Abstract
Campylobacteriosis is the most prevalent bacterial foodborne gastroenteritis affecting humans in the European Union. Human cases are mainly due to Campylobacter jejuni or Campylobacter coli, and contamination is associated with the handling and/or consumption of poultry meat. In fact, poultry constitutes the bacteria's main reservoir. A promising way of decreasing the incidence of campylobacteriosis in humans would be to decrease avian colonization. Poultry vaccination is of potential for this purpose. However, despite many studies, there is currently no vaccine available on the market to reduce the intestinal Campylobacter load in chickens. It is essential to identify and characterize new vaccine antigens. This study applied the reverse vaccinology approach to detect new vaccine candidates. The main criteria used to select immune proteins were localization, antigenicity, and number of B-epitopes. Fourteen proteins were identified as potential vaccine antigens. In vitro and in vivo experiments now need to be performed to validate the immune and protective power of these newly identified antigens.
1. Introduction
Campylobacter is the leading cause of human bacterial gastroenteritis in Europe [1]. It has been estimated that 9 million people are affected each year, costing around €2.4 billion. C. jejuni is responsible for approximately 90% of cases, and C. coli is responsible for 10%. Other species can cause human campylobacteriosis but are more rarely involved [2]. Most infections are not severe, leading to gastroenteritis symptoms, but they can cause extraintestinal manifestations such as reactive arthritis, Guillain-Barré syndrome (GBS), or inflammatory bowel disease (IBD) [3]. In some cases, infection can even lead to death. Human contaminations are mainly associated with handling and/or consuming poultry meat [1]. Domestic and wild birds constitute the bacteria's main reservoir, carrying up to 109 CFU·g−1 of Campylobacter intestinally. In poultry flocks, natural colonization occurs in 2- to 3-week-old chicks by horizontal contamination from the environment [4], and birds remain infected until slaughter.
Decreasing avian colonization would appear to be an effective strategy for reducing the incidence of human campylobacteriosis. In 2013, Romero-Barrios et al. estimated that a reduction in Campylobacter cecal colonization from 2 to 3 log10 units could lead to a 100% reduction in the risk of human disease [5]. Along with the implementation of biosecurity, hygiene, and nutritional measures in flocks, poultry vaccination is one way of reducing avian intestine colonization by Campylobacter [6]. Several vaccine prototypes have already been tested with variable results. These include whole-cell, subunit, or microorganism-vectored vaccines. Globally, whole-cell vaccines have not been efficient in decreasing Campylobacter intestinal loads despite the induction of a specific immune response [7–10]. Among subunit vaccines, flagellin—described as the immunodominant antigen of Campylobacter—has been tested and proved to be able to induce an immune response but this was not necessarily correlated with any decrease in chicken gut colonization [9, 11–13]. Furthermore, because of its weak homology across Campylobacter strains, flagellin-based vaccines do not induce cross-protection, making these vaccines inefficient in combatting all C. jejuni strains [14]. Other antigens such as CjaA [15]—a periplasmic protein—or CadF, FlpA, CmeC [16], and Dsp proteins [17] involved in Campylobacter adherence during colonization have also been trialed as subunit vaccines. In the same way, total outer membrane proteins [18] or fusion proteins [16] have also been tested. Another strategy is to deliver vaccine antigens by vectors such as attenuated bacterial strains. Salmonella enterica serovar Typhimurium [15, 19–21] and Eimeria tenella [22] have been evaluated as a vector for C. jejuni CjaA delivery. For example, in 2004, Wyszyńska et al. [19] indicated that chickens orally immunized with a virulent Salmonella strain carrying the Campylobacter CjaA gene develop a strong specific antibody response, and birds were protected from colonization after a homologous C. jejuni challenge. Recently, the same team was unable to confirm these results [21]. Other antigens were tested in the same way, including Omp18/CjaD, ACE393 [20], Dsp [17], Peb1A, GlnH, and ChuA [15]. Some of these experimental studies gave promising results, combining both the induction of a humoral immune response and a decrease in Campylobacter intestinal colonization in poultry, but experimentation has not yet been followed up. So, despite much research, no anti-Campylobacter vaccine aiming to reduce bacterial colonization in the poultry gut is yet available.
Identifying new potential vaccine antigens is one way of speeding up the development of new vaccines. Reverse vaccinology—a recent approach first described by Rappuoli in the early 2000s [23]—is used to predict antigens through the development of genomics and bioinformatic tools such as genome sequencing. This strategy is different from Hoppe et al. approach, where they identified novel immunodominant proteins by in vitro screening of mRNA of C. jejuni [24]. The following selection criteria are of particular importance for the reverse vaccinology approach. To be potentially good candidates, the selected proteins must be surface-exposed and able to be recognized by the immune system. Proteins with adhesin capacities are known to be involved in bacterial pathogenicity and invasion, so adhesins or adhesin-like proteins appear as good vaccine targets. The transmembrane helix number is also an important criterion. Indeed, it is difficult to purify proteins with more than one transmembrane helix, and it seems wise to exclude these proteins from the selection process [25]. Individual antigenicity and B-epitope density (the ratio between the number of B-epitopes and the protein length) need to be assessed as described by Oprea's study aimed at developing a vaccine against S. aureus endocarditis [26]. Although a few studies are describing innate intestinal inflammations and gut mucosa lesions upon Campylobacter jejuni infection (like in [27]), these bacteria are mostly described as a commensal organism for poultry [28]. In the avian intestinal tract, intensive Campylobacter multiplication occurs in the mucus layer of the epithelial cells. In this way, antigens need to induce a humoral immune response to neutralize and eliminate Campylobacter from the avian intestinal gut. The induction of a cytotoxic cellular response may not be a selection criterion since Campylobacter multiplication in intestinal epithelial cells of chickens was not clearly highlighted [28]. Anyway, bioinformatic tools aiming at predicting T epitopes for avian vaccines are still poorly developed, limiting the reverse vaccinology analysis in this goal. Finally, to provide cross-protection and avoid autoimmune response, it is essential that vaccine candidates are common to many pathogenic strains and do not mimic host proteins [25].
Our research identifies new potential vaccine antigens against Campylobacter using the reverse vaccinology strategy to develop an avian vaccine which could impact the incidence of human campylobacteriosis.
2. Material and Methods
2.1. Bacterial Strain
The highly virulent Campylobacter jejuni subsp. jejuni 81-176 strain was chosen for this in silico analysis. Its genome is available on the NCBI website under accession number CP000538.1 and listed in the Vaxign program used below.
2.2. OMP and Extracellular Protein Preselection
Vaxign (http://www.violinet.org/vaxign/index.php) was used to shortlist proteins with potential as vaccine candidates due to their cellular localization, probability of having adhesin-like characteristics, and number of transmembrane helixes [25]. Vaxign is a web-based pipeline dedicated to vaccine design and integrating several bioinformatic programs. Subcellular localization is predicted using PSORTb2.0 [29]. The probability of adhesin characteristics is predicted by SPAAN software [30] and the transmembrane helix topology is predicted by HMMTOP [31] using a hidden Markov model.
Campylobacter jejuni subsp. jejuni 81-176 is available in the Vaxign database of over 350 listed genomes, along with nine other Campylobacter genomes. Extracellular and outer membrane proteins having an adhesin probability score > 0.51 and either 1 or 0 transmembrane helixes were preselected.
2.3. Protein Antigenicity
VaxiJen v2.0 (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) was used to predict protein antigenicity. This software uses the physicochemical properties of proteins to predict their antigenicity from FASTA-submitted amino acid sequences. This feature is characterized according to an antigenic score. Proteins with an antigenic score > 0.5 were selected as described by Doytchinova and Flower [32].
2.4. Epitope B Prediction
BCPreds software (http://ailab.ist.psu.edu/bcpred/) was used to identify B-cell epitopes in FASTA-submitted amino acid sequences. This program provides two methods based on different algorithms: the amino acid pair (AAP) antigenicity method [33] and the BCPreds method using string kernels [34]. These methods predict antigenic linear nonoverlapping 20-mer epitopes from the whole antigen. Each preselected protein was analyzed and B-cell epitopes with a score >0.8 were accepted (specificity > 80%). The selected epitopes were again submitted to VaxiJen software to check their individual antigenicity and those having an antigenic score >0.5 were selected. Furthermore, for each protein and each algorithm, the ratio of B-epitopes to protein length was calculated to assess B-epitope density.
2.5. BLAST
In order to assess conservation of the selected proteins in the different Campylobacter strains, tblastn analyses were performed for each amino acid sequence against both C. jejuni and C. coli whole genomes available on the NCBI site on the day of analysis (February 9, 2016): 93 for C. jejuni and nine for C. coli. The identity percentage was set to 80% and the minimum query coverage was set to 50%. The amount and percentage of sharing among the available genomes were determined. The proteins with a sharing percentage lower than 80% (i.e., about the value for the flagellin) were eliminated from the protein shortlist.
A blastp analysis was also performed to ensure that the host Gallus gallus does not express the selected proteins. The identity percentage was set to 50% and the minimum query coverage was set to 50%.
3. Results
The reverse vaccinology protocol applied here and results are summarized in Figure 1.
Figure 1.
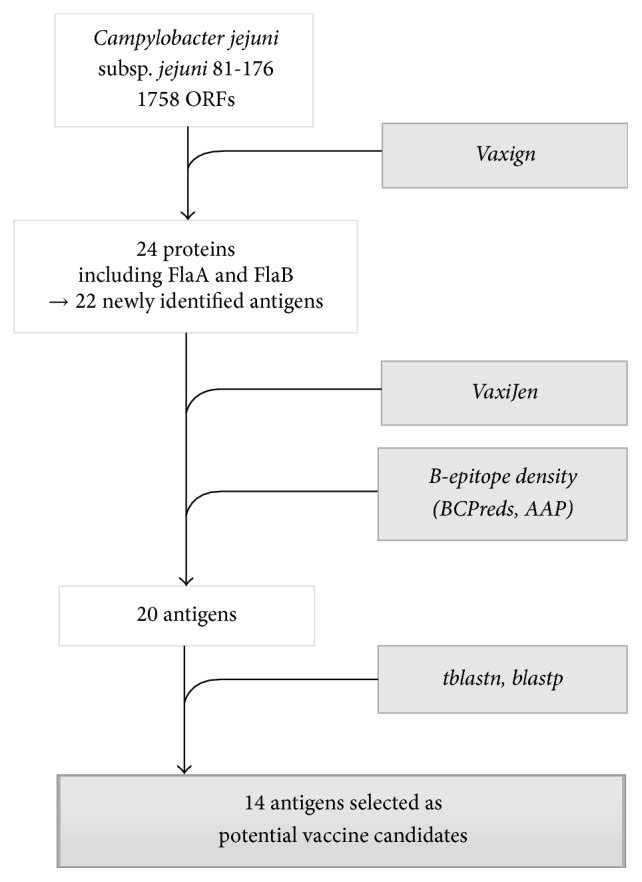
Summary of the reverse vaccinology protocol applied to Campylobacter jejuni for the selection of vaccine candidates.
3.1. Protein Preselection
The Vaxign server was used to preselect vaccine candidates. Of the 1758 ORFs encoded by the C. jejuni 81-176 genome, only 24 were identified as potential vaccine antigens according to the applied criteria (localization, adhesion features, and number of transmembrane helixes) (Table 1). Of these 24 identified ORFs, we found the two known flagellins A and B, which means that 22 new potential antigens were selected at this step.
Table 1.
Potential vaccine candidates selected by the Vaxign program. Localization and length were obtained by the Vaxign program, antigenic score was obtained by the VaxiJen program, and the number of B-cell epitopes was obtained from both BCPreds and AAP methods. The calculated ratio between the number of B-epitopes and protein length is also shown. The two candidates eliminated because of a low antigenic score are shown in italics.
| Protein accession | Description | ID | Localization | Length (aa) | Vaxijen score | BCPreds B-epitopes | AAP B-epitopes | ||
|---|---|---|---|---|---|---|---|---|---|
| N | Ratio N/length | N | Ratio N/length | ||||||
| YP_001000996.1 | Flagellin B | FlaB | Extracellular | 576 | 0.7650 | 10 | 0.017 | 11 | 0.019 |
| YP_001000997.1 | Flagellin A | FlaA | Extracellular | 576 | 0.8185 | 11 | 0.019 | 12 | 0.021 |
|
| |||||||||
| YP_001001371.1 | Flagellar hook protein | FlgE | Extracellular | 838 | 0.7659 | 16 | 0.019 | 18 | 0.021 |
| YP_001000562.1 | Flagellin protein family | Extracellular | 750 | 0.6965 | 15 | 0.020 | 15 | 0.020 | |
| YP_001000248.1 | Flagellar capping protein | FliD | Extracellular | 642 | 0.7021 | 11 | 0.017 | 11 | 0.017 |
| YP_001000204.1 | Putative periplasmic protein | OMP | 553 | 0.6702 | 11 | 0.021 | 11 | 0.021 | |
| YP_001000654.1 | Putative periplasmic protein | OMP | 553 | 0.6702 | 11 | 0.021 | 11 | 0.021 | |
| YP_999769.1 | Flagellar hook protein | FlgE-1 | Extracellular | 545 | 0.6567 | 10 | 0.018 | 12 | 0.022 |
| YP_001001115.1 | Flagellar hook-associated protein | FlgK | Extracellular | 608 | 0.5836 | 11 | 0.018 | 10 | 0.016 |
| YP_001000153.1 | TonB-dependent receptor, putative, degenerate | OMP | 704 | 0.5437 | 12 | 0.017 | 9 | 0.013 | |
| YP_001000945.1 | N-Acetylmuramoyl-L-alanine amidase | OMP | 659 | 0.6475 | 9 | 0.014 | 11 | 0.017 | |
| YP_001001027.1 | Serine protease | OMP | 1121 | 0.5268 | 9 | 0.008 | 11 | 0.010 | |
| YP_001000437.1 | Putative OMP | OMP | 508 | 0.6122 | 9 | 0.018 | 6 | 0.012 | |
| YP_999838.1 | Hypothetical protein | OMP | 400 | 0.6809 | 5 | 0.013 | 9 | 0.023 | |
| YP_999817.1 | Hypothetical protein | OMP | 315 | 0.7827 | 6 | 0.019 | 7 | 0.022 | |
| YP_001000383.1 | Flagellar basal body L-ring protein | FlgH | OMP | 232 | 0.6978 | 6 | 0.026 | 4 | 0.017 |
| YP_001000935.1 | Major OMP | PorA | OMP | 424 | 0.6051 | 5 | 0.012 | 5 | 0.012 |
| YP_001001008.1 | Phospholipase A | PldA | OMP | 329 | 0.5819 | 4 | 0.012 | 3 | 0.009 |
| YP_001001257.1 | TonB-dependent heme receptor | ChuA | OMP | 702 | 0.6213 | 7 | 0.010 | 5 | 0.007 |
| YP_001000615.1 | Hypothetical protein | Extracellular | 294 | 0.5498 | 3 | 0.010 | 3 | 0.010 | |
| YP_001000663.1 | Surface-exposed lipoprotein | JlpA | OMP | 372 | 0.6642 | 2 | 0.005 | 3 | 0.008 |
| YP_001000261.1 | Hypothetical protein | OMP | 309 | 0.5149 | 2 | 0.006 | 3 | 0.010 | |
| YP_001000503.1 | Hypothetical protein | Extracellular | 444 | 0.4603 | / | / | / | / | |
| YP_001000297.1 | Major antigenic peptide | PEB4 | OMP | 273 | 0.4511 | / | / | / | / |
3.2. Protein Selection according to Antigenicity and Number of B-Epitopes
To refine the selection, the 22 preselected proteins were submitted to the VaxiJen server for antigenicity prediction. Antigenicity scores ranged from 0.4511 to 0.7827. This step allowed the elimination of two proteins with an antigenicity score lower than 0.5 (YP_001000503.1 and YP_001000297.1) (Table 1). The VaxiJen software indicated that all other candidates were antigenic (score > 0.5).
Each antigenic protein was assessed in terms of B-epitopes using BCPreds and AAP algorithms, and each B-epitope was studied for its antigenicity. Table 1 summarizes the number of B-epitopes predicted for each protein and each algorithm as well as the ratio between the number of B-epitopes and protein length.
3.3. Conservation of the Selected Proteins in the Sequenced C. jejuni and C. coli Strains
tblastn analyses were performed in order to assess the individual sharing of the preselected proteins among C. jejuni and C. coli strains. As shown in Table 2, all the proteins were shared with available C. coli strains except YP_001001027.1, which was also poorly shared with the available C. jejuni strains (6%). This protein was therefore removed from the list of potential vaccine antigens. Of the remaining 19 shortlisted proteins, five—YP_001001371.1, YP_001000248.1, YP_001000204.1, YP_001000654.1 and YP_001000615.1—were removed from the candidate list because of poor sharing among C. jejuni strains (<80%).
Table 2.
Potential vaccine candidates selected after blast analysis. tblastn analyses were performed for each amino acid sequence against both C. jejuni and C. coli whole genomes available on the NCBI site. The amount and percentage of sharing among the available genomes were determined. A blastp analysis was also performed against the host Gallus gallus. The six candidates eliminated because of poor sharing among Campylobacter strains are shown in italics.
| Protein accession | Description | ID | Sharing among C. jejuni strains | Sharing among C. coli strains |
Similarity in Gallus gallus | ||
|---|---|---|---|---|---|---|---|
| N/93 | % | N/9 | % | ||||
| YP_001000996.1 | Flagellin B | FlaB | 77 | 83 | 9 | 100 | No |
| YP_001000997.1 | Flagellin A | FlaA | 75 | 81 | 9 | 100 | No |
|
| |||||||
| YP_001001371.1 | Flagellar hook protein | FlgE | 15 | 16 | 8 | 89 | No |
| YP_001000562.1 | Flagellin protein family | 93 | 100 | 9 | 100 | No | |
| YP_001000248.1 | Flagellar capping protein | FliD | 38 | 41 | 9 | 100 | No |
| YP_001000204.1 | Putative periplasmic protein | 2 | 2 | 9 | 100 | No | |
| YP_001000654.1 | Putative periplasmic protein | 2 | 2 | 9 | 100 | No | |
| YP_999769.1 | Flagellar hook protein | FlgE-1 | 93 | 100 | 9 | 100 | No |
| YP_001001115.1 | Flagellar hook-associated protein | FlgK | 93 | 100 | 9 | 100 | No |
| YP_001000153.1 | TonB-dependent receptor, putative, degenerate | 90 | 97 | 9 | 100 | No | |
| YP_001000945.1 | N-Acetylmuramoyl-L-alanine amidase | 93 | 100 | 9 | 100 | No | |
| YP_001001027.1 | Serine protease | PEB4 | 6 | 6 | 1 | 11 | No |
| YP_001000437.1 | Putative OMP | 89 | 96 | 6 | 67 | No | |
| YP_999838.1 | Hypothetical protein | 93 | 100 | 9 | 100 | No | |
| YP_999817.1 | Hypothetical protein | 92 | 99 | 9 | 100 | No | |
| YP_001000383.1 | Flagellar basal body L-ring protein | FlgH | 93 | 100 | 9 | 100 | No |
| YP_001000935.1 | Major OMP | PorA | 81 | 87 | 9 | 100 | No |
| YP_001001008.1 | Phospholipase A | PldA | 92 | 99 | 9 | 100 | No |
| YP_001001257.1 | TonB-dependent heme receptor | ChuA | 93 | 100 | 9 | 100 | No |
| YP_001000615.1 | Hypothetical protein | 64 | 69 | 9 | 100 | No | |
| YP_001000663.1 | Surface-exposed lipoprotein | JlpA | 93 | 100 | 9 | 100 | No |
| YP_001000261.1 | Hypothetical protein | 92 | 99 | 9 | 100 | No | |
Table 2 also shows that none of the proteins are expressed by Gallus gallus.
3.4. Final Selection
Table 3 shows the final selection of potential vaccine candidates after the whole bioinformatic analysis process. Fourteen candidates were selected. Of these, three are extracellular proteins whereas the others are outer membrane proteins. Four flagellar proteins were identified and several were not characterized and designated as hypothetical proteins.
Table 3.
Potential vaccine candidates selected after the whole bioinformatic analysis process including Vaxign and VaxiJen programs, BCPreds and AAP algorithms, and blast analyses. Of 1758 ORFs encoded by C. jejuni, strain 81-176 genome, 14 proteins were selected as vaccine candidates.
| Protein accession | Description | Localization | ID |
|---|---|---|---|
| YP_001000562.1 | Flagellin protein family | Extracellular | |
| YP_999769.1 | Flagellar hook protein | Extracellular | FlgE-1 |
| YP_001001115.1 | Flagellar hook-associated protein | Extracellular | FlgK |
| YP_001000153.1 | TonB-dependent receptor, putative, degenerate | OMP | |
| YP_001000945.1 | N-Acetylmuramoyl-L-alanine amidase | OMP | |
| YP_001000437.1 | Putative OMP | OMP | |
| YP_999838.1 | Hypothetical protein | OMP | |
| YP_999817.1 | Hypothetical protein | OMP | |
| YP_001000383.1 | Flagellar basal body L-ring protein | OMP | FlgH |
| YP_001000935.1 | Major OMP | OMP | PorA |
| YP_001001008.1 | Phospholipase A | OMP | PldA |
| YP_001001257.1 | TonB-dependent heme receptor | OMP | ChuA |
| YP_001000663.1 | Surface-exposed lipoprotein | OMP | JlpA |
| YP_001000261.1 | Hypothetical protein | OMP |
4. Discussion
In the last decades, advances in genomics, genome sequencing, and annotation, coupled with the development of bioinformatic tools has revolutionized vaccine development strategy. Reverse vaccinology allows vaccines to be designed even for noncultivable pathogens; genome availability is the only factor enabling in silico analysis or not. All the proteins are targeted even if only transiently expressed or scarce during infection. Furthermore, this strategy considerably reduces the time needed to develop new vaccines [35]. Reverse vaccinology was first successfully applied to the development of a vaccine against B serogroup Neisseria meningitidis [36]. Despite available prophylactic vaccines based on capsular polysaccharides (CPS) for four N. meningitidis serogroups (A, C, W, and Y), the development of a capsular vaccine against serogroup B was not possible because of CPS mimicry of polysialic acid in human cells. In silico analysis identified three proteins (fHbp, NadA, and NHBA) which were combined with outer membrane vesicles containing known antigen PorA and led to the European licensure of the 4CMenB vaccine in 2013 [37]. This strategy was then applied to several other pathogens such as herpes simplex viruses using the Vaxign program [38], Staphylococcus aureus for the in silico characterization of ten surface-exposed proteins [26], Mycobacterium tuberculosis with the identification of six novel antigen candidates to improve the tuberculosis vaccine [39], or Streptococcus pneumonia with the bioinformatic assessment of 13 protein targets [40]. The antigenicity and efficiency of the potential candidates selected in these last in silico studies have not yet been tested in vitro or in vivo.
Until now, and despite many studies, conventional development of a vaccine against Campylobacter in poultry has not led to an efficient vaccine in terms of immunogenicity and protection. Since 2005, Campylobacter has been and remains today the leading cause of bacterial foodborne gastroenteritis in Europe [1]. As poultry vaccination is one of the potential ways of reducing the incidence of human campylobacteriosis, it is important to pursue efforts to test new vaccine antigens. Reverse vaccinology is a suitable strategy to this end.
This in silico study predictively identified new vaccine antigens against Campylobacter. The reference C. jejuni ATCC (American Type Culture Collection) strain 81-176 was chosen for antigen prediction because of its high virulence in human diseases (namely, the chicken vaccine strategy is to prevent human infections). Even if this strain is not a good colonizer for chickens, this strain has been successfully used in several poultry experiments with high colonization levels [18]. Moreover, the reverse vaccinology aims to identify shared proteins among many Campylobacter strains (here more than 100 strains). Thereby, other more avian colonizer Campylobacter strains should be used for in vivo challenge experiments to evaluate the effectiveness of the proteins found by the bioinformatics analysis of the C. jejuni 81-176 genome.
Based on their cellular localization, adhesin-like properties, antigenicity, B-epitope density, and conservation among Campylobacter strains, 14 proteins were selected. It was decided to eliminate proteins with a sharing percentage lower than the flagellin sharing percentage. It has already been observed that flagellin could not be used as a vaccine candidate because of poor sharing among Campylobacter strains and the lack of cross-protection [14]. The known vaccine antigens of flagellins A and B were also identified alongside potential antigens using the same criteria. This strengthens the validity of the bioinformatic protocol used, because the flagellin has already been described and used as the immune-dominant antigen of Campylobacter [9, 11, 12]. However, it is important to keep in mind that the identified proteins were selected on the basis of predictions by various algorithms. Only in vitro and more in vivo experiments will confirm or refute the proteins' immune power. In terms of antigen ranking, proteins with a high antigenicity score and B-epitope density seem to be the best vaccine candidates and should therefore be evaluated for in vivo immunogenicity as a priority. Indeed, it has already been demonstrated that a high epitope density significantly enhances antigenicity and immunogenicity [41]. This strategy, being based on genome analysis, does not take into consideration lipid and saccharide antigens, which could also have immune properties. Concerning Campylobacter, capsule polysaccharides are not targeted through the reverse vaccinology protocol, although they could be immunogenic [42].
Several of the identified proteins had already been characterized and were mainly associated with Campylobacter virulence [43]. This is the case for three selected flagellar proteins—FlgE-1, FlgK, and FlgH—involved in Campylobacter motility, essential for bacteria survival in the gastrointestinal tract. These proteins were recently tested in vitro along with other flagellar proteins [44]. The first two were immunostained by more than 70% of tested sera from chickens older than 5-6 weeks; the third one was immunostained by 50% of the tested sera. The present in silico analysis is in line with these in vitro results, leading us to consider these three flagellar proteins as a potential vaccine antigen. However, no in vivo assessment is available yet. The FliD flagellar protein was similarly tested in vitro and was observed to react strongly to sera from chickens over 4 weeks of age [45]. In the present analysis, this flagellar protein was eliminated from the shortlist because of poor sharing with other Campylobacter strains (41%). Moreover, the ChuA protein—involved in the iron uptake system—had already been tested in an avian vaccine experiment using attenuated Salmonella as a vector [15] and did not significantly reduce cecal Campylobacter counts. Furthermore, the major PorA outer membrane protein was tested in vivo in a mouse model [46]. Mice vaccinations led to significantly higher antibody levels in serum and intestinal lavage fluids. A decrease in C. jejuni colonization levels was also observed after a heterologous challenge. Phospholipase A (PldA) and lipoprotein JlpA are involved in Campylobacter adhesion since it has been demonstrated that mutations of pldA impair the ability of C. jejuni to colonize cecum [47] and since Jin et al. highlighted the interaction of JlpA with a surface-exposed protein of epithelial cells [48].
To conclude, reverse vaccinology—a powerful tool for identifying new vaccine antigens—allowed 14 candidates to be selected for the development of a vaccine against Campylobacter in poultry. Several antigens identified as potential vaccine candidates are currently under in vitro and in vivo investigations to evaluate their immunogenicities and protective potentials against Campylobacter in chickens.
Acknowledgments
This work has received funding from the European Union's Seventh Framework Program for research, technological development and demonstration under Grant Agreement no. 605835.
Competing Interests
The authors declare that there is no conflict of interests regarding publication of this paper.
References
- 1.EFSA. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA Journal. 2015;13(1):p. 3991. doi: 10.2903/j.efsa.2018.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gillespie I. A., O'Brien S. J., Frost J. A., et al. A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerging Infectious Diseases. 2002;8(9):937–942. doi: 10.3201/eid0809.010817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janssen R., Krogfelt K. A., Cawthraw S. A., van Pelt W., Wagenaar J. A., Owen R. J. Host-pathogen interactions in Campylobacter infections: the host perspective. Clinical Microbiology Reviews. 2008;21(3):505–518. doi: 10.1128/cmr.00055-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahin O., Luo N., Huang S., Zhang Q. Effect of Campylobacter-specific maternal antibodies on Campylobacter jejuni colonization in young chickens. Applied and Environmental Microbiology. 2003;69(9):5372–5379. doi: 10.1128/AEM.69.9.5372-5379.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romero-Barrios P., Hempen M., Messens W., Stella P., Hugas M. Quantitative microbiological risk assessment (QMRA) of food-borne zoonoses at the European level. Food Control. 2013;29(2):343–349. doi: 10.1016/j.foodcont.2012.05.043. [DOI] [Google Scholar]
- 6.Meunier M., Guyard-Nicodème M., Dory D., Chemaly M. Control strategies against Campylobacter at the poultry production level: biosecurity measures, feed additives and vaccination. Journal of Applied Microbiology. 2016;120(5):1139–1173. doi: 10.1111/jam.12986. [DOI] [PubMed] [Google Scholar]
- 7.Rice B. E., Rollins D. M., Mallinson E. T., Carr L., Joseph S. W. Campylobacter jejuni in broiler chickens colonization and humoral immunity following oral vaccination and experimental infection. Vaccine. 1997;15(17-18):1922–1932. doi: 10.1016/S0264-410X(97)00126-6. [DOI] [PubMed] [Google Scholar]
- 8.Glünder G., Spiering N., Hinz K. Investigations on parenteral immunization of chickens with a Campylobacter mineral oil vaccine. In: Nagy B., Mulder R., editors. Proceedings of the International Congress of the World Veterinary Poultry Association; 1997; Budapest, Hungary. pp. 247–253. [Google Scholar]
- 9.Widders P. R., Thomas L. M., Long K. A., Tokhi M. A., Panaccio M., Apos E. The specificity of antibody in chickens immunised to reduce intestinal colonisation with Campylobacter jejuni . Veterinary Microbiology. 1998;64(1):39–50. doi: 10.1016/s0378-1135(98)00251-x. [DOI] [PubMed] [Google Scholar]
- 10.Ziprin R. L., Hume M. E., Young C. R., Harvey R. B. Inoculation of chicks with viable non-colonizing strains of Campylobacter jejuni: evaluation of protection against a colonizing strain. Current Microbiology. 2002;44(3):221–223. doi: 10.1007/s00284-001-0088-3. [DOI] [PubMed] [Google Scholar]
- 11.Khoury C. A., Meinersmann R. J. A genetic hybrid of the Campylobacter jejuni flaA Gene with LT-B of Escherichia coli and assessment of the efficacy of the hybrid protein as an oral chicken vaccine. Avian Diseases. 1995;39(4):812–820. doi: 10.2307/1592418. [DOI] [PubMed] [Google Scholar]
- 12.Huang J.-L., Yin Y.-X., Pan Z.-M., et al. Intranasal immunization with chitosan/pCAGGS-flaA nanoparticles inhibits Campylobacter jejuni in a White Leghorn model. Journal of Biomedicine and Biotechnology. 2010;2010:8. doi: 10.1155/2010/589476.589476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meunier M., Guyard-Nicodème M., Vigouroux E., Poezevara T., Beven V. Sequential Optimization of an Avian Vaccine Protocol Against Campylobacter. 2015. [Google Scholar]
- 14.de Zoete M. R., van Putten J. P. M., Wagenaar J. A. Vaccination of chickens against Campylobacter. Vaccine. 2007;25(30):5548–5557. doi: 10.1016/j.vaccine.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Buckley A. M., Wang J., Hudson D. L., et al. Evaluation of live-attenuated Salmonella vaccines expressing Campylobacter antigens for control of C. jejuni in poultry. Vaccine. 2010;28(4):1094–1105. doi: 10.1016/j.vaccine.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 16.Neal-McKinney J. M., Samuelson D. R., Eucker T. P., Nissen M. S., Crespo R., Konkel M. E. Reducing campylobacter jejuni colonization of poultry via vaccination. PLoS ONE. 2014;9(12) doi: 10.1371/journal.pone.0114254.e114254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Theoret J. R., Cooper K. K., Zekarias B., et al. The Campylobacter jejuni dps homologue is important for In vitro biofilm formation and cecal colonization of poultry and may serve as a protective antigen for vaccination. Clinical and Vaccine Immunology. 2012;19(9):1426–1431. doi: 10.1128/cvi.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Annamalai T., Pina-Mimbela R., Kumar A., et al. Evaluation of nanoparticle-encapsulated outer membrane proteins for the control of Campylobacter jejuni colonization in chickens. Poultry Science. 2013;92(8):2201–2211. doi: 10.3382/ps.2012-03004. [DOI] [PubMed] [Google Scholar]
- 19.Wyszyńska A., Raczko A., Lis M., Jagusztyn-Krynicka E. K. Oral immunization of chickens with avirulent Salmonella vaccine strain carrying C. jejuni 72Dz/92 cjaA gene elicits specific humoral immune response associated with protection against challenge with wild-type Campylobacter. Vaccine. 2004;22(11-12):1379–1389. doi: 10.1016/j.vaccine.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Layton S. L., Morgan M. J., Cole K., et al. Evaluation of Salmonella-vectored Campylobacter peptide epitopes for reduction of Campylobacter jejuni in broiler chickens. Clinical and Vaccine Immunology. 2011;18(3):449–454. doi: 10.1128/CVI.00379-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Łaniewski P., Kuczkowski M., Chrzastek K., et al. Evaluation of the immunogenicity of Campylobacter jejuni CjaA protein delivered by Salmonella enterica sv. Typhimurium strain with regulated delayed attenuation in chickens. World Journal of Microbiology and Biotechnology. 2014;30(1):281–292. doi: 10.1007/s11274-013-1447-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark J. D., Oakes R. D., Redhead K., et al. Eimeria species parasites as novel vaccine delivery vectors: anti-Campylobacter jejuni protective immunity induced by Eimeria tenella-delivered CjaA. Vaccine. 2012;30(16):2683–2688. doi: 10.1016/j.vaccine.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Rappuoli R. Reverse vaccinology. Current Opinion in Microbiology. 2000;3(5):445–450. doi: 10.1016/S1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- 24.Hoppe S., Bier F. F., von Nickisch-Rosenegk M. Rapid identification of novel immunodominant proteins and characterization of a specific linear epitope of Campylobacter jejuni . PLoS ONE. 2013;8(5) doi: 10.1371/journal.pone.0065837.e65837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Y., Xiang Z., Mobley H. L. Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development. Journal of Biomedicine and Biotechnology. 2010;2010:15. doi: 10.1155/2010/297505.297505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oprea M., Antohe F. Reverse-vaccinology strategy for designing T-cell epitope candidates for Staphylococcus aureus endocarditis vaccine. Biologicals. 2013;41(3):148–153. doi: 10.1016/j.biologicals.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Humphrey S., Chaloner G., Kemmett K., et al. Campylobacter jejuni is not merely a commensal in commercial broiler chickens and affects bird welfare. mBio. 2014;5(4) doi: 10.1128/mbio.01364-14.e01364-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Deun K., Pasmans F., Ducatelle R., et al. Colonization strategy of Campylobacter jejuni results in persistent infection of the chicken gut. Veterinary Microbiology. 2008;130(3-4):285–297. doi: 10.1016/j.vetmic.2007.11.027. [DOI] [PubMed] [Google Scholar]
- 29.Gardy J. L., Laird M. R., Chen F., et al. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21(5):617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- 30.Sachdeva G., Kumar K., Jain P., Ramachandran S. SPAAN: a software program for prediction of adhesins and adhesin-like proteins using neural networks. Bioinformatics. 2005;21(4):483–491. doi: 10.1093/bioinformatics/bti028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Käll L., Krogh A., Sonnhammer E. L. L. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Research. 2007;35(2):W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doytchinova I. A., Flower D. R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 2007;8, article 4 doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J., Liu H., Yang J., Chou K.-C. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids. 2007;33(3):423–428. doi: 10.1007/s00726-006-0485-9. [DOI] [PubMed] [Google Scholar]
- 34.El-Manzalawy Y., Dobbs D., Honavar V. Predicting linear B-cell epitopes using string kernels. Journal of Molecular Recognition. 2008;21(4):243–255. doi: 10.1002/jmr.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sette A., Rappuoli R. Reverse vaccinology: developing vaccines in the era of genomics. Immunity. 2010;33(4):530–541. doi: 10.1016/j.immuni.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pizza M., Scarlato V., Masignani V., et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287(5459):1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 37.Andrews S. M., Pollard A. J. A vaccine against serogroup B Neisseria meningitidis: dealing with uncertainty. The Lancet Infectious Diseases. 2014;14(5):426–434. doi: 10.1016/s1473-3099(13)70341-4. [DOI] [PubMed] [Google Scholar]
- 38.Xiang Z., He Y. Genome-wide prediction of vaccine targets for human herpes simplex viruses using Vaxign reverse vaccinology. BMC Bioinformatics. 2013;14(supplement 4, article S2) doi: 10.1186/1471-2105-14-s4-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monterrubio-López G. P., González-Y-Merchand J. A., Ribas-Aparicio R. M. Identification of novel potential vaccine candidates against tuberculosis based on reverse vaccinology. BioMed Research International. 2015;2015:16. doi: 10.1155/2015/483150.483150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Argondizzo A. P. C., da Mota F. F., Pestana C. P., et al. Identification of proteins in Streptococcus pneumoniae by reverse vaccinology and genetic diversity of these proteins in clinical isolates. Applied Biochemistry and Biotechnology. 2015;175(4):2124–2165. doi: 10.1007/s12010-014-1375-3. [DOI] [PubMed] [Google Scholar]
- 41.Liu W., Chen Y.-H. High epitope density in a single protein molecule significantly enhances antigenicity as well as immunogenicity: a novel strategy for modern vaccine development and a preliminary investigation about B cell discrimination of monomeric proteins. European Journal of Immunology. 2005;35(2):505–514. doi: 10.1002/eji.200425749. [DOI] [PubMed] [Google Scholar]
- 42.Guerry P., Poly F., Riddle M., Maue A. C., Chen Y.-H., Monteiro M. A. Campylobacter polysaccharide capsules: virulence and vaccines. Frontiers in Cellular and Infection Microbiology. 2012;2:p. 7. doi: 10.3389/fcimb.2012.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolton D. J. Campylobacter virulence and survival factors. Food Microbiology. 2015;48:99–108. doi: 10.1016/j.fm.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 44.Yeh H.-Y., Hiett K. L., Line J. E. Reactions of chicken sera to recombinant Campylobacter jejuni flagellar proteins. Archives of Microbiology. 2015;197(2):353–358. doi: 10.1007/s00203-014-1062-3. [DOI] [PubMed] [Google Scholar]
- 45.Yeh H.-Y., Hiett K. L., Line J. E., Seal B. S. Characterization and antigenicity of recombinant campylobacter jejuni flagellar capping protein FliD. Journal of Medical Microbiology. 2014;63(4):602–609. doi: 10.1099/jmm.0.060095-0. [DOI] [PubMed] [Google Scholar]
- 46.Islam A., Raghupathy R., Albert M. J. Recombinant porA, the major outer membrane protein of Campylobacter jejuni, provides heterologous protection in an adult mouse intestinal colonization model. Clinical and Vaccine Immunology. 2010;17(11):1666–1671. doi: 10.1128/cvi.00255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ziprin R. L., Young C. R., Byrd J. A., et al. Role of Campylobacter jejuni potential virulence genes in cecal colonization. Avian Diseases. 2001;45(3):549–557. doi: 10.2307/1592894. [DOI] [PubMed] [Google Scholar]
- 48.Jin S., Song Y. C., Emili A., Sherman P. M., Chan V. L. JlpA of Campylobacter jejuni interacts with surface-exposed heat shock protein 90α and triggers signalling pathways leading to the activation of NF-κB and p38 MAP kinase in epithelial cells. Cellular Microbiology. 2003;5(3):165–174. doi: 10.1046/j.1462-5822.2003.00265.x. [DOI] [PubMed] [Google Scholar]


