Abstract
The discovery of alterations in the EGFR and ALK genes, amongst others, in NSCLC has driven the development of targeted-drug therapy using selective tyrosine kinase inhibitors (TKIs). To optimize the use of these TKIs, the discovery of new biomarkers for early detection and disease progression is mandatory. These plasma-isolated exosomes can be used as a non-invasive and repeatable way for the detection and follow-up of these biomarkers. One ml of plasma from 12 NSCLC patients, with different mutations and treatments (and 6 healthy donors as controls), were used as exosome sources. After RNAse treatment, in order to degrade circulating miRNAs, the exosomes were isolated with a commercial kit and resuspended in specific buffers for further analysis. The exosomes were characterized by western blotting for ALIX and TSG101 and by transmission electron microscopy (TEM) analysis, the standard techniques to obtain biochemical and dimensional data of these nanovesicles. Total RNA extraction was performed with a high yield commercial kit. Due to the limited miRNA-content in exosomes, we decided to perform retro-transcription PCR using an individual assay for each selected miRNA. A panel of miRNAs (30b, 30c, 103, 122, 195, 203, 221, 222), all correlated with NSCLC disease, were analyzed taking advantage of the remarkable sensitivity and specificity of Real-Time PCR analysis; mir-1228-3p was used as endogenous control and data were processed according to the formula 2-ΔΔct 13. Control values were used as baseline and results are shown in logarithmic scale.
Keywords: Medicine, Issue 111, Exosomes, miRNAs, NSCLC, Liquid biopsy, Cancer, Biomarker
Introduction
The low survival rate in NSCLC (non-small cell lung cancer) patients is mainly due to the limited efficacy of treatments in advanced disease and poor early detection1. Activating mutations in the EGFR gene, that have been discovered in a subpopulation of NSCLC patients, are known to confer sensitivity to tyrosine kinase inhibitors (TKIs). Unfortunately, the majority of EGFR mutated NSCLC patients develop TKI resistance2. Especially in this context, a correct diagnosis is mandatory. In general, due to localization of tumors, obtaining biopsy tissue in NSCLC isn't always feasible. Therefore, several studies are ongoing that use non-invasive methods to obtain these biological data. Exosomes are described as liquid biopsy components that could be investigated in order to obtain diagnostic and prognostic values with non-invasive techniques3.
Exosomes are nanovesicles (40 - 100 nm diameter) of endocytic origin that are released by different cell types both in physiological and pathological conditions4. They can carry protein, lipids, mRNA and microRNAs. Moreover, several studies suggest a pleiotropic role of tumor derived exosomes, which can influence growth and survival of tumor cells, angiogenesis, stromal and extracellular matrix remodeling and drug resistance5.
MicroRNAs (miRNAs) are short non-coding RNA that are involved in post-transcriptional regulation, binding the 3′-UTR of the target mRNAs and leading the mRNA to degradation or to a non-translation6. Selective sorting of miRNAs into exosomes has been described. In this context, recently was demonstrated, in vitro and in vivo, a selective packaging of miR-21 (a well-known miRNA with oncogenic effects) in exosomes released by chronic myelogenous leukemia cell lines after curcumin treatment7.
The exosomal miRNA profile is similar to the miRNA profile of the primary tumor and this feature may be exploited in early diagnosis and prognosis3. Specifically, there is a possibility to characterize, by real-time PCR, selected miRNAs correlated with the molecular profile of disease, e.g., EGFR mutations among others8.
Here, the analysis of a selected panel of NSCLC-correlated miRNAs8-9is described that were isolated from exosomes released in the blood of NSCLC patients. After obtaining the inform consent report from the patients, plasma of 12 NSCLC patients and 6 healthy controls, were analyzed. The exosome isolation was performed with commercial kit according to the manufacturer's protocol. Before exosome isolation, plasma samples were treated with RNAse, in order to degrade contaminant circulating miRNAs. We decided to perform this analysis with a commercial kit due to the limited standardization of other techniques (e.g., ultracentrifugation) which is especially important when working with clinical samples. This kit includes a Proteinase K treatment, which will degrade the RNAse, and thus decreases the risk to degrade exosomal miRNAs after exosome lysis. MicroRNA analysis was performed by sensitive and specific Real-Time PCR analysis; mir-1228-3p was used as an endogenous control and data were normalized according to the formula 2-ΔΔct. Control values are used as baseline and results are shown in a logarithmic scale.
Protocol
1. Exosome Isolation
Note: The isolation of exosomes from plasma samples, was performed with a commercial kit, according to manufacturer's protocol and by adding RNAse treatment: (all these steps must be performed with personal and environment protective equipments and following specific legal procedure for manipulation of biological samples).
Take 1 ml of plasma of each sample, stored at -80 °C, and place it on ice.
Centrifuge the sample at 2,000 × g for 20 min at RT; this passage is mandatory in order to remove cells and debris.
Transfer the supernatant containing the clean plasma to a new 1.5 ml tube.
Centrifuge the new tube at 10,000 × g for 20 min at RT.
Transfer the supernatant containing the clean plasma to a new 1.5 ml tube.
Add 100 ng/ml of RNAse to the supernatant and incubate the tube at 37 °C for 10 min in order to degrade circulating RNAs.
Transfer all the treated plasma to a new 2 ml tube and add 0.5 volume of 1x PBS.
Vortex the sample in order to mix the solutions.
Add 0.05 volumes of Proteinase K solution (enclose in the kit) to the sample.
Vortex the sample and then incubate at 37 °C for 10 min.
Add 0.2 volume of exosome precipitation buffer to the sample and mix the solutions by inversion.
Incubate the sample at 2 °C to 8 °C for 30 min and then centrifuge the sample at 10,000 × g for 5 min at RT.
Aspirate and discard the supernatant. The pellet contains the isolated exosomes.
- Add selected buffer to the exosome pellet in order to resuspend the exosomes. The selection of the buffer depends on the downstream analysis planned, in this case:
- Add 100 µl of 1x PBS in order to perform TEM analysis.
- Add 346.5 µl of specific lysis buffer for RNA extraction (from commercial kit) (CAUTION: chemical hazard).
- Add 100 µl of lysis buffer for protein extraction (Tris-HCl 50 mM pH 7.6 - NaCl 300 mM - Triton X-100 0.5% - PMSF 1 mM - Leupeptin/Aprotinin 10 µg/ml) in order to perform Western Blotting analysis. (CAUTION: chemical hazards).
Store the resuspended exosomes at −20 °C.
2. Exosome Characterization by Western Blot Analysis
Note: In order to characterize the isolated nanovesicles, western blotting with antibodies against ALIX and TSG101 (well-known exosomal markers) is recommended.
After resuspension of the exosome pellet in Lysis buffer, place it on ice for 1 hr in order to dissolve the exosomal membrane.
Centrifuge the lysate at 12,000 g for 10 min and transfer the supernatant to a new tube without disturbing the pellet.
Quantify the total exosomal protein content by Bradford Assay or other methods and load 50 µg of proteins per well in a Tris/Glycine SDS - 8% Polyacrylamide gel. (CAUTION: chemical hazards).
Separate the proteins (and an appropriate molecular weight marker) in 8% SDS-PAGE gel electrophoresis with running buffer (25 mM Tris, 192 mM glycine, 0.1% SDS) for 1 hr and 30 min at 120 V. (CAUTION: chemical hazards).
Transfer the protein to a nitrocellulose membrane by electroblotting with 4 °C cold transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol) for 1 hr at constant 50 V. (CAUTION: chemical hazards).
Block the membrane for 1 hr in slow agitation with 5% non-fat dry Milk in 1x TBS -Tween 0.1%.
Wash the membrane 4 times for 5 min in fast agitation with 1x TBS - Tween 0.1%.
Add the primary antibodies against ALIX and TSG101 (one per membrane) in appropriate buffer (accordingly to the antibody datasheet from the manufacturer) and incubate the membrane O/N in slow agitation.
Wash the membrane 5 times for 5 min in fast agitation with 1x TBS - Tween 0.1% in order to remove excessive antibody.
Add the appropriate secondary antibody in appropriate buffer (accordingly to the antibody datasheet from the manufacturer) and incubate the membrane for 1 hr in slow agitation.
Wash the membrane 5 times for 5 min in fast agitation with 1x TBS - Tween 0.1% in order to remove excessive antibody.
Detect the selected protein by chemiluminescence analysis 11.
3. Exosome Characterization by TEM Analysis
Note: In order to visualize the isolated exosomes, TEM (Transmission Electron Microscopy) analysis was performed.
Place ~10 µg (previously quantified by Bradford assay) of sample of intact exosomes resuspended in 1x PBS on a parafilm and put on the sample drop a formvar carbon coated nickel grid for 1 hr.
Wash the grid 3 times by positioning it on three 40 µl 1x PBS drops.
Wash the grid 5 times by positioning it on five 30 µl ultrapure water drops.
Fix the sample by adding a drop of 2.5% glutaraldehyde (CAUTION: chemical hazard) for 10 min.
Wash the grid 3 times by positioning it on three 40 µl 1x PBS drops.
Add a drop of 2% uranyl acetate (CAUTION: chemical hazards) and put the grid on top of it for 15 min in order to contrast the sample.
Embed the sample on a drop of 0.13% methylcellulose (CAUTION: chemical hazards) and 0.4% uranyl acetate and incubate the grid on top of the drop for 10 min.
Gently remove liquid excess with an absorbing paper and dry the grid for 5 min with the coated side up.
Examine the grid by transmission electron microscope12.
4. Exosomal RNA Extraction and Quantification
Note: We decided to perform the total RNA extraction from exosomal samples using a commercial kit, according to manufacturer's protocol: (CAUTION: chemical hazards, review the manufacturer's protocol and guidelines).
After resuspension of the exosome pellet in 346.5 µl of lysis buffer, add 3.5 µl of β-Mercaptoethanol (CAUTION: chemical hazards) and vortex vigorously; this passage is mandatory in order to destroy the lipid membranes.
Add the lysate to the washing column in a collection tube and centrifuge at 11,000 × g for 1 min. After centrifugation, discard the column and transfer the filtrate to a new 1.5 ml RNase-free tube.
Add 350 µl of 70% Ethanol (CAUTION: chemical hazards) and mix by vortexing for 5 sec.
Load the lysate in the resolving column and centrifuge for 30 sec at 8,000 × g. After centrifugation, discard the flowthrough and transfer the column to a new collection tube.
Load 350 µl of desalting buffer and centrifuge at 11,000 × g for 1 min. After centrifugation, discard the flowthrough and return the column into the same collection tube.
Add 95 µl of DNase I reaction mix (10 µl of reconstituted DNase I + 90 µl of DNase buffer, for each sample) on the silica membrane of the column and incubate at RT for 15 min at RT. (CAUTION: chemical hazard)
Add 200 µl of washing buffer (CAUTION: chemical hazard) to the column and centrifuge at 11,000 × g for 1 min. Place the column into a new collection tube.
Add 600 µl of washing buffer to the column and centrifuge at 11,000 × g for 1 min. After centrifugation, discard flowthrough and place the column into the same collection tube.
Add 250 µl of washing buffer to the column and centrifuge at 11,000 × g for 2 min. Place the column into a 1.5 ml RNAse-free tube.
Elute the RNA in 40 µl of RNAse-free water and centrifuge at 11,000 × g for 1 min, in order to have a high RNA concentration in this small volume.
Immediately place eluted RNA on ice to prevent potential degradation or store at -80 °C.
Quantify the RNA content. Note: Here, the RNA quantization was performed by using 1 µl of sample through spectrophotometer.
5. Retrotrascription of Exosomal miRNAs
Note: Due to the described limited quantity of miRNAs content in exosomes, it was decided to perform the reverse transcription of selected miRNAs through a commercial kit using an individual assay for each miRNA, according to manufacturer's protocol. For each sample, 8 miRNAs correlated to NSCLC status were selected (miR-30b; miR-30c; miR-103; miR-122; miR-195; miR-203; miR-221; miR-222; and miR-1228-3p as endogenous control).
Prepare the Reverse transcription master mix (RT mix). For each reaction prepare 7 µl of RT mix, as described in Table 1 (click here to download). (CAUTION: chemical hazard) Note: In this experiment, 9 different RT mix were prepared for each sample.
Mix gently and store the RT mix on ice. Add 7 µl of RT mix per reaction tube.
Add 5 µl of sample RNA (10 µg of total RNA) per reaction tube and mix gently.
Add 3 µl of the 5x reverse transcription primers (CAUTION: chemical hazard) from each assay set into the corresponding reaction tube.
Mix slowly, close the tube and incubate on ice for 5 min.
Load the reaction tube into the thermal cycler and start the reverse transcription reaction using the following parameter values (HOLD at 16 °C for 30 min; HOLD at 42 °C for 30 min; HOLD at 85 °C for 5 min; HOLD ∞ 4 °C).
6. Real Time PCR Analysis of Exosomal miRNAs
Note: This analysis was performed using a commercial kit for Real Time PCR (qPCR), with three biological and technical replicates per sample.
For each reaction, prepare 17.67 µl of Real Time PCR mix (10 µl of PCR Master Mix + 7.67 µl of Nuclease-free water), mix it and put on ice. (CAUTION: chemical hazard)
Add 1 µl, for each PCR tube, of 20x qPCR primers (CAUTION: chemical hazard).
Place 18.67 µl of the qPCR mix in each well-plate.
Add 1.33 µl of the reverse transcription product into the correspondent well-plate.
Close the plate and spin it (300 g x 5 sec).
Insert the plate in a Real Time PCR system and start the run using the following PCR conditions: HOLD at 95°C for 10 min; 40 cycles of 15 sec at 95°C and 60 sec at 60°C. Process and normalize data according to the formula 2-ΔΔct 13.
Representative Results
A total of 12 plasma of NSCLC patients and 6 healthy controls were processed with commercial kit for exosome isolation from plasma. Each sample was processed by Western Blot analysis in order to evaluate the exosomal markers ALIX (96 kDa) and TSG101 (43 kDa). An example of these results is shown for 3 samples in Figure 1, indicating that exosome isolation from plasma samples is feasible with this protocol.
In order to biophysically characterize exosomal samples, TEM analysis was performed. TEM have shown that the nanovesicles obtained by this isolation protocol have a size average in the range of exosomal size, between 40 - 100 nm (Figure 2).
After exosome isolation and characterization, we investigated their miRNAs content. We selected 8 specific miRNAs, known to be deregulated in NSCLC. As internal control, we used miR-1228 that has been previously validated as a stable and efficient endogenous control in different tumor types10. According to these results miR-1228 is a reliable and stable endogenous control. When the expression level of the 8 selected miRNA between healthy donors and lung cancer patients were compared, it was found that these miRNAs are strongly deregulated in the analyzed clinical samples. In Figure 3 the expression pattern of the 8 selected miRNAs is shown on a logarithmic scale for the 12 samples analyzed (PAD1-12). Not all miRNAs were detectable in every sample and due to the limited amount of samples we cannot perform statistical analysis.
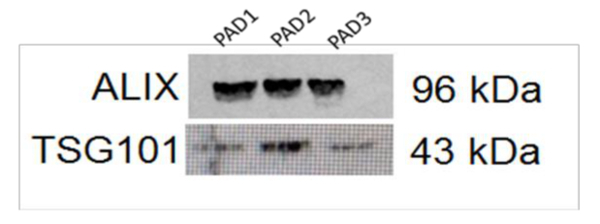 Figure 1. Western Blot Analysis in 3 Exosomal Samples for ALIX and TSG101. The Western Blot shows the presence of the well-known exosomal markers, ALIX and TSG-101, in the analyzed samples. Please click here to view a larger version of this figure.
Figure 1. Western Blot Analysis in 3 Exosomal Samples for ALIX and TSG101. The Western Blot shows the presence of the well-known exosomal markers, ALIX and TSG-101, in the analyzed samples. Please click here to view a larger version of this figure.
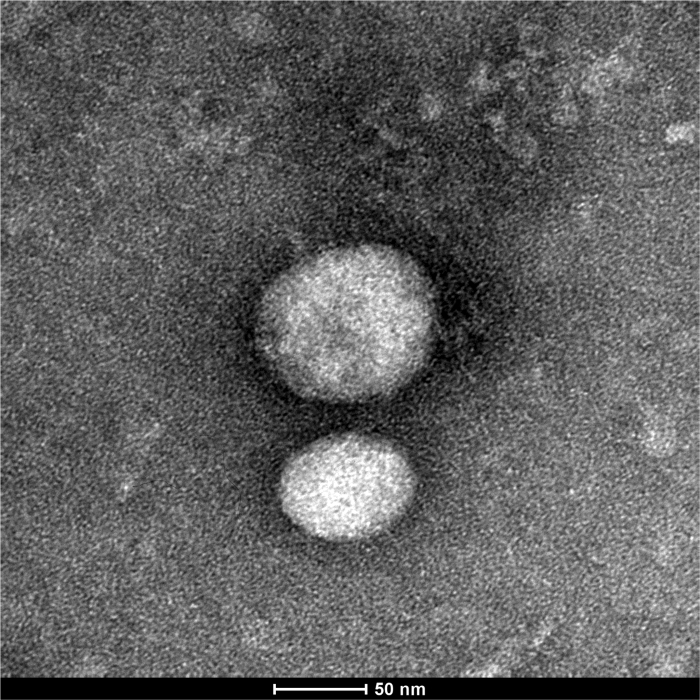 Figure 2. TEM Analysis of Exosomal Samples. These results demonstrate that the isolated nanovesicles have a diameter average, between 40 - 100 nm, inside the exosomal range. Please click here to view a larger version of this figure.
Figure 2. TEM Analysis of Exosomal Samples. These results demonstrate that the isolated nanovesicles have a diameter average, between 40 - 100 nm, inside the exosomal range. Please click here to view a larger version of this figure.
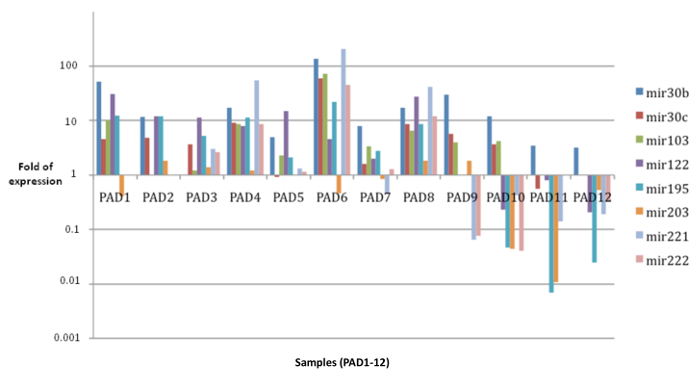 Figure 3. Expression Profile of 8 Selected miRNAs in Exosomal Samples. These data, shown in logarithmic scale, demonstrate that through this protocol is feasible to see up- or down-regulation of selected exosomal miRNAs from clinical NSCLC plasma patients (PAD1-12), leading to potential discovery of exosomal biomarkers of NSCLC. The data are shown as fold of expression relative to healthy controls. Please click here to view a larger version of this figure.
Figure 3. Expression Profile of 8 Selected miRNAs in Exosomal Samples. These data, shown in logarithmic scale, demonstrate that through this protocol is feasible to see up- or down-regulation of selected exosomal miRNAs from clinical NSCLC plasma patients (PAD1-12), leading to potential discovery of exosomal biomarkers of NSCLC. The data are shown as fold of expression relative to healthy controls. Please click here to view a larger version of this figure.
Discussion
With this combined protocol it was possible to perform an exosomal miRNA analysis from plasma of NSCLC patients. These data may reflect the disease status, but this needs to be confirmed by further study.
The protocol used in this article was based on a commercial kit for exosome isolation. The reason was that the other known isolation procedures (e.g., density gradient ultracentrifugation) require more manual procedures, leading to a limited standardization. Nevertheless, one of the limitations of this protocol is that in comparison with density gradient ultracentrifugation for exosome isolation, density gradient ultracentrifugation remains a very useful technique in exosome isolation that exploits the specific density of exosomes (between 1.13 - 1.19 g/ml). A potential modification is a mix of both procedures, which might be a gold standard in this field.
The possibility to process clinical samples with RNase treatment has given us pure samples without contamination of circulating miRNAs. However, the presence of RNase enzymes in the samples could effect the amount of exosomal miRNA after exosome lysis. For this reason we chose a kit with Proteinase K treatment, in order to eliminate circulating contaminant proteins and to degrade RNase enzymes.
After the standardization of all isolation procedures, the future characterization and miRNA analysis of these data can form a useful tool to detect biomarkers in a non-invasive way during initial diagnosis and follow-up.
Disclosures
All authors declare no competing financial interests.
Acknowledgments
This study was also financed by MOCA (Multidisciplinair Oncologisch Centrum Antwerpen) grant 2014.
References
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics. Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Rolfo C, et al. Novel therapeutic strategies for patients with NSCLC that do not respond to treatment with EGFR inhibitors. Cancer Treat Rev. 2014;40:990–1004. doi: 10.1016/j.ctrv.2014.05.009. [DOI] [PubMed] [Google Scholar]
- Rolfo C, et al. Liquid biopsies in lung cancer: the new ambrosia of researchers. Biochim Biophys Acta. 2014;1846:539–546. doi: 10.1016/j.bbcan.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- Fontana S, Saieva L, Taverna S, Alessandro R. Contribution of proteomics to understanding the role of tumor-derived exosomes in cancer progression: state of the art and new perspectives. Proteomics. 2013;13(10-11):1581–1594. doi: 10.1002/pmic.201200398. [DOI] [PubMed] [Google Scholar]
- Taverna S, et al. Exosomal shuttling of miR-126 in endothelial cells modulates adhesive and migratory abilities of chronic myelogenous leukemia cells. Mol Cancer. 2014;11 doi: 10.1186/1476-4598-13-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna S, et al. Curcumin inhibits in vitro and in vivo chronic myelogenous leukemia cells growth: a possible role for exosomal disposal of miR-21. Oncotarget. 2015. [DOI] [PMC free article] [PubMed]
- Garofalo M, et al. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008;27:3845–3855. doi: 10.1038/onc.2008.6. [DOI] [PubMed] [Google Scholar]
- Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10(1):42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- Hu J, et al. Human miR-1228 as a stable endogenous control for the quantification of circulating microRNAs in cancer patients. Int J Cancer. 2014;135(5):1187–1194. doi: 10.1002/ijc.28757. [DOI] [PubMed] [Google Scholar]
- Alessandro R, et al. Effects of carboxyamidotriazole on in vitro models of imatinib-resistant chronic myeloid leukemia. J Cell Physiol. 2008;215:111–121. doi: 10.1002/jcp.21290. [DOI] [PubMed] [Google Scholar]
- Zhao Y, et al. Exosomes Derived from Human Umbilical Cord Mesenchymal Stem Cells Relieve Acute Myocardial Ischemic Injury. Stem Cells Int. 2015;2015 doi: 10.1155/2015/761643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoko T, et al. Up-regulation of MDR'1and induction of doxorubicin resistance by histone deacetylase inhibitor depsipeptide (FK228) and ATRA in acute promyelocytic leukemia cells. Blood. 2006;107(4):1546–1554. doi: 10.1182/blood-2004-10-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]


