Abstract
Methanogens have gained much attention for their metabolic product, methane, which could be an energy substitute but also contributes to the greenhouse effect. One factor that controls methane emission, reversible protein phosphorylation, is a crucial signaling switch, and phosphoproteomics has become a powerful tool for large-scale surveying. Here, we conducted the first phosphorylation-mediated regulation study in halophilic Methanohalophilus portucalensis FDF1T, a model strain for studying stress response mechanisms in osmoadaptation. A shotgun approach and MS-based analysis identified 149 unique phosphoproteins. Among them, 26% participated in methanogenesis and osmolytes biosynthesis pathways. Of note, we uncovered that protein phosphorylation might be a crucial factor to modulate the pyrrolysine (Pyl) incorporation and Pyl-mediated methylotrophic methanogenesis. Furthermore, heterologous expression of glycine sarcosine N-methyltransferase (GSMT) mutant derivatives in the osmosensitive Escherichia coli MKH13 revealed that the nonphosphorylated T68A mutant resulted in increased salt tolerance. In contrast, mimic phosphorylated mutant T68D proved defective in both enzymatic activity and salinity tolerance for growth. Our study provides new insights into phosphorylation modification as a crucial role of both methanogenesis and osmoadaptation in methanoarchaea, promoting biogas production or reducing future methane emission in response to global warming and climate change.
Methane is considered to be an energy substitute for petroleum1,2, but the emission of biologically produced methane is also a critical factor in the greenhouse effect and results in extreme climate events3,4. Methanogenic archaea are exceptional in the unusual type of metabolism that they exhibit, having the ability to gain energy from reducing CO, CO2, formate, methanol, methylamines, or acetate as energy and carbon sources for growth5,6. Thus, methanogens have received much attention because they play a pivotal role in both the recycling of carbon compounds to useful resources and the maintenance of the global carbon flux on Earth3,6. This dual role underscores the importance of regulating both the methanogenesis process and the stress response to environmental changes for methane emission.
Methanogens can utilize three types of methanogenic pathways for energy conservation: CO2-reduction, aceticlastic reactions, and methyl-group reduction (methylotrophic) pathways7. The order Methanosarcinales has the widest substrate range for methanogenesis. They are widely distributed in marine and freshwater sediments, anaerobic sewage digestors, and animal gastrointestinal tracts7,8. The methanogen used in this study, Methanohalophilus portucalensis FDF1T, belongs to the Methanosarcinales order and is cultivated from a naturally hypersaline environment9. The strain FDF1T utilizes only methanol, monomethylamine (MMA), dimethylamine (DMA), and trimethylamine (TMA) as carbon and energy sources for growth through the methylotrophic methanogenesis pathway. Intriguingly, a non-canonical amino acid, pyrrolysine (Pyl), can be incorporated by an in-frame amber codon (UAG) of three distinct methylamine methyltransferases, TMA methyltransferase (mttB), DMA methyltransferase (mtbB), and MMA methyltransferase (mtmB), to initiate methanogenesis from methylamines10,11. The utilization of Pyl is restricted to methyltransferases or other Pyl-containing proteins existing only in a few methanogenic archaea and bacteria. The importance of Pyl in methylamine-dependent methanogenesis was shown when deletion of the amber suppressor tRNAPyl (pylT) gene in Methanosarcina acetivorans disabled methane production from precursor MMA, DMA, or TMA12. Despite the functional roles of the critical components involved in the methanogenesis pathway that have been reported, there is still surprisingly little known about their regulatory networks in methanogens.
Importantly, methane produced by halophilic methanogens contributes significantly to the carbon mineralization in marine and hypersaline environments where large amounts of the greenhouse gas methane are stored13. Furthermore, TMA in these habitats is an important methane precursor for the methylotrophic methanogens, since TMA precursors are constantly provided by the degradation of osmolytes, like glycine betaine (betaine), for osmoregulation to cope with high external salinity14,15. Betaine is a type of quaternary ammonium compound that can equilibrate unbalanced osmolarity and is a common osmoprotectant in prokarya and eukarya16,17,18,19. The model methanogen M. portucalensis FDF1T has been reported to possess the capability for de novo synthesis of betaine through three-step methylation from precursor glycine via glycine sarcosine N-methyltransferase (GSMT) and sarcosine dimethylglycine N-methyltransferase (SDMT)20,21,22. It is known that intracellular salt concentration regulates the expression of GSMT and SDMT in strain FDF1T, and the amount of monovalent ions modulates the activity of rate-limited enzyme GSMT21. Furthermore, drought and salt tolerance in the arabidopsis model also increased in response to heterogeneously expressed GSMT and SDMT from strain FDF1T 23. Despite these studies on key regulatory factors contributing to osmoadapation, there is still little evidence explaining how strain FDF1T has an immediate on-off switch in the betaine synthesis process to respond to environmental changes.
Reversible protein phosphorylation is the most common cellular mechanism to regulate many physiological and adaptational processes. The first identified methanogen phosphoprotein with a known function was the methyltransferase activation protein from Methanosarcina barkeri, which converts methanol to methane via the methylotrophic methanogenesis pathway24. Furthermore, the archaeal two-component system was found to be involved in the regulation of methanogenesis in Methanosaeta harundinacea25. As genome sequences continue to accumulate, it is apparent that methanoarchaea also possess protein kinases and phosphatases, but only a few of them and their protein substrates have been defined by basic biochemical and genetic approaches26,27,28. Therefore, this report will be of particular use in delineating the physiological processes for methanogenesis and salt stress tolerance by displaying the global phosphorylation network from M. portucalensis FDF1T.
In this study, we provided a genome-wide, shotgun and phosphorylation site-specific investigation in a halophilic methanogen through MS-based systematic phosphoproteomic analysis. Both Ser/Thr/Tyr and His/Asp phosphorylation-based signaling systems were involved in diverse biological processes, especially in the single-carbon energy metabolism for methane production and osmolyte biosynthetic pathways. Although a previous study in the E. coli system had uncovered the ability to overcome hyper-salinity stress via expression of the osmolyte betaine synthesizing enzymes, GSMT and SDMT, from strain FDF1T 21, we further undertook a phosphosite mutagenesis analysis of GSMT (MPF_0823) to clarify its enzymatic activity and how its osmoregulatory function could be modulated by phosphorylation modification.
Results
Establishment of the phosphoproteome from M. portucalensis FDF1T
The phosphorylation-mediated cellular signaling network in methylotrophic halophilic M. portucalensis FDF1T was acquired through high accuracy LC-MS/MS analysis to derive a global and site-specific phosphoproteomic data set. In order to analyze a comprehensive phosphoproteome, protein extracts prepared from mid-exponential phase cultures were subjected to a combination of gel-free and gel-based approaches. In addition, TiO2-based HAMMOC was applied for efficient phosphopeptide enrichment29,30, and then analyzed in duplicates, as summarized in Supplementary Fig. 1. In total, we identified 308 unique phosphopeptides originating from 149 phosphoproteins with high confidence and a FDR of less than 1.0%. The distribution of class I phosphosites31 on Ser, Thr, Tyr, Asp, and His with a probability higher than 75.0% is listed in Supplementary Table 1. Following the typical protocols to prepare phosphopeptides, 11.8% of the phosphosites we identified were His and Asp residues. These phosphosites are thought to be mediated through two-component systems. Figure 1a exemplifies the manually annotated MS/MS spectra (full results provided in Supplementary Fig. 2), depicting the sequence VLDVApTGTGFNSVR from GSMT carrying one phosphorylated site on Thr-68. Detailed information about all identified phosphopeptides and matched MS data are listed in Supplementary Table 1.
Figure 1. A representative MS/MS spectrum and classification of the identified phosphoproteins in M. portucalensis FDF1T.
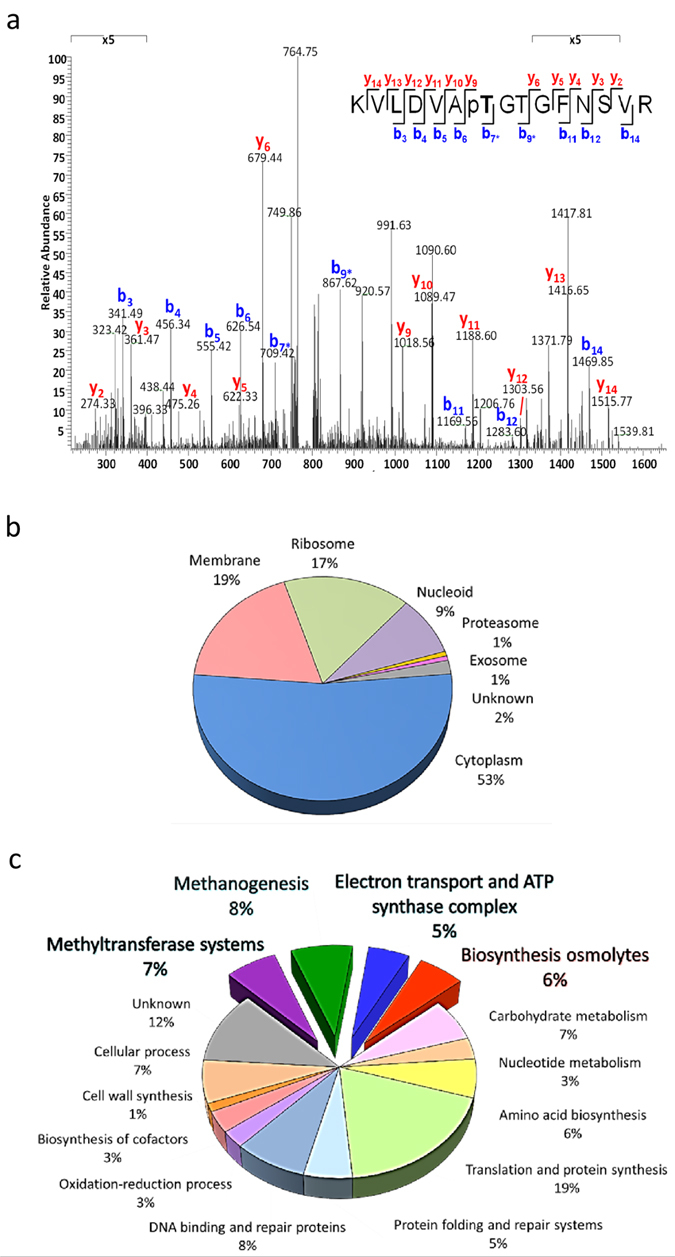
(a) The MS/MS spectrum acquired in the Orbitrap mass spectrometer for a threonine-phosphorylated peptide (VLDVApTGTGFNSVR), in which one phosphorylated site on Thr-68 was identified from GSMT (MPF_0823). Sequence-informative fragmentation ions are summarized on the peptide sequences and the matched “y” and “b” ions are annotated in red and blue, respectively. Phosphorylation site–specific ions are indicated with “p”. Fragment ion signals corresponding to additional neutral loss of NH3 are indicated by asterisks. The identified phosphoproteins were grouped by (b) cellular locations and (c) biological function based on GO terms. The biological functions related to methanogenesis and osmoadaptation are highlighted as bold blue and orange, respectively. Hypothetical proteins are grouped into the “Unknown” category and the percental distribution is given.
Classification of Methanoarchaeal Phosphoproteins
The identified phosphoproteins in M. portucalensis FDF1T were categorized by cellular localization and biological function to gain insight into their functional roles (Fig. 1b,c). Various data mining techniques using GO annotations provided useful information on the different classes, as summarized in Supplementary Table 2. Of the 149 identified phosphoproteins, 146 phosphoproteins were successfully categorized into different cellular compartments, including cytoplasm (53%), membrane (19%), ribosome (17%), nucleoid (9%), proteasome (1%), and exosome (1%) (Fig. 1b and Supplementary Table 2). As for biological function, 88.7% of the identified phosphoproteins could be assigned to 14 functional classes (Fig. 1c and Supplementary Table 1). Among them, many phosphoproteins were involved in pathways responsible for the control of key physiological processes, such as replication, transcription, translation, DNA repair systems, thermosome, proteasome, chaperone systems, osmoadaptation, and S-layer protein synthesis. To facilitate the integration of the phosphoproteome data set and uncover the functional relevance of the identified phosphoproteins, they were mapped into their complex physiological pathways as shown in Supplementary Fig. 3.
Methylotropic halophilic methanogen M. portucalensis FDF1T can metabolize TMA via methyltransferase systems to provide electrons for reducing additional molecules to methane, thereby generating the electrochemical gradient for ATP synthesis32,33. Notably, 20% of the total identified phosphoproteins were classified in methane production for energy gain categories, such as methyltransferase systems (7%), methanogenesis (8%), and electron transport and the ATP synthase complex (5%) (Fig. 1c). Moreover, the proteins involved in osmolyte biosynthesis (6%) were associated with phosphorylation-mediated regulation, which could be essential for M. portucalensis FDF1T to accumulate osmolytes in order to balance cell turgor under hypersaline conditions (Fig. 1c). The novel observation of these unique phosphorylation events advances our understanding of signal transduction in methanogenic archaea, especially for single carbon metabolisms in methanogenesis and osmotic adaptation, suggesting their functions in controlling numerous intracellular signaling and regulatory pathways.
Phosphorylation in the methylotrophic methanogenesis pathway
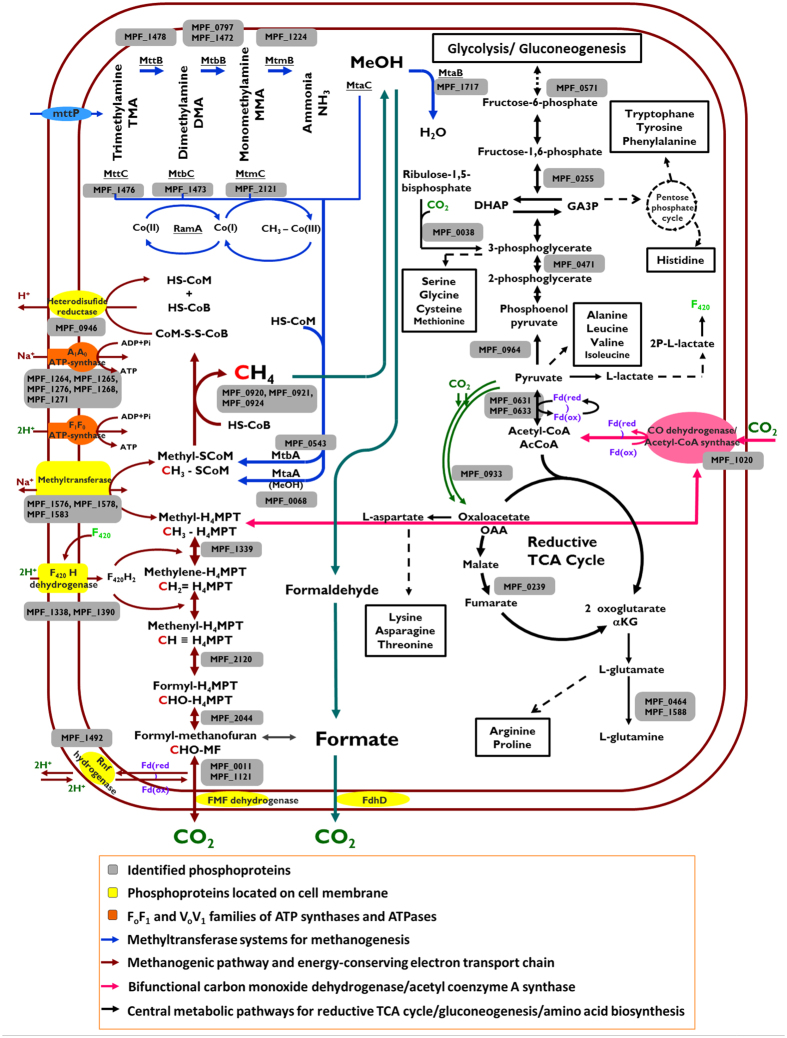
In this study, the only carbon and nitrogen source for M. portucalensis FDF1T growth was TMA. Ten methyltransferases initiating methylotrophic methanogenesis to convert methylamines to methyl groups were phosphorylated (Supplementary Table 1). Also phosphorylated were the proteins participating in the last methane-forming step leading up to an overall transfer of the methyl group from TMA to methane, including the methyl-CoM reductase McrAGB complex (MPF_0920, 0921, and 0924) and heterodisulfide reductase HdrD (MPF_0946). In energy-yielding processes generating a proton gradient for driving ATP synthesis, we found that five distinct ATP synthases, two membrane-bound subunits from the F420H2 dehydrogenase complex, and one electron transport RnfC could be phosphorylated (Supplementary Table 1). Likewise, we found ten phosphoproteins in carbohydrate metabolic processes such as glycolysis, gluconeogenesis, and the reductive tricarboxylic acid cycle. Figure 2 is a schematic drawing of the phosphorylation events, illustrating in greater detail these phosphoproteins participating in TMA utilization for methanogenesis and energy metabolism. These unprecedented findings suggest that protein phosphorylation globally regulates the initiation of methyl transfer reactions for the metabolism of TMA, which produces methane and generates a sufficiently positive redox potential.
Figure 2. Schematic illustration of phosphorylation events in methanogenesis from TMA metabolic pathways in M. portucalensis FDF1T.
Integrated phosphoproteome data mapped into methanogenic and housekeeping pathways for cells to utilize TMA, produce methane, and gain energy, as shown in different color arrows. The carbon atoms of the methanogenesis are labeled in red “C”. The shaded boxes with gene IDs (MPF_numbers) show the phosphorylated enzymes in this study.
Pyrrolysine in phosphorylated methylamine methyltransferases
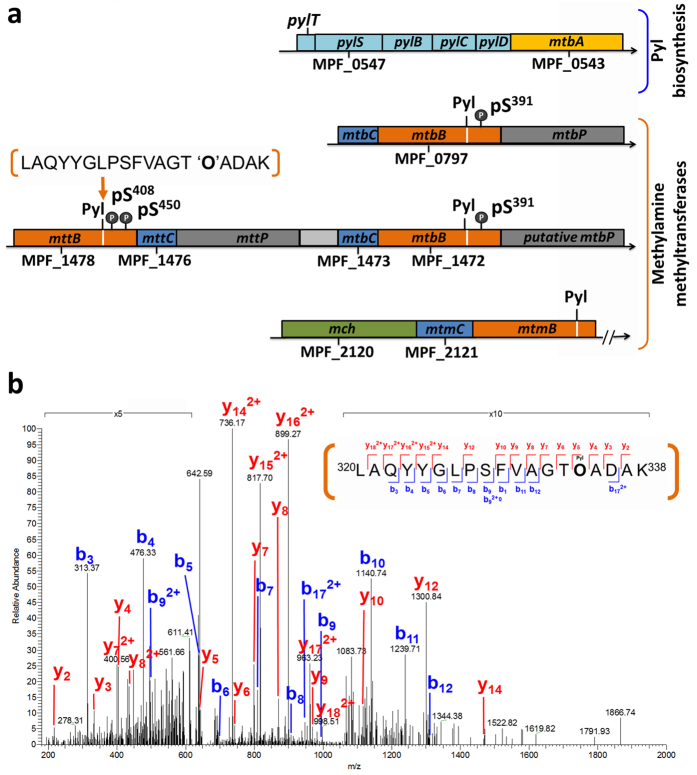
Interestingly, several phosphorylation sites were located in the C-terminal sequence downstream of the amber (UAG) codon, which encodes pyrrolysine (Pyl) during protein synthesis (Fig. 3a and Supplementary Fig. 4a). The peptide with the amber (UAG)-encoded pyrrolysyl-residue incorporated into MttB (MPF_1478) was measured by mass spectrometry as shown in Fig. 3b and Supplementary Table 4, demonstrating that Pyl can be naturally synthesized and incorporated into proteins. Furthermore, the pylTSBCD genes for Pyl synthesis are located within a cluster that contains the gene for methylamine-specific methyltransferase MtbA (MPF_0343) (Fig. 3a). Intriguingly, the pyrrolysyl-tRNA synthetase (MPF_0547/PylS) identified for Pyl-tRNAPyl formation included multiple phosphosites at Ser-390/Ser-391 and possibly Tyr-384/Thr-385 on the C-terminal tail of the predicted structure (Supplementary Fig. 4b), which is the core-binding surface for the tRNAPyl acceptor helix34,35,36. Taking into account the importance of Pyl being present in the active site of monomethylamine methyltransferase MtmB in M. barkeri37, our findings imply that the formation of the Pyl-tRNAPyl may be regulated by protein phosphorylation, thereby influencing the enzymatic integrity of methylamine methyltransferases.
Figure 3. Manual annotation and MS/MS analysis reveal Pyl incorporation at amber codons and downstream phosphorylation.
(a) The completely sequenced M. portucalensis genome revealed the operon (cyan) that synthesizes Pyl and incorporates it into methyltransferases. Most databases truncate methylamine methyltransferases at the amber codon, but our analysis indicated the incorporation of a Pyl residue (white line) and downstream phosphorylation on Ser-408/Ser-450 and Ser-391 in four methyltransferases (orange). (b) LC-MS/MS analysis confirmed the Pyl incorporation in a tryptic peptide (residues 320–338). The pyrrolysine residue “O” with a mass of 237.3 Da was detected at position 334 of TMA methyltransferase MttB (MPF_1478).
Phosphorylation in salt stress response
Unlike bacteria, most archaea lack rigid outer envelopes, like the peptidoglycan, for salt resistance38. They instead rely on strategies like accumulating small molecular osmolytes to overcome the high turgor pressure. M. portucalensis FDF1T synthesizes glycine betaine de novo as a preferred osmolyte for turgor adjustment21. We found that most proteins participating in the betaine uptake and synthesis pathways were phosphorylated (Supplementary Table 1), including the glycine betaine BtaABC transporter (Supplementary Fig. 3) and enzymes involved in the methionine transmethylation cycle and betaine biosynthesis (Supplementary Fig. 5). Because GSMT was a protein known to be the rate-limiting enzyme in the production of intermediate substrate sarcosine for further betaine synthesis21, we investigated the phosphorylation of GSMT. Previous research had uncovered only four phosphorylated serine residues in the GSMT ortholog in rat hepatocytes, Glycine N-methyltransferase (GNMT)39. We identified one of those phosphosites in GSMT as well as nine more. The multiple phosphosites observed on GSMT included six unambiguous phosphosites with a localization probability ≥75% (Ser-46, Thr-68, Thr-70, Tyr-169, Ser-178, and Ser-179) and four putative residues scoring <75% (Ser-74, Asp-170, Asp-174, and Tyr-177) (Supplementary Table 1). Mammalian GNMT exhibits strong structural similarity with the highly phosphorylated M. portucalensis GSMT (MpGSMT) (Supplementary Fig. 6a), suggesting that there are more phosphorylation sites to be uncovered in GNMT, which may underscore the importance of GNMT to a further extent than the four previously identified serine residues. To better understand how cells maintain osmotic balance via protein phosphorylation, we focused on phosphorylated MpGSMT and examined the roles of the identified phosphorylation sites.
The effects of MpGSMT phosphothreonine on methyltransferase activity and salt stress response
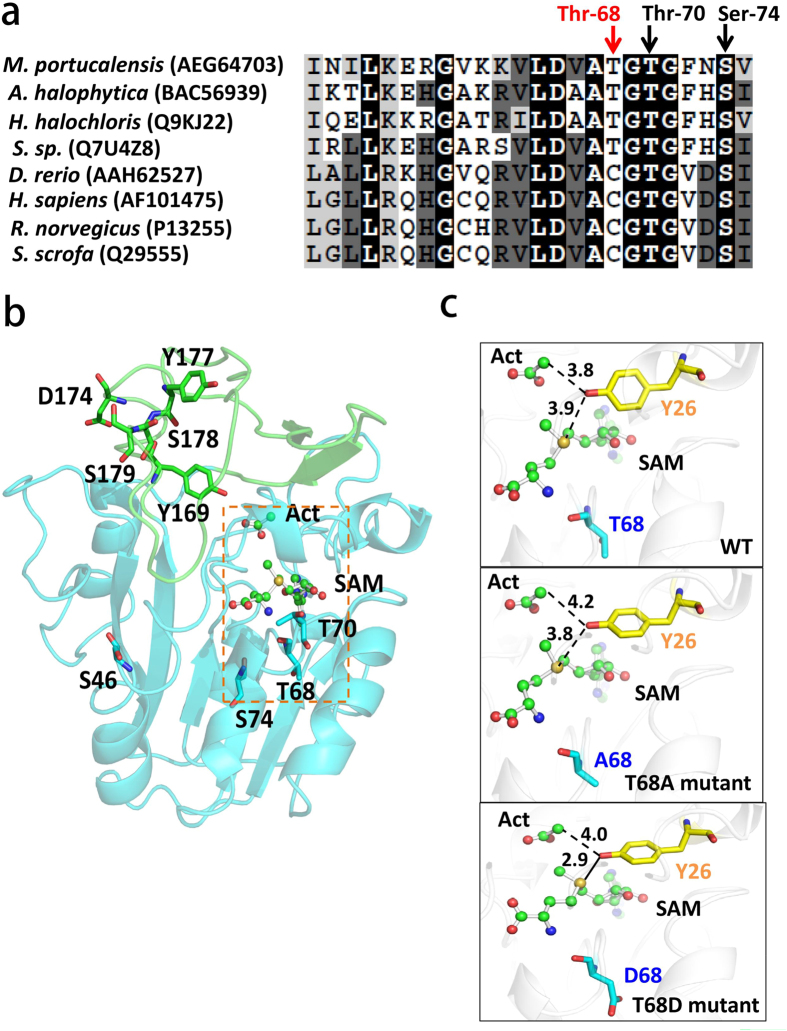
Sequence alignment and predicted MpGSMT structure indicate that the first four phosphorylated sites (Ser-46, Thr-68, Thr-70, and Ser-74) are located in the SAM binding region, and the remaining phosphosites (Tyr-169, Asp-170, Asp-174, Tyr-177, Ser-178, and Ser-179) are located in the lid structure for substrate glycine or sarcosine binding (Fig. 4a,b). Notably, phosphosites Thr-68 and Thr-70 are situated in a highly conserved GxG motif of class I methyltransferases for the interaction with the methionine group of SAM40 (Fig. 4a). Intriguingly, phosphosite Thr-68 was highly conserved in prokaryotic GSMT, but the residue was replaced with a nonphosphorylatable Cys in eukaryotic GNMT, hinting at the possible regulation of MpGSMT mediated by threonine phosphorylation. Based on energy minimization of the catalytic environment of MpGSMT modeling structures (Fig. 4c), the distance between phenolic hydroxyl Tyr-26 and the sulfur atom on SAM indicate suitable geometry to form a hydrogen bond with the potential to inhibit the enzymatic reaction in the T68D phospho-mimicking mutant, but not in the other two modeling structures (WT and T68A).
Figure 4. The transmethylation activity of MpGSMT was regulated via Thr phosphorylation.
(a) Partial sequence alignment of the MpGSMT with its homologous regions from prokaryotic GSMT and eukaryotic GNMT. The amino acid sequences of GSMT were captured from M. portucalensis FDF1T, Aphanothece halophytica, Halorhodospira halochloris, and Synechococcus sp. WH8102, and the sequences of GNMT from Danio rerio, Homo sapiens, Rattus norvegicus, and Sus scrofa. The conserved residues are highlighted in black, while the strongly similar residues are shown in gray background. Arrows indicate the phosphorylated residue Thr-68, Thr-70, and Ser-74 located in SAM binding motif. The red arrow marks the position of phospho-residue that regulate activity in MpGSMT. (b) The phosphorylation sites mapped to MpGSMT predicted structure according to the rat GNMT (PDB: 1nbh) as template, which was solved with SAM and acetate mimicking glycine in catalytic pocket. The lid moiety (167–219) was shown in green ribbon and central domain containing SAM binding motif (43–166 and 220–263) was marked in blue. The phosphosites were highlighted in sticks. (c) An expanded view of catalytic pocket of wild-type GSMT, T68A, and T68D mutants. The Tyr-26 located near the SAM-binding site was labeled in yellow stick. The distances between Tyr-26 and SAM were labeled in dashed lines. The possible hydrogen bond was denoted in solid line with the distance of 2.9 Å.
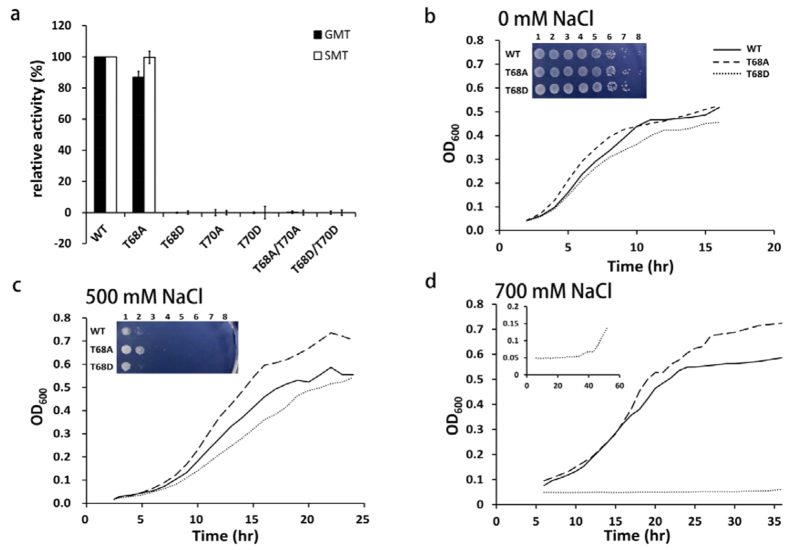
We therefore examined the transmethylation activities by site-specific mutagenesis to determine the essential phosphorylated-threonine residues to be Thr-68 and Thr-70. While WT- and T68A-MpGSMT retained their GMT and SMT enzymatic activities, transmethylation activities were almost completely abrogated in all dephospho- (T70A and T68A/T70A) and phospho-mimetic isoforms (T68D, T70D, and T68D/T70D) (Fig. 5a). These results implied that Thr-68, but not Thr-70, could have a regulatory role for M. portucalensis in response to betaine biosynthesis through phosphorylation/de-phosphorylation. Additionally, circular dichroism (CD) analysis showed that the secondary structures of T68A and T68D mutant proteins had no conformational change (Supplementary Table 5), indicating that these point mutations did not affect their protein structure.
Figure 5. Phosphosite mutation of MpGSMT affects methyltransferase activity and growth rate at elevated osmolarity.
(a) The relative GMT and SMT activities of recombinant WT MpGSMT and various mutant forms. The methyltransferase activities were performed by modified acid-washed charcoal method under 1.0 M of KCl with 0.5 M glycine or sarcosine as the substrate. All the data points were averaged by triplicate experiments and displayed as percentages relative to wild-type MpGSMT. The effect of salt adaptation was assayed by the growth of E. coli MKH13 containing WT or mutant (T68A and T68D) MpGSMT co-expressing with SDMT in M9 minimal media supplemented with (b) 0 mM, (c) 500 mM, or (d) 700 mM NaCl. Growth rate was monitored by measuring 600 nm optical densiometry (OD600) over 24 h at 37 °C. E. coli strain MKH13 without a recombinant GSMT21 had the same growth rate as the T68D mutant. The effect of salt shock shown in the inset of (b,c) was measured by drop tests of serious dilution from 100 to 10−7 of overnight cultures on agar plates, but cells failed to grow on agar plate with (d) 700 mM NaCl. Insect in (d) indicates the growth curve of T68D mutant with a large time-scale over 48 hours.
To further examine the biological effects of MpGSMT isoforms on osmoadaptation, we measured the growth of the osmosensitive strain E. coli MKH13 with heterologous expression of Mpgsmt-sdmt (WT) and mutant Mpgsmt-sdmt (T68A- or T68D-GSMT) genes. Media containing 0 mM, 500 mM or 700 mM NaCl were prepared in solid form for salt shock and liquid form for salt adaptation growth tests. The growth patterns of E. coli MKH13 with WT-, T68A- or T68D-GSMT were similar under the non-saline condition (Fig. 5b). Interestingly, E. coli MKH13 expressing T68A-GSMT had a fast-growth phenotype, adapting faster in 500 mM and 700 mM NaCl, while the strain expressing T68D-GSMT displayed a slow-growth phenotype (Fig. 5c,d). In other words, expression of T68A-GSMT in E. coli MKH13 conferred higher tolerance to salt stress than the expression of WT-GSMT (Fig. 5c,d). Conversely, the strain expressing T68D-GSMT required a longer lag period (45 hours) to overcome 700 mM NaCl salt stress (Fig. 5d). Similarly, the salt shock growth tests using solid media showed that the strain expressing T68A-GSMT possessed higher salt stress tolerance as NaCl concentration increased from 0 to 500 mM NaCl (Fig. 5c). However, none of the constructs could protect the cells against salt shock on solid media with 700 mM NaCl. Taken altogether, our results support the hypothesis that Thr-68 phosphorylation in MpGSMT could regulate the dual catalytic activities of GMT and SMT in GSMT while playing a leading role in osmoprotection.
Discussion
Microbial methanogenesis is important in the carbon cycle, impacting climate change or contributing to renewable energy. A fair amount of research attention in methanogenic ecosystems or biosynthetic functions has therefore approached this issue7,41,42,43. In this study, we provided additional insights into the unique metabolic pathways and environmental adaptation correlated with functionally meaningful phospho-regulatory events in the halophilic methanogen M. portucalensis FDF1T. Through further investigation by site-directed mutagenesis, we clarified the role of MpGSMT phosphorylation in regulating methyltransferase activities and salt tolerance.
The 149 phosphoproteins identified in this study approximately 7.0% of all the ORFs encoded in M. portucalensis FDF1T genome (nearly 2131 encoded proteins), compared with 2.5% of the halophilic archaeon Halobacterium salinarum44. Both archaeal phosphoprotemic analyses revealed that phosphorylated proteins were involved in almost every cellular process, though unique physiological traits such as methanogenesis and osmolyte biosynthesis were uncovered in M. portucalensis (Fig. 1c and Supplementary Fig. 3). We observed that many phosphoproteins were classified as translation and protein repair (Fig. 1c, Supplementary Fig. 3 and Supplementary Table 1), possibly promoting protein homeostasis to adapt to fluctuations in external osmotic pressure. This phenomenon is similarly observed in eukaryotes to buffer normal fluctuations in cellular state and maintain cellular homeostasis via phosphorylation-mediated regulation of translation or chaperone proteins45.
Methanogens in the Methanosarcinaceae family have been found in a wide variety of anaerobic environments where methane is produced from the widest substrate range; methylamines and methanol are generally used to generate energy46. The enzymes involved in methylotrophic methanogenesis and the coupled electron transfer were phosphorylated (Fig. 2), suggesting that methylotrophic methanogenesis might be globally regulated by protein phosphorylation. On the other hand, the formation of a multienzyme complex is known to be essential for demethylating substrates. All of the identified phosphosites from MtmC, MtbC, MttC, MtaA, and MtbA were located within the C-terminal domain with a Rossmann fold structure in C subunits or within the N-terminal segment capping the active center in A subunits (Supplementary Fig. 7), which is the interface between an A subunit and the core complex of the BC subunits47,48. Likewise, some phosphosites on MtaB, MtmB, and MttB were located in the helical layer, which surrounds the TIM barrel structure to interact with the partner C subunit and potentially the A subunit48. Taken together, these findings suggest that phosphorylation may influence the stability of methyltransferase quaternary structural complexes, and in turn alter their enzymatic activities in methylotrophic methanogenesis, but further biological validation is required to confirm the significance of our findings.
Notably, phosphorylated MttB was found to possess an in-frame amber codon with Pyl, and the pyrrolysyl-tRNA synthetase PylS (MPF_0547) for Pyl-tRNAPyl formation and Pyl incorporation was found to be phosphorylated (Fig. 3a and Supplementary Fig. 4). Building upon the known importance of Pyl incorporation in methanogenesis37,49, our results not only demonstrated that M. portucalensis FDF1T is a Pyl-utilizing archaea but also suggested the possibility that protein phosphorylation plays an upstream regulatory role in facilitating methylamine metabolism.
Osmolyte biosynthesis pathways and protein refolding processes are energy consuming systems and are required to respond immediately to osmotic stresses50. Twelve units of ATP are required to generate one SAM, and three units of SAM are capable of synthesizing one unit of the osmolyte betaine, consuming 36 units of ATP in total51,52. From a bioenergetic point of view, phospho-regulatory mechanisms might provide a more energy-efficient strategy to maintain osmotic equilibrium by rapidly switching protein activity on or off. Results from the enzymatic activities and salinity tolerance assay in E. coli MKH13 (Fig. 5) demonstrated that the Thr-68-dephosphorylated MpGSMT was constitutively active in adapting to osmotic fluctuations.
Furthermore, we present the substrate-assisted catalysis model using MpGSMT_WT, T68A, and T68D mutants to illustrate that the phosphorylation-simulated mutant T68D has the potential to form a hydrogen bond between the hydroxyl group of Y26 and the sulfur atom of SAM (Fig. 4c). Because the position of residue Y26 in the predicted MpGSMT structure is a highly conserved homology of the Y21 residue of GNMT (PDB:1nbh) (Supplementary Fig. 6a), the catalytic center of both methyltransferases may be similar53,54. Accordingly, we further propose a transmethylation mechanism for the nonphosphorylated MpGSMT through the electron transferring from Tyr-26 to attack the amine group of glycine, followed by the methyl group attacking the sulfur atom of SAM, which may lead to methyl transfer from SAM to glycine, thus generating methylated glycine (sarcosine) and S-adenosylhomocysteine (SAH) (Supplementary Fig. 6b). Conversely, we speculate that phosphorylation on Thr-68 shortens the distance between Y26 and SAM (Fig. 4c) and in turn retards the transmethylation reaction, as evidenced by the T68D mutant causing an inactive form of MpGSMT (Fig. 5a). These results indicate that threonine phosphorylation negatively regulates MpGSMT for energy conservation.
It should be noted that GNMT is considered a tumor suppressor of human hepatocellular carcinoma, and the position of the phosphorylated residues in the GNMT tertiary structure is likely to affect the protein’s conformation and activity55. For instance, phosphosite Ser-71 of GNMT may affect the microenvironmental net charge of SAM binding pocket, and Ser-182 could modulate the tetramer switching to the dimer39. More surprising is the fact that phosphorylated GNMT significantly increases enzyme activity39, in contrast to phosphorylated MpGSMT, which presented a methyltransferase-inactive phenotype (Fig. 5a). Intriguingly, the phosphosites Ser-74 and Asp-174 identified in MpGSMT corresponding to rat GNMT Ser-71 and Ser-182 were evolutionarily conserved (Supplementary Fig. 8), while the quaternary structure of MpGSMT is regulated via potassium ion concentration but not mediated by protein phosphorylation21. Furthermore, the highly conserved phosphosite Thr-68 in prokaryotic GSMT was replaced by Cys-65 in eukaryotic GNMT (Fig. 4a), revealing that GNMTs in eukarya possessing constitutive activity might be due to the non-phosphorylatable residue Cys-65. It may therefore be regulated through another mechanism modulating the ratio of SAM/SAH in cells. Collectively, our recent findings revealed a tight correlation between the phospho-regulation and methyltransferase activities of MpGSMT, especially as an energy-efficient strategy in response to osmotic regulation.
In conclusion, we present the global phosphorylation-mediated cellular signaling networks in halophilic methanogen M. portucalensis FDF1T to highlight the importance of phospho-regulation in methanogenesis and the betaine biosynthesis process. In particular, we demonstrated that the FDF1T is a pyrrolysine-utilizing archaea and hypothesized that methane production via the methyltrophic methanogenesis may be regulated by protein phosphorylation in both protein translation and the methylamine metabolic process. In addition, this is the first study to elucidate the importance of Thr phosphorylation in prokaryotic GSMT, and clarifying that Thr-68 phosphorylation plays a distinct role in enzymatic activity and salt tolerance. This report advances our knowledge of the underlying mechanisms in the modulation of methanogenesis and osmotic adaptation and potentiates improved methane yields or minimized methane emission, both crucial in addressing global warming and climate change.
Methods
Growth conditions and protein extraction
The bacterial strains used for cloning and heterologous expression were E. coli DH5α and E. coli BL21(DE3)RIL (Novagen, Madison, WI), respectively. The archaeal strain Methanohalophilus portucalensis strain FDF1T (=DSM 7471)9, was used in this study to derive the phosphoproteome.
M. portucalensis FDF1T were cultured in medium containing 120 g L−1 NaCl and 20 mM trimethylamine as a sole carbon and energy source, as described in the literature 56. The harvested mid-exponential phase (OD540 at 0.5) cell pellets were resuspended in fresh lysis buffer (25 mM ammonium bicarbonate, PhosSTOP phosphatase inhibitor mixture tablets (Roche), 6 M urea, and 2 M thiourea) and disrupted by sonication on ice. The protein concentration was quantified by the Bradford protein assay (Bio-Rad).
Phosphopeptide preparation and NanoLC-MS/MS Analysis
In order to compile a comprehensive phosphoproteome data set, the total protein was treated with trypsin in both gel-based and gel-free processes following procedures described in the literature57. Phosphopeptides from the tryptic peptides were enriched by custom-made HAMMOC tips, which were prepared using 0.5 mg TiO2 beads (GL Sciences, Tokyo, Japan) packed into 10-μL C8-StageTips, as described previously29,30.
The peptide mixtures were analyzed by online nanoflow liquid chromatography tandem mass spectrometry (LC-MS/MS) on a nanoAcquity system (Waters, Milford, MA) coupled to an LTQ-Orbitrap Velos hybrid mass spectrometer (Thermo Scientific) equipped with a PicoView nanospray interface (New Objective). Detailed detection conditions are described in Supplementary Information.
MS/MS database search and phosphorylation site analysis
All MS and MS/MS raw data were analyzed using MaxQuant software (version 1.4.1.2) (http://www.maxquant.org/)58 with the built-in search engine Andromeda59 for phosphopeptide identification and phosphorylation site analysis. The protein sequences for the MS/MS database search consisted of an in-house draft genome sequence of M. portucalensis strain FDF1T with all 2, 131 protein sequences and a well-annotated genome of M. mahii DSM 521932, which shares 99.8% sequence identity with M. portucalensis strain FDF1T and contains 1, 987 sequences (data download from the NCBI Reference Sequence database on December 3, 2013). The protein-encoding genes from the M. portucalensis strain FDF1T genome sequence were previously predicted by Glimmer 2.1360, GeneMark 2.4, and GeneMark.hmm 2.161 and annotated with the RefSeq Microbial Genomes database62 using BLASTP in standard settings (E-value < 10−5, identity >40%, and matched length >30%). The detailed search criteria are described in Supplementary Information. The identified phosphoproteins matched to protein sequences in M. portucalensis strain FDF1T were reported and listed in Supplementary Table 1. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium63 via the PRIDE partner repository with the dataset identifier PXD002024. The detailed bioinformatics analyses used to further classify each identified protein are described in Supplementary Information, and all results were compiled into a data set provided in Supplementary Table 2.
Cloning, expression, and purification of recombinant GSMT and mutant proteins
The Mpgsmt (GenBank: AEG64703) (EC 2.1.1.156) was cloned into the pET28a expression vector (Novagen)21 and then used as a template for site-directed mutagenesis64. Residues Thr-68 and Thr-70 of MpGSMT were substituted with Ala to mimic a non-phosphorylated state, and with Asp to mimic a phosphorylated state. The specific primers are listed in Supplementary Table 3. The pET28a-Mpgsmt construct and its mutant derivatives were transformed into E. coli BL21(DE3)RIL for heterologous expression as described in literature21.
Methyltransferase activity assay
MpGSMT exhibiting GMT and SMT activities were determined in the forward direction, measuring sarcosine and dimethylglycine formation from glycine (Sigma) and sarcosine (Sigma) substrates. A modified acid-washed charcoal method was used to detect methyltransferase activities following the standard protocol described in literature21,65.
Salt tolerance test
The plasmid pUHE21-Mpgsmt-sdmt co-expressing MpGSMT/SDMT was initially introduced into the osmolyte uptake mutant E. coli MKH13 as described in literature21. This plasmid was also used as a template for the point mutation of MpGSMT at residue Thr-68 to Ala or Asp for mimicking dephosphorylation and phosphorylation, respectively. The MKH13 cells harboring plasmid pUHE21-Mpgsmt-sdmt were defined as wild type (WT), while cells carrying the plasmid with a mutated version of Mpgsmt at Thr-68 were indicated as mutant T68A or T68D. For growth tests, three E. coli strains (WT, T68A, and T68D) were grown in M9 minimal medium without NaCl for at least three generations, and then subcultured in different NaCl concentrations in solid and liquid medium to investigate the influence of salt shock and salt adaptation, respectively. To assess the effects of salt shock, the overnight cultures of the three strains were diluted to OD600 of 1.0 using M9 medium without NaCl, and then used for further serial dilution to be dotted in equal volumes of each dilution, ranging from 100 to 10−7, on M9 agar plates containing 0, 500, or 700 mM NaCl. All the conditions were carried out in triplicate and incubated at 37 °C. To assess the effects of salt adaptation, the three strains were acclimated in M9 liquid medium containing 0, 500, or 700 mM NaCl for three generations. The cells in 0 mM NaCl were inoculated at an initial OD600 of 0.01 for growth curves. In order to shorten the lag period prior to initiating growth in high concentrations of 500 or 700 mM NaCl, the initial OD600 value was fixed at 0.03. The growth rates of each strain in different salt conditions were determined in duplicate experiments.
Additional Information
How to cite this article: Wu, W.-L. et al. Phosphoproteomic analysis of Methanohalophilus portucalensis FDF1T identified the role of protein phosphorylation in methanogenesis and osmoregulation. Sci. Rep. 6, 29013; doi: 10.1038/srep29013 (2016).
Supplementary Material
Acknowledgments
This work was supported in part by the Ministry of Education, Taiwan, R.O.C. under the ATU plan (to M.C.L.) and the postdoc fellowship has been supported by grant NSC 100-2321-B-005-005-MY3 and MOST 103-2113-M-001-029-MY3 from the Ministry of Sciences and Technology, Taiwan, ROC (to S.J.L.). The postdoc fellowship to W.L.W. was supported by Central Academic Advisory Committee at Academia Sinica. Proteomic mass spectrometry analyses were performed by the Core Facilities for Protein Structural Analysis located at the Institute of Biological Chemistry, Academia Sinica, and supported by a National Science Council grant (NSC100-2325-B-001-029) and the Academia Sinica. Portions of this work were also supported by a grant (AS-102-SS-A19) from Sustainability Science Research Program. We would like to thank Dr. Jiahn-Haur Liao and Dr. Cindy Lee for their advice.
Footnotes
Author Contributions M.-C.L. and S.-H.W. conceived and directed the project execution; W.-L.W. and J.-T.Y. performed phosphoproteomic experiments, data processing, and metabolic reconstruction; S.-J.L. prepared FDF1T samples and performed genome sequence and mutant analysis; S.-Y.L. and C.-H.K. provided support for computational analysis tools and gene prediction and annotation; C.-C.C. performed LC-MS/MS analysis; J.C. carried out homology modeling; W.-L.W., S.-J.L., M.-C.L. and S.-H.W. wrote the initial draft of the paper.
References
- De Vrieze J., Hennebel T., Boon N. & Verstraete W. Methanosarcina: the rediscovered methanogen for heavy duty biomethanation. Bioresour Technol 112, 1–9 (2012). [DOI] [PubMed] [Google Scholar]
- Ferry J. G. Fundamentals of methanogenic pathways that are key to the biomethanation of complex biomass. Curr Opin Biotechnol 22, 351–357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschke S. et al. Three decades of global methane sources and sinks. Nat Geosci 6, 813–823 (2013). [Google Scholar]
- Thauer R. K., Kaster A. K., Seedorf H., Buckel W. & Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nature reviews Microbiology 6, 579–591 (2008). [DOI] [PubMed] [Google Scholar]
- Reeve J. N. Molecular biology of methanogens. Annual review of microbiology 46, 165–191 (1992). [DOI] [PubMed] [Google Scholar]
- Deppenmeier U. The unique biochemistry of methanogenesis. Progress in nucleic acid research and molecular biology 71, 223–283 (2002). [DOI] [PubMed] [Google Scholar]
- Liu Y. & Whitman W. B. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Annals of the New York Academy of Sciences 1125, 171–189 (2008). [DOI] [PubMed] [Google Scholar]
- Hedderich R. & Whitman W. Physiology and biochemistry of the methane-producing Archaea. In: Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. (eds). The Prokaryotes Springer, New York. pp 1050–1079 (2006). [Google Scholar]
- Boone D. R. et al. Isolation and characterization of Methanohalophilus portucalensis sp-nov and DNA reassociation study of the genus Methanohalophilus. Int J Syst Bacteriol 43, 430–437 (1993). [Google Scholar]
- Borrel G. et al. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC genomics 15, 679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston M. A., Jiang R. & Krzycki J. A. Functional context, biosynthesis, and genetic encoding of pyrrolysine. Current opinion in microbiology 14, 342–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra A. et al. Characterization of a Methanosarcina acetivorans mutant unable to translate UAG as pyrrolysine. Molecular microbiology 59, 56–66 (2006). [DOI] [PubMed] [Google Scholar]
- Kvenvolden K. A. Methane hydrate - a major reservoir of carbon in the shallow geosphere. Chem Geol 71, 41–51 (1988). [Google Scholar]
- King G. M. Methanogenesis from methylated amines in a hypersaline algal mat. Applied and environmental microbiology 54, 130–136 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G. M. Metabolism of trimethylamine, choline, and glycine betaine by sulfate-reducing and methanogenic bacteria in marine sediments. Applied and environmental microbiology 48, 719–725 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. H. & Murata N. Glycinebetaine protects plants against abiotic stress: mechanisms and biotechnological applications. Plant Cell Environ 34, 1–20 (2011). [DOI] [PubMed] [Google Scholar]
- Chen T. H. & Murata N. Glycinebetaine: an effective protectant against abiotic stress in plants. Trends Plant Sci 13, 499–505 (2008). [DOI] [PubMed] [Google Scholar]
- Roesser M. & Muller V. Osmoadaptation in bacteria and archaea: common principles and differences. Environmental microbiology 3, 743–754 (2001). [DOI] [PubMed] [Google Scholar]
- Sleator R. D. & Hill C. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS microbiology reviews 26, 49–71 (2002). [DOI] [PubMed] [Google Scholar]
- Lai M. C., Wang C. C., Chuang M. J., Wu Y. C. & Lee Y. C. Effects of substrate and potassium on the betaine-synthesizing enzyme glycine sarcosine dimethylglycine N-methyltransferase from a halophilic methanoarchaeon Methanohalophilus portucalensis. Research in microbiology 157, 948–955 (2006). [DOI] [PubMed] [Google Scholar]
- Lai S. J. & Lai M. C. Characterization and regulation of the osmolyte betaine synthesizing enzymes GSMT and SDMT from halophilic methanogen Methanohalophilus portucalensis. PloS one 6, e25090 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. Y., Lai M. C., Lai S. J. & Lee Y. C. Characterization of osmolyte betaine synthesizing sarcosine dimethylglycine N-methyltransferase from Methanohalophilus portucalensis. Archives of microbiology 191, 735–743 (2009). [DOI] [PubMed] [Google Scholar]
- Lai S. J., Lai M. C., Lee R. J., Chen Y. H. & Yen H. E. Transgenic Arabidopsis expressing osmolyte glycine betaine synthesizing enzymes from halophilic methanogen promote tolerance to drought and salt stress. Plant molecular biology 85, 429–441 (2014). [DOI] [PubMed] [Google Scholar]
- Daas P. J. et al. Purification and properties of an enzyme involved in the ATP-dependent activation of the methanol:2-mercaptoethanesulfonic acid methyltransferase reaction in Methanosarcina barkeri. J Biol Chem 271, 22339–22345 (1996). [DOI] [PubMed] [Google Scholar]
- Li J., Zheng X., Guo X., Qi L. & Dong X. Characterization of an archaeal two-component system that regulates methanogenesis in Methanosaeta harundinacea. PloS one 9, e95502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennelly P. J. Archaeal protein kinases and protein phosphatases: insights from genomics and biochemistry. Biochem J 370, 373–389 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Potts M. & Kennelly P. J. The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiol Rev 22, 229–253 (1998). [DOI] [PubMed] [Google Scholar]
- Kennelly P. J. Protein Ser/Thr/Tyr Phosphorylation in the Archaea. J Biol Chem (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N. et al. Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol Cell Proteomics 6, 1103–1109 (2007). [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Mann M. & Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2, 1896–1906 (2007). [DOI] [PubMed] [Google Scholar]
- Olsen J. V. et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 (2006). [DOI] [PubMed] [Google Scholar]
- Spring S. et al. The genome sequence of Methanohalophilus mahii SLP(T) reveals differences in the energy metabolism among members of the Methanosarcinaceae inhabiting freshwater and saline environments. Archaea 2010, 690737 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross R. L. & Muller V. The evolution of A-, F-, and V-type ATP synthases and ATPases: reversals in function and changes in the H+/ATP coupling ratio. FEBS letters 576, 1–4 (2004). [DOI] [PubMed] [Google Scholar]
- Nozawa K. et al. Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality. Nature 457, 1163–1167 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. M. et al. Structure of Desulfitobacterium hafniense PylSc, a pyrrolysyl-tRNA synthetase. Biochem Biophys Res Commun 374, 470–474 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa T. et al. Crystallographic studies on multiple conformational states of active-site loops in pyrrolysyl-tRNA synthetase. J Mol Biol 378, 634–652 (2008). [DOI] [PubMed] [Google Scholar]
- Hao B. et al. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science 296, 1462–1466 (2002). [DOI] [PubMed] [Google Scholar]
- Kandler O. & Konig H. Cell wall polymers in Archaea (Archaebacteria). Cell Mol Life Sci 54, 305–308 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luka Z. et al. Identification of phosphorylation sites in glycine N-methyltransferase from rat liver. Protein science : a publication of the Protein Society 15, 785–794 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. L. & McMillan F. M. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Current opinion in structural biology 12, 783–793 (2002). [DOI] [PubMed] [Google Scholar]
- Youngblut N. D. et al. Genomic and phenotypic differentiation among Methanosarcina mazei populations from Columbia River sediment. The ISME journal 9, 2191–2205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J. L., Patel B. K. & Ollivier B. Taxonomic, phylogenetic, and ecological diversity of methanogenic Archaea. Anaerobe 6, 205–226 (2000). [DOI] [PubMed] [Google Scholar]
- Costa K. C. & Leigh J. A. Metabolic versatility in methanogens. Current opinion in biotechnology 29, 70–75 (2014). [DOI] [PubMed] [Google Scholar]
- Aivaliotis M. et al. Ser/Thr/Tyr protein phosphorylation in the archaeon Halobacterium salinarum–a representative of the third domain of life. PloS one 4, e4777 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda S., Gorman A. M., Hori O. & Samali A. Cellular stress responses: cell survival and cell death. Int J Cell Biol 2010, 214074 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Haridon S., Chalopin M., Colombo D. & Toffin L. Methanococcoides vulcani sp. nov., a marine methylotrophic methanogen that uses betaine, choline and N,N-dimethylethanolamine for methanogenesis, isolated from a mud volcano, and emended description of the genus Methanococcoides. International journal of systematic and evolutionary microbiology 64, 1978–1983 (2014). [DOI] [PubMed] [Google Scholar]
- Hoeppner A. et al. Structure of the corrinoid:coenzyme M methyltransferase MtaA from Methanosarcina mazei. Acta Crystallogr D Biol Crystallogr 68, 1549–1557 (2012). [DOI] [PubMed] [Google Scholar]
- Hagemeier C. H., Krer M., Thauer R. K., Warkentin E. & Ermler U. Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex. Proc Natl Acad Sci USA 103, 18917–18922 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul L., Ferguson D. J. Jr. & Krzycki J. A. The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons. Journal of bacteriology 182, 2520–2529 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. Bioenergetic aspects of halophilism. Microbiology and molecular biology reviews: MMBR 63, 334–348 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson D. Cellular energy metabolism and its regulation. Academic Press, New York, 72–75 (1977). [Google Scholar]
- Nyyssola A., Kerovuo J., Kaukinen P., von Weymarn N. & Reinikainen T. Extreme halophiles synthesize betaine from glycine by methylation. The Journal of biological chemistry 275, 22196–22201 (2000). [DOI] [PubMed] [Google Scholar]
- Velichkova P. & Himo F. Methyl transfer in glycine N-methyltransferase. A theoretical study. J Phys Chem B 109, 8216–8219 (2005). [DOI] [PubMed] [Google Scholar]
- Takata Y. et al. Catalytic mechanism of glycine N-methyltransferase. Biochemistry 42, 8394–8402 (2003). [DOI] [PubMed] [Google Scholar]
- Yen C. H., Lin Y. T., Chen H. L., Chen S. Y. & Chen Y. M. The multi-functional roles of GNMT in toxicology and cancer. Toxicology and applied pharmacology 266, 67–75 (2013). [DOI] [PubMed] [Google Scholar]
- Lai M. C., Sowers K. R., Robertson D. E., Roberts M. F. & Gunsalus R. P. Distribution of compatible solutes in the halophilic methanogenic archaebacteria. Journal of bacteriology 173, 5352–5358 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W. L. et al. Phosphoproteomic analysis reveals the effects of PilF phosphorylation on type IV pilus and biofilm formation in Thermus thermophilus HB27. Molecular & cellular proteomics: MCP 12, 2701–2713 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. & Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- Cox J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10, 1794–1805 (2011). [DOI] [PubMed] [Google Scholar]
- Delcher A. L., Bratke K. A., Powers E. C. & Salzberg S. L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky M., Mills R., Besemer J. & Lomsadze A. Prokaryotic gene prediction using GeneMark and GeneMark.hmm. Curr Protoc Bioinformatics Chapter 4, Unit4 5 (2003). [DOI] [PubMed] [Google Scholar]
- Pruitt K. D., Tatusova T. & Maglott D. R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35, D61–65 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J. A. et al. ProteomeXchange provides globally co-ordinated proteomics data submission and dissemination. Nature biotechnology 32, 4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Baumann U. & Reymond J. L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Research 32 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook R. J. & Wagner C. Glycine N-methyltransferase is a folate binding-protein of rat-liver cytosol. P Natl Acad Sci-Biol 81, 3631–3634 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.