Abstract
The mechanism underlying impaired learning and memory in Alzheimer’s disease is not fully elucidated. The phosphorylation of cyclic-AMP response element binding protein (pCREB) in the hippocampus is thought to be a critical initiating step in the formation of long-term memories. Here, we tested CRE-driven gene expression following learning in mice harboring the familial Alzheimer’s disease-linked APPswe/PS1ΔE9 mutations using CRE-β galactosidase reporter. We show that young adult APPswe/PS1ΔE9 mice exhibit impaired recognition memory and reduced levels of pCREB, and its cofactors CREB binding protein (CBP) and p-300 following a learning task, compared to their wild type littermate counterparts. Impairments in learning-induced activation of CREB in these mice are manifested by reduced CRE-driven gene transcription. Importantly, expression of the CRE-driven immediate early gene, Egr-1 (Zif268) is decreased in the CA1 region of the hippocampus. These studies implicate defective CREB-dependent plasticity in the mechanism underlying learning and memory deficits in Alzheimer’s disease.
Keywords: Alzheimer’s disease, CBP, CRE, CREB, hippocampal plasticity, learning and memory
INTRODUCTION
Cognitive decline and deficits in learning and memory are the primary characteristics of Alzheimer’s disease (AD), a progressive neurodegenerative disease. The mechanisms underlying these cognitive deficits are not fully understood. cAMP response element binding protein (CREB) is a key transcription factor involved in learning and memory (for review see [1]). Following neuronal stimulation, CREB phosphorylation at Ser133 induces the transcription of genes that play a crucial role in the initiation of long-term memory [2] and in the consolidation and retrieval of spatial and fear memories [3]. CREB binding protein (CBP) and p300 are CREB’s transcriptional coactivator paralogs that are recruited following CREB phosphorylation at Ser133 and are necessary for the initiation of gene transcription. CBP deficient mice show a strong defect in enriched environment (EE)-induced neurogenesis as well as impairments in EE- mediated enhancement of spatial navigation and pattern separation [4]. Impaired CREB activation has detrimental effects on learning and memory [2], including compromised spatial reference memory [5], a type of memory known to be highly impaired in AD patients. Levels of CREB and pCREB are reduced in the aging human brain, as well as in postmortem brain samples of AD patients[6, 7]. Likewise, level and activity of Type I adenylyl cyclase is reduced in AD [8, 9]. These results suggest that a decrease in CREB signaling may play a role in AD [7, 10, 11]. Nevertheless, information in mouse models of familial AD (FAD) is controversial [12–15]. Specifically, studies in FAD mouse models expressing mutant forms of amyloid-β protein precursor (AβPP) and/or presenilin (PS) reveal loss of CREB function and decreased levels of pCREB [12, 13, 16–19]. Reduced CREB levels were attributed to Aβ toxicity [20–23]. However, other studies report increased CREB expression and activity, leading to a gain of transcriptional function in PS1 mutant variants, Tg2576 mice and in the 3XTg-AD mice [13, 21, 24]. Here we show that steady state levels of pCREB and its coactivators, CREB binding protein (CBP) and p-300, were reduced in FAD-linked APPswe/PS1ΔE9 mice compared to wild type littermates. Importantly, following learning, pCREB and CBP levels were significantly lower in the hippocampus of APPswe/PS1ΔE9 mice compared to their wild type littermates. Moreover, impaired performance of APPswe/PS1ΔE9 mice in a contextual fear-conditioning test correlated with lower levels of CBP and pCREB when compared to wild type mice. Using a reporter mouse co-expressing CRE-LacZ and APPswe/PS1ΔE9 mice (CRE-AD), we show that following learning or novel experience, levels of β-galactosidase in the hippocampus of CRE-AD mice were dramatically lower when compared to levels in the CRE/wild type counterparts. Finally, levels of the CRE-driven immediate early gene Zif268 (Egr-1) were not increased in the hippocampus of APPswe/PS1ΔE9 mice following learning tasks as they were in their wild type littermates. Taken together, these results strongly suggest that impairments in CREB signaling may underlie learning and memory impairments in FAD.
METHODS
Mice
All animal experiments were approved by the University of Illinois at Chicago Institutional Animal Care and Use Committee (IACUC). APPswe/PS1ΔE9 have been previously described [25]. CRE-AD mice were generated by breeding a CRE-LacZ male transgenic mouse (previously described [26] and generously provided by Dr. Daniel Storm, University of Washington) with a female APPswe/PS1ΔE9 mouse. Animals were maintained in standard conditions (14/10 h light/dark cycle) with full access to food and water ad libitum. Female APPswe/PS1ΔE9 and their non-transgenic littermates were used for fear conditioning experiments. CRE and CRE-AD mice were used for novel object recognition experiments. Each mouse was used for only a single behavioral task and then immediately sacrificed. To control for time-dependent expression of CREB signaling, the time between the test and sacrifice was recorded and mice were time-matched between genotype for comparison.
Environmental enrichment
Two- to three-month old mice were either exposed to an enriched environment for one week or were maintained in standard housing conditions as described previously [27] with the following modification: the mice remained in the cage for an entire week, and were removed immediately prior to sacrifice.
Contextual fear conditioning
Three-month-old APPswe/PS1ΔE9 or nontransgenic female mice were placed into a 17.8 × 17.8 × 30.5 cm chamber with two clear Plexiglass walls, two metal walls, and a stainless steel grid floor (Coulbourn Instruments). The chamber was located inside an opaque isolation cabinet. The mice remained in the cage for 148 s before receiving a single 0.75 mA foot shock lasting 2 s. The mice remained in the cage for another 30 s after the shock before being returned to their home cage. 24 h later the mice were returned to the chamber for 5 min. Freezing was measured using a digital video camera mounted above the chamber and FreezeFrame software (Actimetrics). Percentage freezing time was analyzed using FreezeView software (Actimetrics). For statistical analysis, a two tailed, paired t-test was used ( *p < 0.05).
Novel object recognition
Novel object recognition was performed as previously described [28]. Three-month-old female CRE-LacZ, APPswe/PS1ΔE9 and CRE-LacZ/APPswe/PS1ΔE9 mice were habituated to the experimental box (24 × 17 × 11 inches) for 5 min on two consecutive days. On the third day the mice were familiarized to two identical objects placed in the center of the box for 5 min. On the fourth day the mice were placed in a box with one familiar object and one novel object. The mice were recorded by video on the familiarization and novel object days. The videos were analyzed by counting the number of frames the mice spent exploring each object. “Exploring” was defined as pointing the nose at the object or touching the object with the front paws. The time was determined by the frame rate of the video. Mice that explored for fewer than 7 s on either the familiarization or novel object day were excluded from the analysis. Statistical analysis was performed by a two-tailed, paired t-test ( *p < 0.05).
Nuclear protein extraction and western blot
Nuclear protein from mouse hippocampus was extracted using CelLytic NuCLEAR Extraction Kit (Sigma NXTRACT) per the manufacturer’s instructions. The sample preparations were supplemented with protease and phosphatase inhibitors (2 mM sodium orthovanadate, 50 nM RR microcystin, 50 nM okadaic acid, 100 nM K52a, 100 nM staurosporine, 1 μL/100 μL buffer mammalian protease inhibitor cocktail II (Sigma P8430), 1 μL/100 μL buffer potassium phosphatase cocktail II (Milipore 524625), 1 mM PMSF). The concentration of protein was determined by BCA assay (ThermoScientific). The samples were run on a 5% (CBP and p-300) or 7.5% (all other proteins) Tris-glycine gel and transferred to a 0.45 μM nitrocellulose membrane (Biorad) at 100 mV for 2 h. Membranes were blocked for 1 h in 5% milk in TBST (TBS +0.1% Tween-20) supplemented with the phosphatase inhibitors 1 mM sodium orthovanadate and 10 mM sodium fluoride. The primary antibodies (pCREB 1:1000 Cell Signaling, Total CREB 1:1000 Cell Signaling, CBP 1:2000 Santa Cruz, p-300 1:1000 Santa Cruz, PKA 1:1000 Santa Cruz, PKC 1:1000 Santa Cruz, CaMKIV 1:500 eBioscience, pERK 1:1000 Santa Cruz, Total ERK 1:1000 Santa Cruz, Lamin B 1:500 Abcam) were incubated at 4°C overnight with shaking in 2% BSA in TBST with phosphatase inhibitors. The membranes were rinsed 3 times for 15 min each time in TBS-T with phosphatase inhibitors and then placed in the appropriate secondary (Rabbit HRP 1:10,000 Promega) for 2 h at room temperature with shaking. The membranes were rinsed again 3 times for 15 min each time and then imaged using an ECL chemiluminesence substrate (GE Healthcare). Relative levels of protein were quantified using densiometric measurements from ImageJ software. For statistical analysis, a two-tailed, unpaired t-test was used. The ratio of phosphorylated protein to total protein was determined for each mouse followed by averaging the ratios for each group.
PKA activity assay
The PKA kinase activity assay was performed according to the manufacturer’s instructions (Enzo) using 0.2 μg of total hippocampal protein. Absorbance was measured at 450 nm on a DTX 880 MultimodeDetector (Beckman Coulter). For statistical analysis, a two-tailed, unpaired t-test was used.
X-gal staining
Mice were perfused with ice cold PBS, and the right hemisphere fixed in 4% PFA for 4 h. The hemispheres were then transferred to a 30% sucrose solution for 16 h, after which they were frozen in OCT and stored at –80°C. Sections of 20 μm thickness were prepared using a cryostat. X-gal staining was performed as previously described [29]. Briefly, slides were washed for 30 min twice at room temperature in a PBS buffer containing 2 mM MgCl2, followed by a 30 min wash at room temperature in a PBS buffer containing 2 mM MgCl2, 0.02% Nonidet P40, and 0.01% sodium deoxycholate. The slides were then incubated overnight at 37°C in a PBS buffer containing 2 mM MgCl2, 0.02% Nonidet P40, 0.01% sodium deoxycholate, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, and 0.6 mg/mL 5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside (X-gal) (Sigma B4252). Slides were then rinsed in PBS, and mounted inPVA-DABCO.
Immunofluorescent staining
For immunofluorescent staining, 50 μm floating sagittal sections were prepared on a microtome (Leica SM2000 R). The slides were blocked in TBS containing 5% Normal Donkey Serum (Jackson) and 0.25% Triton X-100. The sections were then incubated with primary antibodies pCREB (1:400 Cell Signaling 9197) or Egr-1 (1:250 Santa Cruz) at 4°C for 72 h with gentle shaking. The sections were rinsed 3 times in TBS for 5 min each and then incubated with donkey anti-rabbit Cy3 (1:250 Jackson Immunoresearch) for 2 h at room temperature in the dark. The sections were rinsed in TBS 3 times for 5 min each. Finally, the sections were counterstained for DAPI, then mounted using an aqueous mounting medium (PVA-DABCO) and stored at 4°C. Sections were imaged on a Ziess LSM 510 confocal microscope. For quantification of Egr-1 immunoflourescence, the area of interest (CA1 region) was traced in ImageJ software and the mean intensity was quantified. The mean intensity of the background fluorescence was also measured and subtracted from the value for the region of interest. For statistical analysis, an unpaired, two-tailed t-test was used ( *p < 0.05).
RESULTS
Basal levels of CREB signaling components are reduced in the nuclear fraction of the hippocampus of APPswe/PS1ΔE9 mice
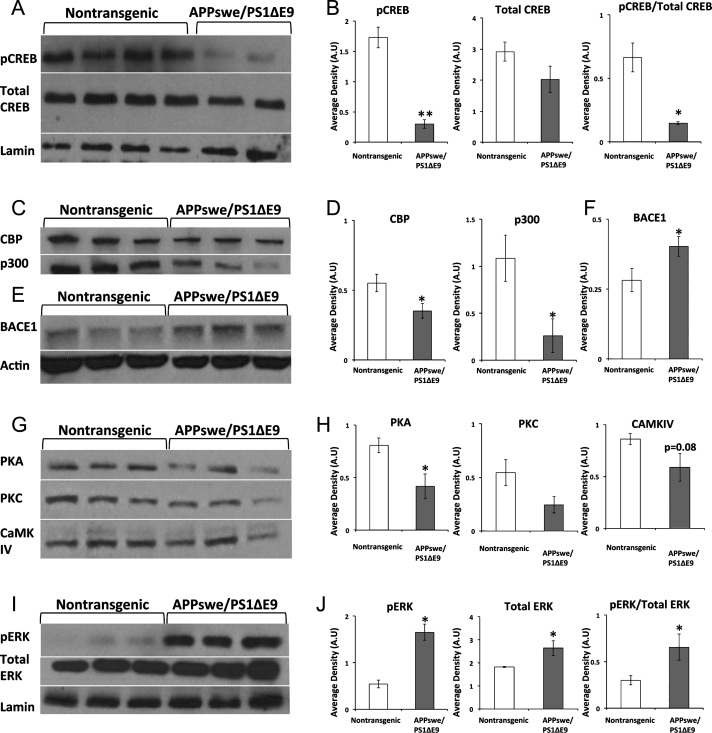
First, we examined whether basal levels of CREB and pCREB were altered in the hippocampus of young adult APPswe/PS1ΔE9 mice. Using Western blot analysis of nuclear fractions prepared from hippocampal protein extract of 3-month-old APPswe/PS1ΔE9 mice, we observed that expression levels of total CREB were comparable in the hippocampus of APPswe/PS1ΔE9 and wild type mice (Fig. 1A-B). However, levels of pCREB were reduced in the hippocampus of APPswe/PS1ΔE9 compared to wild type mice. Next, we asked whether CREB’s transcriptional coactivator paralogs, CBP and p-300, were also altered. We observed that protein levels of both CBP and p-300 were reduced in the hippocampus of APPswe/PS1ΔE9 mice (Fig. 1C-D). These results indicate a reduction in the availability of critical CREB signaling components in the hippocampus of adult APPswe/PS1ΔE9 mice. To start to understand the mechanism underlying these deficits, we first examined BACE1 levels in the brains of these mice. Elevated BACE1 levels have been shown to reduce CREB phosphorylation, PKA activity and cAMP levels [17]. Levels of BACE1 were significantly increased in the hippocampus of APPswe/PS1ΔE9 compared to wild type mice (Fig. 1E-F). Next, we asked whether reduced levels of CREB phosphorylation could result from alterations in the kinases that phosphorylate CREB. Although a multitude of kinases phosphorylate CREB in vitro, thus far, only protein kinase A (PKA), protein kinase C (PKC), the calcium/calmodulin-dependent protein kinases CaMKII and CaMKIV, the mitogen- and stress-activated protein kinase (MSK), and the90 kDa ribosomal S6 kinase (RSK) have been shown to activate CREB-dependent transcription in vivo [30]. Levels of PKA were significantly reduced in the hippocampus of APPswe/PS1ΔE9 compared to wild type mice (Fig. 1G-H).
Fig.1.
CREB signaling components are differentially expressed in the nuclear fraction of the hippocampus of APPswe/PS1ΔE9 mice.A) Representative blot of pCREB and Total CREB in 3-month-old female non-transgenic (n = 8) and APPswe/PS1ΔE9 (n = 3) mice. B) ImageJ analysis of pCREB normalized to Lamin (p = 0.001), total CREB normalized to Lamin (p = 0.150), ratio of pCREB/Total CREB (p = 0.025). C) Representative blot of CBP and p300 in 3-month-old female non-transgenic (n = 4) and APPswe/PS1ΔE9 mice (n = 3). D) ImageJ analysis of CBP normalized to Lamin (p = 0.04), p300 normalized to Lamin (p = 0.05). E) Representative blot of BACE1 expression in total hippocampal homogenate in 3-month-old non-transgenic (n = 8) and APPswe/PS1ΔE9 (n = 6) mice. F) ImageJ analysis of BACE1 normalized to actin (p = 0.05). G) Representative blot of PKA, PKC, and CaMKIV in 3-month-old female non-transgenic (n = 4) and APPswe/PS1ΔE9 (n = 3) mice. H) ImageJ analysis of PKA normalized to Lamin (p = 0.03), PKC normalized to Lamin (p = 0.1), CaMKIV normalized to Lamin (p = 0.08).I) Representative blot of pERK and Total ERK in 3-month-old female non-transgenic (n = 4) and APPswe/PS1ΔE9 (n = 3) mice. J) ImageJ analysis of pERK normalized to Lamin (p = 0.001), total ERK normalized to Lamin (p = 0.03), and the ratio of pERK to total ERK (p = 0.04).
Similarly, expression levels of PKC and CaMKIV were reduced in hippocampal extracts of APPswe/PS1ΔE9 compared to wild type mice, although this difference was not statistically significant (Fig. 1G-H). Next, we examined levels of ERK and pERK, which activate MSK and RSK. Interestingly, we observed that levels of pERK and Total ERK, as well as the ratio of pERK to Total ERK were significantly increased in the brains of APPswe/PS1ΔE9 compared to wild type mice (Fig. 1I-J). Taken together, these results suggest that the reduction in PKA expression, and the somewhat reduced levels of PKC and CaMKIV, may underlie the downregulated levels of pCREB. Further, enhanced ERK activation may not be sufficient to overcome the reduction in the level of the other kinases. However, it was not clear whether these alterations would result in functional deficits in learning and memory in young APPswe/PS1ΔE9 mice.
Impairments in CREB signaling are linked to deficits in associative learning in APPswe/PS1 ΔE9 mice, and are apparent in brain regions that process learning and memory
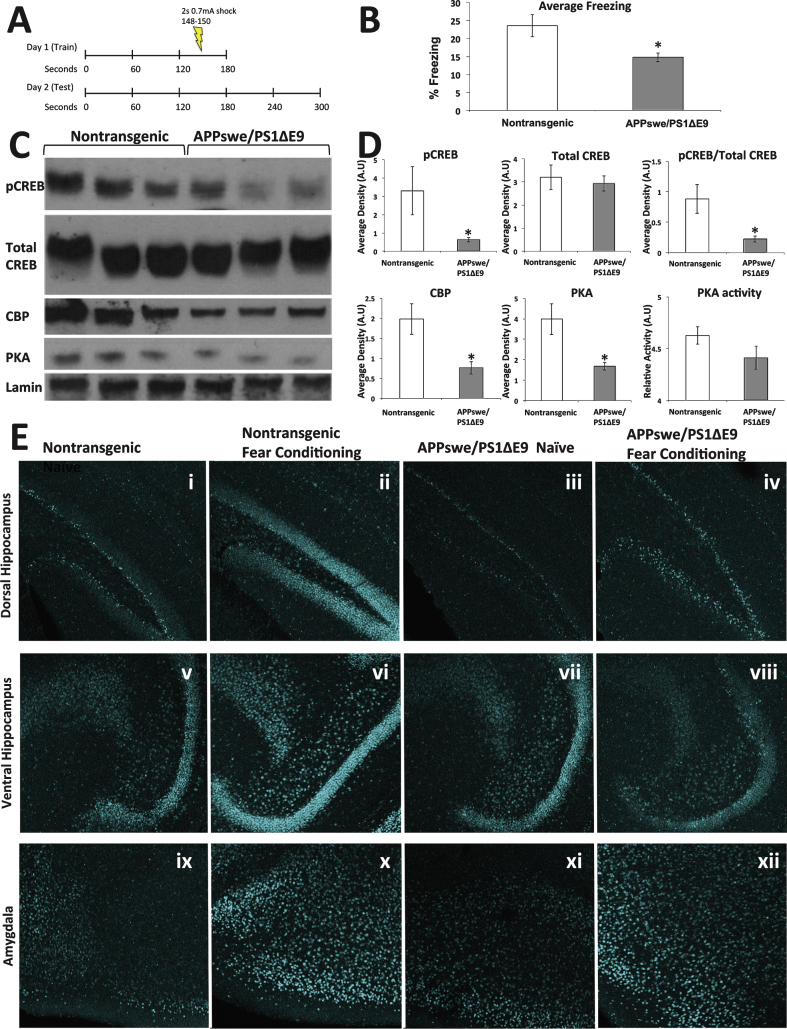
CREB and CBP deficient mice have been previously shown to exhibit impairments in contextual fear conditioning [31–33]. Thus, to associate learning ability and CREB impairment in FAD, we tested the performance of APPswe/PS1ΔE9 mice in a contextual fear conditioning paradigm and examined CREB signaling in the brain following the task (Fig. 2A). We observed no differences in mobility prior to the shock, and no differences in the magnitude of the response to the shock between the wild type and APPswe/PS1ΔE9 mice (Supplementary Fig. 1). However, wild type mice froze significantly more than APPswe/PS1ΔE9 mice when re-introduced to the context 24 h later (Fig. 2B). Analysis of the nuclear fraction of the hippocampus revealed that the APPswe/PS1ΔE9 mice, again, exhibited significantly lower levels of pCREB and CBP, as well as significantly lower levels of PKA expression and a trending reduction in PKA activity (Fig. 2C-D). To define the regional effect of fear conditioning on CREB activation, we immunolabeled brain sections of these mice. We observed that levels of pCREB were lower in the dorsal (Fig. 2E i,iii) but not ventral (Fig. 2E v, vii) hippocampus of naive APPswe/PS1ΔE9 mice compared to their naïve wild type counterparts, suggesting that compromised CREB signaling is particularly apparent in the hippocampal area thought to be involved in learning and memory, rather than mood and emotion. That said, lack of activation of pCREB following the learning task was apparent in both the dorsal (Fig. 2E ii, iv) and ventral (Fig. 2E vi, viii) hippocampus. In contrast, we could not detect any difference in either basal or learning-activated levels of pCREB expression in the amygdala between wild type and APPswe/PS1ΔE9 mice (Fig. 2E ix-xii), again supporting the hypothesis that impairments in pCREB following fear conditioning are apparent in neuronal populations implicated in explicit memory.
Fig.2.
Memory impairments in 3-month-old female APPswe/PS1ΔE9 mice correspond with deficits in hippocampal CREB signaling. A) Experimental paradigm. Long-term memory consolidation was tested by a contextual fear conditioning paradigm. Mice were placed in the box for 2 min and 28 s before receiving a 2 s shock (0.7 mA). The mice remained in the cage for another 30 s and were then removed. 24 h later the mice were placed in the cage for 5 min and average freezing time was measured. B) Average freezing for 3-month-old female mice (non-transgenic, n = 14; APPswe/PS1ΔE9, n = 11; p = 0.026). C) Representative blot of pCREB, total CREB, and CBP expression levels following contextual fear conditioning training. D) ImageJ analysis of pCREB expression normalized to Lamin B (non-transgenic, n = 7; APPswe/PS1ΔE9, n = 7; p = 0.06), Total CREB expression normalized to Lamin B (p = 0.68), the ratio of pCREB expression to Total CREB (p = 0.02), CBP expression normalized to Lamin B (p = 0.01), PKA expression normalized to Lamin (p = 0.04), and PKA activity (p = 0.14). E) pCREB expression in the ventral and dorsal hippocampus and amygdala of nontransgenic and APPswe/PS1ΔE9 mice either naïve or following fear conditioning.
Reduced CRE-driven gene transcription following environmental enrichment in APPswe/PS1ΔE9 mice.
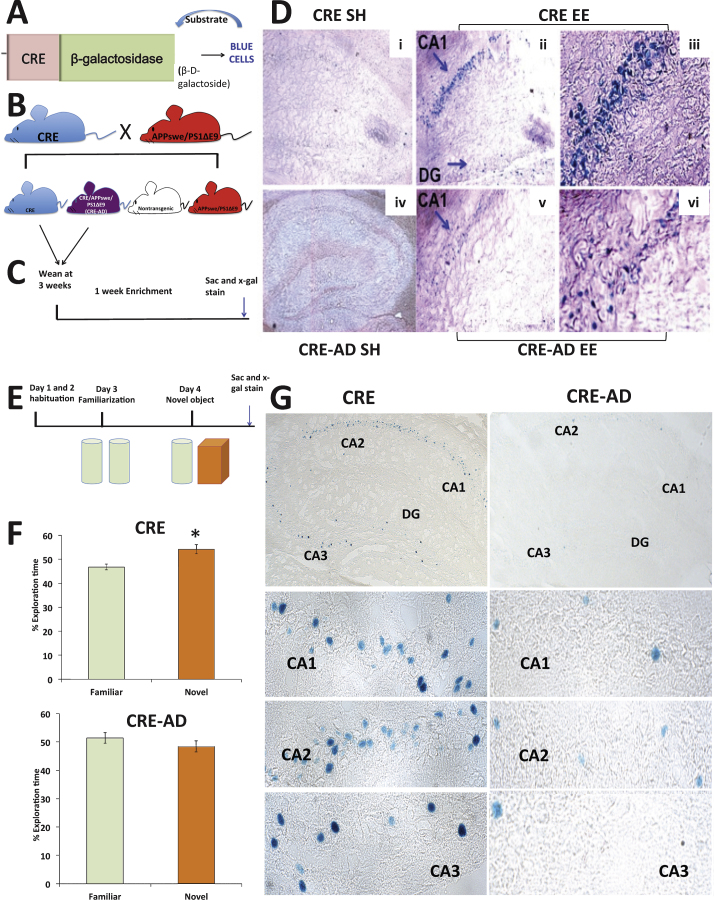
CRE-driven gene transcription is increased in the CA1 and CA3 regions of the hippocampus following fear conditioning [34]. Our observation that levels of CBP and pCREB remain lower in APPswe/PS1ΔE9 mice following learning in the fear conditioning task indicates that CREB signaling is nonresponsive to a stimulus, and suggests that the transcription of CRE-driven genes required for synaptic plasticity and long-term memory, such as Egr-1 (Zif 268), may be impaired [35–39]. To this end, we decided to use environmental enrichment (EE) for the enhancement of CRE-driven gene transcription, to get a maximal effect of experience on brain plasticity, compared to the confined effect of a specific learning task. We have previously shown that in contrast to wild type mice, pCREB is not upregulated in the hippocampus of young adult APPswe/PS1ΔE9 mice following EE [40]. Following up on this observation, we measured the amount of CBP in the nuclear fraction of the hippocampus. Indeed, we observed that following EE, CBP levels were increased in the hippocampus of young adult wild type mice (Supplementary Fig. 2A-B), but not in their APPswe/PS1ΔE9 counterparts (Supplementary Fig. 2C-D). To directly address whether CRE-driven gene transcription is impaired in FAD under these conditions, we crossed reporter CRE-LacZ mice that express β-galactosidase driven by the CRE promoter (Fig. 3A, generously provided by Dr. Daniel Storm [26, 29]) with the APPswe/PS1ΔE9 mouse (Fig. 3B), and allowed the mice to experience EE (Fig. 3 C). Abundant X-gal staining was detected throughout the hippocampus following EE in the CRE reporter mouse, particularly in the CA1 region (Fig. 3D i-iii). In contrast, X-gal positive cells in the hippocampus of the CRE-AD mice were scarce (Fig. 3D iv-vi), suggesting that EE-induced CRE-driven gene expression is defective in the APPswe/PS1ΔE9 mouse. This observation indicates that, in the APPswe/PS1ΔE9 mouse, the CREB pathway is not responding to a stimulus known to enhance neuroplasticity and memory function, resulting in reduced CRE-driven gene transcription.
Fig.3.
Novel object recognition (NOR) and CRE-driven gene expression are impaired in 3-month-old APPswe/PS1ΔE9 mice. A) Schematic of CRE-reporter. B) Breeding scheme of CRE-reporter mouse with APPswe/PS1ΔE9 mice to generate the CRE-AD mouse. C) Enrichment timeline scheme. D) CRE-gene transcription is increased following EE in CRE mice (i-iii) but is not increased following EE in CRE-AD mice (iv-vi). E) Schematic of NOR experimental design. F) CRE reporter mice significantly prefer a novel object over a familiar object (n = 9, p = 0.04), while CRE-AD mice do not show a preference for a novel object over a familiar object (n = 10, p = 0.4). G) X-gal staining in the hippocampus of CRE or CRE-AD mice following NOR (30 min post-task).
CRE- driven gene transcription is increased following a novel object recognition task in the hippocampus of CRE-LacZ mice, but not CRE-AD mice
To address the association between deficits in CREB signaling and memory impairment in a specific learning task, we asked whether CRE-driven gene transcription is impaired in the hippocampus of FAD mice following the novel object recognition task (NOR, Fig. 3E). Deficits in this task have been previously observed in CREB and CBP deficient mice [41]. For this purpose, we subjected a separate group of CRE and CRE-AD mice to NOR. We observed no difference in preference for the two similar objects on the familiarization day (Supplementary Fig. 3). However, the CRE mice significantly preferred the novel object the next day, whereas the CRE-AD mice did not (Fig. 3F). Following the task we observed a clear expression of CRE-gene transcription in the hippocampus of the CRE mice (Fig. 3 G). In the CRE-AD mice, however, CRE-gene transcription was markedly reduced (Fig. 3 G). Taken together, these experiments suggest that reduced expression of pCREB and CBP in the hippocampus is manifested by impairments in CRE-based gene transcription and may underlie deficits in hippocampus-dependent tasks.
Egr-1 expression is not increased following learning in hippocampus of APPswe/PS1ΔE9 mice
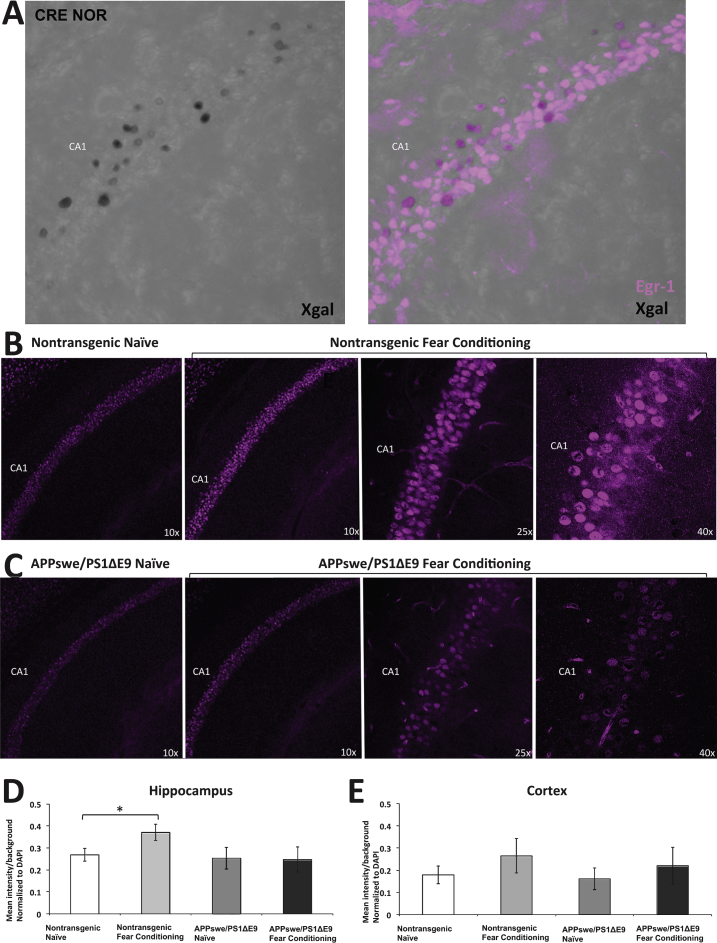
To further establish the impairment in CRE-driven transcription following stimulation, we examined the protein expression level of Egr-1, a CRE-driven immediate early gene implicated in learning-induced brain plasticity [42]. Egr-1 has been shown to be one of many CREB-dependent transcription factors important for training-induced long-term memory [43]. First, we examined Egr-1 expression following learning in the hippocampus of CRE and CRE-AD mice that were subject to the NOR task. We observed colocalization between X-gal and Egr-1 in the hippocampus (Fig. 4A). Notably, there were more cells that expressed Egr-1 than were positive for X-gal. This may be attributed to differences in activation time of the different genes following learning, as well as to the processing time of the β-galactosidase activity. Second, we examined Egr-1 expression in the hippocampus of APPswe/PS1ΔE9 or nontransgenic mice that were subject to fear conditioning. We observed that Egr-1 expression was increased following learning in the hippocampus of nontransgenic mice (Fig. 4B), but not in the hippocampus of APPswe/PS1ΔE9 mice (Fig. 4 C). Quantification of Egr-1 expression in the CA regions of the hippocampus revealed that Egr-1 was significantly increased following fear conditioning in nontransgenic mice, but not in the APPswe/PS1ΔE9 mice (Fig. 4D). No differences in Egr-1 expression were observed in the cortex (Fig. 4E). These results strongly suggest that the upregulation of Egr-1, and possibly other important CRE-driven genes, is compromised following learning in the hippocampus of APPswe/PS1ΔE9 mice, leading to deficient brain plasticity and memory formation.
Fig.4.
Egr-1 is increased in non-transgenic mice, but not in APPswe/PS1ΔE9 mice, following contextual fear conditioning. A) X-gal staining overlaps with Egr-1 staining in the CA1 region of the CRE mouse following NOR. B) Egr-1 expression in naïve non-transgenic mice (n = 4) and following fear conditioning (n = 4). C) Egr-1 expression in naïve APPswe/PS1ΔE9 mice (n = 3) and following fear conditioning (n = 3). D) ImageJ analysis average intensity of Egr-1 staining relative to background in the hippocampus. E) ImageJ analysis average intensity of Egr-1 staining relative to background in the cortex.
DISCUSSION
This study provides several novel aspects concerning the role of CREB deficits in learning and memory in FAD mouse models. First, we show that basal levels of pCREB, CBP, and p-300 are reduced in the nuclear fraction of the hippocampus of APPswe/PS1ΔE9 mice. This observation is in line with postmortem analysis of pCREB expression in the hippocampus of AD patients [7]. Lower levels of basal pCREB and CBP could indicate a reduction in the availability of these critical factors for learning and memory. Reduced levels of PKA, and possibly PKC and CaMKIV (the latter two were observed as non-significant trends), may suggest lower activity of these kinases, and, in concert with increased BACE1 activity, may play a role in reduced levels of basal pCREB. Recent evidence suggests that PKA-CREB signaling may also reduce expression of tau [44]. The negative regulation of CREB via BACE1-PKA signaling [17] provides an Aβ-independent, alternative mechanism for memory impairments in AD. BACE1 activity increases with aging and in sporadic and familial cases of AD.Perhaps increased BACE1 activity may partially account for poor cognitive performance even in the presence of relatively mild neuropathology. While PKA expression was significantly reduced, difference in PKA activity was not statistically significant. However, it is not clear what extent of activity loss is required for a significant effect on the level of pCREB. In addition, PKA activity levels in this study were measured in young adults, and may decrease further as a function of mouse age. Interestingly, we observed increases in pERK, total ERK, and the ratio of pERK to total ERK in APPswe/PS1ΔE9 mice. This observation is interesting in light of the fact that ERK signaling has been previously reported to be abnormally increased in AD [45]. It has been suggested that increased ERK signaling may even result in accelerated neurodegeneration [46]. On the other hand, ERK was reported to lead to activation of CREB via RSK and MSK (for review see [30]). Our observations support an increase in ERK and pERK expression in the APPswe/PS1ΔE9.Nevertheless, this increase does not result in normal CREB phosphorylation.
Second, we show that learning or EE-induced CRE-driven gene transcription is deficient in APPswe/PS1ΔE9 mice. While levels of pCREB, CBP, and CRE-driven gene transcription are significantly increased following learning or EE in wild type or CRE-reporter mice, this activation is deficient in the APPswe/PS1ΔE9 mice. Clearly, these impairments are not a result of age or disease progression, as they are detected as early as 2-3 months of age in this study. This suggests that enhancement of plasticity by experience in EE may not be sufficient for a complete rescue of cognitive impairmentsin AD. Interestingly, some studies have failed to observe memory impairments in APPswe/PS1ΔE9 mice even as late as 6 months of age [47]. One reason for this difference may be the use of males in their analysis, as cognitive decline occurs more slowly in males in this mouse model. In support of our observations, others have shown memory deficits at 3.5 months of age in females [48]. It is also important to note that we designed our experiments based on paradigms previously used for the examination of memory deficits in CREB or CBP deficient mice [32, 33, 41]. The experimental design may have profound effects on behavioral outcomes [31].
Third, we show that impairments in CREB signaling correlate with deficits in the formation of long-term memory in contextual fear conditioning and novel object recognition paradigms. While contextual fear conditioning depends on the amygdala as well as the hippocampus, we observed that deficits in pCREB were mainly localized to the dorsal hippocampus, and no differences were detected in the amygdala of wild type and APPswe/PS1ΔE9 mice. To verify this observation, we used two learning tasks as well as EE, in part to ensure that our results were not task-specific, and in part to test the association between impairments in CREB signaling and the involvement of the hippocampus and amygdala. Taken together, these results lead us to conclude that the impairments in performance on learning tests result from deficient CREB signaling in memory pathways in the hippocampus.
Fourth, we generated a novel mouse model by crossing the CRE-reporter mouse with the FAD-linked APPswe/PS1ΔE9 mice. This allowed us to visualize CRE-driven gene transcription during the formation of long-term memories. We not only observed a lack of preference for the novel object in our CRE-AD mice, we also observed reduced CRE-driven gene transcription throughout the hippocampus immediately following the NOR task. These results profoundly demonstrate that impairments in CRE-driven gene expression are associated with (and may lead to) deficits in long-term memory formation.
Fifth, we investigated the expression of the CRE-driven gene Egr-1 following contextual fear conditioning. We showed overlap between X-gal and Egr-1 staining in the CA regions of the hippocampus, although not every Egr-1 + cell was X-gal positive. A few explanations for this observation are possible. First, X-gal staining is dependent on the activity of the galactosidase enzyme, which begins to decay during tissue processing. Time of tissue processing was carefully controlled across groups, but the interval of processing may have resulted in some decay of enzymatic activity. Second, X-gal expression following stimulation is transient, and may be returning to baseline by the time the epitope for Egr-1 is fully expressed. However, the fact that we do see overlap between X-gal and Egr-1 staining further supports that the CRE-driven gene expression observed following learning results in the transcription of factors important for the formation of long-term memories. The observation that Egr-1 levels are not upregulated in the hippocampus of APPswe/PS1ΔE9 mice following learning further supports our result that CRE-based gene transcription is impaired in these mice.
Our study suggests impairments in CREB signaling on several levels, which may suggest several potential mechanisms. Impairments in CREB in FAD have been attributed to both amyloid-dependent [6, 49–51] and independent [13, 17] mechanisms. Female APPswe/PS1ΔE9 mice exhibit increased levels of Aβ42 as well as oligomeric Aβ with onset of deposition around 4-5 months of age [52, 53] and we cannot exclude the possibility of an effect of Aβ on CREB expression. We observed increased BACE1 expression in the hippocampus of the APPswe/PS1ΔE9 mice at 3 months of age. This observation is in line with previous observations [54, 55]. Interestingly, BACE1 has been previously shown to modulate CREB signaling independent of amyloid, offering another potential mechanism underlying CREB deficits [17]. In addition, several studies suggest that PS1 itself regulates CREB expression and function [13, 24, 56, 57]. It is yet to be elucidated whether one or more pathways is sufficient or necessary for the rescue of CREB and memory deficit.
In summary, our results indicate that the expression of critical components of CREB signaling are reduced and nonresponsive to learning stimuli in APPswe/PS1ΔE9 mice, resulting in impaired transcription of genes important for long-term memory formation, and leading to deficits in long-term memory.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Dr. Daniel Storm, The University of Washington, Seattle, for providing the CRE-β-galactosidase mice. This study was supported by NIA AG033570 (OL) and the PECTS Award CCTS Pre-doctoral Education Clinical and Translational Scientists Program (NB).
Authors’ disclosures available online (http://j-alz.com/manuscript-disclosures/15-0650r1).
Appendix
The supplementary table and figure are available in the electronic version of this article: http://dx.doi.org/10.3233/JAD-150650.
REFERENCES
- [1]. Alberini CM, Chen DY (2012) Memory enhancement: consolidation, reconsolidation and insulin-like growth factor 2. Trends Neurosci 35, 274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Lee YS, Silva AJ (2009) The molecular and cellular biology of enhanced cognition. Nat Rev Neurosci 10, 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Kim R, Moki R, Kida S (2011) Molecular mechanisms for the destabilization and restabilization of reactivated spatial memory in the Morris water maze. Mol Brain 4, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. Lopez-Atalaya JP, Ciccarelli A, Viosca J, Valor LM, Jimenez-Minchan M, Canals S, Giustetto M, Barco A (2011) CBP is required for environmental enrichment-induced neurogenesis and cognitive enhancement. EMBO J 30, 4287–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Mizuno M, Yamada K, Maekawa N, Saito K, Seishima M, Nabeshima T (2002) CREB phosphorylation as a molecular marker of memory processing in the hippocampus for spatial learning. Behav Brain Res 133, 135–141. [DOI] [PubMed] [Google Scholar]
- [6]. Pugazhenthi S, Wang MR, Pham S, Sze CI, Eckman CB (2011) Downregulation of CREB expression in Alzheimer’s brain and in A beta-treated rat hippocampal neurons. Mol Neurodegener 6, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Yamamoto-Sasaki M, Ozawa H, Saito T, Rosler M, Riederer P (1999) Impaired phosphorylation of cyclic AMP response element binding protein in the hippocampus of dementia of the Alzheimer type. Brain Res 824, 300–303. [DOI] [PubMed] [Google Scholar]
- [8]. Yamamoto M, Ozawa H, Saito T, Frolich L, Riederer P, Takahata N (1996) Reduced immunoreactivity of adenylyl cyclase in dementia of the Alzheimer type. Neuroreport 7, 2965–2970. [DOI] [PubMed] [Google Scholar]
- [9]. Yamamoto M, Ozawa H, Saito T, Hatta S, Riederer P, Takahata N (1997) Ca2+/CaM-sensitive adenylyl cyclase activity is decreased in the Alzheimer’s brain: ossible relation to type I adenylyl cyclase. J Neural Transm 104, 721–732. [DOI] [PubMed] [Google Scholar]
- [10]. Cowburn RF, Oneill C, Ravid R, Alafuzoff I, Winblad B, Fowler CJ (1992) Adenylyl cyclase activity in postmortem human brain – evidence of altered g-protein mediation in Alzheimer’s disease. J Neurochem 58, 1409–1419. [DOI] [PubMed] [Google Scholar]
- [11]. Schnecko A, Witte K, Bohl J, Ohm T, Lemmer B (1994) Adenylyl-cyclase activity in Alzheimer’s disease brain - stimulatory and inhibitory signal-transduction pathways are differently affected. Brain Res 644, 291–296. [DOI] [PubMed] [Google Scholar]
- [12]. Saura CA, Choi SY, Beglopoulos V, Malkani S, Zhang D, Shankaranarayana Rao BS, Chattarji S, Kelleher RJ 3rd, Kandel ER, Duff K, Kirkwood A, Shen J (2004) Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration. Neuron 42, 23–36. [DOI] [PubMed] [Google Scholar]
- [13]. Marambaud P, Wen PH, Dutt A, Shioi J, Takashima A, Siman R, Robakis NK (2003) A CBP binding transcriptional repressor produced by the PS1/epsilon-cleavage of N-cadherin is inhibited by PS1 FAD mutations. Cell 114, 635–645. [DOI] [PubMed] [Google Scholar]
- [14]. Francis YI, Stephanou A, Latchman DS (2006) CREB-binding protein activation by presenilin 1 but not by its M146L mutant. Neuroreport 17, 917–921. [DOI] [PubMed] [Google Scholar]
- [15]. Francis YI, Diss JK, Kariti M, Stephanou A, Latchman DS (2007) p300 activation by Presenilin 1 but not by its M146L mutant. Neurosci Lett 413, 137–140. [DOI] [PubMed] [Google Scholar]
- [16]. Caccamo A, Maldonado MA, Bokov AF, Majumder S, Oddo S (2010) CBP gene transfer increases BDNF levels and ameliorates learning and memory deficits in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci U S A 107, 22687–22692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]. Chen YM, Huang XW, Zhang YW, Rockenstein E, Bu GJ, Golde TE, Masliah E, Xu HX (2012) Alzheimer’s beta-Secretase (BACE1) Regulates the cAMP/PKA/CREB Pathway Independently of beta-Amyloid. J Neurosci 32, 11390–11395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18]. Nishimoto I, Okamoto T, Matsuura Y, Takahashi S, Okamoto T, Murayama Y, Ogata E (1993) Alzheimer amyloid protein precursor complexes with brain GTP-binding protein G(o). Nature 362, 75–79. [DOI] [PubMed] [Google Scholar]
- [19]. Wang RS, Zhang YW, Sun P, Liu RZ, Zhang X, Zhang X, Xia K, Xia JH, Xu HX, Zhang ZH (2006) Transcriptional regulation of PEN-2, a key component of the gamma-secretase complex, by CREB. Mol Cell Biol 26, 1347–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Dineley KT, Kayed R, Neugebauer V, Fu Y, Zhang W, Reese LC, Taglialatela G (2010) Amyloid-beta oligomers impair fear conditioned memory in a calcineurin-dependent fashion in mice. J Neurosci Res 88, 2923–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Dineley KT, Westerman M, Bui D, Bell K, Ashe KH, Sweatt JD (2001) Beta-amyloid activates the mitogen-activated protein kinase cascade via hippocampal alpha7 nicotinic acetylcholine receptors: In vitro and in vivo mechanisms related to Alzheimer’s disease. J Neurosci 21, 4125–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22]. Ma QL, Harris-White ME, Ubeda OJ, Simmons M, Beech W, Lim GP, Teter B, Frautschy SA, Cole GM (2007) Evidence of Abeta- and transgene-dependent defects in ERK-CREB signaling in Alzheimer’s models. J Neurochem 103, 1594–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Espana J, Valero J, Minano-Molina AJ, Masgrau R, Martin E, Guardia-Laguarta C, Lleo A, Gimenez-Llort L, Rodriguez-Alvarez J, Saura CA (2010) beta-amyloid disrupts activity-dependent gene transcription required for memory through the CREB coactivator CRTC1. J Neurosci 30, 9402–9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Muller M, Cardenas C, Mei L, Cheung KH, Foskett JK (2011) Constitutive cAMP response element binding protein (CREB) activation by Alzheimer’s disease presenilin-driven inositol trisphosphate receptor (InsP3R) Ca2+ signaling. Proc Natl Acad Sci U S A 108, 13293–13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Jankowsky JL, Slunt HH, Ratovitski T, Jenkins NA, Copeland NG, Borchelt DR (2001) Co-expression of multiple transgenes in mouse CNS: a comparison of strategies. Biomol Eng 17, 157–165. [DOI] [PubMed] [Google Scholar]
- [26]. Impey S, Mark M, Villacres EC, Poser S, Chavkin C, Storm DR (1996) Induction of CRE-mediated gene expression by stimuli that generate long-lasting LTP in area CA1 of the hippocampus. Neuron 16, 973–982. [DOI] [PubMed] [Google Scholar]
- [27]. Hu YS, Xu P, Pigino G, Brady ST, Larson J, Lazarov O (2010) Complex environment experience rescues impaired neurogenesis, enhances synaptic plasticity, and attenuates neuropathology in familial Alzheimer’s disease-linked APPswe/PS1 Delta E9 mice. FASEB J 24, 1667–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. Singh P, Thakur MK (2014) Reduced recognition memory is correlated with decrease in DNA methyltransferase1 and increase in histone deacetylase2 protein expression in old male mice. Biogerontology 15, 339–346. [DOI] [PubMed] [Google Scholar]
- [29]. Barth AL, McKenna M, Glazewski S, Hill P, Impey S, Storm D, Fox K (2000) Upregulation of cAMP response element-mediated gene expression during experience-dependent plasticity in adult neocortex. J Neurosci 20, 4206–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Lonze BE, Ginty DD (2002) Function and regulation of CREB family transcription factors in the nervous system. Neuron 35, 605–623. [DOI] [PubMed] [Google Scholar]
- [31]. Kogan JH, Frankland PW, Blendy JA, Coblentz J, Marowitz Z, Schutz G, Silva AJ (1997) Spaced training induces normal long-term memory in CREB mutant mice. Curr Biol 7, 1–11. [DOI] [PubMed] [Google Scholar]
- [32]. Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, Silva AJ (1994) Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell 79, 59–68. [DOI] [PubMed] [Google Scholar]
- [33]. Barrett RM, Malvaez M, Kramar E, Matheos DP, Arrizon A, Cabrera SM, Lynch G, Greene RW, Wood MA (2011) Hippocampal focal knockout of CBP affects specific histone modifications, long-term potentiation, and long-term memory. Neuropsychopharmacology 36, 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34]. Impey S, Smith DM, Obrietan K, Donahue R, Wade C, Storm DR (1998) Stimulation of cAMP response element (CRE)-mediated transcription during contextual learning. Nat Neurosci 1, 595–601. [DOI] [PubMed] [Google Scholar]
- [35]. Alberini CM, Ghirardi M, Huang YY, Nguyen PV, Kandel ER (1995) A molecular switch for the consolidation of long-term memory: cAMP-inducible gene expression. Ann N Y Acad Sci 758, 261–286. [DOI] [PubMed] [Google Scholar]
- [36]. Hu SC, Chrivia J, Ghosh A (1999) Regulation of CBP-mediated transcription by neuronal calcium signaling. Neuron 22, 799–808. [DOI] [PubMed] [Google Scholar]
- [37]. Schwaninger M, Blume R, Kruger M, Lux G, Oetjen E, Knepel W (1995) Involvement of the Ca(2+)-dependent phosphatase calcineurin in gene transcription that is stimulated by cAMP through cAMP response elements. J Biol Chem 270, 8860–8866. [DOI] [PubMed] [Google Scholar]
- [38]. Shieh PB, Hu SC, Bobb K, Timmusk T, Ghosh A (1998) Identification of a signaling pathway involved in calcium regulation of BDNF expression. Neuron 20, 727–740. [DOI] [PubMed] [Google Scholar]
- [39]. Wilson BE, Mochon E, Boxer LM (1996) Induction of bcl-2 expression by phosphorylated CREB proteins during B-cell activation and rescue from apoptosis. Mol Cell Biol 16, 5546–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40]. Hu YS, Long N, Pigino G, Brady ST, Lazarov O (2013) Molecular mechanisms of environmental enrichment: impairments in Akt/GSK3 beta, neurotrophin-3 and CREB signaling. Plos One 8, e64460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Valor LM, Pulopulos MM, Jimenez-Minchan M, Olivares R, Lutz B, Barco A (2011) Ablation of CBP in forebrain principal neurons causes modest memory and transcriptional defects and a dramatic reduction of histone acetylation but does not affect cell viability. J Neurosci 31, 1652–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42]. Jones MW, Errington ML, French PJ, Fine A, Bliss TV, Garel S, Charnay P, Bozon B, Laroche S, Davis S (2001) A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat Neurosci 4, 289–296. [DOI] [PubMed] [Google Scholar]
- [43]. Lakhina V, Arey RN, Kaletsky R, Kauffman A, Stein G, Keyes W, Xu D, Murphy CT (2015) Genome-wide functional analysis of CREB/long-term memory-dependent transcription reveals distinct basal and memory gene expression programs. Neuron 85, 330–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44]. Liu HL, Jin XX, Yin XM, Jin NN, Liu F, Qian W (2015) PKA-CREB signaling suppresses tau transcription. J Alzheimers Dis 46, 239–248. [DOI] [PubMed] [Google Scholar]
- [45]. Perry G, Roder H, Nunomura A, Takeda A, Friedlich AL, Zhu X, Raina AK, Holbrook N, Siedlak SL, Harris PL, Smith MA (1999) Activation of neuronal extracellular receptor kinase (ERK) in Alzheimer disease links oxidative stress to abnormal phosphorylation. Neuroreport 10, 2411–2415. [DOI] [PubMed] [Google Scholar]
- [46]. Subramaniam S, Zirrgiebel U, von Bohlen und Halbach O, Strelau J, Laliberte C, Kaplan DR, Unsicker K (2004) ERK activation promotes neuronal degeneration predominantly through plasma membrane damage and independently of caspase-3. J Cell Biol 165, 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47]. Savonenko A, Xu GM, Melnikova T, Morton JL, Gonzales V, Wong MP, Price DL, Tang F, Markowska AL, Borchelt DR (2005) Episodic-like memory deficits in the APPswe/PS1dE9 mouse model of Alzheimer’s disease: relationships to beta-amyloid deposition and neurotransmitter abnormalities. Neurobiol Dis 18, 602–617. [DOI] [PubMed] [Google Scholar]
- [48]. Zhang W, Bai M, Xi Y, Hao J, Liu L, Mao N, Su CJ, Miao JT, Li ZY (2012) Early memory deficits precede plaque deposition in APPswe/PS1dE9 mice: Involvement of oxidative stress and cholinergic dysfunction. Free Radic Biol Med 52, 1443–1452. [DOI] [PubMed] [Google Scholar]
- [49]. Rosa E, Fahnestock M (2015) CREB expression mediates amyloid beta-induced basal BDNF downregulation. Neurobiol Aging 36, 2406–2413. [DOI] [PubMed] [Google Scholar]
- [50]. Vitolo OV, Sant’Angelo A, Costanzo V, Battaglia F, Arancio O, Shelanski M (2002) Amyloid beta-peptide inhibition of the PKA/CREB pathway and long-term potentiation: Reversibility by drugs that enhance cAMP signaling. Proc Natl Acad Sci U S A 99, 13217–13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51]. Tong LQ, Thornton PL, Balazs R, Cotman CW (2001) beta-Amyloid-(1-42) impairs activity-dependent cAMP-response element-binding protein signaling in neurons at concentrations in which cell survival is not compromised. J Biol Chem 276, 17301–17306. [DOI] [PubMed] [Google Scholar]
- [52]. Lazarov O, Lee M, Peterson DA, Sisodia SS (2002) Evidence that synaptically released beta-amyloid accumulates as extracellular deposits in the hippocampus of transgenic mice. J Neurosci 22, 9785–9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53]. Lazarov O, Robinson J, Tang YP, Hairston IS, Korade-Mirnics Z, Lee VM, Hersh LB, Sapolsky RM, Mirnics K, Sisodia SS (2005) Environmental enrichment reduces Abeta levels and amyloid deposition in transgenic mice. Cell 120, 701–713. [DOI] [PubMed] [Google Scholar]
- [54]. Li W, Yu J, Liu Y, Huang XJ, Abumaria N, Zhu Y, Huang X, Xiong WX, Ren C, Liu XG, Chui DH, Liu GS (2014) Elevation of brain magnesium prevents synaptic loss and reverses cognitive deficits in Alzheimer’s disease mouse model. Mol Brain 7, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55]. Wang Y, Tang XC, Zhang HY (2012) Huperzine A alleviates synaptic deficits and modulates amyloidogenic and nonamyloidogenic pathways in APPswe/PS1dE9 transgenic mice. J Neurosci Res 90, 508–517. [DOI] [PubMed] [Google Scholar]
- [56]. Bonds JA, Kuttner-Hirshler Y, Bartolotti N, Tobin MK, Pizzi M, Marr R, Lazarov O (2015) Presenilin-1 dependent neurogenesis regulates hippocampal learning and memory. Plos One 10, e0131266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57]. Watanabe H, Smith MJ, Heilig E, Beglopoulos V, Kelleher RJ, Shen J (2009) Indirect regulation of presenilins in CREB-mediated transcription. J Biol Chem 284, 13705–13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The supplementary table and figure are available in the electronic version of this article: http://dx.doi.org/10.3233/JAD-150650.