Abstract
IMPORTANCE
Advances have been made in identifying genetic susceptibility loci for autoimmune diseases, but evidence is needed regarding their association with prognosis and treatment response.
OBJECTIVE
To assess whether specific HLA-DRB1 haplotypes associated with rheumatoid arthritis (RA) susceptibility are also associated with radiological severity, mortality, and response to tumor necrosis factor (TNF) inhibitor drugs.
DESIGN, SETTING, AND PARTICIPANTS
The Norfolk Arthritis Register (NOAR; 1691 patients and 2811 radiographs; recruitment: 1989–2008; 2008 as final follow-up) was used as a discovery cohort and the Early Rheumatoid Arthritis Study (421 patients and 3758 radiographs; recruitment: 1986–1999; 2005 as final follow-up) as an independent replication cohort for studies of radiographic outcome. Mortality studies were performed in the NOAR cohort (2432 patients; recruitment: 1990–2007; 2011 as final follow-up) and studies of treatment response in the Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate cohort (1846 patients enrolled at initiation of TNF inhibitor; recruitment: 2006–2010; 2011 as final follow-up). Longitudinal statistical modeling was performed to integrate multiple radiograph records per patient over time. All patients were from the United Kingdom and had self-reported white ancestry.
EXPOSURES
Sixteen HLA-DRB1 haplotypes defined by amino acids at positions 11, 71, and 74.
MAIN OUTCOMES AND MEASURES
Radiological outcome using the Larsen score (range: 0 [none] to 200 [severe joint damage]) and erosions of the hands and feet on radiographs, all-cause mortality, and treatment response measured by change in Disease Activity Score based on 28 joint counts and European League Against Rheumatism (EULAR) response.
RESULTS
Patients with RA and valine at position 11 of HLA-DRB1 had the strongest association with radiological damage (OR, 1.75 [95% CI, 1.51–2.05], P = 4.6E-13). By year 5, the percentages of patients with erosions of the hands and feet were 48% of noncarriers (150/314) of valine at position 11, 61% of heterozygote carriers (130/213), and 74% of homozygote carriers (43/58). Valine at position 11 also was associated with higher all-cause mortality in patients with inflammatory polyarthritis (hazard ratio, 1.16 [95% CI, 1.03–1.31], P = .01) (noncarriers: 319 deaths in 1398 patients over 17 196 person-years, mortality rate of 1.9% per year; carriers: 324 deaths in 1116 patients in 13 208 person-years, mortality rate of 2.5% per year) and with better EULAR response to TNF inhibitor therapy (OR, 1.14 [95% CI, 1.01–1.30], P = .04) (noncarriers: 78% [439/561 patients] with moderate or good EULAR response; heterozygote carriers: 81% [698/866]; and homozygote carriers: 86% [277/322]). The risk hierarchy defined by HLA-DRB1 haplotypes was correlated between disease susceptibility, severity, and mortality, but inversely correlated with TNF inhibitor treatment response.
CONCLUSIONS AND RELEVANCE
Among patients with RA, the HLA-DRB1 locus, which is associated with disease susceptibility, was also associated with radiological severity, mortality, and treatment response. Replication of these findings in other cohorts is needed as a next step in evaluating the role of HLA-DRB1 haplotype analysis for management of RA.
Like many autoimmune diseases, the success in identifying genetic loci associated with rheumatoid arthritis (RA) susceptibility has not informed clinical practice. The largest RA genetic susceptibility effect is conferred by the HLA locus,1 and studies conducted in the 1980s identified multiple RA risk alleles within the HLA-DRB1 gene, encoding a similar amino acid motif at positions 70 through 74, leading to the “shared epitope” hypothesis.2 The shared epitope is associated with the development of anticitrullinated protein antibodies and has been consistently associated with markers of severe disease, such as radiological joint damage3,4 and mortality in patients with RA.5,6 However, the epitope has not shown a consistent association with treatment response.7–10
Amino acid positions 11, 71, and 74 within HLA-DRB1 are the major determinants of the association with RA susceptibility11 because no residual association at other HLA-DRB1 amino acid positions was observed after conditioning on these 3 positions. These 3 positions define 16 HLA-DRB1 haplotypes that can be ranked in a hierarchy based on the risk they confer and better model the association at HLA-DRB1 than the shared epitope alone. We hypothesized that these markers of disease susceptibility are also markers of disease severity and treatment response to tumor necrosis factor (TNF) inhibitor drugs. In this study, we tested their association with multiple measures of RA severity (radiological damage and mortality) and with response to TNF inhibitor drugs.
Methods
Patients and Cohorts
The Norfolk Arthritis Register (NOAR) was used as a discovery cohort and the Early Rheumatoid Arthritis Study (ERAS) as an independent replication cohort for studies of radiographic outcome. Mortality studies were performed in the NOAR cohort and studies of treatment response in the Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate (BRAGGSS) cohort. All patients were from the United Kingdom and had self-reported white ancestry thus avoiding spurious associations caused by population stratification. To compare the odds ratios (ORs) for disease severity with susceptibility and the ORs for treatment response with susceptibility, we recalculated the ORs for susceptibility using 9585 cases and 33 742 controls (described in eMethods in Supplement 1).
Radiographic Outcome: NOAR and ERAS Cohorts
We used the NOAR and ERAS cohorts to test association with radiographic outcome. NOAR is a primary care–based inception cohort of patients recruited since 1989 presenting with at least 2 swollen joints for at least 4 weeks (inflammatory polyarthritis) and followed up prospectively for 20 years or less.12,13 Patients with inflammatory polyarthritis who satisfied the 1987 American College of Rheumatology criteria14 during follow-up were classified as having RA. Radiographs of the hands and feet were performed during the first 10 years of follow-up and were scored using the Larsen method15 (described previously16,17).
Briefly, a score ranging from 0 (no joint damage) to 5 (complete destruction) is assigned to the small joints of the hands and feet. The individual joint scores are summed to attain a Larsen score per patient. The presence of a joint erosion was defined as a cortical break of 2 mm or larger and was assigned a score of 2 or greater. The Larsen score ranges from 0 to 200; a higher score indicates a more severe level of damage. Radiographs were read independently by 2 medically qualified observers, who underwent a specific training in radiograph reading and who were blinded to the sequence. Disagreement on the erosion status was settled by arbitration by a third investigator. All patients were recruited following informed consent and with ethical approval from the Norwich research ethics committee.
The ERAS cohort represents an independent group of patients recruited from rheumatology outpatient clinics in 9 districts of England between 1986 and 1999.17,18 Entry criteria included a clinical diagnosis of RA, symptoms for less than 2 years, and no prior treatment with disease-modifying antirheumatic drugs. Radiographs of the hands and feet were performed yearly and scored using the Larsen technique for the first 9 years of follow-up. Patients were recruited with informed consent and ethical approvals from the West Hertfordshire local research ethics committee and the Caldicott Guardian.
Mortality: NOAR Cohort
Patients in the NOAR cohort who were recruited between 1990 and 2007 were included if they had genetic and all-cause mortality data (provided by the UK Office for National Statistics) and censoring was applied on June 30, 2011. Data ascertainment and validation have been described.6,19,20 Briefly, mortality data were verified according to the UK Statistics Authority Code of Practice for Official Statistics21 to ensure quality regarding relevance, accuracy, timeliness, accessibility, and comparability.
Treatment Response to TNF Inhibitor Drugs: BRAGGSS
The BRAGGSS cohort has been described.22,23 Patients with RA starting treatment with a TNF inhibitor drug (either infliximab, etanercept, or adalimumab) were recruited between 2006 and 2010. However, patients who had been treated previously with TNF inhibitor drugs or other biologic agents also were included. Blood samples were collected prior to treatment and patients were followed up prospectively to assess response at a single time point between 3 and 6 months after therapy. A multicenter ethics committee (COREC 04/Q1403/37) approved the study and all patients provided informed consent. Further details appear in the eMethods in Supplement 1.
Genotyping
A 4-digit HLA typing corresponds to the determination of the full and unambiguous amino acid sequence of the HLA protein (eg, HLA-DRB1*04:01, *04:04). A 2-digit typing refers to the determination of the first 2 digits (eg, HLA-DRB1*04). The HLA-DRB1 alleles starting with the same 2 digits (eg, *04:01, *04:02, *04:03) will share some amino acid sequence similarities, but are not identical. Two-digit typing therefore does not systematically allow the unambiguous determination of the amino acid carried by a patient at a specific position. Six-digit typing (eg, HLA-DRB1*04:01:01) is used to characterize alleles that differ only by synonymous nucleotide substitutions (silent or noncoding substitutions). The amino acid sequence of all 6-digit alleles sharing the same initial 4 digits is therefore identical. The HLA typing was performed using a semiautomated, reverse dot-blot method.24 Amino acids at positions 11, 71, and 74 of HLA-DRB1 were assigned (eMethods and eTables 1 and 2 in Supplement 1). The 4-digit HLA-DRB1 shared epitope alleles have been listed previously.25
In addition, all samples available in 2010 from the NOAR and BRAGGSS cohorts with a diagnosis of RA and of sufficient DNA quality were genotyped using a single-nucleotide polymorphism microarray (Illumina Infinium Immunochip) and imputed at the amino acid resolution as described previously.26–28 In NOAR, 1490 patients had an available 2-, 4- or 6-digit HLA-DRB1 typing determined experimentally by the reverse dot-blot method and 881 from imputation of the HLA region (Immunochip). In 680 overlapping samples, the concordance was 91.8% at the 4-digit level, 96.5% at the 2-digit level, and 96.7% for amino acid positions 11, 71, and 74 (eTable 3 in Supplement 1).
Statistical Analysis
Radiographic outcome was assessed by the presence of erosions of the hands and feet and by the Larsen score. Radiographic outcome was modeled longitudinally to enhance power17 by integrating multiple records per patient and by incorporating more patients into the model than a cross-sectional study because radiographs were not performed systematically at every time point.
The presence of erosions of the hands and feet was treated as a longitudinal binary variable and modeled using a generalized estimating equation (GEE) model with logit-link function and an exchangeable within-subject correlation structure. Measures of association for GEE models are given as ORs and 95% CIs.
The Larsen score (as a longitudinal continuous nonnormally distributed outcome variable) was fitted using a generalized linear latent and mixed model (GLLAMM29–31) with discrete random effects and 3 latent classes. The measures of association for GLLAMM are given as a change in Larsen score. Technical details are provided in the eMethods in Supplement 1.
Adjustment for age, disease duration, and the square of them was performed systematically for all GEE and GLLAMM analyses to allow for a quadratic relationship between radiographic outcome and disease duration and age. We have previously reported a nonlinear relationship between these covariates and RA outcome in these cohorts,17,32 and have demonstrated the appropriateness of fitting a quadratic term.
Mortality
The association between genetic factors and all-cause mortality was assessed using Cox proportional hazard models. A forward-stepwise approach showed a significant and independent association with mortality for the nongenetic cardiovascular risk factors of sex, obesity, and use of antihypertensive drugs. These were subsequently included as covariates in the study of genetic factors. Measures of association are reported as hazard ratios (HRs).
Treatment Response to TNF Inhibitor Drugs
Treatment response was defined as a change in Disease Activity Score based on 28 joint counts (DAS28)23 (change in DAS28 = DAS28 at 3 to 6 months - DAS28 at baseline) or European League Against Rheumatism (EULAR) response (0 for no, 1 for moderate, 2 for good response).33 Treatment response was assessed at 1 time point between 3 and 6 months after treatment initiation. The DAS28 score ranges from 0 to 10; a higher score indicates a higher level of disease activity. A change in DAS28 of greater than 0.6 constitutes a clinically meaningful change.9 The EULAR response categories are based on this threshold.33
Association with genotypes was tested with linear regression for change in DAS28 or ordinal logistic regression for EULAR response. A forward-stepwise approach showed a significant and independent association of the following nongenetic predictors of response to TNF inhibitor drugs, which were subsequently included as covariates: sex, concurrent treatment with disease-modifying antirheumatic drugs, pretreatment DAS28 score, and the pretreatment Health Assessment Questionnaire disability index (a self-reported, patient-oriented measure of functional disability [score range: 0–3]; a higher score indicates a higher level of disability).34 Further details appear in the eMethods in Supplement 1.
Model Definition and Corrections for Multiple Testing
For all analyses, an additive model of association was used by creating a numerical variable (0, 1, 2) for the number of alleles carried by a patient. An allele is either a specific amino acid at a specific position or a haplotype. A haplotype refers to a combination of amino acids coded by DNA positions inherited independently of each other. The statistical effects are reported for 1 copy of every allele. We systematically tested the association of individual amino acids, individual positions, and individual haplotypes with all outcome measures. Model construction is explained in detail in the eMethods in Supplement 1.
Uncorrected 2-sided P values are presented. The Benjamini-Hochberg false discovery rate (0.05) method was used to correct for multiple testing in univariable and multivariable analyses. P values that remained significant when this correction was applied are indicated.
To test for differences between ORs, the linear combination βhaplotype A minus βhaplotype B, in which βhaplotype A is log (ORhaplotype A) and βhaplotype B is log(ORhaplotype B), was calculated with its standard error and a P value for the difference in association. The same approach was used for the HRs.
Statistical analysis was performed using Stata version 12.1 (StataCorp).
Results
Radiographic Outcome: Univariable Analysis of Amino Acid Positions 11, 71, and 74 of HLA-DRB1 in NOAR
At the time of the analysis, NOAR comprised 4293 patients with inflammatory polyarthritis and 24 093 follow-up visits. The 1691 patients with inflammatory polyarthritis (comprising 1363 patients with RA) with available genetic data and at least 1 radiograph were included in the study of radiographic outcome (Table 1). Of all amino acids tested at HLA-DRB1 listed in Table 2, valine at position 11 (outside the shared epitope) showed the strongest association with erosive disease in patients with inflammatory polyarthritis (OR, 1.75 [95% CI, 1.52–2.01], P = 8.7E-15) and was associated with an increase in Larsen score of 1.47 (95% CI, 0.85–2.10, P = 3.8E-06) compared with the carriers of nonvaline amino acids. Results for the subset of patients with RA were similar for valine at position 11 (OR, 1.75 [95% CI, 1.51–2.05], P = 4.6E-13). The crude unadjusted data appear in Table 3 and in the eTable in Supplement 2.
Table 1.
Cohort Characteristics for the Norfolk Arthritis Register (NOAR), Early Rheumatoid Arthritis Study (ERAS), and Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate (BRAGGSS)
| Cohort Characteristics | NOAR | ERAS | BRAGGSS |
|---|---|---|---|
| No. of patients successfully genotyped with radiological outcome or treatment response data by yeara | 1691 | 421 | 1846 |
| 1 | 1038 | 421 | NA |
| 5 | 719 | 373 | NA |
| 10 | 194 | 256 | NA |
| No. of radiographs included in the study | 2811 | 3758 | NA |
| Recruitment period | 1989–2008 | 1986–1999 | 2006–2010 |
| Final follow-up date | 2008 (radiographs) 2011 (mortality) |
2005 | 2011 |
| Satisfy ACR criteria for RA during follow-up, No. (%)b | 1363 (81) | 404 (96) | NA |
| Female sex, No. (%) | 1143 (68) | 276 (66) | 1414 (77) |
| Age, median (IQR), yc | 56 (44–67) | 56 (45–65) | 57 (50–64) |
| Duration of symptoms at baseline | |||
| Median (IQR), mo | 6 (3–12) | 7 (4–12) | NA |
| 90% percentile (maximum), y | 2 (5) | 1.7 (2) | NA |
| Duration of follow-upd | 8 (0–20) | 11 (1–15) | 4.1 (3.0–6.0) |
| Mortality and genetic data, No./total (%) | |||
| Obese | 511/2432 (21) | NA | NA |
| Taking antihypertensive drugs | 270/2432 (11) | NA | NA |
| Mean No. of radiographs/patient (range) | 1.7 (1–5) | 12 (2–17) | NA |
| Ever tested positive, No./total (%)e | 549/1583 (35) | 370/421 (88) | 984/1184 (83) |
| ≥1 Copy of shared epitope, No./total (%) | 1030/1648 (62) | 297/395 (75) | NA |
| Health Assessment Questionnaire disability index score, median (IQR)f | 0.875 (0.250–1.625) | 0.875 (0.125–1.500) | 2.0 (1.6–2.4) |
| Disease Activity Score, median (IQR)g | |||
| At baseline | NA | NA | 6.5 (5.8–7.2) |
| At 3–6 mo | NA | NA | 4.0 (2.9–5.1) |
| EULAR response at 3–6 mo, No. (%) | |||
| None | NA | NA | 357 (19) |
| Moderate | NA | NA | 941 (51) |
| Good | NA | NA | 548 (30) |
| Concurrent treatment with disease-modifying antirheumatic drugs by year, No./total (%) | |||
| Baseline | 484/1691 (29) | NA | 1393/1845 (76) |
| 1 | 727/1601 (45) | NA | NA |
| 3 | NA | 339/402 (84) | NA |
| 5 | 597/1320 (45) | 323/372 (87) | NA |
| 10 | 295/705 (42) | NA | NA |
| 20 | 47/123 (38) | NA | NA |
| Treatment with TNF inhibitor | |||
| Infliximab | NA | NA | 613 (33.2) |
| Etanercept | NA | NA | 650 (35.2) |
| Adalimumab | NA | NA | 582 (31.5) |
| Larsen score by year, median (IQR) | NA | NA | NA |
| 5 | 6 (0–23) | 10 (2–27) | NA |
| 9 | NA | 25 (7–48) | NA |
| 10 | 34 (15–56) | NA | NA |
| Erosive disease by year, No./total (%)h | |||
| 5 | 332/719 (46) | 275/372 (74) | NA |
| 10 | 176/194 (91) | 220/256 (86) | NA |
Abbreviations: EULAR, European League Against Rheumatism; IQR, interquartile range; NA, not available or not relevant; RA, rheumatoid arthritis; TNF, tumor necrosis factor.
NOAR and ERAS collected radiological outcome data and BRAGGSS collected treatment response data. Patients in NOAR underwent radiography only at baseline and years 1, 2, 5, and 10.
The 1987 American College of Rheumatology (ACR) criteria for RA applied cumulatively over the entire duration of follow-up in NOAR.
At symptom onset in NOAR and ERAS and at baseline in BRAGGSS (baseline means at initiation of treatment with biologic agents).
Median (range) in years for NOAR and ERAS and median (IQR) in months for BRAGGSS.
For anticitrullinated protein antibodies in NOAR and BRAGGSS or rheumatoid factor in ERAS.
At year 5 in NOAR and ERAS and at baseline in BRAGGSS. Scores range from 0 to 3; a higher score indicates a higher level of functional disability.
Score range from 0 to 10; a higher score indicates a higher level of disease activity.
Defined as the presence of at least 1 erosive joint (Larsen score ≥2; cortical break ≥2 mm). The Larsen score15 ranges from 0 to 200; a higher score indicates a more severe level of joint damage.
Table 2.
Univariable Analysis of the Association Between Amino Acids Tested at HLA-DRB1 and Erosive Disease in the Norfolk Arthritis Register (NOAR) and the Early Rheumatoid Arthritis Study (ERAS)
| Position | Amino Acida | No.b | Joint Erosions of the Hands and Feet, OR (95% CI)c | P Value | Change in Larsen Score (95% CI)d | P Value |
|---|---|---|---|---|---|---|
| Inflammatory polyarthritis in NOAR (1691 patients, 2811 radiographs)e | ||||||
| 11 | Valine | 779 | 1.75 (1.52 to 2.01) | 8.7E-15f | 1.47 (0.85 to 2.10) | 3.8E-06f |
| 11 | Leucine | 456 | 1.08 (0.90 to 1.32) | .39 | −0.09 (−0.97 to 0.79) | .84 |
| 11 | Aspartic acid | 52 | 1.27 (0.77 to 2.10) | .35 | 2.63 (0.58 to 4.69) | .01f |
| 11 | Proline | 347 | 0.82 (0.66 to 1.01) | .07 | −0.58 (−1.48 to 0.33) | .21 |
| 11 | Glycine | 357 | 0.89 (0.72 to 1.09) | .27 | 0.39 (−0.48 to 1.25) | .38 |
| 11 | Serine | 937 | 0.64 (0.55 to 0.73) | 6.9E-11f | −1.42 (−2.00 to −0.85) | 1.1E-06f |
| 71 | Lysine | 821 | 1.14 (0.98 to 1.32) | .08 | −0.34 (−0.98 to 0.30) | .29 |
| 71 | Arginine | 1154 | 1.13 (0.99 to 1.30) | .07 | 0.97 (0.39 to 1.55) | 1.0E-03f |
| 71 | Alanine | 298 | 0.81 (0.65 to 1.02) | .08 | −1.10 (−2.09 to −0.12) | .03 |
| 71 | Glutamic acid | 261 | 0.62 (0.48 to 0.79) | 1.3E-04f | −0.99 (−1.96 to −0.03) | .04 |
| 74 | Alanine | 1480 | 1.27 (1.11 to 1.48) | 7.20E-04f | 0.64 (0.07 to 1.21) | .03f |
| 74 | Glutamic acid | 185 | 0.90 (0.68 to 1.19) | .44 | −0.03 (−1.11 to 1.06) | .96 |
| 74 | Arginine | 404 | 0.73 (0.60 to 0.89) | 1.5E-03f | −1.34 (−2.13 to −0.55) | 9.3E-04f |
| 74 | Glutamine | 353 | 0.89 (0.72 to 1.11) | .28 | 0.40 (−0.46 to 1.26) | .36 |
| 74 | Leucine | 72 | 1.00 (0.62 to 1.63) | .99 | −0.50 (−2.18 to 1.19) | .56 |
| Rheumatoid arthritis in NOAR (1363 patients, 2384 radiographs)e | ||||||
| 11 | Valine | 661 | 1.75 (1.51 to 2.05) | 4.6E-13f | 1.81 (1.08 to 2.55) | 1.2E-06f |
| 11 | Leucine | 359 | 1.11 (0.90 to 1.36) | .35 | 0.28 (−0.74 to 1.29) | .59 |
| 11 | Aspartic acid | 42 | 1.39 (0.79 to 2.44) | .25 | 2.91 (0.50 to 5.32) | .02f |
| 11 | Proline | 284 | 0.80 (0.64 to 1.01) | .06 | −0.56 (−1.81 to 0.70) | .39 |
| 11 | Glycine | 284 | 0.90 (0.71 to 1.13) | .36 | 0.57 (−0.63 to 1.76) | .35 |
| 11 | Serine | 726 | 0.63 (0.54 to 0.73) | 3.0E-10f | −1.91 (−2.57 to −1.24) | 2.0E-08f |
| 71 | Lysine | 679 | 1.19 (1.01 to 1.39) | .04 | −0.36 (−1.10 to 0.38) | .34 |
| 71 | Arginine | 942 | 1.13 (0.97 to 1.31) | .10 | 1.08 (0.39 to 1.77) | 2.1E-03f |
| 71 | Alanine | 246 | 0.81 (0.63 to 1.04) | .10 | −1.19 (−2.49 to 0.12) | .07 |
| 71 | Glutamic acid | 201 | 0.54 (0.41 to 0.71) | 1.4E-05f | −1.35 (−2.52 to −0.18) | .02f |
| 74 | Alanine | 1194 | 1.28 (1.11 to 1.51) | 1.20E-03f | 0.88 (0.21 to 1.55) | .01f |
| 74 | Glutamic acid | 145 | 0.87 (0.64 to 1.19) | .37 | −0.32 (−1.58 to 0.94) | .62 |
| 74 | Arginine | 324 | 0.74 (0.60 to 0.91) | 5.0E-03f | −1.60 (−2.53 to −0.67) | 7.9E-04f |
| 74 | Glutamine | 283 | 0.89 (0.70 to 1.12) | .30 | 0.45 (−0.66 to 1.57) | .43 |
| 74 | Leucine | 56 | 0.90 (0.53 to 1.57) | .73 | −1.03 (−3.00 to 0.94) | .31 |
| Rheumatoid arthritis in ERAS (421 patients, 3758 radiographs)e,g | ||||||
| 11 | Valine | 242 | 1.39 (1.10 to 1.75) | 5.77E-03f | (n = 219)b 0.82 (0.21 to 1.43) |
8.77-03f |
| 11 | Leucine | 99 | 1.44 (0.95 to 2.19) | .09 | (n = 92)b 1.78 (0.76 to 2.80) |
6.52E-04f |
| 11 | Aspartic acid | 9 | 0.28 (0.06 to 1.26) | .10 | (n = 8)b −1.29 (−4.08 to 1.49) |
.36 |
| 11 | Proline | 83 | 0.95 (0.64 to 1.42) | .80 | (n = 77)b 0.43 (−0.48 to 1.33) |
.36 |
| 11 | Glycine | 71 | 0.70 (0.42 to 1.16) | .17 | (n = 66)b −0.28 (−1.36 to 0.79) |
.61 |
| 11 | Serine | 194 | 0.57 (0.42 to 0.78) | 4.20E-04f | (n = 170)b −2.31 (−3.06 to −1.55) |
2.12E-09f |
| 71 | Lysine | 225 | 1.31 (0.97 to 1.78) | .08 | (n = 207)b −0.59 (−1.36 to 0.18) |
.13 |
| 71 | Arginine | 249 | 0.79 (0.58 to 1.07) | .12 | (n = 226)b 0.69 (−0.11 to 1.48) |
.09 |
| 71 | Alanine | 60 | 1.07 (0.69 to 1.66) | .76 | (n = 56)b 1.37 (0.31 to 2.44) |
.01f |
| 71 | Glutamic acid | 18 | 0.58 (0.24 to 1.37) | .21 | (n = 17)b −0.68 (−3.04 to 1.67) |
.57 |
| 74 | Alanine | 371 | 1.41 (1.02 to 1.93) | .03 | (n = 340)b 0.99 (0.25 to 1.74) |
9.27E-03f |
| 74 | Glutamic acid | 18 | 1.25 (0.55 to 2.87) | .60 | (n = 14)b −0.45 (−2.79 to 1.88) |
.70 |
| 74 | Arginine | 90 | 0.87 (0.58 to 1.31) | .51 | (n = 84)b −1.43 (−2.43 to −0.43) |
5.00E-03f |
| 74 | Glutamine | 69 | 0.68 (0.41 to 1.15) | .15 | (n = 65)b 0.08 (−1.05 to 1.20) |
.89 |
| 74 | Leucine | 17 | 0.67 (0.27 to 1.63) | .38 | (n = 13)b −4.53 (−7.07 to −1.99) |
4.70E-04f |
The comparison group is all other amino acids at this position listed in the Table. The amino acids listed herein represent the exhaustive list of all possible amino acids at positions 11, 71, or 74 in the white population. The strongest association signal with radiological outcome mapped to valine 11.
Indicates the number of homozygote and heterozygote patients for that specific amino acid with available radiograph and genetic data.
Tested with a generalized estimating equation model.
Tested with a generalized latent and linear mixed model. The Larsen score ranges from 0 to 200; a higher score indicates a more severe level of joint damage.
The total number of patients with available radiographs and an available genotype located at the position of at least 1 of the 3 positions studied; however, only a subset of those will have been successfully assigned a genotype at every position.
Remains significant with a false discovery rate of 0.05 after adjustment with the Benjamini-Hochberg method.
Number of patients is different for different measures of radiographic outcome because erosion status and the Larsen score have been determined independently of each other.
Table 3.
Crude Unadjusted Data for Number of Copies of Valine at Position 11 and Radiographic Joint Erosions of the Hands and Feet in Patients With Inflammatory Polyarthritis in the Norfolk Arthritis Registera
| No. of Copies of Valine at Position 11b | No./Total (%) of Patients With Inflammatory Polyarthritis and Radiographic Joint Erosions of the Hands and Feet by Year | ||||
|---|---|---|---|---|---|
| Baseline | 1 | 2 | 5 | 10 | |
| 0 | 116/283 (41) | 189/553 (34) | 43/182 (24) | 156/396 (39) | 77/84 (92) |
| 1 | 106/181 (59) | 188/386 (49) | 44/129 (34) | 133/255 (52) | 73/84 (87) |
| 2 | 28/49 (57) | 64/99 (65) | 16/30 (53) | 43/68 (63) | 26/26 (100) |
Even though patients with rheumatoid arthritis in general show a higher proportion of joint erosions than patients with inflammatory polyarthritis, the association with genotypes was similar (crude unadjusted data for all amino acids and all positions for rheumatoid arthritis and inflammatory polyarthritis appear in the eTable in Supplement 2).
The strongest association signal with radiological outcome mapped to valine at amino acid position 11.
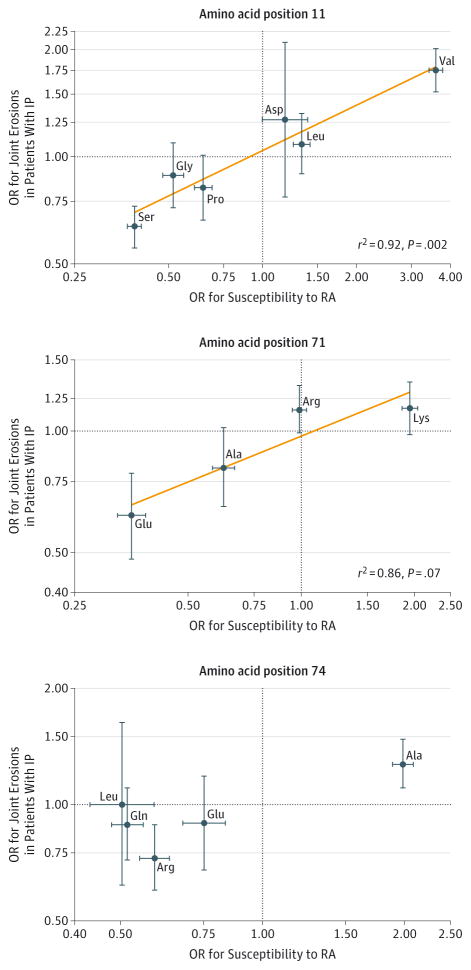
The association of valine at amino acid position 11 was independent of the shared epitope (eTable 5 in Supplement 1). Conversely, serine at amino acid position 11 was associated with a decreased risk of erosions of the hands and feet (OR, 0.64 [95% CI, 0.55 to 0.73], P = 6.9E-11) and change in Larsen score (−1.42 [95% CI, −2.00 to −0.85], P = 1.1E-06) (Table 2). We performed a linear regression of the ORs for severity of inflammatory polyarthritis on the ORs for susceptibility to RA and observed a significant alignment of amino acids at position 11 (r2 = 0.92, P = .002; Figure 1). However, the ORs were systematically smaller for radiographic outcome compared with susceptibility. Similar results were obtained for amino acid positions 71 and 74. Univariable analysis of positions showed that amino acid positions 71 and 74 were also significantly associated with radiographic outcome (eTable 6 in Supplement 1).
Figure 1. Linear Correlation Between Odds Ratios (ORs) for Rheumatoid Arthritis (RA) Susceptibility and ORs for Severity of RA From Univariable Analysis.
The reference group for every amino acid comprised noncarriers of that specific amino acid. Similar results were obtained for RA severity (instead of inflammatory polyarthritis [IP]) vs anticitrullinated protein antibody–positive RA. The orange line in the top 2 panels was fitted by linear regression. Horizontal and vertical error bars indicate 95% CIs. Ala indicates alanine; Arg, arginine; Asp, aspartic acid; Gln, glutamine; Glu, glutamic acid; Gly, glycine; Leu, leucine; Lys, lysine; Pro, proline; Ser, serine; Val, valine. More information appears in eTable 4 in Supplement 1 and in Raychaudhuri et al.11
Radiographic Outcome: Haplotype and Multivariable Analysis in NOAR
In multivariable analysis, Larsen score in patients with inflammatory polyarthritis was associated with amino acid positions 11 (P = 6.79E-03), 71 (P = 5.67E-03), and 74 (P = .04) independently of each other. No association was observed with shared epitope (an amino acid motif at positions 70–74; P = .34), indicating that a statistical model including amino acid positions 11, 71, and 74 completely superseded the shared epitope model.
Given that the associations at amino acid positions 11, 71, and 74 were independent of each other, we explored the association of the corresponding haplotypes. With 6 different amino acids at position 11, 4 at 71, and 5 at 74, 120 different haplotypes were theoretically possible. However, only 16 haplotypes were found in NOAR, matching those reported in our susceptibility study.11 The 16 haplotypes and their multivariable association results are presented for RA in Table 4 and for inflammatory polyarthritis in eTable 7 in Supplement 1. The shared epitope corresponded to the 3 first haplotypes in Table 4 (VKA, VRA, and LRA) and was therefore fully defined by (or collinear with) the 16-haplotype model.
Table 4.
Multivariable Analysis of 16 Haplotypes and Their Association With Radiographic Outcome in Patients With Rheumatoid Arthritis (RA) in the Norfolk Arthritis Register
| Haplotype Namea | Amino Acid by Positionb
|
No. of Patients With RA
|
GEEd
|
GLLAMMe
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 71 | 74 | Heterozygote Carriers (n = 1832) | Homozygote Carriers (n = 1832) | Haplotype Frequency in RA, %c | Joint Erosions, OR (95% CI) (n = 1249) | P Value | Change in Larsen Score (95% CI) (n = 1249)f | P Value | |
| VKAg | Val | Lys | Ala | 541 | 55 | 17.8 | 1.82 (1.35 to 2.46) | 8.85E-05h | 2.07 (0.69 to 3.44) | 3.28E-03h |
|
| ||||||||||
| VRAg | Val | Arg | Ala | 316 | 14 | 9.4 | 1.84 (1.31 to 2.59) | 4.00E-04h | 3.31 (1.64 to 4.98) | 1.00E-04h |
|
| ||||||||||
| LRAg | Leu | Arg | Ala | 458 | 31 | 14.2 | 1.48 (1.05 to 2.08) | .03 | 1.52 (−0.10 to 3.14) | .07 |
|
| ||||||||||
| PRA | Pro | Arg | Ala | 32 | 1 | 0.9 | 1.05 (0.56 to 1.97) | .88 | 3.37 (0.06 to 6.69) | .05 |
|
| ||||||||||
| VRE | Val | Arg | Glu | 81 | 6 | 2.5 | 1.55 (0.92 to 2.64) | .10 | 1.60 (−0.67 to 3.88) | .17 |
|
| ||||||||||
| DRE | Asp | Arg | Glu | 60 | 2 | 1.7 | 1.26 (0.68 to 2.32) | .47 | 3.21 (0.38 to 6.04) | .03 |
|
| ||||||||||
| VEA | Val | Glu | Ala | 12 | 0 | 0.3 | 0.24 (0.05 to 1.19) | .08 | −2.46 (−7.41 to 2.48) | .33 |
|
| ||||||||||
| SKA | Ser | Lys | Ala | 25 | 0 | 0.7 | 0.88 (0.33 to 2.39) | .80 | 0.01 (−3.70 to 3.72) | 1.00 |
|
| ||||||||||
| PAAg,i | Pro | Ala | Ala | 339 | 25 | 10.6 | 1 [Reference] | 0 [Reference] | ||
|
| ||||||||||
| GRQg | Gly | Arg | Gln | 383 | 24 | 11.8 | 1.04 (0.75 to 1.45) | .82 | 1.08 (−0.47 to 2.62) | .17 |
|
| ||||||||||
| SRAg | Ser | Arg | Ala | 171 | 6 | 5.0 | 1.07 (0.71 to 1.60) | .75 | 1.38 (−0.48 to 3.24) | .15 |
|
| ||||||||||
| SRE | Ser | Arg | Glu | 67 | 1 | 1.9 | 0.42 (0.23 to 0.77) | .01h | −1.38 (−3.79 to 1.03) | .26 |
|
| ||||||||||
| LEA | Leu | Glu | Ala | 32 | 1 | 0.9 | 0.59 (0.26 to 1.36) | .22 | −3.07 (−7.47 to 1.32) | .17 |
|
| ||||||||||
| SRL | Ser | Arg | Leu | 74 | 0 | 2.0 | 1.02 (0.54 to 1.92) | .96 | −0.10 (−2.41 to 2.21) | .93 |
|
| ||||||||||
| SKRg | Ser | Lys | Arg | 407 | 32 | 12.9 | 0.92 (0.67 to 1.28) | .64 | −0.18 (−1.68 to 1.31) | .81 |
|
| ||||||||||
| SEAg | Ser | Glu | Ala | 236 | 17 | 7.4 | 0.75 (0.51 to 1.09) | .13 | 0.26 (−1.41 to 1.93) | .76 |
Abbreviations: GEE, generalized estimating equation; GLLAMM, generalized latent and linear mixed model; OR, odds ratio.
Derived from Table 1 in the article by Raychaudhuri et al.11
Ala indicates alanine; Arg, arginine; Asp, aspartic acid; Gln, glutamine; Glu, glutamic acid; Gly, glycine; Leu, leucine; Lys, lysine; Pro, proline; Ser, serine; Val, valine.
Calculated as (No. of heterozygote carriers + 2 × No. of homozygote carriers)/(2 × 1832). Haplotype frequency was assessed in the entire RA population in the Norfolk Arthritis Register cohort with available haplotype (1832 patients), including individuals without available radiological assessment.
Population-averaged model for the presence of erosive disease (longitudinal dummy variable). The haplotype group P value is 2.70E-11.
The haplotype group P value is 1.22E-06.
The Larsen score (longitudinal continuous variable) ranges from 0 to 200; a higher score indicates a more severe level of joint damage. The association study for joint erosions of the hands and feet or Larsen score was performed on samples with available haplotype and radiological data (1249 patients with RA). This number is smaller than the 1363 patients with RA described in Table 1 because some have missing genotypes at 1 or 2 of the 3 positions used to construct haplotypes.
Frequency greater than 5%; cumulatively, these haplotypes represent 89% of the RA population.
Remains significant with a false discovery rate of 0.05 after adjustment with the Benjamini-Hochberg method.
Used as the reference group; previously reported to be the most frequent haplotype in control samples.11
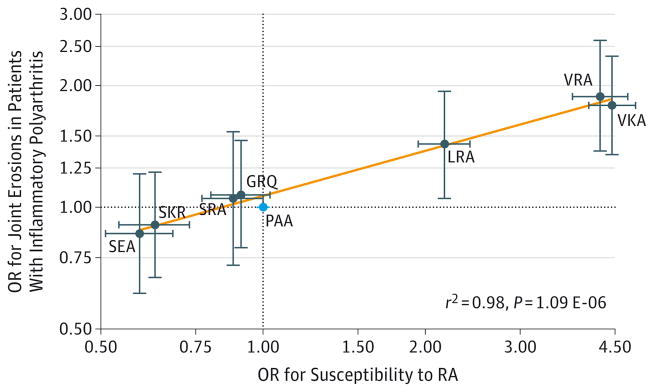
The PAA haplotype was used as the reference because it was the most frequent haplotype in control samples.11 The VKA and VRA haplotypes, which were significantly associated with an increased risk of developing RA,11 also were significantly associated with an increased risk of radiographic damage (Table 4). The same haplotype risk hierarchy established for susceptibility was fully conserved with that seen for radiographic severity (Figure 2). We observed a significant alignment for the 16-haplotype model (r2 = 0.98 for the 8 haplotypes with a frequency of ≥5% associated with erosions of the hands and feet vs susceptibility; P = 1.09E-06).
Figure 2. Eight Haplotypes With Frequency of 5% or Greater Associated With Odds Ratios (ORs) for Joint Erosions and ORs for Susceptibility to Rheumatoid Arthritis (RA).
Severity correlates with susceptibility in this Figure. The reference group is the PAA haplotype (cyan dot). Similar results were obtained for RA vs anticitrullinated protein antibody–positive RA. The orange line was fitted by linear regression. Horizontal and vertical error bars indicate 95% CIs. The odds ratios for susceptibility are from Table 1 of the article by Raychaudhuri et al.11
The genetic risk score35 for severity (calculated solely with the 16 HLA-DRB1 haplotypes) also was highly associated with the genetic risk score for susceptibility (r2 = 0.57, P < 1.0E-300; eFigure in Supplement 1). The 16-haplotype model was associated with Larsen score (P = 3.73E-06) as was the shared epitope (P = 1.10E-06); however, only the 16-haplotype model remained significantly associated (P = 1.74E-03) when tested together with the shared epitope (P = .99). Mediation analysis detected a signific ant direct assoc iation of the 16-haplotype model on radiographic outcome, independently of the anticitrullinated protein antibody status (eResults in Supplement 1).
Radiographic Outcome: Replication in ERAS
To replicate associations with radiographic outcome, we tested a second cohort of patients with early-stage RA (Table 1). At the time we conducted the study, the ERAS cohort had 1399 patients with phenotypic information; however, a DNA sample was only available for 421 patients. The demographic and clinical characteristics of this subgroup were similar to those of the entire ERAS cohort.36 The same pattern in NOAR was seen in ERAS (Table 2); valine at amino acid position 11 was associated with erosions of the hands and feet and Larsen score. Position 11 as a whole was associated with erosions of the hands and feet (P = 5.05E-05) and with Larsen score (P = 9.71E-13), and remained significant after adjustment for the shared epi-tope (3.43E-06), which was then no longer associated with RA outcome (P = .69). The 16-haplotype model was associated with Larsen score (P = 7.42E-25) and completely removed the association of the shared epitope.
Comparison With Shared Epitope
We formally tested the incremental explanatory power of the 16-haplotype model compared with the shared epitope to model radiographic damage and found the goodness of fit of the 16-haplotype model to be significantly better. The log likelihood (a measure of the goodness of fit of a model) for GLL AMM (L arsen score) in RA was −8483 for the 16-haplotype model, which was greater than −9016 for the shared epitope. The higher the log likelihood, the better the model fit. This difference in the goodness of fit was statistically significant (P = .03) when compared using the likelihood ratio test (eResults in Supplement 1).
Mortality Study in NOAR
All-cause mortality also was tested to confirm that HLA-DRB1 haplotypes associated with an increased risk of radiographic damage were also associated with an increased risk of other measures of severe disease outcome. Of 2432 patients with data and DNA available, 642 patients died during a median duration of follow-up of 12 years (interquartile range, 8–17 years). Valine at amino acid position 11 was again significantly associated with higher all-cause mortality in patients with inflammatory polyarthritis (HR, 1.16 [95% CI, 1.03–1.31], P = .01) (noncarriers: 319 deaths in 1398 patients over 17 196 person-years, mortality rate of 1.9% per year; carriers: 324 deaths in 1116 patients in 13 208 person-years, mortality rate of 2.5% per year).
Restricting the analysis to the subset of patients with RA showed the same pattern. When the 16-haplotype model was tested, the same hierarchy was observed. All-cause mortality in patients with inflammatory polyarthritis was increased for carriers of the VKA haplotype (mortality rates for noncarriers vs carriers: 1.9% vs 2.5% per year; HR, 1.15 [95% CI, 0.99–1.33], P = .08), whereas the SEA haplotype was associated with decreased mortality (mortality rates for noncarriers vs carriers: 2.2% vs 1.5% per year; HR, 0.76 [95% CI, 0.60–0.97], P = .03). The difference in HRs of 0.37 (95% CI, 0.10–0.64) was statistically significant (P = 8.3E-03), indicating that the same risk hierarchy observed for radiographic damage also applied to other measures of disease severity.
Treatment Response to TNF Inhibitor Drugs in BRAGGSS
We also tested for an association between HLA-DRB1 amino acid positions 11, 71, and 74 and treatment response to TNF inhibitor drugs in a third independent cohort of 1846 patients from the BRAGGSS cohort with available genotype and treatment response data (Table 1). We found no significant association for change in DAS28 with anticitrullinated protein antibodies (P = .15) or the shared epitope (P = .21); similar results were found for the EULAR response (P = .55 and P = .08, respectively). However, valine at amino acid position 11 was significantly associated with a better EULAR response (OR, 1.14 [95% CI, 1.01–1.30], P = .04) (noncarriers: 78% [439/561 patients] with moderate or good EULAR response; heterozygote carriers: 81% [698/866 patients]; and homozygote carriers: 86% [277/322 patients]).
The crude unadjusted data for treatment response and haplotypes appear in Table 5. The VKA haplotype was significantly associated with a better EULAR response (OR, 1.23 [95% CI, 1.06–1.43], P = .007; Table 6). The OR shows the increased chance of moving from one EULAR response category to the next per copy of the VKA haplotype. This translates to an OR of 2.32 for a good EULAR response vs no response from a VKA haplotype homozygote patient vs a noncarrier (assuming an additive model). The association of the VKA haplotype was independent of known factors (DAS28, Health Assessment Questionnaire disability index, disease-modifying antirheumatic drugs, and sex) associated with good response.
Table 5.
Crude Unadjusted Measures of Treatment Response in the Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate Cohort of 1846 Patients
| SEA Haplotype (n = 1736) | VKA Haplotypea
|
|||||
|---|---|---|---|---|---|---|
| 0 Copies
|
1 Copy
|
2 Copies
|
||||
| Change in DAS28 | EULAR Response | Change in DAS28 | EULAR Response | Change in DAS28 | EULAR Response | |
| 0 copies | −2.43 (1.54) | 1.06 (0.70) | −2.49 (1.49) | 1.15 (0.69) | −2.58 (1.43) | 1.16 (0.63) |
|
| ||||||
| 1 copy | −2.23 (1.54) | 0.98 (0.67) | −2.52 (1.40) | 1.33 (0.66) | ||
|
| ||||||
| 2 copies | −1.75 (1.88) | 0.80 (1.10) | ||||
Abbreviations: DAS28, Disease Activity Score based on 28 joint counts; EULAR, European League Against Rheumatism.
Data are expressed as mean (SD). Change in DAS28 and EULAR response are measures of treatment response. Scores for DAS28 range from 0 to 10 (a higher score indicates a higher level of disease activity) and EULAR response range from 0 (no response), 1 (moderate response), to 2 (good response). A negative value for change in DAS28 indicates treatment response and corresponds to a value greater than 0 for EULAR response. There are empty cells because the carriage of the 2 haplotypes is mutually exclusive.
Table 6.
Adjusted Measures of Treatment Response in the Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate Cohort
| Markera | Change in DAS28b
|
EULAR Responsec
|
||
|---|---|---|---|---|
| Coefficient (95% CI) | P Value | OR (95% CI) | P Value | |
| VKA haplotyped | −0.12 (−0.23 to −0.01) | 3.25E-02 | 1.23 (1.06 to 1.43) | .007 |
|
| ||||
| SEA haplotyped | 0.17 (−0.08 to 0.42) | 1.80E-01 | 0.96 (0.68 to 1.35) | 8.08E-01 |
|
| ||||
| DAS28 | −0.60 (−0.67 to −0.53) | 1.71E-57 | 0.92 (0.84 to 1.02) | 1.29E-01 |
|
| ||||
| Health Assessment Questionnaire (HAQ) disability indexe | 0.40 (0.27 to 0.52) | 5.35E-10 | 0.61 (0.51 to 0.73) | 3.34E-08 |
|
| ||||
| Disease-modifying antirheumatic drugsf | −0.45 (−0.61 to −0.30) | 1.03E-08 | 1.77 (1.43 to 2.18) | 1.11E-07 |
|
| ||||
| Sexg | 0.22 (0.06 to 0.38) | 5.93E-03 | 0.76 (0.62 to 0.95) | 1.44E-02 |
Abbreviations: DAS28, Disease Activity Score based on 28 joint counts; OR, odds ratio.
Haplotypes were the main variables, whereas DAS28, HAQ disability index, disease-modifying antirheumatic drugs, and sex were the covariates. The reference group for the regressions was the group of patients without a copy of the VKA or SEA haplotype.
Multivariable linear regression was performed. Regression coefficients are expressed in DAS28 units and are given for an increase of 1 unit for every variable. For example, for every point in HAQ at treatment initiation, a patient will on average experience an increase in DAS28 by 0.40 during treatment, but a decrease by 0.12 for every copy of the VKA haplotype, and the magnitude of these associations are independent of any other parameters tested. DAS28 at baseline (ie, at initiation of treatment with tumor necrosis factor inhibitor therapy) is strongly associated with treatment response as measured by change in DAS28, but is not associated with European League Against Rheumatism (EULAR) response in this Table, reflecting the fact that EULAR response is a composite measure of change in DAS28 and absolute DAS28.
Multivariable ordinal logistic regression was performed. The ORs for EULAR response are given for an increase of 1 unit for every variable.
The VKA haplotype was chosen because it has been shown to have the largest magnitude of association with an increased susceptibility and severity in rheumatoid arthritis. The SEA haplotype was chosen because it has been shown to have the largest magnitude of association with a decreased susceptibility and severity in rheumatoid arthritis (Table 4 and Figure 2; and Table 1 of article by Raychaudhuri et al11).
Measured at baseline; functional disability measure with a score range from 0 to 3; a higher score indicates a higher level of disability.
Concurrent treatment was coded as 1 (no concurrent treatment was coded as 0 and serves as the reference group).
Female was coded as 1 (male was coded as 0 and serves as reference group).
The risk hierarchy of the SEA haplotype with regard to the VKA haplotype was significantly conserved. The difference in change in DAS28 between 1 copy of the VKA haplotype and 1 copy of the SEA haplotype was significant (difference in change in DAS28 of 0.29 [95% CI, 0.04–0.54], P = .03). Because this difference in change in DAS28 is reported for 1 copy of each haplotype, it means that a homozygote carrier of the VKA haplotype will have a DAS28 score of 0.58 units lower on average between 3 and 6 months after therapy than a homozygote carrier of the SEA haplotype. The 6 variable model presented in Table 6 explained 15% of the variance in treatment response. When all other haplotypes were included in the analysis, the risk hierarchy also was significantly conserved (Figure 3 and Figure 4).
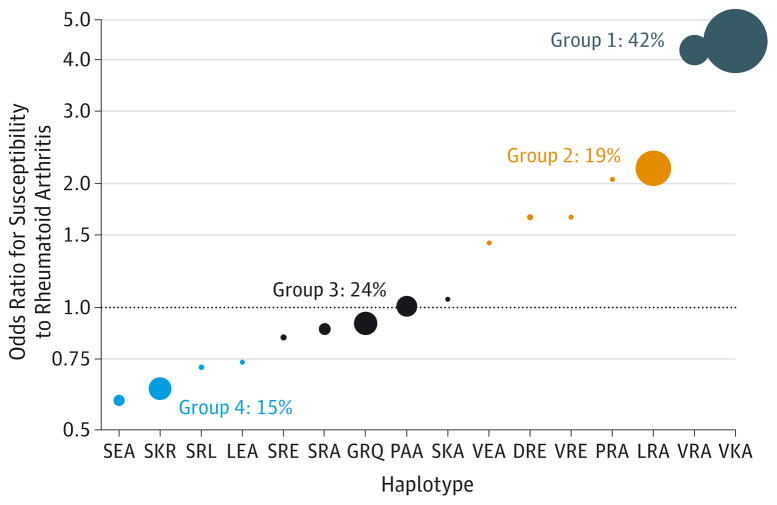
Figure 3. Definition of 4 Haplotype Groups by 16 Haplotypes Defined by Amino Acid Positions 11, 71, and 74 of HLA-DRB1.
The size of the filled circle representing a haplotype is proportional to its frequency in the Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate (BRAGGSS) cohort; the smallest point represents frequencies below 1%. The allocation of haplotypes to groups was performed to group rare haplotypes with frequent ones having similar odds ratios. If the PAA haplotype is set as the reference, then haplotypes associated with a decreased risk to develop rheumatoid arthritis are represented below the dashed line, whereas haplotypes associated with an increased risk are above it. Of the 1846 patients from the BRAGGSS cohort presented in Table 1, 1819 had nonmissing genotypes at the 3 positions used to construct the haplotypes. Group 1 comprises 894 heterozygote patients (1 copy of a group 1 haplotype; eg, VKA or VRA) and 314 homozygote patients (2 copies of VKA or 2 copies of VRA, 1 copy of VKA and VRA). Group 2 comprises 594 heterozygote patients and 46 homozygote patients. Group 3 comprises 715 heterozygote patients and 80 homozygote patients. Group 4 comprises 480 heterozygote patients and 37 homozygote patients. The haplotype frequency presented was calculated as: (No. of heterozygote carriers + 2 × No. of homozygote carriers)/(2 × 1819). The nomenclature for haplotype names is presented in Table 4.
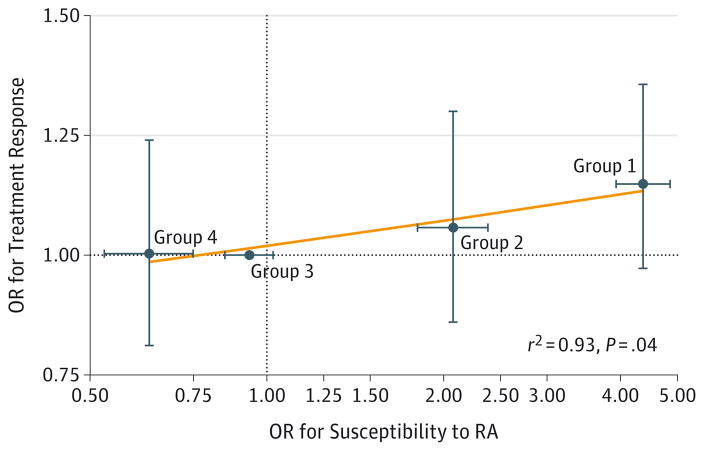
Figure 4. Correlation Between Odds Ratios (ORs) for Susceptibility to Rheumatoid Arthritis (RA) and ORs for Treatment Response in BRAGGSS Cohort.
Treatment response correlates with susceptibility in this Figure. Horizontal and vertical error bars indicate 95% CIs. The orange line was fitted by linear regression. Haplotype groups are defined in Figure 3. The OR for a haplotype group for the association with RA susceptibility has been calculated as the weighted average of the OR of the individual haplotypes belonging to the group; haplotype frequency was used as the weight. The OR for the association with treatment response has been calculated using a multivariable ordinal logistic regression of European League Against Rheumatism response on the haplotype groups. Every patient in the Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate (BRAGGSS) cohort carries either 0, 1, or 2 copies of a haplotype classified as a group 1, 2, 3, or 4 haplotype. The multivariable ordinal logistic regression includes all 4 haplotype groups in the same model and the following markers of treatment response as covariates: sex, concurrent treatment with disease-modifying antirheumatic drugs, Disease Activity Score based on 28 joint counts, and Health Assessment Questionnaire disability index score at treatment initiation (baseline). The Disease Activity Score ranges from 0 to 10; a higher score indicates a higher level of disease activity. Health Assessment Questionnaire measures functional disability and ranges from 0 to 3; a higher score indicates a higher level of disability.
Discussion
To our knowledge, this is the first study to explore the associations of 16 HLA-DRB1 haplotypes defined by amino acids at positions 11, 71, and 74 with RA severity, mortality, and treatment response. We found that the same RA susceptibility risk variants also were associated with these outcome measures. We have reproduced previously published results regarding the association of the shared epitope on disease severity,4 mortality,5,6 and its lack of association with treatment response.10,37 The classification11 of 16 haplotypes appears to supersede the previous shared epitope model, which classified patients with RA into 2 groups of patients positive for the shared epitope or negative for it.
Amino acid position 11 (outside of the shared epitope) was associated with the development of radiographic damage in patients with inflammatory polyarthritis and RA. This was independent of the classic shared epitope. Valine at this position represents what we believe is the strongest single genetic association with radiographic damage identified to date. The association was confirmed at genome-wide thresholds and was replicated in an independent cohort. Every valine carried was associated with a higher Larsen score (1.8 Larsen units); therefore, homozygosity at this locus corresponds to a Larsen score that is higher by 3.6 Larsen units than the one of nonvaline carriers, and is clinically relevant (the minimal clinically important difference is 2.3).38
Valine at amino acid position 11 also was associated with mortality and treatment response. The HLA-DRB1 positions 11, 71, and 74 were associated independently of each other and formed 16 haplotypes that were hierarchically associated with radiographic damage, all-cause mortality, and treatment response to TNF inhibitor drugs. The valine-containing VKA haplotype was associated with RA susceptibility (OR, 4.44 [95% CI, 4.02-4.91), joint erosions in patients with RA (OR, 1.82 [95% CI, 1.35–2.46]), and a better response to TNF inhibitor drugs (OR, 1.23 [95% CI, 1.06–1.43]). Although this is a modest increase for treatment response, it should be recognized that it refers to the likelihood of moving from the EULAR category of none to a moderate response, or from a moderate to a good response for every copy of the VKA haplotype carried by the patient compared with noncarriers.
The 16-haplotype model provided significantly more information than the currently used shared epitope classification and modeled the association of the HLA-DRB1 locus with RA severity significantly better. The association of HLA-DRB1 did not appear to be exclusively mediated by anticitrullinated protein antibodies because we identified a significant direct effect of HLA-DRB1 on radiographic outcome (eResults in Supplement 1), but no association of anticitrullinated protein antibodies with treatment response.
There are some strengths and limitations of the study. The availability of a large prospective cohort of patients with inflammatory polyarthritis or RA recruited at disease onset in the NOAR allowed us to determine 3 associations independent of each other within the HLA-DRB1 gene. The ERAS replication cohort provided independent validation of the results. The lack of available replication cohorts for other measures of disease outcome (mortality and treatment response) is a limitation and results for those outcomes require replication in independent cohorts. However, a strength of the present study is the comparison of association of the same genetic markers across various measures of disease severity and outcome. This allowed us to test the hypothesis that at least some genetic markers of autoimmune disease susceptibility, outcome, and treatment response are shared.
The study design did not allow us to answer the question regarding whether the increased genetic risk of developing severe and lethal disease was outweighed by the higher probability of responding to treatment because we did not have a cohort with concomitantly available severity and treatment response data.
Conclusions
Among patients with RA, the HLA-DRB1 locus, which is associated with disease susceptibility, was also associated with radiological severity, mortality, and treatment response. Replication of these findings in other cohorts is needed as a next step in evaluating the role of HLA-DRB1 haplotype analysis for management of RA.
Supplementary Material
Acknowledgments
Funding/Support: This work was funded by core programme grant 20385 from Arthritis Research UK, by the National Institute for Health Research Manchester Musculoskeletal Biomedical Research Unit, and by the Innovative Medicines Initiative Be The Cure (contract 115142-2). At the start of this project, Dr Viatte was supported by research grant PASMP3_134380 from the Swiss Foundation for Medical-Biological Scholarships, managed by the Swiss National Science Foundation. That grant is financed by a donation from Novartis to the Swiss Foundation for Medical-Biological Scholarships. Dr Raychaudhuri is supported by the Arthritis Foundation, the Doris Duke Foundation, and National Institutes of Health grants 1R01AR063759-01A1, 1R01AR062886-01, and 5U01GM092691.
Footnotes
Role of the Funder/Sponsor: The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Disclaimer: The views expressed are those of the authors and not necessarily those of the National Health Service, the National Institute for Health Research, or the UK Department of Health.
Additional Contributions: We acknowledge the Early Rheumatoid Arthritis Study clinicians and research nurses: Josh J. Dixey (New Cross Hospital, Wolverhampton, England), C. Solymossy (St Albans City Hospital, St Albans, England), Paul Davies and Lynn Hill (Chelmsford, England), Jo Devlin, Paul Emery, and Lynn Waterhouse (Birmingham, England), Helen Tate (Grimsby, England), Cathy Boys (Basingstoke, England), Dora White (Medway, England), Helen Dart (Oswestry, England), Sue Stafford (Winchester, England), John Winfield (Sheffield, England), Annie Seymour (St Albans, England). Study coordinators and data managers: Cathy Mayes and Marie Hunt (St Albans, England). We also acknowledge the Norfolk Arthritis Register clinicians, research nurses, metrologists, and the database management team in Manchester, England, and the contributions of local general practitioners and rheumatologists in Norwich, England. We also acknowledge the Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate members, clinicians (Kimme L. Hyrich, Ann W. Morgan, Anthony G. Wilson, John D. Isaacs, and Anne Barton), research nurses, patients, and the database management team in Manchester, England (also see appendix A of article by Plant et al23). We also acknowledge the assistance given by the University of Manchester IT Services and use of the Computational Shared Facility.
Author Contributions: Dr Viatte had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Viatte, Plant, Thomson, Symmons, Worthington, Wilson, Raychaudhuri, Barton.
Acquisition, analysis, or interpretation of data: Viatte, Plant, Han, Fu, Yarwood, Symmons, Young, Hyrich, Morgan, Isaacs, Raychaudhuri, Barton.
Drafting of the manuscript: Viatte, Han, Fu, Wilson, Raychaudhuri, Barton.
Critical revision of the manuscript for important intellectual content: Viatte, Plant, Fu, Yarwood, Thomson, Symmons, Worthington, Young, Hyrich, Morgan, Isaacs, Raychaudhuri, Barton.
Statistical analysis: Viatte, Plant, Han, Fu, Yarwood, Hyrich, Raychaudhuri, Barton.
Obtained funding: Symmons, Worthington, Hyrich, Morgan, Raychaudhuri, Barton.
Administrative, technical, or material support: Thomson, Young, Morgan, Barton.
Study supervision: Thomson, Symmons, Worthington, Raychaudhuri, Barton.
Conflict of Interest Disclosures: The authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Dr Hyrich reported receiving speaker’s fees from Abbvie and honoraria from Pfizer. Dr Raychaudhuri reported receiving personal fees from Johnson & Johnson, Pfizer, and Novartis. Dr Barton reported receiving consultancy fees and grant support from Eli-Lilly, Pfizer, and Abbvie. No other disclosures were reported.
References
- 1.Viatte S, Plant D, Raychaudhuri S. Genetics and epigenetics of rheumatoid arthritis. Nat Rev Rheumatol. 2013;9(3):141–153. doi: 10.1038/nrrheum.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. Arthritis Rheum. 1987;30(11):1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 3.Bukhari M, Thomson W, Naseem H, et al. The performance of anti-cyclic citrullinated peptide antibodies in predicting the severity of radiologic damage in inflammatory polyarthritis. Arthritis Rheum. 2007;56(9):2929–2935. doi: 10.1002/art.22868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viatte S, Barton A. The role of rheumatoid arthritis genetic susceptibility markers in the prediction of erosive disease. Eur Musculoskeletal Rev. 2012;7(2):102–107. [Google Scholar]
- 5.Ajeganova S, Andersson ML, Frostegård J, Hafström I. Disease factors in early rheumatoid arthritis are associated with differential risks for cardiovascular events and mortality depending on age at onset. J Rheumatol. 2013;40(12):1958–1966. doi: 10.3899/jrheum.130365. [DOI] [PubMed] [Google Scholar]
- 6.Farragher TM, Goodson NJ, Naseem H, et al. Association of the HLA-DRB1 gene with premature death, particularly from cardiovascular disease, in patients with rheumatoid arthritis and inflammatory polyarthritis. Arthritis Rheum. 2008;58(2):359–369. doi: 10.1002/art.23149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Criswell LA, Lum RF, Turner KN, et al. The influence of genetic variation in the HLA-DRB1 and LTA-TNF regions on the response to treatment of early rheumatoid arthritis with methotrexate or etanercept. Arthritis Rheum. 2004;50(9):2750–2756. doi: 10.1002/art.20469. [DOI] [PubMed] [Google Scholar]
- 8.Cui J, Stahl EA, Saevarsdottir S, et al. Genome-wide association study and gene expression analysis identifies CD84 as a predictor of response to etanercept therapy in rheumatoid arthritis. PLoS Genet. 2013;9(3):e1003394. doi: 10.1371/journal.pgen.1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plant D, Bowes J, Potter C, et al. Genome-wide association study of genetic predictors of anti-tumor necrosis factor treatment efficacy in rheumatoid arthritis identifies associations with polymorphisms at seven loci. Arthritis Rheum. 2011;63(3):645–653. doi: 10.1002/art.30130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plant D, Wilson AG, Barton A. Genetic and epigenetic predictors of responsiveness to treatment in RA. Nat Rev Rheumatol. 2014;10(6):329–337. doi: 10.1038/nrrheum.2014.16. [DOI] [PubMed] [Google Scholar]
- 11.Raychaudhuri S, Sandor C, Stahl EA, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44(3):291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plant D, Thomson W, Lunt M, et al. The role of rheumatoid arthritis genetic susceptibility markers in the prediction of erosive disease in patients with early inflammatory polyarthritis. Rheumatology (Oxford) 2011;50(1):78–84. doi: 10.1093/rheumatology/keq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Symmons DP, Silman AJ. The Norfolk Arthritis Register (NOAR) Clin Exp Rheumatol. 2003;21(5 suppl 31):S94–S99. [PubMed] [Google Scholar]
- 14.Arnett FC, Edworthy SM, Bloch DA, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31(3):315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 15.Larsen A, Dale K, Eek M. Radiographic evaluation of rheumatoid arthritis and related conditions by standard reference films. Acta Radiol Diagn (Stockh) 1977;18(4):481–491. doi: 10.1177/028418517701800415. [DOI] [PubMed] [Google Scholar]
- 16.Solymossy C, Dixey J, Utley M, et al. Larsen scoring of digitized X-ray images. Rheumatology (Oxford) 1999;38(11):1127–1129. doi: 10.1093/rheumatology/38.11.1127. [DOI] [PubMed] [Google Scholar]
- 17.Viatte S, Plant D, Lunt M, et al. Investigation of rheumatoid arthritis genetic susceptibility markers in the early rheumatoid arthritis study further replicates the TRAF1 association with radiological damage. J Rheumatol. 2013;40(2):144–156. doi: 10.3899/jrheum.121034. [DOI] [PubMed] [Google Scholar]
- 18.James D, Young A, Kulinskaya E, et al. Orthopaedic intervention in early rheumatoid arthritis. Rheumatology (Oxford) 2004;43(3):369–376. doi: 10.1093/rheumatology/keh059. [DOI] [PubMed] [Google Scholar]
- 19.Farragher TM, Plant D, Flynn E, et al. Association of a rheumatoid arthritis susceptibility variant at the CCL21 locus with premature mortality in inflammatory polyarthritis patients. Arthritis Care Res (Hoboken) 2010;62(5):676–682. doi: 10.1002/acr.20208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ibrahim I, McAllister K, Plant D, et al. Investigation of an interleukin-6 receptor gene polymorphism (rs2228145) as a predictor of cardiovascular mortality in inflammatory polyarthritis. Ann Rheum Dis. 2014;73(4):787–788. doi: 10.1136/annrheumdis-2013-204330. [DOI] [PubMed] [Google Scholar]
- 21.Statistics and Registration Service Act 2007.
- 22.Hyrich KL, Watson KD, Silman AJ, et al. Predictors of response to anti-TNF-alpha therapy among patients with rheumatoid arthritis. Rheumatology (Oxford) 2006;45(12):1558–1565. doi: 10.1093/rheumatology/kel149. [DOI] [PubMed] [Google Scholar]
- 23.Plant D, Prajapati R, Hyrich KL, et al. Replication of association of the PTPRC gene with response to anti-tumor necrosis factor therapy in a large UK cohort. Arthritis Rheum. 2012;64(3):665–670. doi: 10.1002/art.33381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomson W, Harrison B, Ollier B, et al. Quantifying the exact role of HLA-DRB1 alleles in susceptibility to inflammatory polyarthritis. Arthritis Rheum. 1999;42(4):757–762. doi: 10.1002/1529-0131(199904)42:4<757::AID-ANR20>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 25.Viatte S, Plant D, Bowes J, et al. Genetic markers of rheumatoid arthritis susceptibility in anti-citrullinated peptide antibody negative patients. Ann Rheum Dis. 2012;71(12):1984–1990. doi: 10.1136/annrheumdis-2011-201225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eyre S, Bowes J, Diogo D, et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet. 2012;44(12):1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia X, Han B, Onengut-Gumuscu S, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One. 2013;8(6):e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yarwood A, Han B, Raychaudhuri S, et al. A weighted genetic risk score using all known susceptibility variants to estimate rheumatoid arthritis risk. Ann Rheum Dis. 2015;74(1):170–176. doi: 10.1136/annrheumdis-2013-204133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rabe-Hesketh S, Skrondal A, Pickles A. Generalized multilevel structural equation modelling. Psychometrika. 2004;69:167–190. [Google Scholar]
- 30.Rabe-Hesketh S, Skrondal A, Pickles A. Maximum likelihood estimation of limited and discrete dependent variable models with nested random effects. J Econom. 2005;128:301–323. [Google Scholar]
- 31.Rabe-Hesketh S, Skrondal A. Classical latent variable models for medical research. Stat Methods Med Res. 2008;17(1):5–32. doi: 10.1177/0962280207081236. [DOI] [PubMed] [Google Scholar]
- 32.Lee JC, Espéli M, Anderson CA, et al. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell. 2013;155(1):57–69. doi: 10.1016/j.cell.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Gestel AM, Prevoo ML, van’t Hof MA, et al. Development and validation of the European League Against Rheumatism response criteria for rheumatoid arthritis. Arthritis Rheum. 1996;39(1):34–40. doi: 10.1002/art.1780390105. [DOI] [PubMed] [Google Scholar]
- 34.Barra L, Pope JE, Payne M. Real-world anti-tumor necrosis factor treatment in rheumatoid arthritis, psoriatic arthritis, and ankylosing spondylitis. J Rheumatol. 2009;36(7):1421–1428. doi: 10.3899/jrheum.081122. [DOI] [PubMed] [Google Scholar]
- 35.Karlson EW, Chibnik LB, Kraft P, et al. Cumulative association of 22 genetic variants with seropositive rheumatoid arthritis risk. Ann Rheum Dis. 2010;69(6):1077–1085. doi: 10.1136/ard.2009.120170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norton S, Sacker A, Dixey J, Done J, Williams P, Young A. Trajectories of functional limitation in early rheumatoid arthritis and their association with mortality. Rheumatology (Oxford) 2013;52(11):2016–2024. doi: 10.1093/rheumatology/ket253. [DOI] [PubMed] [Google Scholar]
- 37.Potter C, Hyrich KL, Tracey A, et al. Association of rheumatoid factor and anti-cyclic citrullinated peptide positivity, but not carriage of shared epitope or PTPN22 susceptibility variants, with anti-tumour necrosis factor response in rheumatoid arthritis. Ann Rheum Dis. 2009;68(1):69–74. doi: 10.1136/ard.2007.084715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruynesteyn K, van der Heijde D, Boers M, et al. Determination of the minimal clinically important difference in rheumatoid arthritis joint damage of the Sharp/van der Heijde and Larsen/Scott scoring methods by clinical experts and comparison with the smallest detectable difference. Arthritis Rheum. 2002;46(4):913–920. doi: 10.1002/art.10190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.