Abstract
Mesenchymal stem/stromal cells (MSC) are promising candidates for use in cell-based therapies. In most cases, therapeutic response appears to be cell-dose dependent. Human term placenta is rich in MSC and is a physically large tissue that is generally discarded following birth. Placenta is an ideal starting material for the large-scale manufacture of multiple cell doses of allogeneic MSC. The placenta is a fetomaternal organ from which either fetal or maternal tissue can be isolated. This article describes the placental anatomy and procedure to dissect apart the decidua (maternal), chorionic villi (fetal), and chorionic plate (fetal) tissue. The protocol then outlines how to isolate MSC from each dissected tissue region, and provides representative analysis of expanded MSC derived from the respective tissue types. These methods are intended for pre-clinical MSC isolation, but have also been adapted for clinical manufacture of placental MSC for human therapeutic use.
Keywords: Developmental Biology, Issue 112, Placenta, human, term, third trimester, maternal, mesenchymal stem cell, stromal cell
Introduction
Mesenchymal stem/stromal cells (MSC) are emerging as a promising candidate for use in cell-based therapies 1. Most applications appear to target MSC-mediated tissue repair or immune regulation 2. In many of these applications, allogeneic MSC may be as effective as autologous MSC 3. The use of allogeneic MSC has the economic advantage of being compatible with the large-scale manufacture of multiple cell doses from a single source tissue 4 to treat many patients.
Historically, pre-clinical and clinical studies have utilized MSC-derived from the bone marrow 4. Bone marrow is generally collected from the iliac crest of a volunteer donor. This process is invasive, and only a small volume of marrow (~20 ml) is collected through a single puncture. Generating clinically meaningful numbers of MSC requires extensive in vitro expansion. Cell potency decreases with passage number 3, creating a paradox where the theoretical number of cells required for clinical efficacy is increased the more the cell population is expanded. In contrast to bone marrow aspirates, term placenta is a physically large starting tissue (typically 500-750 g 4), which can be harvested aseptically during cesarean section with no risk to the donor. MSC derived from placenta have long-term proliferation 5 and immunomodulatory capacity 6, superior to bone marrow-derived MSC. In a previous study, we demonstrated that a single term placenta contained sufficient MSC for the manufacture of up to 7,000 clinical doses 4. These characteristics make placenta an ideal source tissue for the manufacture of allogeneic MSC.
The placenta is a fetomaternal organ consisting of both fetal and maternal tissue 7, and thus MSC of fetal or maternal origin can be, theoretically, isolated. The following references provide detailed information on the development and pathology, as well as microscopic and macroscopic examination of the human placenta and adnexa 8,9. The placenta proper is comprised largely of fetal blood vessels and secretory and supporting cells called trophoblasts, making up the chorionic villi covered by the chorion frondosum (plate) 8. The branched placental villi are bathed in maternal blood delivered from the uterine spiral arteries, enabling nutrient, hormone and gas exchange between fetus and mother. The placenta is anchored to the endometrium via maternal decidual stromal cells and fetal extravillious trophoblasts are interspersed in extracellular matrix 8. The villi converge onto the fetal chorionic plate where they form the umbilical cord 8.
An outcome of the first International Workshop on Placental-Derived Stem Cells (2008) was an appreciation of the need to standardize the isolation and characterization of cells from human term placenta 10. Because of the anatomy of the placenta, dissection of the different tissues, isolation of MSC and anticipated culture outcomes can be overwhelming for newcomers to the field. In this protocol, the harvest of placental chorionic tissues, followed by MSC isolation and expansion is thoroughly detailed. MSC characterization via flow cytometry and in vitro differentiation are considered routine 5,11-13, and thus only briefly detailed here.
As highlighted in a recent systematic literature review 14, MSC obtained from the placental chorionic villi are generally assumed to be fetal. Although, only 18% of studies examined the origin of the MSC obtained, and of those, only half of the studies reported fetal MSC and the other half reported maternal or mixed MSC populations. Each of the three tissue components described herein (chorionic villi, chorionic plate and decidua basalis) are composed primarily of the fetal membrane/villi, and a small proportion of uterine-derived maternal cells, which remain attached to the delivered placenta. We provide data demonstrating that isolating MSC from the maternal side of the placenta, rather than the fetal side of the placenta, as we have previously reported 5,11, is a more appropriate starting material if maternal MSC are desired. This protocol also describes the use of XY FISH to validate fetal or maternal contribution to cell cultures. While this is a standard protocol from the manufacturer, this analysis is often neglected and its importance underestimated 14.
Protocol
The Human Research Ethics Committees at Mater Health Services, Royal Brisbane and Women's Hospital, Queensland University of Technology and the University of Queensland approved the research and collection of human placenta samples used in the study. All protocols complied with national research guidelines. Patients provided informed written consent for the use of tissue for research purposes.
Third trimester placentas were obtained from healthy mothers following routine Caesarean section (CS) births at term from the above-mentioned hospitals in Brisbane, Australia. Male discordant pregnancies for term samples were utilized in this study to distinguish fetal from maternal cells. Fetal gender had been determined by ultrasound prior to birth and/or visual inspection of the neonate at birth by clinical staff. X and Y chromosome fluorescence in situ hybridization (FISH) was utilized to further validate gender and fetal or maternal origin of tissue preserved from the original placentas.
1. Prior to Harvest, Prepare the Following
Note: When making up enzyme solutions, the specific requirements and concentrations provided by the manufacturer for each product should be followed. Enzymes are often provided as a crude mix of proteins, therefore similar enzymes from different companies are likely to have different activities/concentrations. For this reason the manufacturer's advice should be acknowledged and solution preparation may have to be modified accordingly.
Prepare or purchase 1.5 to 2 L total sterile Hank's Balanced Salt Solution (HBSS) for this protocol.
Prepare collagenase I stock solution by weighing out collagenase type I and dissolve 1 mg/ml in HBSS and vortex gently to ensure complete dissolution. Determine the volume of HBSS (containing calcium and magnesium) required to bring collagenase solution to 100 units (U)/µl (1,000x stock solution) and then filter using a 0.2 µm syringe filter. Aliquot 1 ml volumes into 1.5 ml tubes and store at -20 °C until needed.
Prepare dispase stock solution by dissolving non-sterile dispase at 10 mg/ml in Dulbecco's phosphate-buffered saline (DPBS) without calcium or magnesium. Further dilute with DPBS without calcium or magnesium to a final concentration of 2.4 U/ml. Filter sterilize through a 0.2 µm syringe filter. Aliquot 1 ml volumes in 1.5 ml tubes and store at -20 °C until needed.
Prepare deoxyribonuclease (DNase)-I stock solution by dissolving DNase-I at 10 mg/ml in 0.15 M NaCl. Filter thorugh a 0.2 µm syringe filter. Aliquot 1 ml volumes in 1.5 ml tubes and store at -20 °C until needed.
Prepare the working digestion solution by combining 100 U/ml collagenase type I, I.5 µg/ml DNase I and 2.4 U/ml dispase in serum-free DMEM. Freshly defrost and dilute enzyme stocks immediately prior to use. Approximately, a 1:1 ratio of digestion solution volume to tissue volume is required (e.g.10 ml of tissue, measured in a 50 ml tube, requires 10 ml of digestion solution).
Prepare a solution of 4% paraformaldehyde (PFA) in PBS and adjust to pH 7.4 15. Store 25 mL aliquots at -20 °C for long term or at 4 °C for 1 week.
Prepare MSC culture medium from DMEM-Low Glucose (DMEM-LG) supplemented with 1x antibiotic and antimycotic solution and 10% non-heat inactivated fetal bovine serum. MSC-grade FBS can be purchased from many FBS suppliers.
Autoclave sterilize dissection equipment in advance as per institutional guidelines. These include scissors, forceps, scalpels, and a large metal dissection tray. Collect additional sterile tissue culture plasticware and consumables (e.g. scalpel blades, 50 ml and 15 ml tubes, 25 ml, 10 ml and 5 ml pipettes, and 75 or 175 cm2 cell culture flasks).
Use standard primary cell culture laboratory equipment: a class II biosafety cabinet suitable for primary cell isolation, cell culture incubator set at 5% CO2 and 37 °C, a rocker in a 37 °C incubator, and a centrifuge with a capacity for 50 ml tubes.
Use appropriate personal protective equipment: a laboratory coat, 2 pairs of gloves (at all times), safety glasses, and close-toed shoes as per institutional guidelines.
Prepare appropriate decontamination solutions for human blood specimens as per institutional guidelines and liquid biological waste containers in advance.
2. Isolation of the Placental MSC
- Preparation of the Placenta
- Dress in the required personal protective equipment. Set up the biological safety cabinet with required materials, solutions and waste containers to carry out the procedure up to the enzymatic digestion (section 2.5.6).
- Obtain a placenta with informed consent and ethical approval. Collect the placenta via cesarean section under aseptic surgical conditions, and package in a sterile bag or bucket for transport to the laboratory.
- During transport and prior to dissection, store the placenta at room temperature or 4 °C. Note: Tissue can be stored for up to 6 hr without affecting cell culture performance.
- Limit the time between placenta collection and tissue processing where possible. This recommendation is based on practical issues. In some instances, we have delayed the cell isolation process for up to 6 hr after birth. In these instances placentas have been kept at room temperature or 4 °C (in the lab or at the collection center). No detectable differences were observed, above general donor variation, in the MSC obtained from placentas when they were stored at 4 °C or at room temperature.
- Place the container with placenta in the biological safety cabinet. Open the container and transfer the placenta into a sterile tray.
- Open out the fetal membranes (amniotic sac or amnion-chorionic laeve) 8. Become orientated with the parts of the placenta. The fetal side has the umbilical cord insertion, which is in general, centrally inserted in the placenta. The maternal side (lined with decidua basalis), which is opposite to the fetal side, has obvious cotyledons (or lobes).
- To begin the dissection, orientate the placenta with the umbilical cord facing upwards in the sterile tray. Note: This protocol is designed to yield sufficient cells to seed into one T175 flask from each of the three tissue types. Each digest begins with approximately 10 g of tissue. This is a similar starting number of flasks as MSC cultures initiated from a 20 ml bone marrow aspirate. If more cells are desired, then more tissue can be harvested and the protocol can be scaled linearly. The sizes of tissue to be harvested are only indicative of what is practical with the placental tissue. There are few defining features in the placental villi, which can be used as a reference point. The indicated dimensions serve as an estimate and the umbilical cord is used as a landmark on the fetal surface.
- Dissection of the Decidual (D) Tissue
- Flip the placenta over so the maternal surface (decidua basalis) is facing up. Ensure that the fetal side with the umbilical cord insertion point is facing down.
- Cut pieces of 0.5 cm thickness from the maternal side of the placenta (containing decidua basalis tissue).
- Place tissue pieces into a Petri dish containing HBSS during the dissection to keep the tissue pieces hydrated.
- Transfer sufficient tissue to fill a tube up to the 10 ml mark in a 50 ml tube (this is approximately 10 g of tissue).
- Dissection of the Chorionic Plate (CP) Tissue
- Mechanically remove of the amniotic membrane from the fetal surface of the placenta (not the amniotic sac), leaving the chorionic frondosum (chorionic plate) intact.
- Cut pieces ~1 cm wide by 0.5 cm deep from the chorionic plate. Harvest the chorionic plate from the region nearest to the umbilical cord, away from the edge of the placenta.
- Place tissue pieces into a petri dish containing HBSS during the dissection to keep them hydrated.
- Transfer sufficient tissue to fill a 50 ml tube up to the 10 ml mark (~10 g of tissue).
- Dissection of the Chorionic Villi (CV) Tissue
- Dissect CV tissue ~1 cm2 x 0.5-1 cm deep from placental tissue where the CP has already been removed. Again, harvest CV from the region closest to the umbilical cord, away from the edge of the placenta. Try to stay at least 1 cm away from the maternal side of the placenta (containing decidua basalis tissue); the goal is to harvest tissue from the interior of the placental villi tissue.
- Place tissue pieces into a Petri dish containing HBSS during the dissection to keep them hydrated.
- Transfer sufficient tissue to fill a 50 ml tube up to the 10 ml mark (~10 g of tissue).
- Mincing and Enzymatic Digestion of the D, CP, and CV Placental Tissues
- As there are ~10 g of D, CP or CV tissues in three different 50 ml tubes, fill each tube with approximately 40 ml of HBSS and repeatedly invert the tube in order to wash the tissue (repeat for ~10 sec).
- Decant the supernatant and repeat this wash step 2-3 times until the solution is largely free from blood. Use a 10 ml pipette or long tweezers to ensure the tissue pieces are not lost out of the tube(s) when decanting the supernatant.
- Return tissue pieces to a 10 cm petri dish with minimal liquid transfer. Chop/mince the tissue into fine pieces of approximately 1-5 mm3 with scissors or razor blade.
- Transfer the minced tissues back into appropriately labeled (D, CP or CV) 50 ml tubes.
- Add freshly prepared digest media in at least a 1:1 ratio with the tissue (e.g. 10 ml of tissue plus 10 ml of digest media). Replace the cap on each tube and invert the tubes several times to mix.
- Incubate the tissues in the tubes with digest media for 1-2 hours at 37 °C. A humidified atmosphere and 5% CO2 are not necessary for this step. Note: Rapid shaking is required in order to efficiently dissociate cells from the tissue and maximize MSC yield. Limited yield will be evident by very few cells attached to the culture flask at 48 hr and a first passage time longer than 1-2 weeks. The incubation time and shaking method is dependent on the 37 °C incubator available in the laboratory and may require optimization.
- For an incubator with a rapid and controllable shaking mechanism, place tissue and digest medium in a 50 ml tube or sterile conical flask, place in incubator and set shaking speed to 250 rpm. This will result in a complete digest of the tissue in 1 hr.
- Alternatively, for an incubator with a gentle rocking or no rocking mechanism, manually shake the tube rapidly and vigorously by hand for 10 sec every 30 min for 1.5 to 2 hr. The enzyme incubation period is a suitable time to discard waste, clear the biosafety cabinet of unnecessary items, change laboratory coat if soiled and prepare for the next part of the procedure.
- Collection of Mononuclear Cells and Plating into Flasks Note: From the previous step, there are three 50 ml tubes (D, CP or CV) with digesting tissue pieces. When the tissue digest solution evolves a cloudy appearance and white blood vessels are obvious in the tissue, the digestion is complete.
- Add 30 ml of MSC media containing FBS to each tube to inactivate the enzymes contained in the digestion solution.
- To separate the mononuclear cells from the large undigested debris, pulse centrifuge the 50 ml tube. Briefly, allow the centrifuge to reach 340 xg for 5 sec and then stop. This will force the large debris to pellet at the bottom of the tube, while leaving the mononuclear cells in liquid suspension. Note: The mononuclear cells will remain in the liquid phase if the tube is only briefly (pulsed) centrifuged. If centrifugation is extended, the mononuclear cells will also pellet at the bottom of the tube with the tissue pieces, resulting in undesirable cell loss.
- Collect the supernatant, containing the mononuclear cells, and transfer this into a new 50 ml tube with a pipette.
- Add 30 ml of media or HBSS to the remaining placental tissue debris and shake vigorously. This will bring remaining detached mononuclear cells into suspension and enable a second harvest of putative MSC from the tissue.
- Pulse centrifuge the tube a second time, and again transfer the supernatant to a new 50 ml tube. Repeat this step a third time to maximize the collection efficiency of mononuclear cells from each tissue source.
- Pool the supernatants from each of the individual tissue types into two 50 ml tubes. This will yield 6 tubes total (2 tubes of each D, CP and CV). Centrifuge each tube for 5 min at 340 x g. This step will pellet the mononuclear cells to the bottom of the tubes.
- Carefully decant or pipette off the supernatant into the waste. It is unnecessary to try to eliminate every drop of supernatant. Note: The cell pellet will be fragile due to a large number of red blood cells. Be careful not to accidentally discard the pellet if you are decanting directly from the 50 ml tube.
- After removing most of the supernatant, gently flick the tube several times with a finger to dislodge the cell pellet. Combine the pellets, so that there is a single tube for each D, CP or CV tissue and resuspend each in 35 ml of MSC media.
- Optional: Filter the mononuclear fraction through a 100 µm mesh cell strainer set up in a 50 ml tube to remove cell clumps or fibrous material. Note: Care is required as the filters can easily clog up and over-fill. Additionally, air bubbles can obstruct filters from draining. If this occurs, slowly lift the filter out of the tube to allow air to flow under the filter, wash filter through with cell culture media or exchange to a new filter. Excluding the filtering step does not appear to alter yield or quality of the subsequent MSC cultures.
- Optionally, a red blood cell (RBC) lysis or density gradient centrifugation can be carried out at this stage 15. However, our experience is that RBC do not interfere with MSC attachment or proliferation and therefore RBC lysis is not necessary. Mononuclear cells could be counted at this step if RBC removal was carried out 15. Although, the vast majority of cells at this stage will not be MSC, but be fetal origin trophoblasts, endothelial cells and hematopoietic cells, as well as maternal origin hematopoietic cells and decidual stromal cells.
- Transfer the cell suspension for each D, CP or CV tissue into a single T175 flask and culture in a humidified incubator at 37 °C and 5% CO2.
- Post Isolation-cell Expansion
- At 48 hr following the initial isolation, remove the medium from each of the flasks (D, CP and CV) and replace with 35 ml of fresh MSC media. The spent medium and detached cells can be discarded, as putative MSC are presumed to be attached to the tissue culture flask 16. Return flasks to the incubator for a further 24 hr.
- Following an additional 24 hr of culture, wash the flasks twice with DPBS (25 ml) to remove debris and RBCs. Swirl the liquid around the bottom of the flask to dislodge RBCs. Use a pipette to remove liquid and debris. Add 35 ml of MSC media to the flasks and return the flasks to the incubator.
- Replace media twice per week, and incubate cultures until the cell monolayers are 80- 90% confluent. This initial expansion takes 4-14 days, depending on the quality of the tissue, amount of starting material and efficiency and time of incubation with the digest solution.
3. Subculture of pMSCs
When cultures are 80-90% confluent, wash twice with 20 ml 1x HBSS or DPBS and discard washes. Add trypsin-substitute to each flask (i.e. use 5 ml for a T175 flask).
Incubate the flasks for 5 min at 37 °C to liberate MSC from the tissue culture surface. Tap the sides of the flask every ~2 min to facilitate detachment.
When cells are detached from the surface and in a single cell suspension, wash the cells from the flask with MSC media and collect the cells in tubes. The MSC media will dilute and deactivate the trypsin-substitute (use ~15 ml of MSC media per T175 flask).
Transfer the contents of each tissue culture flask to a 50 ml tube.
Centrifuge tubes at 340 x g for 5 min to pellet the cells.
Discard the supernatant and resuspend the cell pellets in 1 ml of MSC media.
Dilute 10 µl of cell suspension in 10 µl trypan blue. Count cells using a hemocytometer and calculate the total number of cells 15.
Transfer cells to new flasks at 1,150 cells/cm2 in fresh MSC media and incubate in a tissue culture incubator. At this seeding density, seed 200,000 MSC into each new T175 flask in 35 ml of MSC media. Feed cells twice per week until 80-90% confluent. Seed subsequent passages at 1,150 cells/cm2 or 200,000 MSC into each T175 flask.
When the Passage 1 (P1) or P2 cells are confluent, cryopreserve the majority of the cells for future use, and reseed only one T175 flask for further propagation and characterization. The propagated cells can be used at P3 or P4 for mesodermal differentiation assays and cell characterization via flow cytometry. Note: In early passages, there may be very few sparsely separated colonies, but the local cell density within each colony may be very high. This is not ideal, as the local dense packing of the cells will result in contact inhibition, delaying growth and reducing culture performance. We recommend when dense colonies are observed, the cells should be harvested and reseeded into a new flask. This redistribution process provides cells with room to proliferate, and overall culture performance will be improved.
To cryopreserve MSC, collect ~1 x 106 cells in 1 ml of 90% FCS and 10% dimethylsulphoxide (DMSO) and freeze using standard protocols. Note: In the representative results section, there are descriptions of MSC characterization using flow cytometry, mesodermal differentiation and XY FISH analysis to determine the gender of the three different (D, CP and CV) expanded cell populations. In our study, placentas were from male babies; thus all fetal cells could be distinguished as male (Y chromosome) and all maternal cells could be distinguished as female (X chromosome only).
Representative Results
The placental MSC isolation procedure is summarized in Figure 1. The three areas of the placental anatomy from which MSC were isolated are highlighted in Figure 2. These are the maternal decidua, as well as the largely fetal tissues of the chorionic plate and chorionic villi. Many text books, articles and online resources detail the development and functional role of the various placental tissues (please see reference 8).
Morphology of cultures 48 hr after isolation and removal of the tissue debris. Following 48 hr of culture, the MSCs will have attached to the tissue culture plastic, while RBCs and most other cellular debris will not have attached. At this time, the medium must be replaced with 35 ml of fresh culture medium. Prior to this medium exchange, it is difficult to make precise observations because of the large numbers of RBCs, which will obstruct visual assessment. The appearance of the culture supernatant may vary substantially between placenta donors. This variation can be seen visually in Example 1 and 2 (Figure 3A). These two MSC isolations, which appeared to be very different, were performed simultaneously from two different placentas. However, once washed, both cultures appeared similar (see washed example 3, Figure 3A).
Under a microscope, after the 48 hr media exchange, only a few cells will be attached to the flask (Figure 3B), and the cells will appear significantly different from more extensively expanded MSCs. Some debris and RBCs will be visible as floating or attached clumps and these will not interfere with the growth of the MSCs. The longer fibroblastic cells adhered to the bottom of the flask are likely to be a mix of cells including MSC, hematopoietic, trophoblastic or endothelial cells. Again, the non-MSC cells do not compromise the MSC cultures, as these cells will not generally survive more than 1-2 passages in the MSC culture conditions. Despite some variation in the initial appearance of cultures, the subsequent expansion results are generally consistent.
Morphology of MSC cultures over time. In this representative example, seven days following isolation, small fibroblastic MSC colonies were visible, although non-MSC cells could also be seen as round or loosely attached cells (Figure 4A). The attached cells are what was originally termed a "colony unit forming-fibroblast" (CFU-F) 17 and later termed MSC 18. Thirteen days following isolation, fibroblastic MSC colonies were large (Figure 4B). Generally, the monolayer will be 80-90% confluent at this point and the cells should be passaged. From passage 2 onwards, the placental MSC monolayer will develop the characteristic whirlpool-like morphology at confluence (Figure 4C). Unless the cells are passaged at low density, the CFU-F formation will no longer be observed. At low density, placental-derived MSC have a smaller, squarer appearance than adult bone marrow-derived MSC 1. While placental-derived MSC and bone marrow-derived MSC exhibit similar proliferation rates, the placental-derived cells are less prone to rapid senescence 5.
Characterization of placental MSC in vitro. Each expanded cell population must be characterized to ensure that it conforms to standard MSC criteria 16, including (1) plastic adherence (2) the presence of mesenchymal surface markers and the absence of hematopoietic surface markers, and (3) the capacity to undergo mesodermal differentiation.
Placental MSCs are plastic adherent. As shown in Figure 4, the MSCs in culture are plastic adherent and have a fibroblastic-like morphology; this validates that the cells meet the first criteria that define MSC 16.
Placental MSC display mesenchymal surface markers. The second defining MSC characteristic is the presence of mesenchymal surface markers and the absence of hematopoietic surface markers 16. As there is no single marker capable of definitively identifying an MSC, panels of markers are generally utilized in conjunction with flow cytometry analysis to identify cells that are mesenchymal, but not hematopoietic. In the representative data set provided here (Figure 5A), we evaluated cell expression of mesenchymal markers CD73, CD105, CD90, CD146, and CD44, the hematopoietic markers CD45 and CD34, and HLA-DR, as well as the endothelial marker CD31. All of the antibodies utilized in this placental MSC characterization process are listed in the Table of Materials/Equipment. Staining was performed as per the manufactures instructions, with analysis methods described here 19.
The cells were positive for the mesenchymal markers CD73, CD105, CD44, and negative for the cell surface markers CD45, CD34, HLA-DR and CD31, as expected 5,20. Approximately, 37% and 57% of cells in our representative data set were positive for CD90 and CD146 21, respectively. Both CD90 and CD146 are commonly utilized MSC markers 21. MSC cell surface marker profiles may be different depending on MSC tissue source, medium composition, or passage number 22. In our many years of experience, we have not observed long-term contamination of placental-derived MSC with non-mesenchymal cells following 1-2 passages 5,11.
Placental MSC display mesenchymal differentiation potential By definition, MSC must possess in vitro mesodermal differentiation capacity 5,13. Mesodermal differentiation potential is commonly assessed through either tri- or bi-lineage differentiation assays. Bi-lineage assays generally assess osteogenic and adipogenic differentiation capacity, while tri-lineage assays additionally assess chondrogenic differentiation capacity. In the representative results presented here, we show that the expanded MSC populations form both calcium deposits, indicative of osteogenic differentiation, and lipid vacuoles, indicative of adipogenesis (Figure 5B).
To characterize the MSC populations reported here, we seeded cells into 24 culture wells at 6 x 104 cells in 1 ml of induction medium. The medium components are listed in the Table of Materials/Equipment. While induction medium formulations are common in the literature, there is considerable variability in published formulations. For this reason we briefly list our induction medium formulations and staining approaches here. Osteogenic induction medium contained DMEM-HG, 10% FBS, 1x antibiotic antimycotic solution, 10 mM β-glycerol phophate, 100 nM dexamethasone, and 50 μM L-ascorbic acid 2-phosphate. Adipogenic induction medium contained DMEM-HG, 10% FBS, 1x antibiotic antimycotic solution, 10 μg/ml insulin, 100 nM dexamethasone, 200 μM indomethacin, and 500 μM 3-isobutyl-1-methyl xanthine. Here, cultures were maintained in a 37 °C, 5% CO2 incubator and cultured for 14 days. Induction medium was exchanged twice per week over the culture period. Following 14 days of induction, cultures were characterized for either bone-like matrix (osteogenesis) or lipid vacuoles (adipogenesis) as per our previous publication 5. Osteogenic calcium matrix deposition analysis was achieved by first aspirating off the medium, fixing the cultures with 4% paraformaldehyde for 20 min, washing the monolayer with DPBS, and then staining with Alizarin Red S according to the manufactures instructions. Adipogenic induction was assessed by aspirating off the medium, fixing the cultures with 4% paraformaldehyde for 20 min, washing the monolayer, and staining with Oil Red O solution according to the manufactures instructions. Stained calcium deposits and oil vacuoles were then visualized with a light microscope, and images saved for future reference.
In previous publications, we have characterized the differentiation potential of placental-derived MSC more extensively 4,5. Placental-derived MSC osteogenesis is similar to bone marrow-derived MSC, while adipogenesis is generally less efficient in placental-derived MSC 5. We do not routinely carry out chondrogenic differentiation for several reasons, although this has been previously reported by us for placental-MSC 5. Firstly, while mesodermal differentiation capacity is a defining characteristic of MSC, it is likely of secondary importance 23-25, especially where the therapeutic benefit is likely to be derived from the MSC paracrine secretions 26. Secondly, although Dominici et al. proposed minimal criteria for the clinical production of human adult bone marrow-derived MSC 16, more recent studies indicate MSC from different niches have different inherent properties and differentiation capabilities 5,13,27-32. In fact, Parolini et al. proposed that placenta-derived MSC should differentiate into "one or more mesodermal" lineages rather than all three lineages 10. Finally, many MSC studies exclude chondrogenic differentiation as it occurs through a similar intracellular signaling pathway as osteogenesis (TGFβ family pathway) 33-35.
Placental MSC are maternal in origin using this method of culture, despite the anatomical location of the starting material. Many publications assume that cells isolated from the fetal chorion yield fetal MSC upon culture 14. However, as we have reported previously 5, all cultures derived from fetal chorion, using this protocol, rapidly become enriched for maternal MSC as would intuitively be expected for the maternal decidual MSC cultures. In these representative results we used placental tissue from male babies so that it was possible to easily delineate the fetal and maternal cell contribution in the expanded cell populations. For these studies we used the XY FISH kit listed in the Table of Materials/Equipment, and followed the manufacturers instructions.
In the representative data provided here, the cultures derived from maternal decidua were ~90% maternal cells (XX) and ~10% fetal cells (XY) at passage 0 (Figure 6). By passage 2, the cell populations derived from the maternal tissues were ~100% maternal cells (XX), and fetal cells (XY) were undetectable. This suggests that physical dissection of maternal tissue led to the enrichment of maternal cells in the subsequent cultures. However, it is critical to consider the culture outcomes from the fetal chorionic villi and chorionic plate-derived cultures. At passage 0, both fetal chorionic villi and chorionic plate derived cultures were ~85% XY, or of fetal origin, indicating that targeted dissection enriched for fetal cells (Figure 6). At passage 0, both cultures contained ~15% maternal cell (XX) contamination. Surprisingly, at passage 2, both fetal cultures were populated with ~100% maternal cells (XX), and fetal cells (XY) were no longer detectable. XY FISH analysis reveals that maternal cells (XX chromosomes) rapidly and consistently takeover the cultures derived from the fetal chorionic tissues. This is a critical culture observation that is often overlooked 5. The detail of this analysis is included in this protocol because it demonstrates the very important observation that maternal cells rapidly populate all cultures when DMEM supplemented with 10% FBS is used without additional factors designed to support the fetal-derived populations.
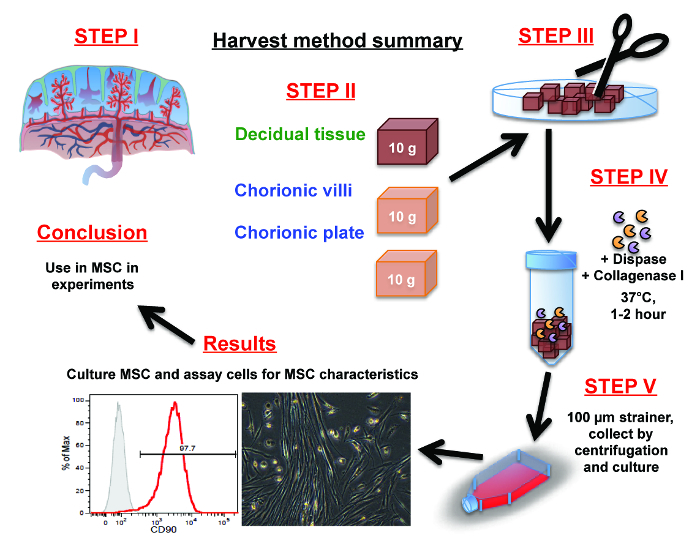 Figure 1: Summary of placental MSC isolation procedure. (Step 1) Orientate yourself with the placenta anatomy. (Step 2) Manually dissect 10 g of tissue from either the decidua, chorionic villi or the chorionic plate using scissors. (Step 3) Mince the dissected portions of decidua, chorionic villi or the chorionic plate tissues into fine pieces with scissors or a scalpel. (Step 4) Liberate cells from the fine pieces via a 1-2 hr digestion in dispase and collagenase I. (Step 5) Separate the cells from the fibrous tissue by pulse centrifugation and/or washing them through a cell strainer. Collect and resuspend cells in culture medium and placed into culture flasks. Please click here to view a larger version of this figure.
Figure 1: Summary of placental MSC isolation procedure. (Step 1) Orientate yourself with the placenta anatomy. (Step 2) Manually dissect 10 g of tissue from either the decidua, chorionic villi or the chorionic plate using scissors. (Step 3) Mince the dissected portions of decidua, chorionic villi or the chorionic plate tissues into fine pieces with scissors or a scalpel. (Step 4) Liberate cells from the fine pieces via a 1-2 hr digestion in dispase and collagenase I. (Step 5) Separate the cells from the fibrous tissue by pulse centrifugation and/or washing them through a cell strainer. Collect and resuspend cells in culture medium and placed into culture flasks. Please click here to view a larger version of this figure.
Mesenchymal stromal cells (MSC) will be selected based on their propensity for plastic adherence and capacity to survive and proliferate in the culture medium. Finally, in the results section, the expanded MSC can be characterized and stored for use in future experiments.
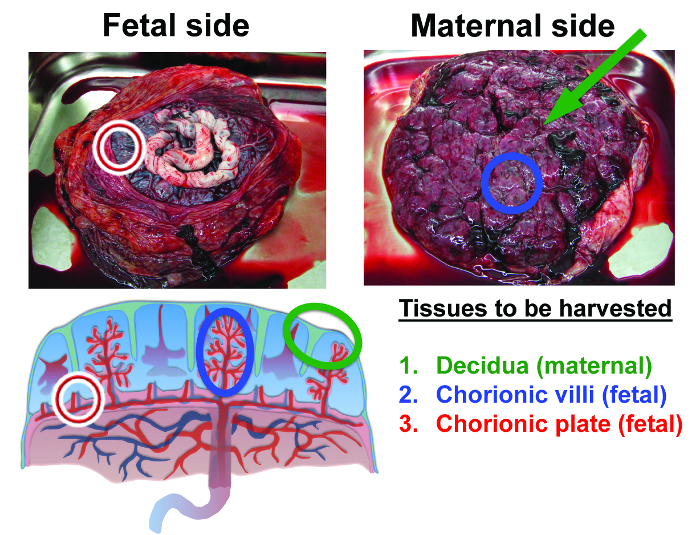 Figure 2: Anatomy of the human term placenta and tissues isolated in this procedure. The first tissue to be harvested is maternal decidua. Decidua is tissue that remains as a thin layer on the surface of the placenta after it is shed from the uterine wall (decidua is identified by the green markers). The second tissue that will be harvested from the interior of the placenta is the fetal chorionic villi (blue markers). The third tissue to be harvested is fetal chorionic plate (red markers) (adapted from reference 36). Please click here to view a larger version of this figure.
Figure 2: Anatomy of the human term placenta and tissues isolated in this procedure. The first tissue to be harvested is maternal decidua. Decidua is tissue that remains as a thin layer on the surface of the placenta after it is shed from the uterine wall (decidua is identified by the green markers). The second tissue that will be harvested from the interior of the placenta is the fetal chorionic villi (blue markers). The third tissue to be harvested is fetal chorionic plate (red markers) (adapted from reference 36). Please click here to view a larger version of this figure.
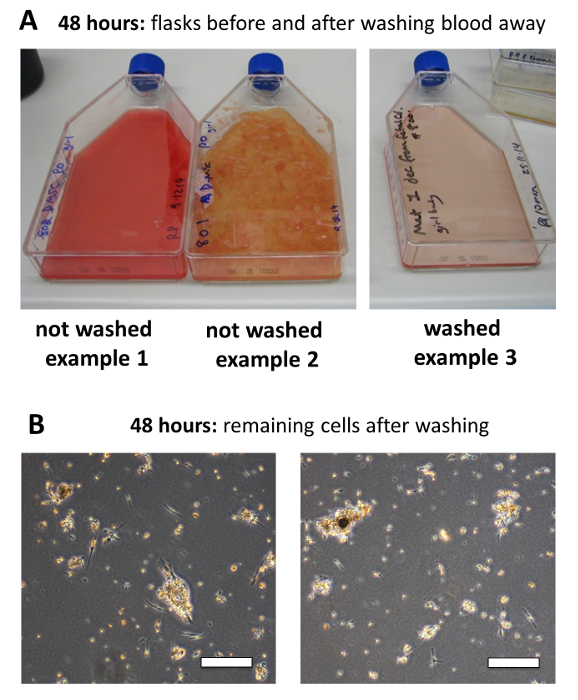 Figure 3: Morphology of cultures 48 hr after isolation and removing the tissue debris. (A) The appearance of the culture supernatant can vary substantially between placenta donors before washing off the debris. Example cultures 1 and 2 demonstrate this variation. These two isolations were performed simultaneously, but from two different placentas. Once washed cultures will be clear of RBCs and tissue debris as shown in example 3. The subsequent expansion results are generally consistent. (B) Following the 48 hr media exchange, only a few cells will be attached to the flask. Scale bar = 200 μm. Please click here to view a larger version of this figure.
Figure 3: Morphology of cultures 48 hr after isolation and removing the tissue debris. (A) The appearance of the culture supernatant can vary substantially between placenta donors before washing off the debris. Example cultures 1 and 2 demonstrate this variation. These two isolations were performed simultaneously, but from two different placentas. Once washed cultures will be clear of RBCs and tissue debris as shown in example 3. The subsequent expansion results are generally consistent. (B) Following the 48 hr media exchange, only a few cells will be attached to the flask. Scale bar = 200 μm. Please click here to view a larger version of this figure.
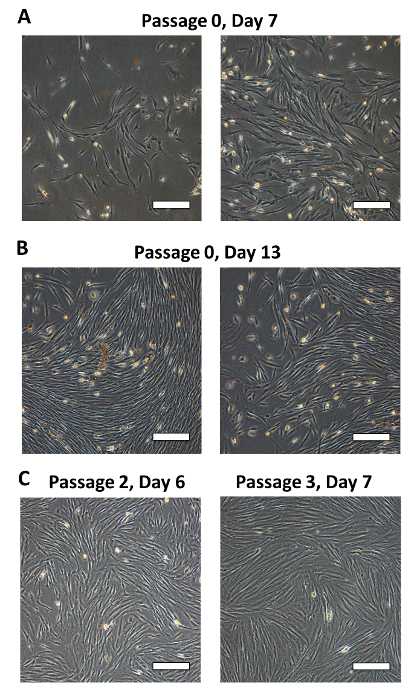 Figure 4: Morphology of MSC cultures over time. (A) Seven days after isolation, small fibroblastic MSC colonies are visible although non-MSC cells will also be present as round or loosely attached cells.(B) 13 days after isolation, fibroblastic MSC colonies are large and often the monolayer of MSC is confluent and ready to passage. (C) From passage 2 onwards, the MSC monolayer will develop a characteristic whirlpool-like morphology at confluence. Scale bar = 200 μm. Please click here to view a larger version of this figure.
Figure 4: Morphology of MSC cultures over time. (A) Seven days after isolation, small fibroblastic MSC colonies are visible although non-MSC cells will also be present as round or loosely attached cells.(B) 13 days after isolation, fibroblastic MSC colonies are large and often the monolayer of MSC is confluent and ready to passage. (C) From passage 2 onwards, the MSC monolayer will develop a characteristic whirlpool-like morphology at confluence. Scale bar = 200 μm. Please click here to view a larger version of this figure.
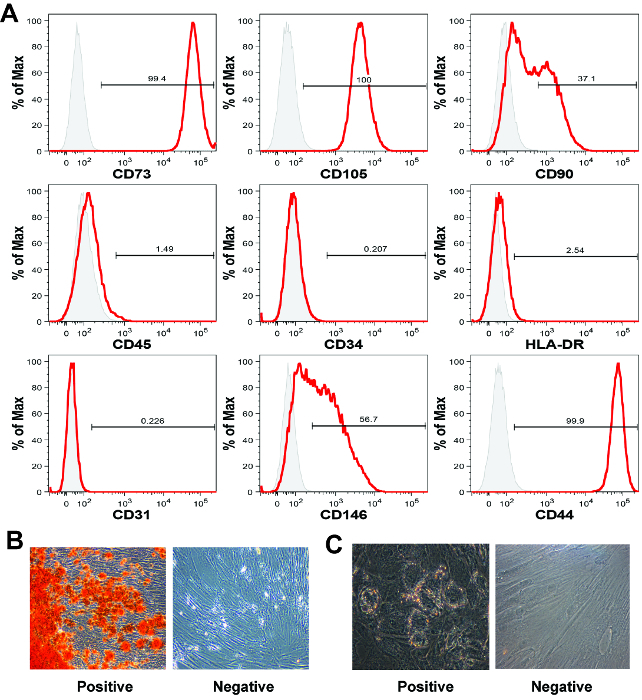 Figure 5: MSC characterization by flow cytometry and mesodermal differentiation. (A) The placental chorionic villi-derived MSC display a classic MSC marker profile by flow cytometry analysis, although for these cell surface markers, expression is similar for all types of human MSC. Each histogram shows the signal intensity (x-axis) versus the normalized cell count on the y-axis (% of Max). In this representative data set the cells were positive for CD73, CD105 and CD44, and negative for the hematopoietic markers CD45 and CD34 and HLA-DR. (B) Placental MSC generally undergo robust osteogenic differentiation, however (C) adipogenic differentiation can be less efficient than with bone marrow-derived MSC. Images in captions B and C were taken at 40X magnification. Please click here to view a larger version of this figure.
Figure 5: MSC characterization by flow cytometry and mesodermal differentiation. (A) The placental chorionic villi-derived MSC display a classic MSC marker profile by flow cytometry analysis, although for these cell surface markers, expression is similar for all types of human MSC. Each histogram shows the signal intensity (x-axis) versus the normalized cell count on the y-axis (% of Max). In this representative data set the cells were positive for CD73, CD105 and CD44, and negative for the hematopoietic markers CD45 and CD34 and HLA-DR. (B) Placental MSC generally undergo robust osteogenic differentiation, however (C) adipogenic differentiation can be less efficient than with bone marrow-derived MSC. Images in captions B and C were taken at 40X magnification. Please click here to view a larger version of this figure.
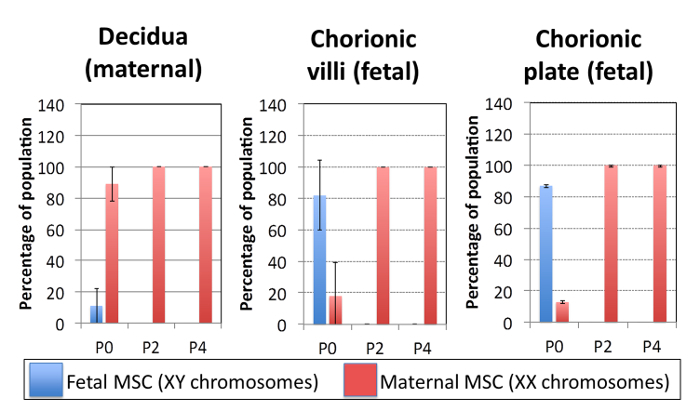 Figure 6: Placental MSC are maternal in origin using this method of culture, despite the anatomical location of the starting material. The plots show quantification of the fetal (male, XY) and maternal (female, XX) cell composition of the placental MSC cultures isolated from decidual, chorionic villi and chorionic plate tissues at every second passage. Maternal cells (XX) rapidly and reproducibly take over the cultures derived from the fetal chorionic tissues. Fetal = male = XY chromosomes detected in an individual cell, maternal = female = XX chromosomes detected in an individual cell. Data presented here was from N = 3 independent donor placentas from male babies, with a minimum of 100 cells stained for XY FISH and counted for each data point. Bars represent averages, and error bars reflect one standard deviation. Please click here to view a larger version of this figure.
Figure 6: Placental MSC are maternal in origin using this method of culture, despite the anatomical location of the starting material. The plots show quantification of the fetal (male, XY) and maternal (female, XX) cell composition of the placental MSC cultures isolated from decidual, chorionic villi and chorionic plate tissues at every second passage. Maternal cells (XX) rapidly and reproducibly take over the cultures derived from the fetal chorionic tissues. Fetal = male = XY chromosomes detected in an individual cell, maternal = female = XX chromosomes detected in an individual cell. Data presented here was from N = 3 independent donor placentas from male babies, with a minimum of 100 cells stained for XY FISH and counted for each data point. Bars represent averages, and error bars reflect one standard deviation. Please click here to view a larger version of this figure.
Discussion
The placenta is a physically large fetomaternal organ, from which fetal or maternal MSC can be isolated 22. Herein, we provided a detailed overview of placental anatomy and instruction on how to specifically dissect decidua (maternal), chorionic villi (fetal), and chorionic plate (fetal) tissue (Step 2.2-2.4). Subsequently, we outlined a robust protocol that enables MSC isolation from each of these three tissues (Step 2.5-2.6). Placental MSC expansion is efficient and cultures appear similar to bone marrow-derived MSC cultures (Figures 3 and 4). Osteogenic differentiation is reliable (Figure 5B), while adipogenic differentiation is generally less efficient (Figure 5C) 5.
Many newcomers to the field will presume that cultures derived from fetal chorionic villi or chorionic plate tissues will be enriched for fetal MSC. However, in our hands fetal cell enrichment is only transient when standard MSC expansion medium such as DMEM-LG + 10% FBS is utilized 5. Here we provide representative results using placental tissues derived from a male baby. By using placenta tissue from a male baby, the fetal cells are readily identifiably as having XY chromosomes, while maternal cells are identifiable as having XX chromosomes. Figure 6 shows XY FISH results for a representative culture. While fetal MSC (XY) are enriched (up to 80%) in the initial cultures derived from fetal chorionic villi or chorionic plate tissues, these same cultures are rapidly overtaken (~100%) by maternal (XX) MSC over the first two passages. In standard medium, composed of DMEM-LG + 10% FBS, the few maternal-derived MSC that contaminate the fetal tissues outcompete the fetal-derived cells in culture.
A critical step outlined in this protocol is an appreciation of the placental anatomy and from where fetal and maternal tissue can be most effectively harvested. As outlined in the representative results section, dissection of fetal tissue does enable transient enrichment for fetal-derived MSC. Improvements in expansion medium formulation, through specific exogenous growth factor medium supplementation should allow selective expansion of the fetal-derived MSC populations, and manufacture a cell product that is enriched for fetal rather than maternal cells (our group is currently developing such medium formulations). The manufacture of fetal MSC populations may have a number of advantages, as fetal MSC are purported to have greater angiogenesis and immunosuppressive properties than equivalent maternal MSC populations 37.
In each of the described isolation protocols, we used approximately 10 g of tissue. A whole placenta is typically 500-750 g, and in previous work we demonstrated that through automated tissue digest and a cell expansion bioreactor processes that it should be possible to manufacture over 7,000 clinical cell doses from a single placenta 4. These numbers highlight the potential suitability of placental-derived MSC in allogeneic MSC therapies, and the significance of this method regardless of MSC origin (fetal or maternal). From a therapeutic perspective, it is most critical that users have a full understanding of the cell product and the capacity to reliably manufacture this cell product. We hope that our video will assist researchers to understand placenta anatomy, isolate MSC from placenta, and anticipate the likely fetal or maternal cell composition of their cultures.
Disclosures
The authors have nothing to disclose.
Acknowledgments
RP was supported by a National Health and Medical Research Council (NHMRC) Postdoctoral Training Fellowship. VS was supported by a University of Queensland International Postgraduate Student scholarship. MRD was supported by the NHMRC and Inner Wheel Australia.
We thank clinical and nursing staff for assisting in patient consent and sample collection. We thank Prof. Nickolas Fisk, Prof. Kerry Atkinson and Dr Rohan Lourie for insightful discussions in obstetrics, feto-placental development and placenta anatomy.
References
- Brooke G, et al. Therapeutic applications of mesenchymal stromal cells. Semin Cell dev Biol. 2007;18:846–858. doi: 10.1016/j.semcdb.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Wei X, et al. Mesenchymal stem cells: a new trend for cell therapy. Acta Phamacol Sin. 2013;34:747–754. doi: 10.1038/aps.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, et al. Immunobiology of mesenchymal stem cells. Cell Death Differ. 2014;21:216–225. doi: 10.1038/cdd.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmins NE, et al. Closed system isolation and scalable expansion of human placental mesenchymal stem cells. Biotechnol Bioeng. 2012;109:1817–1826. doi: 10.1002/bit.24425. [DOI] [PubMed] [Google Scholar]
- Barlow S, et al. Comparison of human placenta- and bone marrow-derived multipotent mesenchymal stem cells. Stem Cell Dev. 2008;17:1095–1107. doi: 10.1089/scd.2007.0154. [DOI] [PubMed] [Google Scholar]
- Jones BJ, Brooke G, Atkinson K, McTaggart SJ. Immunosuppression by placental indoleamine 2,3-dioxygenase: a role for mesenchymal stem cells. Placenta. 2007;28:1174–1181. doi: 10.1016/j.placenta.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Parolini O, et al. Toward cell therapy using placenta-derived cells: disease mechanisms, cell biology, preclinical studies, and regulatory aspects at the round table. Stem Cell Dev. 2010;19:143–154. doi: 10.1089/scd.2009.0404. [DOI] [PubMed] [Google Scholar]
- Benirschke K, Burton GJ, Baergen RN. Pathology of the Human Placenta. 2012. 6 edn.
- Moore K, Persaud T, Torchia M. The developing human: clinically oriented embryology. Elsevier/ Saunders; 2013. [Google Scholar]
- Parolini O, et al. Concise review: isolation and characterization of cells from human term placenta: outcome of the first international Workshop on Placenta Derived Stem Cells. Stem cells. 2008;26:300–311. doi: 10.1634/stemcells.2007-0594. [DOI] [PubMed] [Google Scholar]
- Brooke G, et al. Manufacturing of human placenta-derived mesenchymal stem cells for clinical trials. Brit J Haematol. 2009;144:571–579. doi: 10.1111/j.1365-2141.2008.07492.x. [DOI] [PubMed] [Google Scholar]
- Pelekanos RA, et al. Intracellular trafficking and endocytosis of CXCR4 in fetal mesenchymal stem/stromal cells. BMC cell biology. 2014;15:15. doi: 10.1186/1471-2121-15-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YS, et al. Small molecule mesengenic induction of human induced pluripotent stem cells to generate mesenchymal stem/stromal cells. Stem Cells Transl Med. 2012;1:83–95. doi: 10.5966/sctm.2011-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood CF, et al. High incidence of contaminating maternal cell overgrowth in human placental mesenchymal stem/stromal cell cultures: a systematic review. Stem Cells Transl Med. 2014;3:1305–1311. doi: 10.5966/sctm.2014-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn P, Bork S, Wagner W. Standardized isolation of human mesenchymal stromal cells with red blood cell lysis. Methods Mol Biol. 2011;698:23–35. doi: 10.1007/978-1-60761-999-4_3. [DOI] [PubMed] [Google Scholar]
- Dominici M, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- Friedenstein AJ, et al. Precursors for fibroblasts in different populations of hematopoietic cells as detected by the in vitro colony assay method. Exp Hematol. 1974;2:83–92. [PubMed] [Google Scholar]
- Caplan AI. Mesenchymal stem cells. J Orthop Res. 1991;9:641–650. doi: 10.1002/jor.1100090504. [DOI] [PubMed] [Google Scholar]
- Futrega K, et al. The microwell-mesh: A novel device and protocol for the high throughput manufacturing of cartilage microtissues. Biomaterials. 2015;62:1–12. doi: 10.1016/j.biomaterials.2015.05.013. [DOI] [PubMed] [Google Scholar]
- Liu Y, Goldberg AJ, Dennis JE, Gronowicz GA, Kuhn LT. One-step derivation of mesenchymal stem cell (MSC)-like cells from human pluripotent stem cells on a fibrillar collagen coating. PloS one. 2012;7:33225. doi: 10.1371/journal.pone.0033225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleki M, Ghanbarvand F, Reza Behvarz M, Ejtemaei M, Ghadirkhomi E. Comparison of mesenchymal stem cell markers in multiple human adult stem cells. Int J Stem Cell. 2014;7:118–126. doi: 10.15283/ijsc.2014.7.2.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass R, Kasper C, Bohm S, Jacobs R. Different populations and sources of human mesenchymal stem cells (MSC): A comparison of adult and neonatal tissue-derived MSC. Cell Commun Signal. 2011;9:12. doi: 10.1186/1478-811X-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P, Robey PG, Simmons PJ. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell stem cell. 2008;2:313–319. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P, et al. The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine. Nature med. 2013;19:35–42. doi: 10.1038/nm.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva Meirelles L, Caplan AI, Nardi NB. In search of the in vivo identity of mesenchymal stem cells. Stem cells. 2008;26:2287–2299. doi: 10.1634/stemcells.2007-1122. [DOI] [PubMed] [Google Scholar]
- Caplan AI, Correa D. The MSC: an injury drugstore. Cell Stem Cell. 2011;9:11–15. doi: 10.1016/j.stem.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmeyer H, et al. Mesenchymal Stromal Cell Characteristics Vary Depending on Their Origin. Stem Cell Dev. 2013. [DOI] [PMC free article] [PubMed]
- Guillot PV, Gotherstrom C, Chan J, Kurata H, Fisk NM. Human first-trimester fetal MSC express pluripotency markers and grow faster and have longer telomeres than adult MSC. Stem cells. 2007;25:646–654. doi: 10.1634/stemcells.2006-0208. [DOI] [PubMed] [Google Scholar]
- Covas DT, et al. Multipotent mesenchymal stromal cells obtained from diverse human tissues share functional properties and gene-expression profile with CD146+ perivascular cells and fibroblasts. Exp Hemol. 2008;36:642–654. doi: 10.1016/j.exphem.2007.12.015. [DOI] [PubMed] [Google Scholar]
- Crisan M, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell stem cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Pelekanos RA, et al. Comprehensive transcriptome and immunophenotype analysis of renal and cardiac MSC-like populations supports strong congruence with bone marrow MSC despite maintenance of distinct identities. Stem cell res. 2012;8:58–73. doi: 10.1016/j.scr.2011.08.003. [DOI] [PubMed] [Google Scholar]
- da Silva Meirelles L, Chagastelles PC, Nardi NB. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci. 2006;119:2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- James AW. Review of Signaling Pathways Governing MSC Osteogenic and Adipogenic Differentiation. Scientifica (Cairo) 2013;2013:684736. doi: 10.1155/2013/684736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, et al. Cross-Talking between PPAR and WNT Signaling and Its Regulation in Mesenchymal Stem Cell Differentiation. Curr Stem Cell Res Ther. 2015. [DOI] [PubMed]
- Zhuang H, et al. Molecular Mechanisms of PPAR-gamma Governing MSC Osteogenic and Adipogenic Differentiation. Curr Stem Cell Res Ther. 2015. [DOI] [PubMed]
- Moore K, Persaud T, Torchia M. Before We Are Born: Essentials of Embryology and Birth Defects. Elsevier Health Sciences; 2011. 8 edn. [Google Scholar]
- Zhu Y, et al. Placental mesenchymal stem cells of fetal and maternal origins demonstrate different therapeutic potentials. Stem Cell Res Ther. 2014;5:48. doi: 10.1186/scrt436. [DOI] [PMC free article] [PubMed] [Google Scholar]


