Abstract
Beijing B0/W148, a “successful” clone of Mycobacterium tuberculosis, is widespread in the Russian Federation and some countries of the former Soviet Union. Here, we used label-free gel-LC-MS/MS shotgun proteomics to discover features of Beijing B0/W148 strains that could explain their success. Qualitative and quantitative proteome analyses of Beijing B0/W148 strains allowed us to identify 1,868 proteins, including 266 that were differentially abundant compared with the control strain H37Rv. To predict the biological effects of the observed differences in protein abundances, we performed Gene Ontology analysis together with analysis of protein-DNA interactions using a gene regulatory network. Our results demonstrate that Beijing B0/W148 strains have increased levels of enzymes responsible for long-chain fatty acid biosynthesis, along with a coincident decrease in the abundance of proteins responsible for their degradation. Together with high levels of HsaA (Rv3570c) protein, involved in steroid degradation, these findings provide a possible explanation for the increased transmissibility of Beijing B0/W148 strains and their survival in host macrophages. Among other, we confirmed a very low level of the SseA (Rv3283) protein in Beijing B0/W148 characteristic for all «modern» Beijing strains, which could lead to increased DNA oxidative damage, accumulation of mutations, and potentially facilitate the development of drug resistance.
Mycobacterium tuberculosis (MTB) is the causative agent of tuberculosis (TB) and, according to the Global Tuberculosis Report produced by the World Health Organization (WHO), nine million people had TB in 2014 and 1.5 million died because of the disease1. Of note, 80% of TB cases are concentrated in 22 “high-burden” countries. The Russian Federation belongs to this list and has a relatively high rate of new TB cases (80/100,000 population/year) according to WHO statistics2. Analysis of the MTB population structure in The Russian Federation has defined three main genetic families, Ural, LAM and Beijing3. According to earlier studies, more than 50% of all MTB strains isolated in Russia belong to the Beijing family, and a quarter of them are the Beijing B0/W148 variant4,5.
A recent systematic and critical review summarized various biological and phylogenetic features of the Beijing B0/W148 cluster6. Strains of this cluster possess unique pathogenic properties, including stronger association with multidrug resistance and higher levels of clustering (i.e. higher transmissibility) compared with other Beijing variants, as demonstrated by a meta-analysis of studies from across the former Soviet Union6. Additionally, members of this cluster demonstrate increased virulence in a macrophage model7, although in a mouse model, no increased virulence was observed8. Beijing variant MTB strains have a unique genome organization; recently, we reported large scale chromosomal inversions spanning 350 and 550 kb segments of the chromosome9. The presence of these inversions in Beijing B0/W148 cluster strains was confirmed by PCR, sequencing, and RFLP analysis. In addition, we identified Beijing B0/W148 cluster-specific SNPs. However, the inversions and the SNPs are insufficient to explain the success of the Beijing B0/W148 cluster. Hence, there is a particular interest in studying the proteomes of these pathogens, which will extend the genomic data by allowing detailed analyses of protein abundance, as well as protein-protein interactions.
According to the TubercuList database, a total of 4,018 proteins are encoded in the genome of M. tuberculosis H37Rv strains10. Not long ago the majority of MTB proteomic studies focused on the analysis of protein groups and individual proteins involved in certain processes, for example, in the development of drug resistance11,12,13,14. The constant technological improvements in analysis methods for biomolecules have made it possible to apply discovery driven shotgun proteomics approaches to the investigation of MTB, with a focus on the identification and quantification of the whole proteome of these strains. The most comprehensive proteome of M. tuberculosis Н37Rv was described recently by Schubert et al.15. The authors used discovery-driven mass spectrometry analysis based on extensive off-gel fractionation followed by LC-MS/MS to identify and quantify 3,074 proteins, whereas the implementation of gel-LC-MS/MS for shotgun proteomics allows the identification of about 2,000 proteins16,17. However, relatively few studies have focused on the proteomes of specific genetic families of MTB and only two reports characterizing the proteomes of Beijing family strains have been published. De Souza et al.18 described the proteomic profiles of hypo- and hypervirulent clinical Beijing isolates, whereas de Keijzer et al. disclosed the proteomic features of MTB strains belonging to ancient (atypical) and modern (typical) sublineages of the Beijing family19.
In this study, we have applied a label-free gel-LC-MS/MS shotgun proteomics approach for empirical ‘bottom-up’ exploration of Beijing B0/W148 strains.
Results
Selection of M. tuberculosis strains for proteome analysis
Seven Beijing B0/W148 cluster strains were selected for inclusion in the proteomic study. Whole genome sequencing of five of the seven strains had been performed previously (Table 1). All studied Beijing strains carried the large scale chromosomal inversions, spanning 350 and 550 kb segments of the chromosome, which we described previously9. The laboratory H37Rv strain was used for comparative analysis. Each strain was grown in three biological replicates, independently, to give a total of 24 samples. Bacterial cells were collected in stationary phase, and total proteins were extracted.
Table 1. Description of M. tuberculosis strains.
| Sample | Сlade | SIT № | 24-VNTR* | Ac Numb |
|---|---|---|---|---|
| H37Rv | H37Rv | SIT-451 | 233′226133321242534233552 | NC_000962.3 |
| Sp1** | Beijing | SIT-1 | 223325173533424672454433 | SRX216883 |
| Sp7** | Beijing | SIT-1 | 223325173533424672454433 | SRX216889 |
| Sp10 | Beijing | SIT-1 | 223325173533424672444433 | SRX216892 |
| Sp13 | Beijing | SIT-1 | 223325173533424672454433 | SRX216895 |
| Sp22 | Beijing | SIT-1 | 223325173533424572454433 | SRX216900 |
| Sp27 | Beijing | SIT-1 | 223325173533424572454433 | |
| Sp45 | Beijing | SIT-1 | 223325173533424672454433 |
Comprehensive proteome analysis of M. tuberculosis
For comprehensive proteomic analysis via LC-MS/MS, the proteins from the seven Beijing B0/W148 cluster strains and H37Rv were fractionated by SDS-PAGE, followed by in-gel tryptic digestion and analysis of the resulting peptide mixtures. The combined analysis yielded a total of 1,098,994 MS/MS spectra, of which 366,621 were assigned to unique peptide sequences using two different MS/MS search algorithms (peptide FDR < 1%).
For the H37Rv strain we identified a total of 1,560 proteins with a minimum of two unique peptides in two biological replicates. For the seven Beijing B0/W148 samples 1,868 proteins were identified, of which 1,176 (>60%) were identified in all strains. Identified proteins and peptides are presented in Tables S1–S3.
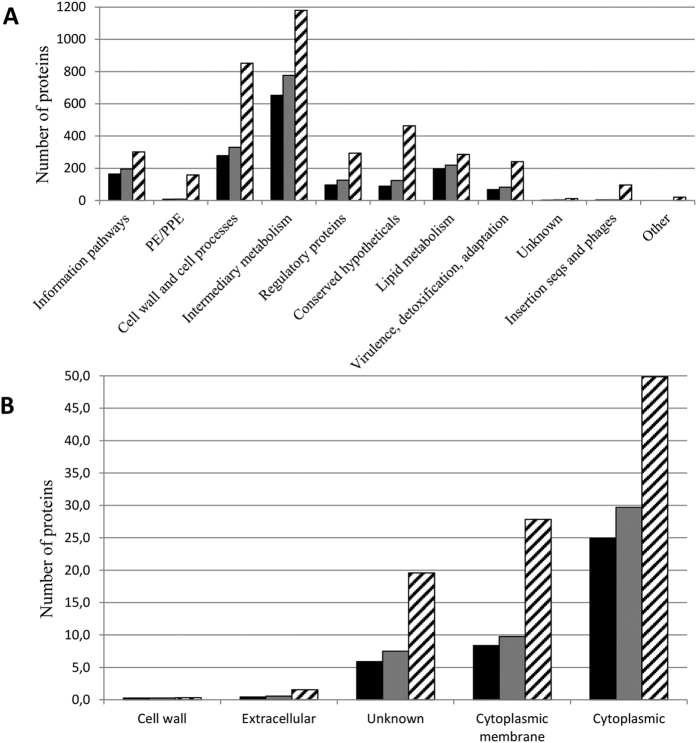
We compared the numbers of identified proteins in H37Rv and the Beijing strains in different functional categories (as defined by TubercuList) and subcellular localizations (as defined by PSORTdb) and did not find any biases between H37Rv and the Beijing B0/W148 strains (Fig. 1A,B).
Figure 1. Functional distribution of the proteins identified by LC-MS/MS.
Proteins present in our MS dataset for H37Rv (black bars) and Beijing B0/W148 (gray bars) and all annotated genes (black/white-banded bars) were categorized by. (A) Functional class categories according to TubercuList v 2.6 (http://tuberculist.epfl.ch/). (B) Localization as given by PSORTdb v 3.0 (http://db.psort.org/).
Qualitative proteome analysis of M. tuberculosis strains
Initially, qualitative proteome analysis was performed to compare proteins identified in the group of Beijing B0/W148 cluster strains with those from H37Rv. To achieve this, we created two lists of proteins; the first included proteins identified in five of seven Beijing B0/W148 cluster strains and the second comprised proteins identified both from H37Rv in our study and in the study of Schubert et al.15.
In this way, we identified 17 proteins characteristic of the Beijing B0/W148 strains that were not detectable in H37Rv. The majority of these were also identified in Beijing strains in a recent report18. In addition, 57 proteins not detectable from the Beijing B0/W148 strains were present in H37Rv (Table S4).
The available WGS dataset for five of the Beijing B0/W148 strains was used to estimate the concordance between genomic and proteomic data. We found genetic changes with potential to explain the presence of 8 of the 17 (47%) of Beijing B0/W148 specific proteins. In H37Rv the upstream region of the Rv2277c, Rv2475c, and Rv3323c genes carries the IS6110 element, which is missing in the Beijing B0/W148 strains and is likely to affect gene expression. In addition, there are three CG repeats present in the Rv2974c upstream region in the H37Rv genome, while the Beijing B0/W148 genomes contain two such repeats. We also detected a single nucleotide insertion in the Rv0976c upstream region and non-synonymous SNPs (nsSNPs) in the Rv0945, Rv1319c, and Rv2351c coding regions of Beijing B0/W148 strains, relative to that of H37Rv.
Among the 57 proteins that were not detectable in the proteomes of Beijing B0/W148 strains but present in the proteome of H37Rv, 33 carried genetic mutations compared to the H37Rv genome. Among these, six genes (Rv0072 (part of RD105), Rv1576c and Rv1586c (part of RD149), Rv1762c (part of RD152), Rv2263 (part of RD181) and Rv2818c (part of RD207)) mapped to chromosome regions showing differences between the two groups of strains20. The absence of two proteins from the Beijing B0/W148 group can be explained by an insertion (Rv0888: 987586 insGG) and a deletion (Rv1997: 2241032 delG) in the coding regions of their respective genes, which both lead to sequence changes causing protein coding frameshifts. In addition, we found changes in the upstream regions of three genes and a further 22 genes carried nsSNPs in their coding regions (Table S4).
Quantitative proteome analysis of M. tuberculosis strains in the Beijing B0/W148 cluster
The abundance of proteins in Beijing B0/W148 cluster strains was compared to that in H37Rv using Progenesis LC-MS software. For this experiment, we limited our analysis to the 1,016 proteins identified in both experimental groups (Tables S5 and S6; Figure S1). In total, we identified 192 proteins with abundances that were significantly different between the two groups (p < 0.05). Among these, 24 were over-represented in the Beijing B0/W148 cluster strains and 168 under-represented (Table S4). Worth noting, we considered all significant alterations in protein abundance, without thresholds for fold change, to allow for maximum identification of differentially abundant proteins.
Genes encoding proteins with significant differences in levels of abundance (n = 192) were matched to known operons of the H37Rv genome using MicrobesOnline Operon Predictions (www.microbesonline.org)21, resulting in the identification of 30 genes in 11 operons. In the majority of cases we observed changes in protein abundances in only one direction (over or under) for genes encoded by the same operon. However, changes in two genes in the same operon, Rv1380 (over) and Rv1384 (under), resulted in opposite changes in the abundances of the corresponding proteins.
We also found insertions (Rv3234c: 3610391 insC), deletions (Rv0927c: 1034211 delTGC; Rv1884: 2094915 delCGTCAG) and 35 nsSNPs in the coding regions of genes encoding proteins with different abundances in the two strains. Additionally, we searched for mutations in transcription initiation sites and transcription factor (TF) binding sites within our genomic data22,23. Mutations in TF binding sites for Rv0169, Rv1129c, Rv1872c and in transcription start sites for Rv0169, Rv1196, Rv1508c, Rv1872c, Rv2627c, Rv2711 were identified (Table S4).
Functional characteristics of differentially abundant proteins
To determine possible cumulative effects of the differentially abundant proteins on the function of mycobacterial cells, we combined the results of qualitative and quantitative proteome analysis. Thus, proteins exclusively identified in or not detectable from the Beijing B0/W148 cluster strains were attributed to the groups of over- or under-represented proteins, respectively; the extended over- and under-represented groups consisted of 41 proteins and 225 proteins, respectively (Table S4, Figure S1).
Proteins were classified according to the Gene Ontology (GO) annotations “biological process” (BP), “cellular component” (CC) and “molecular function” (MF) using the PANTHER classification system (Figure S2A). In case of “molecular function” term both under- and over-represented proteins enriched “oxidoreductase activity” and “transferase activity” sub-categories of GO “catalytic activity” and additionally “transporter activity” category. For “biological process” term over-represented proteins enriched “primary lipid metabolic process”, whereas under-represented proteins were distributed across different categories (Table 2, Figure S2).
Table 2. Enriched functional clusters of differential proteins discussed in the text.
| Gene | Synonym | Reg | Functional_category | Product | log2 fold | p-value |
|---|---|---|---|---|---|---|
| transporter activity (GO:0005215) | ||||||
| fadD15 | Rv2187 | over | Lipid metabolism | long-chain-fatty-acid–CoA ligase FadD15 | 0.64 | 0.049358831 |
| – | Rv2971 | over | Intermediary metabolism and respiration | oxidoreductase | 0.5 | 0.041065417 |
| – | Rv0073 | under | cell wall and cell processes | glutamine ABC transporter ATP-binding protein | ||
| – | Rv0143c | under | cell wall and cell processes | transmembrane protein | ||
| pstB | Rv0933 | under | cell wall and cell processes | phosphate ABC transporter ATP-binding protein PstB | −2.41 | 4.65402E-07 |
| pstS1 | Rv0934 | under | cell wall and cell processes | phosphate ABC transporter substrate-binding lipoprotein PstS | −1.12 | 0.00155862 |
| oppD | Rv1281c | under | cell wall and cell processes | oligopeptide ABC transporter ATP-binding protein OppD | −0.6 | 0.045017563 |
| atpA | Rv1308 | under | Intermediary metabolism and respiration | ATP synthase subunit alpha | −0.46 | 0.001330189 |
| atpD | Rv1310 | under | Intermediary metabolism and respiration | ATP synthase subunit beta | −0.66 | 0.00060944 |
| glnQ | Rv2564 | under | cell wall and cell processes | glutamine ABC transporter ATP-binding protein | −0.76 | 0.00036177 |
| – | Rv2690c | under | cell wall and cell processes | integral membrane protein | ||
| lipid metabolic process (GO:0006629) | ||||||
| fadD15 | Rv2187 | over | Lipid metabolism | long-chain-fatty-acid–CoA ligase FadD15 | 0.64 | 0.049358831 |
| – | Rv2277c | over | Intermediary metabolism and respiration | glycerolphosphodiesterase | ||
| agpS | Rv3107c | over | Lipid metabolism | alkyldihydroxyacetonephosphate synthase | 3.5 | 0.022585521 |
| hsaA | Rv3570c | over | Intermediary metabolism and respiration | flavin-dependent monooxygenase oxygenase subunit HsaA | 1 | 0.043815093 |
| rmlA | Rv0334 | under | Intermediary metabolism and respiration | glucose-1-phosphate thymidylyltransferase | −1.41 | 7.01184E-05 |
| pks6 | Rv0405 | under | Lipid metabolism | membrane bound polyketide synthase | ||
| fadB2 | Rv0468 | under | Lipid metabolism | 3-hydroxybutyryl-CoA dehydrogenase | −1.33 | 2.15157E-07 |
| fadB | Rv0860 | under | Lipid metabolism | fatty oxidation protein FadB | −0.47 | 0.008655458 |
| echA6 | Rv0905 | under | Lipid metabolism | enoyl-CoA hydratase EchA6 | −0.96 | 0.002614821 |
| fadD21 | Rv1185c | under | Lipid metabolism | fatty-acid–CoA ligase FadD21 | −1.35 | 0.004873026 |
| – | Rv3230c | under | Intermediary metabolism and respiration | stearoyl-CoA 9-desaturase electron transfer partner | −1.38 | 0.03233442 |
| dprE2 | Rv3791 | under | Lipid metabolism | decaprenylphosphoryl-D-2-keto erythropentose reductase | −0.85 | 0.003881878 |
| pks2 | Rv3825c | under | Lipid metabolism | phthioceranic/hydroxyphthioceranic acid synthase | −1 | 0.03160682 |
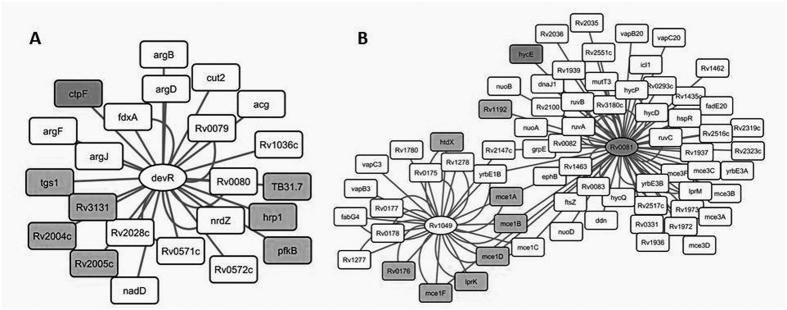
Given the regulon organization of prokaryotic genes and operons, we hypothesized that proteins controlled by a single transcription factor must be assembled into “unidirectional change” groups. To verify this hypothesis we analyzed 266 differentially abundant proteins by mapping them to a gene regulatory network consisting of 65 TFs and 431 genes regulated by these TFs24. Visualization by Cytoscape v 2.8.3 software allowed us to position the proteins in the network. Five and 38 proteins from the over- and under-represented groups, respectively were found to belong to this gene network (Table 3, Fig. 2).
Table 3. Differential proteins identified on the gene regulatory network.
| Synonym | Gene | Reg | TF | Product | Functional_category | log2 fold | p-value |
|---|---|---|---|---|---|---|---|
| Rv0509 | – | over | Rv1353c | glutamyl-tRNA reductase | Intermediary metabolism and respiration | ||
| Rv1464 | – | over | Rv1460 | cysteine desulfurase | Intermediary metabolism and respiration | 1.56 | 0.0489072 |
| Rv3094c | fadE2 | over | Rv0494; Rv3095 | hypothetical protein | Intermediary metabolism and respiration | 9.61 | 0.0386941 |
| Rv3494c | yrbE1A | over | Rv0022c | Mce family protein Mce4 | virulence, detoxification, adaptation | 1.46 | 0.0259927 |
| Rv3570c | mce1A | over | Rv0678; Rv1353c; Rv3574 | flavin-dependent monooxygenase oxygenase subunit HsaA | Intermediary metabolism and respiration | 1 | 0.0438151 |
| Rv0034 | – | under | Rv3249c | hypothetical protein | Intermediary metabolism and respiration | ||
| Rv0035 | – | under | Rv3249c | fatty-acid–CoA ligase FadD34 | Lipid metabolism | ||
| Rv0081 | bioF2 | under | Rv0081 | HTH-type transcriptional regulator | Regulatory proteins | ||
| Rv0087 | acpA | under | Rv0081 | formate hydrogenase HycE | Intermediary metabolism and respiration | ||
| Rv0101 | – | under | Rv0047c; Rv2069; Rv0324 | peptide synthetase Nrp | Lipid metabolism | ||
| Rv0154c | fadD34 | under | Rv1423 | acyl-CoA dehydrogenase FadE2 | Lipid metabolism | −0.59 | 0.0008057 |
| Rv0169 | – | under | Rv0023; Rv0081; Rv0757; Rv1049; Rv3416 | Mce family protein Mce1A | virulence, detoxification, adaptation | −1.44 | 5.763E-06 |
| Rv0170 | – | under | Rv0023; Rv0081; Rv1049; Rv3416 | Mce family protein Mce1B | virulence, detoxification, adaptation | −1.38 | 0.0017153 |
| Rv0172 | sdaA | under | Rv0023; Rv0081; Rv1049 | Mce family protein Mce1D | virulence, detoxification, adaptation | −1.38 | 0.0001941 |
| Rv0173 | glyA2 | under | Rv1049 | Mce family lipoprotein LprK | cell wall and cell processes | −1.29 | 7.012E-05 |
| Rv0174 | - | under | Rv0023; Rv0081; Rv1049 | Mce family protein Mce1F | virulence, detoxification, adaptation | −0.84 | 7.012E-05 |
| Rv0176 | – | under | Rv1049 | Mce associated transmembrane protein | cell wall and cell processes | −1.81 | 0.0175553 |
| Rv0241c | – | under | Rv0238; Rv1049 | 3-hydroxyacyl-thioester dehydratase HtdX | Intermediary metabolism and respiration | −0.86 | 0.0386941 |
| Rv0243 | – | under | Rv0238 | acetyl-CoA acetyltransferase FadA | Lipid metabolism | −0.91 | 0.0166606 |
| Rv0675 | – | under | Rv0674 | enoyl-CoA hydratase EchA5 | Lipid metabolism | −1.1 | 0.0007597 |
| Rv0768 | – | under | Rv0576; Rv1255c | aldehyde dehydrogenase AldA | Intermediary metabolism and respiration | ||
| Rv0824c | hycD | under | Rv0472c | acyl-ACP desaturase DesA | Lipid metabolism | −1.98 | 5.763E-06 |
| Rv0989c | hycP | under | Rv0767c | polyprenyl-diphosphate synthase GrcC | Intermediary metabolism and respiration | ||
| Rv1094 | hycQ | under | Rv0472c | acyl-ACP desaturase DesA | Lipid metabolism | −1.91 | 5.196E-09 |
| Rv1192 | hycE | under | Rv2034; Rv0081 | hypothetical protein | cell wall and cell processes | −1.91 | 0.0006327 |
| Rv1386 | - | under | Rv1033c; Rv2359 | PE family protein PE15 | PE/PPE | ||
| Rv1856c | fcoT | under | Rv1353c | oxidoreductase | Intermediary metabolism and respiration | −1.06 | 0.037951 |
| Rv1997 | ctpF | under | Rv3133c | cation transporter ATPase F | cell wall and cell processes | ||
| Rv2004c | – | under | Rv3133c | hypothetical protein | Regulatory proteins | −1.45 | 0.0004367 |
| Rv2005c | nrp | under | Rv3133c | universal stress protein | virulence, detoxification, adaptation | −1.24 | 0.023446 |
| Rv2029c | gmhB | under | Rv3133c | 6-phosphofructokinase PfkB | Intermediary metabolism and respiration | −3 | 0.0006327 |
| Rv2048c | hddA | under | Rv0767c | polyketide synthase | Lipid metabolism | −1.38 | 0.0393413 |
| Rv2103c | msrA | under | Rv1990c | ribonuclease VapC37 | virulence, detoxification, adaptation | ||
| Rv2410c | – | under | Rv0022c | hypothetical protein | Intermediary metabolism and respiration | −1.42 | 0.0082996 |
| Rv2623 | – | under | Rv3133c | universal stress protein | virulence, detoxification, adaptation | −1.26 | 0.0157466 |
| Rv2626c | ptbB | under | Rv3133c | hypoxic response protein | Regulatory proteins | −1.56 | 0.0216908 |
| Rv3130c | pntAa | under | Rv3133c | diacyglycerol O-acyltransferase | Lipid metabolism | −2.29 | 0.0001161 |
| Rv3131 | pntAb | under | Rv3133c | NAD(P)H nitroreductase | Intermediary metabolism and respiration | −1.69 | 0.0026414 |
| Rv3400 | pntB | under | Rv0135c | hydrolase | Intermediary metabolism and respiration | −0.93 | 0.0451627 |
| Rv3509c | yrbE1B | under | Rv0324 | acetohydroxyacid synthase large subunit | Intermediary metabolism and respiration | −0.66 | 0.0080181 |
| Rv3602c | mce1B | under | Rv1353c | pantothenate synthetase | Intermediary metabolism and respiration | −1.25 | 0.0014439 |
| Rv3825c | mce1C | under | Rv0757 | phthioceranic/hydroxyphthioceranic acid synthase | Lipid metabolism | −1 | 0.0316068 |
Figure 2. Network map of co-regulated groups of genes.
Ellipse and rectangles indicates TF and genes, respectively. Gray color indicates genes which correspond to under-represented proteins. White color indicates genes which correspond to proteins without changes or unidentified proteins. (A) Group of genes bound by TF devR/DosR; (B) Group of genes bound by TFs Rv0081 and Rv1049.
Next, we focused our attention on co-regulated groups of proteins controlled by the same TF and identified a set of five TFs responsible for the regulation of 24 genes. Of these TFs, Rv3133c, Rv1049 and Rv0081 had the most extensive connections and Rv3133c, a key member of the DosR regulon, was associated with eight under-represented proteins, Rv1997, Rv2004c, Rv2005c, Rv2029c, Rv2623, Rv2626c, Rv3130c and Rv3131 (Fig. 2A). Therefore we examined the abundance profiles of proteins belonging to the DosR regulon, which consists of 52 genes and the sensor histidine kinase, dosT15. In this study, we identified 33 of the 53 DosR proteins (62%), 11 of which were under-represented in strains of the Beijing B0/W148 cluster (Figure S1). Most of the 11 under-represented proteins have been proposed to be involved in lipid transport and degradation and are likely to function in the assimilation of exogenous lipids from host cell membranes. Interestingly, no difference in the abundance of DosR transcription factor itself was observed between Beijing B0/W148 strains and H37Rv.
Another group included the TFs Rv1049 and Rv0081, which regulate the synthesis of eight proteins under-represented in Beijing B0/W148 strains (Fig. 2B). Of these eight, six proteins, Rv0169, Rv0170, Rv0172, Rv0173, Rv0174, and Rv0176, are involved in membrane transport of phospholipids and belong to the ABC transporter family. Of note, Rv0081, which was recently proposed to be a hypoxia regulator25,26, is a member of the ArsR/SmtB family of metal-dependent transcriptional regulators and is directly regulated by the response regulators DosR/DevR and MprAB27. Our results indicate that MprA (Rv0981) is under-represented in Beijing B0/W148 cluster strains.
Discussion
According to previous studies, the Beijing genotype represents approximately 50% of MTB strains in East Asia and at least 13% of strains worldwide28. Among them the Beijing B0/W148 clonal cluster, can be distinguished. It is defined as a “successful” Russian clone of MTB6 and is known under different names, initially as B0 or W1485,29, or, more recently, as clade B of the “East European” sublineage3, or the Resistant European Tuberculosis cluster30. The pathobiology, genomic characteristics and phylogeography of the B0/W148 clonal cluster are described in a recent systematic and critical review6, although many questions concerned its pathogenomics remain unclear.
In this study we focused on the specific features of the proteome of the Beijing B0/W148 clonal cluster strains. For this reason, we selected seven МТВ strains of this cluster with a similar VNTR-profile and containing the specific chromosomal rearrangements. The well-studied laboratory H37Rv strain was used for comparison.
For the proteomic analysis we used discovery-driven MS, also known as the shotgun MS approach, aimed at maximizing proteome coverage. The first studies using this method to investigate MTB were published in 200431 and at the time of writing there were 30 such articles in the PubMed database, including two ones focused on the Beijing family. In 2010 Souza et al. applied a label-free gel-LC-MS/MS to identify 1,668 proteins in hyper- and hypovirulent MTB Beijing isolates18. The other study of the Beijing MTB family derived 2,392 proteins using a label-based SCX-LC-MS/MS approach, and described the differences between ancient and modern Beijing strains19. In our study we identified 1,951 proteins for Beijing B0/W148 strains and 1,560 ones for H37Rv, which is comparable to the number of proteins identified in earlier reports.
It is known that different MTB genetic groups can exhibit different features that affect protein extraction. Because we used the same workflow both for Beijing B0/W148 and H37Rv strains, to be sure of its effectiveness we compared the number of identified proteins within functional categories and subcellular localizations for tested groups (Fig. 1A,B). Both groups showed a comparable distribution of proteins across functional categories, in agreement with the results of de Keijzer et al.19, and subcellular localizations. It allows us to conclude that the differences in protein abundance we observed between Beijing B0/W148 and H37Rv strains are independent from our workflow and reflect the true physiological characteristics of pathogens.
For a detailed description of the specific properties of the Beijing B0/W148 cluster, we performed a comparative proteomic analysis of these strains with H37Rv using a combination of qualitative and quantitative proteomic data. In total, 266 differentially abundant proteins were identified, of which 41 were over- and 225 were under-represented, respectively, in Beijing B0/W148 cluster strains.
By analyzing the sequences of available Beijing B0/W148 genomes, we found possible explanations for 47% of the changes revealed by qualitative proteomic analysis. Specifically, we demonstrated that the absence of six proteins was due to the fact they mapped to five deleted regions, a characteristic feature of the whole Beijing family32. Additionally, we identified nsSNPs in coding regions and nucleotide substitutions/IS6110 insertions in the upstream regions of genes encoding 33 proteins that were present or not detectable in the Beijing B0/W148 cluster strains. However, the majority of differentially abundant proteins identified by quantitative proteomic analysis could not be explained by differences in the genomic data.
Two complementary approaches were used to predict the functional effects of the observed features of the Beijing B0/W148 proteome. We first applied GO analysis to identify functional categories enriched for the differentially abundant proteins (n = 266). We believe that the distribution of proteins categorized by the GO annotation “biological processes” is most relevant. Accordingly, we found that a substantial group of differentially abundant proteins (33/266, 12.4%) belong to the “metabolic process” category (GO:008152). In-depth analysis revealed an enrichment for over-represented proteins (4/41, 9.8%) in the GO:0006629 category, “lipid metabolic process”, while under-represented proteins were relatively equally distributed among the categories: “lipid” (GO:0006629), “cellular amino acid” (GO:0006520) and “carbohydrate” (GO:0005975) metabolic processes.
Among the over-represented proteins involved in lipid biosynthesis, the long-chain-fatty-acid-CoA ligase, FadD15, is known to be the one of seven fatty-acid-CoA synthases induced in virulent strains33. Another protein, AgpS, is an alkyl-DHAP synthase that initiates lipid anabolism. Together, these results suggest lipid synthesis is upregulated in Beijing B0/W148 cluster strains. Consistent with this hypothesis, the majority of proteins involved in fatty acid catabolism were under-represented in Beijing B0/W148 cluster strains (Table 2). By contrast, the HsaA oxygenase subunit of the flavin-dependent monooxygenase, encoded by one of 126 genes necessary for survival in macrophages34, was over-represented in Beijing B0/W148 cluster strains. This protein is involved in the catabolism of steroids and could have important effects on the infected host by reducing the local concentration of membrane cholesterol, altering immunoregulatory sterols, and producing novel secondary metabolites35.
We also investigated proteins classified into the “transporter activity” term (GO:0005215), based on GO analysis by “Molecular Function”. Three and 15 “transporter activity” proteins, respectively, were over- and under-represented in the Beijing B0/W148 strains. Five of the under-represented proteins are classified as ABC transporters and, of these, PstB and PstS1 belong to a single operon involved in phosphate import during fasting, which is a characteristic of bacteria inside phagosomes34. In addition, PstS1 is overexpressed during phosphate starvation36.
Another interesting transport protein is CtpF, which was not detectable in the Beijing B0/W148 strains. This protein is a P-type ATPase and potential alkali/alkaline earth metal cation transporter. CtpF is the only P-type ATPase gene that is regulated by the global latency regulator, DosR, and is highly overexpressed under conditions of hypoxia. From the genomic data we determined that the corresponding gene had a single nucleotide deletion (Rv1997: 2241032 delG) leading to a frame shift mutation. This mutation is not specific to Beijing B0/W148 strains, but is rather a general characteristic of the Beijing family.
It should be noted that GO analysis reflects the independent roles of differentially abundant proteins, and does not take into account potentially related changes in the bacterial cell. To estimate the effects of co-regulated proteins, we used a gene regulatory network for MBT from a recently published study by Peterson et al.24. This analysis revealed that there was a decreased representation of 11 proteins from the DosR system in Beijing B0/W148 strains. The MTB DosR system has been well documented; in H37Rv, it is induced under three conditions that inhibit aerobic respiration: hypoxia, NO, or CO26,37–39. It is well known that, during the exponential phase of MTB growth, the levels of DosR transcripts are constitutively higher in Beijing B0/W148 strains than in H37Rv40,41. In contrast, Badillo-López et al. demonstrated that, during hypoxia, the level of DosR is significantly lower in the Beijing strains than in H37Rv42. This is explained by the fact that Beijing strains of MTB implement an alternative response to hypoxic stress than that used by H37Rv42. Our results demonstrated an under-representation of DosR-regulated proteins in the Beijing B0/W148 strains during the stationary growth phase. This might be due to pre-adaptation of bacterial cells to the potentially low levels of oxygen under prolonged cultivation in vitro.
Of interest, the transcription factor Rv0081 of the DosR regulon was not detectable in any Beijing B0/W148 strain. This protein was recently described by Galagan et al. as a regulator hub during the response to hypoxia26 and, together with Rv1049, controls the mce1 operon, consisting of six mce genes (Rv0169–Rv0174), two yrbE genes (Rv0167, Rv0168), and four mce-associated genes (Rv0175–Rv0178). In our study we found that four Mce proteins (Rv0169, Rv0170, Rv0172, and Rv0174), and one Mce-associated protein (Rv0176) were under-represented in the Beijing B0/W148 strains. The proteins encoded by the mce1 operon are thought to be ABC transporters involved in the transport of phospholipids, and are required for mycobacterial survival in macrophages or mouse models of infection36,43, and deletion of the mce1 operon results in a hypervirulent phenotype36. Notably, these proteins were not defined in the GO analysis, highlighting the need to use different approaches to functionally assess proteins and better understand how they work in concert to ensure cell survival.
In earlier study, the efflux pump proteins Rv0341, Rv2688c, Rv3728, are found exclusively in Beijing MTB strains19. In contrast, we found that Rv0341 was present in all strains tested, including H37Rv, and that its abundance was similar between Beijing B0/W148 strains and H37Rv. However, Rv2688 and Rv3728 proteins were not identified in our dataset. Notably, we observed that the Rv3728 gene carried a B0/W148-specific mutation, leading to the formation of an early stop codon, indicating why this protein may be absent from the B0/W148 proteome.
Beijing B0/W148 strains belong to the “modern” Beijing sublineage6, which differ from the “ancient” sublineage by the over-representation of the proteins Rv0450c and Rv3137, and the under-representation of Rv1269c and Rv328319. Also in our data, the abundance of Rv3283 (SseA) was dramatically lower in Beijing B0/W148 strains than in H37Rv (fold change = 0.04). This protein is a predicted thiol-oxidoreductase and, together with the superoxide detoxifying enzyme, SodA (Rv3846) and an integral membrane protein, DoxX (Rv3005c), it forms a membrane-associated oxidoreductase complex (MRC). Loss of any MRC component is correlated with defective recycling of mycothiol, which is a functional analog of glutathione in MTB44. The decreased abundance of Rv3283 (SseA) in «modern» Beijing cells might lead to the accumulation of oxidative cellular damage, caused by reactive oxygen species (ROS).
Taken together, our data suggest that the distinctive proteomic features of Beijing B0/W148 strains are likely to contribute to their enhanced virulence and successful geographical spread. We observed an increased abundance of enzymes responsible for long-chain fatty acid biosynthesis, which coincided with a decrease in proteins responsible for the degradation of these molecules. Mycobacteria can use long chain fatty acids (up to 86–95 carbon atoms in length) to produce mycolic acids and various lipids that are considered to be the major virulence factors of MTB, in particular during the early stage of infection, when bacilli encounter host macrophages. We also observed an increased abundance of the HsaA protein involved in steroid degradation. In the intracellular environment, MTB uses cholesterol as an energy source and for the biosynthesis of the cell wall lipids. The difference we observed in the abundance of HsaA protein could increase the survival of MTB in host macrophages, a known characteristic of Beijing B0/W148 strains7,45. In addition, we observed a decreased abundance of proteins encoded by mce1 operon genes, the deletion of which is known to lead to a hypervirulent phenotype36. Our data also provide a possible basis for the well-known ease with which stains with the Beijing B0/W148 genotype develop drug resistance. We confirmed very low levels of SseA protein in B0/W148 strains, which is likely to lead to an accumulation of ROS, followed by DNA damage. The latter has the potential to generate a wide range of genetic variants, supporting the survival of MTB populations under positive selection, in particular during drug therapy.
Materials and Methods
Mycobacteria cultivation conditions
Eight strains of MTB were used; seven Beijing B0/W148 cluster strains, which were treated as an experimental group, and the control H37Rv strain (Table 1). Mycobacterium tuberculosis strains were grown in Middlebrook 7H11 media with OADC supplement at 35 °C without shaking for 14–16 days to a cell density of 1 ± 2 × 108 cells ml−1. Each strain was grown in three biological replicates. The bacterial cells were washed in Tris-HCl, PBS+2%, Triton-X100 (pH 7.5–8) and incubated at 80 °C for 20 min. Further cells pellet was received by centrifugation at 4,500 g, 4 °С for 15 min and stored at −80 °C until required.
Whole genome sequencing and PCR analysis of Beijing B0/W148 strains
DNA extraction was performed as previously described46. Strains of the Beijing B0/W148-cluster were detected using a PCR assay47. Large chromosomal inversions in the B0/W148 genome were detected as described previously9. Spoligotyping and 24-locus VNTR typing were performed as previously described48,49, as were genome sequencing and SNP calling9.
Protein Extraction from M. tuberculosis
Bacterial cell pellets were resuspended in 50 μL lysozyme and 100 μL 100 mМ TrisHСl pH 7.6, with 3 μL of Protease inhibitor Mix (GE Healthcare, USA). Cells were disrupted using a bead-beating homogenizer (MPBio, FastPrep-24, USA) with 0.5 mm silica-zirconium beads for 4 min, followed by 5 min on ice. For protein solubilization, SDS (Panreac, Spain) was added to the collected suspension to a final concentration of 10%. To reduce disulfide bonds, DTT (BioRad, USA) was added to a final concentration of 100 mM. Samples were then incubated at 60 °C for 30 min, centrifuged at 13,000 g at 4 °С for 5 min, and the supernatant used as protein solution. Protein concentration was measured by the Bradford method using the Bradford Protein Assay Kit (Bio Rad, USA).
Trypsin Digestion
Protein samples (200 μg) were loaded onto a 7.5% SDS-PAGE gel and separated by electrophoresis at 20 mA for 20 min and 40 mA overnight using a PROTEAN II system (Bio-Rad, USA). The gel was stained using a Colloidal Blue Staining Kit (Invitrogen, USA)50. Proteolytic in gel digestion was performed with trypsin (Trypsin Gold, Mass Spectrometry Grade, Promega, USA) as described previously51. Cleavage was stopped by adding 5% formic acid (FA) and peptides were extracted in a solution containing 50% ACN and 5% FA (2v/v), followed by extraction in 75% ACN and 5% FA (2v/v). Peptides were concentrated by Speedvac and dissolved in 20 μL 1% acetic acid. Supernatant peptides were removed and cleaned using C18 Sep-Pak columns (Waters, USA).
LC-MS/MS analysis
Analysis was performed on a TripleTOF 5600+ mass-spectrometer with a NanoSpray III ion source (AB Sciex, Canada) coupled to a NanoLC Ultra 2D+ nano-HPLC system (Eksigent, Singapore). The HPLC system was configured in a trap-elute mode. For a sample loading buffer and buffer A, a mix of 98.9% water, 1% methanol, and 0.1% formic acid (v/v) was used. Buffer B was 99.9% acetonitrile and 0.1% formic acid (v/v). Samples were loaded on a trap column Chrom XP C18, 3 mm, 120 Å, 350 mm × 0.5 mm (Eksigent, Singapore) at a flow rate of 3.5 μl/min over 10 min and eluted through the separation column 3C18-CL-120 (3 mm, 120 Å) 75 mm × 150 mm (Eksigent, Singapore) at a flow rate of 300 nl/min. The gradient was from 5 to 40% of buffer B in 120 min. The column and the pre-column were regenerated between runs by washing with 95% buffer B for 7 min and equilibrated with 5% buffer B for 25 min. Between samples, to ensure the absence of carryover, both the column and the precolumn were thoroughly washed with blank trap-elute gradient, including 5–7 min of 5-95-95-5% waves of buffer B followed by 25 min of equilibration with 5% buffer B.
Mass spectra were acquired in the positive ion mode. Information-dependent mass-spectrometer experiments included one survey MS1 scan followed by 50 dependent MS2 scans. MS1 acquisition parameters were as follows: mass range for analysis and subsequent ion selection for MS2 analysis was 300–1250 m/z, signal accumulation time was 250 ms. Ions for MS2 analysis were selected on the basis of intensity, with a threshold of 200 cps and charge state between 2 and 5. MS2 acquisition parameters were as follows: resolution of quadrupole was set to UNIT (0.7 Da), measurement mass range was 200–1800 m/z, optimization of ion beam focus was set to obtain maximal sensitivity, and signal accumulation time was 50 ms for each parent ion. Collision activated dissociation was performed with nitrogen gas, with collision energy ramping from 25 to 55 V within the 50 ms signal accumulation time. Analyzed parent ions were sent to a dynamic exclusion list for 15 sec, in order to collect the next MS2 spectra of the same compound around its chromatographic peak apex (the minimum peak width throughout the gradient was approximately 30 s).
Protein identification
Raw data (.wiff files) were converted to Mascot Generic Format (.mgf files, peak lists) using the command-line program, AB SCIEX MS Data Converter v.1.3 (AB SCIEX, Framingham, MA, USA) and the “-proteinpilot” parameter. Mascot v. 2.2.07 was used for identification against the Mycobacterium tuberculosis H37Rv sequence database (3,932 amino acid sequences, including 26 contaminant sequences) downloaded from the RefSeq database52 (RefSeq: NC_000962.3). In the Mascot search results, when the significance threshold was set at 0.05, the individual ions score was >11 (Table S7). The peptide false discovery rate (peptide FDR) was calculated using Decoy database analysis. Frequently observed contaminants, such as trypsin, bovine proteins and human keratins, were removed from the results, along with proteins supported by a single unique peptide. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium53 via the PRIDE partner repository with the dataset identifier PXD002542 (Reviewer account details: Username - reviewer44310@ebi.ac.uk and password - KJzyqV1z).
Additionally, for protein identification, .wiff data files were analyzed with ProteinPilotTM (AB Sciex, Canada) software version 4.5, revision 1656, using search algorithm Paragon 4.5.0.0, revision 1654 (AB Sciex, Canada) and a standard set of identification settings to search against the RefSeq database (RefSeq: NC_000962.3), supplemented with sequences of trypsin and common protein contaminants, to give a total of 4298 protein sequences. Peptide identifications were processed with default settings by using the ProGroup algorithm integral to ProteinPilot software. The software algorithm includes any modification listed in UniMod, based on the estimated probability of its occurrence54. The final protein identification list for each sample was obtained by leaving out protein identifications with unused scores below the threshold calculated by the ProteomicS Performance Evaluation Pipeline (PSPEP) algorithm for 1% global FDR from fit (which is defined using protein hits for decoy reversed sequences in the provided database)55. In addition, only identifications for which two or more unique peptides with confidence scores above the threshold calculated by PSPEP software for 1% global FDR were retained.
We used TubercuList version 2.6 (http://tuberculist.epfl.ch/) and PSORTdb v 3.0 (http://db.psort.org/) databases to determine functional categories and localization of the identified proteins.
Label-free protein quantitation
For label-free quantitation, raw MS data files (.wiff files) were imported and processed in Progenesis LC-MS software v.4.1 (Nonlinear Dynamics, Newcastle, UK). The sample of M. tuberculosis H37Rv with the highest number of MS/MS spectra was set as the reference and all other runs were aligned to it. Searches were performed using Mascot Search Engine as described in “Protein identification” section. The results of peptide quantitation were normalized using an iterative median-based normalization as implemented in the Progenesis software. Differences in the abundance of a protein between the three biological replicates of M. tuberculosis H37Rv and all Beijing B0/W148 cluster strains were evaluated using a two-sided unpaired Student’s T-test. P-values < 0.05 were considered statistically significant. Adjusted p-values for multiple tests (q-values) were generated using the Benjamini–Hochberg method56.
Gene ontology analysis
To functionally characterize differentially abundant proteins for biological interpretation, Gene Ontology (GO) analysis was performed. Gene Ontology annotation for H37Rv proteins was obtained from UniProt57 using the ID mapping function (http://www.uniprot.org/uploadlists/). The TopGO R package from Bioconductor was used for GO enrichment analysis58. A two-tailed Fisher’s Exact Test was used to measure the significance of enrichment. Proteins assigned to enriched GO categories (p-value < 0.05) were grouped according to the PANTHER classification system59.
Additional Information
How to cite this article: Bespyatykh, J. et al. Proteome analysis of the Mycobacterium tuberculosis Beijing B0/W148 cluster. Sci. Rep. 6, 28985; doi: 10.1038/srep28985 (2016).
Supplementary Material
Acknowledgments
We are grateful to O. Pobeguts for helping in sample preparation for proteomic analysis. We also thank O. Manicheva and N. Melnikova from Research Institute of Phtisiopulmonology for bacteriological work. This work was partly supported by 14-15-00689 grant of the Russian Science Foundation of the Russian Federation.
Footnotes
Author Contributions J.B., E.S., I.B., I.A. and E.I. wrote the main manuscript text. J.B. and E.S. prepared Figures 1 and 2. I.B. and I.A. prepared supplementary Tables and Figure. J.B. and E.S. conducted genome and proteome analysis. I.B., I.A. and D.A. conducted mass-spectrometry and statistical analysis. V.Z., M.D. and P.Y. cultivated mycobacteria. E.I. and V.M. designed the experiment. All authors reviewed the manuscript.
References
- World Health Organization. Global Tuberculosis Report 20th edn World Health Organization (2015). [Google Scholar]
- World Health Organization. World Health Statistic. World Health Organization (2015).
- Casali N. et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet. 46, 279–286, doi: 210.1038/ng.2878 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afanas’ev M. V. et al. Molecular typing of Mycobacterium tuberculosis circulated in Moscow, Russian Federation. Eur J Clin Microbiol Infect Dis. 30, 181–191, doi: 110.1007/s10096-10010-11067-z (2011). [DOI] [PubMed] [Google Scholar]
- Bifani P. J., Mathema B., Kurepina N. E. & Kreiswirth B. N. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10, 45–52 (2002). [DOI] [PubMed] [Google Scholar]
- Mokrousov I. Insights into the origin, emergence, and current spread of a successful Russian clone of Mycobacterium tuberculosis. Clin Microbiol Rev. 26, 342–360, doi: 310.1128/CMR.00087-00012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasunskaia E. et al. Emerging multidrug resistant Mycobacterium tuberculosis strains of the Beijing genotype circulating in Russia express a pattern of biological properties associated with enhanced virulence. Microbes Infect. 12, 467–475, doi: 410.1016/j.micinf.2010.1002.1008 (2010). [DOI] [PubMed] [Google Scholar]
- Andreevskaia S. N., Chernousova L. N., Smirnova T. G., Larionova E. E. & Kuz’min A. V. [Impact of M. tuberculosis genotype on survival in mice with experimental tuberculosis]. Probl Tuberk Bolezn Legk. 7, 45–50 (2007). [PubMed] [Google Scholar]
- Shitikov E. A. et al. Unusual large-scale chromosomal rearrangements in Mycobacterium tuberculosis Beijing B0/W148 cluster isolates. PLoS One. 9, e84971, doi: 84910.81371/journal.pone.0084971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew J. M., Kapopoulou A., Jones L. M. & Cole S. T. TubercuList–10 years after. Tuberculosis (Edinb). 91, 1–7, doi: 10.1016/j.tube.2010.1009.1008 (2011). [DOI] [PubMed] [Google Scholar]
- Betts J. C. et al. Comparison of the proteome of Mycobacterium tuberculosis strain H37Rv with clinical isolate CDC 1551. Microbiology. 146, 3205–3216 (2000). [DOI] [PubMed] [Google Scholar]
- Mascarello A. et al. Discovery of Mycobacterium tuberculosis protein tyrosine phosphatase B (PtpB) inhibitors from natural products. PLoS One. 8, e77081, doi: 77010.71371/journal.pone.0077081 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattow J. et al. Comparative proteome analysis of culture supernatant proteins from virulent Mycobacterium tuberculosis H37Rv and attenuated M. bovis BCG Copenhagen. Electrophoresis. 24, 3405–3420 (2003). [DOI] [PubMed] [Google Scholar]
- Parandhaman D. K., Sharma P., Bisht D. & Narayanan S. Proteome and phosphoproteome analysis of the serine/threonine protein kinase E mutant of Mycobacterium tuberculosis. Life Sci. 109, 116–126, doi: 110.1016/j.lfs.2014.1006.1013 (2014). [DOI] [PubMed] [Google Scholar]
- Schubert O. T. et al. The Mtb proteome library: a resource of assays to quantify the complete proteome of Mycobacterium tuberculosis. Cell Host Microbe. 13, 602–612, doi: 610.1016/j.chom.2013.1004.1008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena H. P. et al. Comparison of the membrane proteome of virulent Mycobacterium tuberculosis and the attenuated Mycobacterium bovis BCG vaccine strain by label-free quantitative proteomics. J Proteome Res. 12, 5463–5474, doi: 5410.1021/pr400334k (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malen H., De Souza G. A., Pathak S., Softeland T. & Wiker H. G. Comparison of membrane proteins of Mycobacterium tuberculosis H37Rv and H37Ra strains. BMC Microbiol. 11, 18, doi: 10.1186/1471-2180-1111-1118 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza G. A. et al. Using a label-free proteomics method to identify differentially abundant proteins in closely related hypo- and hypervirulent clinical Mycobacterium tuberculosis Beijing isolates. Mol Cell Proteomics. 9, 2414–2423, doi: 2410.1074/mcp.M900422-MCP900200 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Keijzer J., de Haas P. E., de Ru A. H., van Veelen P. A. & van Soolingen D. Disclosure of selective advantages in the “modern” sublineage of the Mycobacterium tuberculosis Beijing genotype family by quantitative proteomics. Mol Cell Proteomics. 13, 2632–2645, doi: 2610.1074/mcp.M2114.038380 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymova M. A. et al. Characterization of extensively drug-resistant Mycobacterium tuberculosis isolates circulating in Siberia. BMC Infect Dis. 14, 478, doi: 10.1186/1471-2334-1114-1478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P. S. et al. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 38, D396–400, doi: 310.1093/nar/gkp1919 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes T. et al. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 5, 1121–1131, doi: 1110.1016/j.celrep.2013.1110.1031 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkarslan S. et al. A comprehensive map of genome-wide gene regulation in Mycobacterium tuberculosis. Sci Data. 2, 150010, doi: 10.1038/sdata.2015.1010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson E. J. et al. A high-resolution network model for global gene regulation in Mycobacterium tuberculosis. Nucleic Acids Res. 42, 11291–11303, doi: 11210.11093/nar/gku11777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas P. et al. The oxidation-sensing regulator (MosR) is a new redox-dependent transcription factor in Mycobacterium tuberculosis. J Biol Chem. 287, 37703–37712, doi: 37710.31074/jbc.M37112.388611 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan J. E. et al. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature. 499, 178–183, doi: 110.1038/nature12337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Bretl D. J., Penoske R. M., Anderson D. M. & Zahrt T. C. Components of the Rv0081-Rv0088 locus, which encodes a predicted formate hydrogenlyase complex, are coregulated by Rv0081, MprA, and DosR in Mycobacterium tuberculosis. J Bacteriol. 193, 5105–5118, doi: 5110.1128/JB.05562-05511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parwati I., van Crevel R. & van Soolingen D. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect Dis. 10, 103–111, doi: 110.1016/S1473-3099(1009)70330-70335 (2010). [DOI] [PubMed] [Google Scholar]
- Narvskaia O. V., Mokrousov I. V., Otten T. F. & Vishnevskii B. I. [Genetic marking of polyresistant Mycobacterium tuberculosis strains isolated in the north-west of Russia]. Probl Tuberk., 3, 39–41 (1999). [PubMed] [Google Scholar]
- De Beer J. L., Kodmon C., van der Werf M. J., van Ingen J. & van Soolingen D. Molecular surveillance of multi- and extensively drug-resistant tuberculosis transmission in the European Union from 2003 to 2011. Euro Surveill. 19(11), 20742. 20734th Congress of the European Society of Mycobacteriology (22013) (2014). [DOI] [PubMed]
- Schmidt F. et al. Complementary analysis of the Mycobacterium tuberculosis proteome by two-dimensional electrophoresis and isotope-coded affinity tag technology. Mol Cell Proteomics. 3, 24–42 (2004). [DOI] [PubMed] [Google Scholar]
- Gagneux S. & Small P. M. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis. 7, 328–337 (2007). [DOI] [PubMed] [Google Scholar]
- Li A. H. et al. Contrasting transcriptional responses of a virulent and an attenuated strain of Mycobacterium tuberculosis infecting macrophages. PLoS One. 5, e11066, doi: 11010.11371/journal.pone.0011066 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan J., Bloom B. R. & Rubin E. J. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci USA 102, 8327–8332 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresen C. et al. A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism. J Biol Chem. 285, 22264–22275, doi: 22210.21074/jbc.M22109.099028 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrellad M. A. et al. Virulence factors of the Mycobacterium tuberculosis complex. Virulence. 4, 3–66, doi: 10.4161/viru.22329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido P. A., Novoa-Aponte L., Villamil N. & Soto C. Y. The DosR dormancy regulator of Mycobacterium tuberculosis stimulates the Na(+)/K (+) and Ca (2+) ATPase activities in plasma membrane vesicles of mycobacteria. Curr Microbiol. 69, 604–610, doi: 610.1007/s00284-00014-00632-00286 (2014). [DOI] [PubMed] [Google Scholar]
- Selvaraj S., Sambandam V., Sardar D. & Anishetty S. In silico analysis of DosR regulon proteins of Mycobacterium tuberculosis. Gene. 506, 233–241, doi: 210.1016/j.gene.2012.1006.1033 (2012). [DOI] [PubMed] [Google Scholar]
- Sivaramakrishnan S. & de Montellano P. R. The DosS-DosT/DosR Mycobacterial Sensor System. Biosensors (Basel). 3, 259–282 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech P., Kolly G. S., Leon-Solis L., Fallow A. & Reed M. B. Massive gene duplication event among clinical isolates of the Mycobacterium tuberculosis W/Beijing family. J Bacteriol. 192, 4562–4570, doi: 4510.1128/JB.00536-00510 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallow A., Domenech P. & Reed M. B. Strains of the East Asian (W/Beijing) lineage of Mycobacterium tuberculosis are DosS/DosT-DosR two-component regulatory system natural mutants. J Bacteriol. 192, 2228–2238, doi: 2210.1128/JB.01597-01509 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badillo-Lopez C. et al. Differential expression of dnaA and dosR genes among members of the Mycobacterium tuberculosis complex under oxic and hypoxic conditions. Int Microbiol. 13, 9–13 (2010). [DOI] [PubMed] [Google Scholar]
- Uchida Y. et al. Accelerated immunopathological response of mice infected with Mycobacterium tuberculosis disrupted in the mce1 operon negative transcriptional regulator. Cell Microbiol. 9, 1275–1283 (2007). [DOI] [PubMed] [Google Scholar]
- Nambi S. et al. The Oxidative Stress Network of Mycobacterium tuberculosis Reveals Coordination between Radical Detoxification Systems. Cell Host Microbe. 17, 829–837, doi: 810.1016/j.chom.2015.1005.1008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardini M. et al. Characteristics of drug-resistant tuberculosis in Abkhazia (Georgia), a high-prevalence area in Eastern Europe. Tuberculosis (Edinb). 89, 317–324, doi: 310.1016/j.tube.2009.1004.1002. Epub 2009 Jun 1017 (2009). [DOI] [PubMed] [Google Scholar]
- van Embden J. D. et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 31, 406–409 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokrousov I. et al. Russian “successful” clone B0/W148 of Mycobacterium tuberculosis Beijing genotype: a multiplex PCR assay for rapid detection and global screening. J Clin Microbiol. 50, 3757–3759, doi: 3710.1128/JCM.02001-02012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply P. et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 44, 4498–4510 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bespyatykh J. A. et al. Spoligotyping of Mycobacterium tuberculosis complex isolates using hydrogel oligonucleotide microarrays. Infect Genet Evol. 26, 41–46, doi: 10.1016/j.meegid.2014.1004.1024 (2014). [DOI] [PubMed] [Google Scholar]
- Neuhoff V., Arold N., Taube D. & Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 9, 255–262 (1988). [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O. & Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 68, 850–858 (1996). [DOI] [PubMed] [Google Scholar]
- Tatusova T., Ciufo S., Fedorov B., O’Neill K. & Tolstoy I. RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 43, 3872, doi: 3810.1093/nar/gkv3278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino J. A. et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 32, 223–226, doi: 210.1038/nbt.2839 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilov I. V. et al. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 6, 1638–1655 (2007). [DOI] [PubMed] [Google Scholar]
- Tang W. H., Shilov I. V. & Seymour S. L. Nonlinear fitting method for determining local false discovery rates from decoy database searches. J Proteome Res. 7, 3661–3667, doi: 3610.1021/pr070492f (2008). [DOI] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300, doi: 10.2307/2346101 (1995). [DOI] [Google Scholar]
- Magrane M. & Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford). 2011, bar009, doi: 10.1093/database/bar1009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A., Rahnenfuhrer J. & Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 22, 1600–1607 (2006). [DOI] [PubMed] [Google Scholar]
- Thomas P. D. et al. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 31, 334–341 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.