Abstract
Histone three lysine 27 (H3K27) methyltransferase enhancer of zeste homolog 2 (EZH2) is a critical epigenetic modifier, which regulates gene transcription through the trimethylation of the H3K27 residue leading to chromatin compaction and gene repression. EZH2 has previously been identified to regulate human bone marrow-derived mesenchymal stem cells (MSC) lineage specification. MSC lineage specification is regulated by the presence of EZH2 and its H3K27me3 modification or the removal of the H3K27 modification by lysine demethylases 6A and 6B (KDM6A and KDM6B). This study used a bioinformatics approach to identify novel genes regulated by EZH2 during MSC osteogenic differentiation. In this study, we identified the EZH2 targets, ZBTB16, MX1, and FHL1, which were expressed at low levels in MSC. EZH2 and H3K27me3 were found to be present along the transcription start site of their respective promoters. During osteogenesis, these genes become actively expressed coinciding with the disappearance of EZH2 and H3K27me3 on the transcription start site of these genes and the enrichment of the active H3K4me3 modification. Overexpression of EZH2 downregulated the transcript levels of ZBTB16, MX1, and FHL1 during osteogenesis. Small interfering RNA targeting of MX1 and FHL1 was associated with a downregulation of the key osteogenic transcription factor, RUNX2, and its downstream targets osteopontin and osteocalcin. These findings highlight that EZH2 not only acts through the direct regulation of signaling modules and lineage-specific transcription factors but also targets many novel genes important for mediating MSC osteogenic differentiation.
Introduction
Osteogenic differentiation of bone marrow mesenchymal stem/stromal cells (MSCs) [1,2] is associated with the repression of adipogenic differentiation [3–5]. This reciprocal relationship between adipogenesis and osteogenesis is modulated by molecules such as Msx2, which promote osteogenesis and suppress adipogenesis [6,7]. Conversely, osteogenic differentiation can be inhibited by the activation of protein kinase A, which upregulates peroxisome proliferator-activated receptor gamma 2 (PPARγ2) and in turn inhibits runt-related transcription factor 2 (RUNX2) and osteopontin (OP), promoting adipogenesis [8]. RUNX2, a key lineage specific transcription factor, and signaling pathway molecules such as WNT and bone morphogenic protein (BMP) play a critical role in dictating MSC osteogenic differentiation [9–13].
Recent investigations have identified the epigenetic regulation of chromatin as a key mechanism in dictating lineage-specific differentiation of MSC. The structure of chromatin can be regulated through methylation, acetylation, phosphorylation, SUMOylation, and ubiquitination of histone tails. Methyltransferases and demethylases are thought to be responsible for the switch between repressive H3K27 domains and active H3K4 domains. Given the diverse array of histone modifications, their various combinations form the “histone code,” which can influence the recruitment of effector proteins and transcription factors to chromatin, determining the functional state of chromatin [14–16].
The epigenetic modifier enhancer of zeste homolog 2 (EZH2) exists within a multisubunit complex known as the polycomb repressor complex 2 (PRC2), which contains suppressor of zeste-12 (SUZ12), yin yang 1 (YY1), and embryonic ectoderm development [17]. EZH2 is a histone three lysine 27 (H3K27) methyltransferase that contains a carboxy-terminal catalytic SET domain responsible for the mono-, di-, or trimethylation modification of H3K27 residues of histone tails [18]. EZH2 H3K7 tri methylation (me3) modification facilitates the recruitment of the PRC1 during chromatin remodeling, compaction of chromatin and gene repression [19–21]. During development, EZH2 is required for neural crest-derived cartilage and bone formation and anterior/posterior skeletal patterning in mice [22,23]. In postnatal skeletal tissue, reports have identified EZH2 as having a role in regulating MSC lineage differentiation. EZH2 was found to promote adipogenesis by disrupting Wnt/β-catenin signaling through the direct repression of pro-osteogenic Wnt genes, Wnt1, -6, -10a, and -10b, in mouse peripheral preadipocytes [24].
More recently, enforced expression of EZH2 in human bone marrow-derived MSC promoted adipogenesis in vitro and inhibited osteogenic differentiation potential in vitro and in vivo [25]. Conversely, inhibition of EZH2 enzymatic activity and knockdown of EZH2 gene expression inhibited adipogenesis and promoted osteogenic differentiation by MSC [25]. EZH2 has been reported to be regulated through phosphorylation at Thr 487 by cyclin-dependent kinase 1 (CDK1). CDK1 phosphorylation suppresses EZH2 activity, promoting MSC osteoblast differentiation in vitro [26]. Chromatin immunoprecipitation (ChIP) analysis has revealed that the presence of EZH2 and its H3K27me3-associated modification is reduced at the transcription start site (TSS) of key osteogenic transcription factor RUNX2 during osteogenic differentiation [25,26]. However, it is still unclear whether EZH2 directly targets other critical genes and pathways during MSC osteogenic differentiation. The present study aimed to identify novel EZH2 targets in osteogenesis through a bioinformatics approach using publically available ChIP-on-chip, ChIP-Seq, and microarray data sets.
Materials and Methods
Bioinformatic analysis
Bioinformatic analysis was performed to assess gene expression, EZH2 occupancy, H3K27, and H3K4 methylation in three independent data sets. These data sets compared primary cultured human bone marrow-derived MSC versus in vitro differentiated osteoblasts (OB) differentiated with dexamethasone 100 nM (Dex) and 10 mM β-glycerophosphate and with the addition of 50 μg/mL ascorbic acid for 28 days (GSE9451 [27]; [26]) or 50 nM ascorbic acid for 16 days (GSE27900 [28]). GSE9451 data set contained three independent MSC lines. These MSC lines were differentiated under osteogenic inductive conditions to generate paired OB for microarray analysis.
ChIP data sets
ChIP-on-chip data for genes bound by EZH2 in undifferentiated MSC and not bound by EZH2 in OB were obtained from Wei et al. [26]. H3K27me3 and H3K4me3 ChIP-Seq data were obtained from GEO (Gene Expression Omnibus, www.ncbi.nlm.nih.gov.geo/, NCBI; data set GSE35573) [28]. A list of all genes identified in either of these studies was generated to generate a results profile for each gene. Data sets were parsed and reformatted using a set of custom Perl Scripts. Gene annotation was retrieved from the Gene Ontology Database (www.geneontology.org).
Microarray data sets
To assess the differential expression of the genes identified in the ChIP data sets, two data sets were used, both conducted on Affymetrix GeneChip Human Genome U133 plus 2.0 arrays. Microarray (Affymetrix U133A) GEO data set GSE9451 [27] was normalized using RMA (RMA Express program http://rmaexpress.bmbolstad.com/). For GSE9451, raw microarray data (.CEL files) were downloaded from ArrayExpress (EMBL-EBI) and were normalized by RMA using the bioconductor package (affy) [29] and R (version 3.03) and log2 transformed. Genes that were not expressed (signal < log2100) in either MSC or OB were excluded from further analysis. Initially, expression of the genes identified differentially expressed genes (adjusted P ≤ 0.05) were initially identified using linear models from microarray data (LIMMA [30–32]) in MultiExperiment Viewer (MeV [33]). Initially, genes that were significantly >2-fold up- or downregulated with OB differentiation in GSE9451 were identified.
Isolation and culture of MSC
Bone marrow aspirates were isolated from the posterior iliac crest of healthy human adult donors, following informed consent (SA Pathology normal bone marrow donor program, Royal Adelaide Hospital Human Ethics Number 940911a). MSC were isolated and cultured using plastic adherence [34]. Culture media: Alpha modified Eagle's medium (α-MEM) (Sigma Aldrich, Inc., St Louis, MO) supplemented with 10% (v/v) fetal calf serum (SAFC Biosciences, Melbourne, VIC), 50 U/mL, 5 μg/mL penicillin, streptomycin (Sigma Aldrich, Inc.), 1 mM sodium pyruvate (Sigma Aldrich, Inc.), 100 μM L-ascorbate-2-phosphate (Wako Pure Chemical industries, Richmond, VA), 2 mM l-glutamine (JRH Biosciences/Sigma, Lenexa, KS), and 10 mM HEPES (Sigma Aldrich, Inc.), and cultured at 37°C, >90% humidity and 5% CO2.
Retroviral transduction overexpression
The human EZH2 coding region of the gene was ligated into pRUF-IRES-GFP retroviral vector using polymerase chain reaction primers to amplify the coding region with Xho1 restriction sites [25]. The pRUF-IRES-GFP-EZH2 construct was transfected into the HEK293T viral packaging cell line together with PGP and VSVG (viral envelope proteins, SBI System Biosciences, Mountain View, CA), and viral supernatant was used for infection of MSCs and sorted by fluorescence-activated cell sorting (FACS: Beckman Coulter Epics Ultra cell sorter, Lane Cove, NSW, AUS). MSC were passaged, and at passage 5 were seeded for osteogenic differentiation assays and RNA assays.
siRNA transfection
Passage 5 MSC were seeded in either 24-well plates (1.9 cm2) at 3 × 104 or 96-well plates (0. 32 cm2) at 3 × 103 cells per well. Early the next day, MSC were treated with 12 pmol siRNA (Life Technologies, Scoresby VIC). Negative control siRNA (4390843), EZH2 #1 (s4916), EZH2 #2 (s4918), FHL1 #1 (n261445), FHL1 #2 (n261446), MX1 #1 (s9100), and MX1 #2 (s9099) siRNAs with Lipofectamine RNAi MAX were used for this study (Invitrogen/Life Technologies, Scoresby VIC, AUS). siRNA were added to cells as specified by the manufacturer. Seventy-two hours post-transfection, media were removed and culture growth media, adipogenic, or osteogenic inductive media were added [25].
In vitro osteogenic differentiation assay
siRNA-treated MSC in 96-well plates were cultured in either noninductive culture media or osteogenic inductive media (α-MEM supplemented with 5% (v/v) FCS, 2 mM l-Glutamine, 1 mM sodium pyruvate, 10 mM HEPES buffer and 50 U/mL penicillin–streptomycin, 100 μM L-ascorbate-2-phosphate, 0.1 μM dexamethasone, and 2.6 mM KH2PO4) [34] for 14 days with media changes twice weekly. Mineral formation by human MSCs under osteogenic differentiation was identified by 1% Alizarin red (PH 4. 3) (Sigma Aldrich, Inc.) staining of hydroxyapatite deposits. Extracellular calcium levels were assessed by Calcium Arsenazo III assay (Thermo Scientific/Life technologies, Scoreby, VIC, AUS) in replicate samples and normalized to DNA content per well by PicoGreen incorporation (Life technologies).
In vitro adipogenic differentiation assay
siRNA-treated MSC in 96-well plates were cultured in either noninductive culture media or adipogenic inductive media (α-MEM supplemented with 10% [v/v] FCS, 2 mM l-Glutamine, 1 mM sodium pyruvate, 10 mM HEPES buffer and 50 U/mL penicillin–streptomycin, 100 μM l- ascorbate-2-phosphate, 0.5 mM methyl isobutyl methylxanthine, 0.5 μM hydrocortisone, and 60 μM indomethacin) [34] for 14 days with media changes twice weekly. Oil Red O (Sigma Aldrich, Inc.) identified lipid formed by human MSC under adipogenic differentiation.
Real-time polymerase chain reaction analysis
Culture expanded MSCs infected with pRUF-IRES-GFP and pRUF-IRES-GFP-EZH2 were seeded at 5.64 × 104 per/well in six-well plates in the presence of either noninductive or osteogenic inductive media as previously described [25]. siRNA-treated MSC were cultured in the presence of noninductive, adipogenic, or osteogenic inductive media in 24-well plates. Total RNA was extracted using TRIzol reagent (Invitrogen/Life Technologies) and converted to cDNA by reverse transcription. Gene expression was assessed by real-time polymerase chain reaction (RT-PCR) amplification using specific primer sets (Table 1), SYBR Green/Rox PCR master mix (Qiagen, Doncaster, Chadstone Centre, VIC, AUS), and Rotor-Gene 6000 Real-Time Thermal Cycler (Corbett Research, Mortlake, NSW, AUS). Changes in gene expression were calculated relative to β-actin using the 2-dCT method [25,35].
Table 1.
Human Real-Time PCR Primers
| Gene | Accession no. | Forward primer 5′-3′ | Reverse primer 5′-3′ |
|---|---|---|---|
| EZH2 | NM_004456.4 (All variants) | gggacagtaaaaatgtgtcctgc | tgccagcaatagatgctttttg |
| RUNX2 | NM_001271893.3 | ctcttgctggtgacattgtc | cccttctctcgacgctgg |
| OP | NM_001040058.1 | atgagagccctcacactcctcg | gtcagccaactcgtcacagtcc |
| OC | NM_199173.5 (All variants) | acatccagtaccctgatgctacag | gtgggtttcagcactctggt |
| β-ACTIN | NM_001101.3| | gatcattgctcctcctga | gtcatagtccgcctagaagcat |
| ZBTB16 | NM_006006.4 (All variants) | ggtcgagcttcctgataac | accgcactgatcacagacaa |
| MX1 | NM_001144925.2 (All variants|) | taccagcgagctcatcacac | catttgtggaactcgtgtcg |
| FHL1 | NM_001159702.2 (All variants) | atgagacctttgtggccaag | cttgttgcacttcacgcaat |
EZH2, methyltransferase enhancer of zeste homolog 2; OP, Osteopontin; OC, Osteocalcin; β-ACTIN, Beta Actin; PCR, polymerase chain reaction.
Western blot analysis
Passage 5 MSC were seeded in either 24-well plates (1.9 cm2) at 3 × 104 or 96-well plates (0. 32 cm2) at 3 × 103 cells per well in duplicate. MSC were treated with siRNA and cultured under osteogenic differentiation conditions for 14 days. Whole cell lysates (40 mg) were separated on SDS gel as previously described [25]. Membranes were probed with anti-EZH2 mouse IgG (Acc2 Cell Signaling Tecnology, Inc./Genesearch Pty. Ltd., Arundel, QLD, AUS; 1/1,000 dilution), anti-MX1 rabbit IgG (Abcam/Sapphire Bioscience Pty. Ltd., Waterloo, NSW, AUS; ab95926, 1/500 dilution), anti-FHL1 rabbit IgG (Abcam/Sapphire Bioscience, ab133661, 1/1,000 dilution), anti-H3K27me3 rabbit IgG (Merk Millipore, Kilsyth, VIC, AUS; 1/1,000 dilution), and anti-β-actin mouse IgG (Cell Signaling Tecnology, Inc., 8H10D10; 1/1,000 dilution). Secondary detection was performed using anti-Rabbit-Alk Phos (Millipore; 1/10,000) and anti-Mouse-Alkphos (Millipore; 1/10,000) antibodies.
BrdU analysis
MSC at passage 5 were seeded at 2 × 103 cells per well in 24-well plates and cultured overnight at 37°C. Cell proliferation ELISA, BrdU protocol, was followed as described in the instruction manual version August 2007 (Roche applied biosciences, Mannheim, GER). On the sixth day, the BrdU reaction was measured on the Ultra Micro plate reader EL808 (Bio-Tek Instruments, Winooski, VT) at 450 nm.
ChIP analysis
Passage 5 MSC were seeded at 2 × 106 cells per flask (75 cm2) and induced under normal growth medium and/or osteogenic differentiation media for 14 days. ChIP was adapted from the Abcam (Abcam, Melbourne, VIC, AUS) crosslinking chromatin immunoprecipitation (X-ChIP) protocol. Briefly, chromatin was cross-linked with a final of 0.75% formaldehyde. Adherent cells were detached using 1 × trypsin EDTA, and the remaining cells were scraped from the bottom of the flask. Cells were lysed with 400 μL of FA lysis buffer (50 mM HEPES KOH pH7.5, 140 mM NaCl, 1 mM EDTA pH8, 1% Triton X-100, 0.1% Sodium deoxycholate, 0.1% SDS, and protease inhibitors). DNA was sheared with probe sonication on ice. Sonicated samples were used for immunoprecipitation [36].
Immunoprecipitation
Antibodies against anti-rabbit H3K27me3 (1 μg, Merck, Millipore 07-449, Bayswater, VIC, AUS, http://www.merck.com.au/), anti-human EZH2 mouse monoclonal (1 μg, Millipore AC22 17-662), anti-human H3K4me3 rabbit polyclonal (1 μg, Abcam ab8580-100), and anti-human IgG rabbit polyclonal control (1 μg Millipore, Bayswater, VIC, AUS) were used for immunoprecipitation. Immunoselected genomic DNA was used for RT-PCR using primers targeting the promoters of genes of interest (Table 2) as previously described [37].
Table 2.
Human ChIP Transcription Start Site Primers
| Gene | Accession no. | Forward primer 5′-3′ | Reverse primer 5′-3′ |
|---|---|---|---|
| GAPDH | NM_002046.5 | cggctactagcggttttacg | aagaagatgcggctgactgt |
| IL2 | NM_000586.3 | attgtggcaggagttgaggt | cagtcagtctttgggggttt |
| INK4A | NM_000077.4 | accccgattcaatttggcag | aaaaagaaatccgcccccg |
| ZBTB16 | NM_006006.4 (All variants) | atctgctgtggcagaacctt | ttctttccttctgggctctg |
| MX1 | NM_001144925.2 (All variants) | tcagcacagggtctgtgagt | gcgcccttgctatgattatg |
| FHL1 | NM_001159702.2 (All variants) | acacagcctccgtgcagt | agggaaagaggggaggaag |
IL2, Interleukin 2.
Statistical analysis
Statistical analysis was carried out using GraphPad Prism 6 (GraphPad Software, La Jolla, CA). Paired student's t-test was used to assess statistical significance for mRNA expression for microarray and adipogenic RT-PCR experiments. Unpaired Student's t-test was used to assess statistical significance for siRNA knockdown of EZH2/MX1 and EZH2/FHL1 experiments. One-way ANOVA with Dunnett's multiple comparison test was used to assess statistical significance for siRNA and BrdU assays. Two-way ANOVA with Sidak's multiple comparison test was used for ChIP and EZH2 overexpression studies.
Results
Identification of novel EZH2 targets during MSC osteogenic differentiation
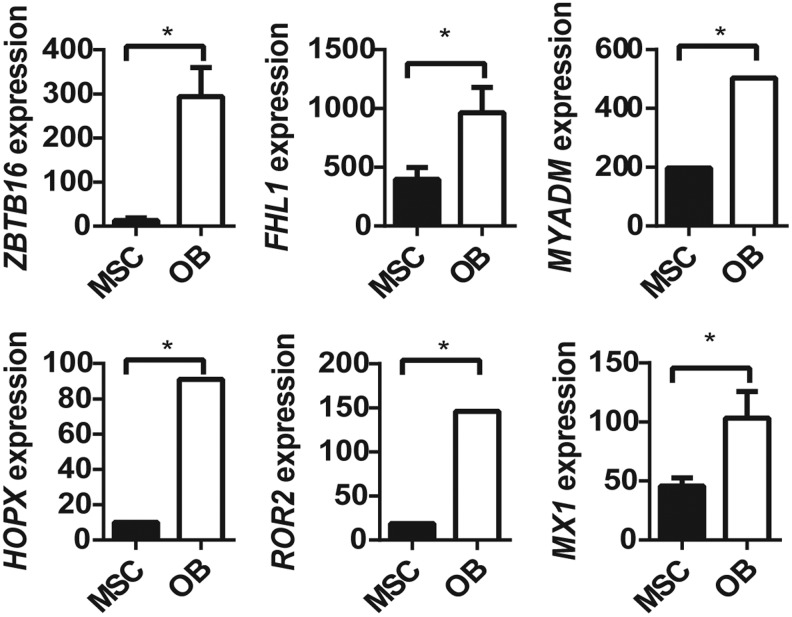
Initially, we examined three independent data sets to identify novel genes that may be switched on during osteogenic differentiation by removal of EZH2 binding and its associated H3K27me3 mark. We identified 99 genes that were reported to be bound by EZH2 in MSC, but not following osteogenic differentiated in vitro [26], and were found to have a loss of the repressive modification H3K27me3 during osteogenic differentiation of MSCs in the independent ChIP-Seq data set GSE35576 [28]. Assessment of differential gene expression in the independent microarray data set GSE9451 [27] identified that, of these 99 genes, 6 genes (ZBTB16, HOPX, ROR2, MYADM, FHL1, MX1) were significantly upregulated (>2-fold change; P ≤ 0.05, LIMMA), while no genes were significantly downregulated, under osteogenic differentiation, when compared with undifferentiated MSC (Fig. 1). Of these, two (ZBTB16, FHL1) gained the activating modification H3K4me3 under osteogenic differentiation, one gene lost the H3K4me3 mark in OB (HOPX), one gene (MYADM) had the H3K4me3 mark in both MSC and OB, and two genes (MX1, ROR2) did not have the H3K4me3 mark in either MSC or OB (Table 3). These findings suggest that ZBTB16, HOPX, ROR2, MYADM, FHL1, and MX1 are potential EZH2 targets that may play a role in regulating osteogenic cell fate determination. Two genes, ZBTB16 and FHL1, were selected for subsequent validation studies that met all the selection criteria. MX1, a known mediator of early mesenchymal differentiation, was also assessed as a positive control.
FIG. 1.
Genes with upregulated expression and loss of EZH2 during osteoblast differentiation associated with loss of H3K27 methylation and/or gain of H3K4 methylation. Genes upregulated expression with loss of EZH2 and H3K27me3 in OB. (A) Microarray (GSE9451) [28] expression of ZBTB16, FHL1, MYADM, HOPX, ROR2, and MX1 in OB compared with MSC was significantly upregulated (*P < 0. 05 paired t-test). EZH2, methyltransferase enhancer of zeste homolog 2; MSC, mesenchymal stem cell.
Table 3.
Genes Regulated by EZH2 in MSC During Osteogenic Differentiation
| GEO: GSE9451 | |||||
|---|---|---|---|---|---|
| Gene symbol | Gene name | H3K27me3 on MSC | H3K4me on OB | Fold change | Adjusted *P |
| ZBTB16 | Zinc finger and BTB domain containing 16 | + | + | 4.6 | 0.0070 |
| FH1 | Four and a half domain 1 | + | + | 1.1 | 0.0141 |
| MYADM | Myeloid-associated differentiation marker | + | + | 1.3 | 0.0011 |
| HOXP | HOP homeobox | + | − | 3.2 | 0.0197 |
| ROR2 | Receptor tyrosine kinase-like orphan receptor 2 | + | − | 3.0 | 0.0007 |
| MX1 | Myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 | + | − | 1.2 | 0.0129 |
Bioinformatics analysis of data-based GEO data set GSE9451 identifying genes bound by EZH2 in MSC, but not in osteoblasts (OB), associated with loss of EZH2 binding and H3K27me3 and/or gain of H3K4me3, during osteogenic differentiation into OB. Fold change is represented as OB/MSC (*P < 0.05). GEO, gene expression omnibus; MSC, mesenchymal stem cell.
Gene expression levels of ZBTB16, HOPX, ROR2, MYADM, FHL1, and MX1 were assessed in MSC cultured under adipogenic inductive conditions (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/scd). Real-time PCR analysis found no statistical difference in the transcript levels for ZBTB16, ROR2, MYADM, FHL1, and MX1 during adipogenic differentiation, when compared to control MSC. However, gene expression of HOPX was significantly increased during adipogenic differentiation when compared to levels expressed in control MSC. These findings suggest that HOPX is potentially important for dual lineage differentiation of MSC. Transient knockdown of ZBTB16, FHL1, and MX1 with two independent siRNA, during MSC adipogenic differentiation, had no effect on lipid formation compared to scramble control.
EZH2 directly represses ZBTB16, MX1, and FHL1 gene expression in undifferentiated MSC
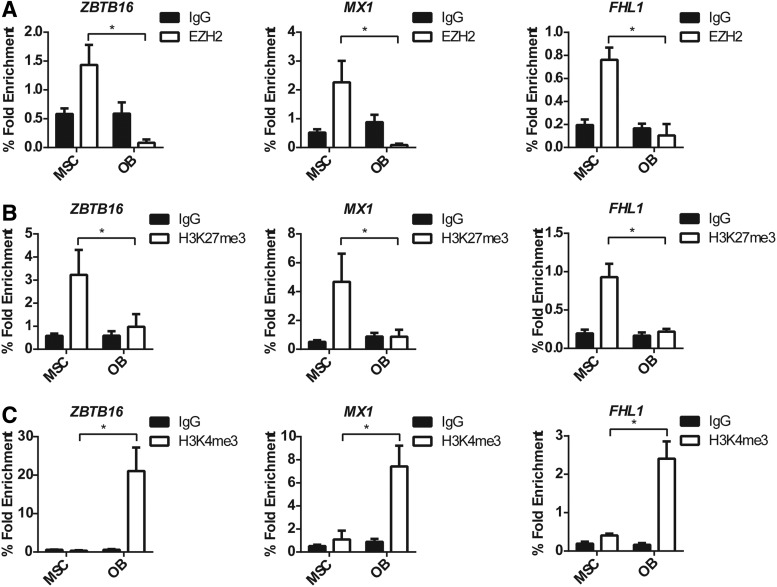
To confirm the bioinformatics analysis, manual ChIP analysis was performed for ZBTB16, FHL1, and MX1, using anti-EZH2, -H3K27me3, -H3K4me3, or IgG control antibodies on genomic DNA isolated from human MSC cultured under normal growth conditions or in osteogenic inductive media. Primers targeting the TSS of ZBTB16, MX1, and FHL1 were used to assess the presence of epigenetic modifying enzymes and modifications. P16 was used as the positive control, and GAPDH was used as the negative control for the EZH2 and H3K27me3 ChIP analysis (Supplementary Fig. S2A, B). GAPDH was used as the positive control and the IL2 was used as the negative control for the H3K4me3 ChIP (Supplementary Fig. S2C).
The TSS of ZBTB16, MX1, and FHL1 were significantly enriched for EZH2 binding in undifferentiated MSC compared with MSC cultured under osteogenic conditions (Fig. 2A). This correlated to a significant enrichment of the gene silencing mark H3K27me3 at the TSS of ZBTB16, MX1, and FHL1 in undifferentiated MSC compared with OB (Fig. 2B). In contrast, the active mark H3K4me3 was present at the TSS of all genes in OB correlating with an increase in gene expression during osteogenesis (Fig. 1). Importantly, EZH2 and the H3K27me3 modification were no longer present at the TSS of ZBTB16, MX1, and FHL1 during osteogenesis.
FIG. 2.
EZH2, H3K27me3, and H3K4me3 ChIP analysis of MSC and osteoblast confirmed modification patterns identified by bioinformatics analysis. Manual ChIP analysis confirms EZH2 regulates ZBTB16, MX1, and FHL1 in osteogenic differentiation. MSC (nondifferentiated) and osteogenic differentiated OB were immunoprecipitated with anti-H3K27me3, anti-EZH2, anti-H3K4me3, and anti-IgG control antibody, and RT-PCR was used to measure enrichment. (A) EZH2 enrichment was present at the transcription start site (TSS) of ZBTB16, MX1, and FHL1 in MSC and was absent in OB. (B) EZH2 H3K27me3 repressive modification was present at the TSS of ZBTB16, MX1, and FHL1 and was removed in OB. (C) H3K4me3 active gene modification was not present on the TSS of ZBTB16, MX1, and FHL1 in MSC; however, the modification was present in OB. Mean values ± SEM, n = 3 independent MSC donors (*P < 0.05, 2-way ANOVA with Sidak's multiple comparison test). Positive and negative ChIP controls are represented in Supplementary data Figure S1. RT-PCR, real-time polymerase chain reaction; SEM, standard error of the mean.
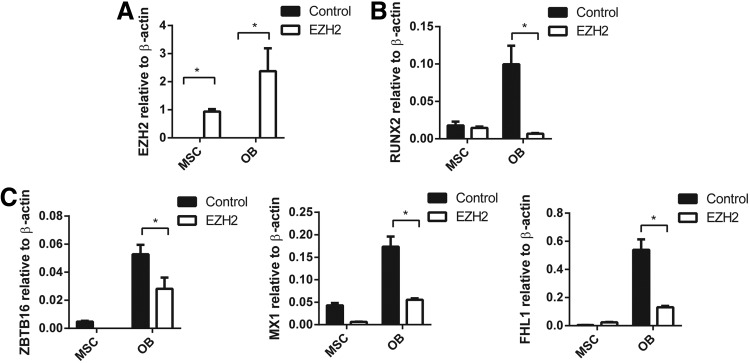
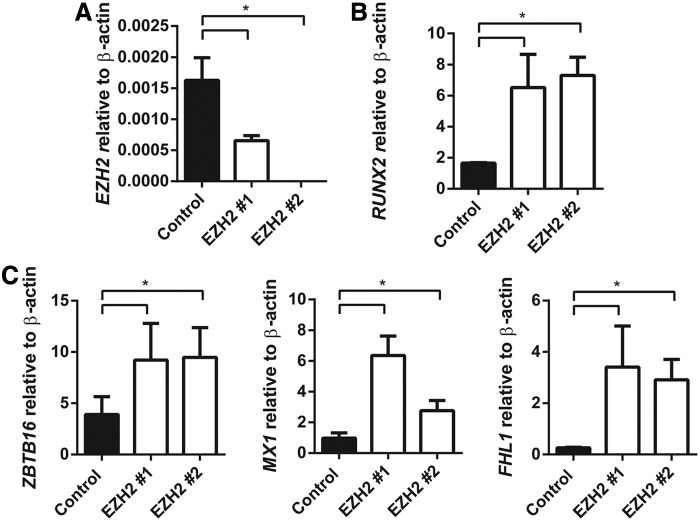
We have previously reported that EZH2 inhibits human MSC osteogenic differentiation in vitro and in vivo [25]. In the present study, the role of EZH2 in regulating ZBTB16, MX1, and FHL1 gene expression was confirmed using stably transduced EZH2 overexpressing MSC lines and vector control MSC (Fig. 3A), cultured under normal growth conditions or osteogenic inductive conditions. EZH2 overexpressing MSC was found to exhibit significantly lower expression levels of the osteogenic master regulatory factor, RUNX2, during osteogenic differentiation when compared with vector control MSC (Fig. 3A, B). Furthermore, EZH2 overexpressing MSC demonstrated repressed gene expression levels of ZBTB16, MX1, and FHL1 following osteogenic induction, when compared with vector control MSC (Fig. 3C). In parallel studies, two independent siRNA were used to knockdown gene expression of EZH2 in MSC cultured under osteogenic inductive conditions (Fig. 4A). Knockdown of EZH2 in MSC increased expression of RUNX2 (Fig. 4B) and significantly increased the expression of ZBTB16, MX1, and FHL1 compared with MSC treated with scramble control siRNA (Fig. 4C).
FIG. 3.
Enforces expression of EZH2 inhibits ZBTB16, MX1, and FHL1 expression in MSC and OB. pRUF-IRES-GFP-EZH2 overexpressing or pRUF-IRES-GFP control MSC were treated with no inductive media (MSC) or osteogenic differentiation media (OB) for 14 days. RT-PCR analysis: (A) Ezh2 overexpression. (B) Expression of RUNX2. (C) Expression of ZBTB16, MX1, and FHL1 was significantly downregulated in EZH2 overexpressing OB compared with control OB. Mean values ± SEM, n = 3 independent MSC donors (*P < 0. 05, 2-way ANOVA with Sidak's multiple comparison test).
FIG. 4.
siRNA knockdown of EZH2 promotes expression of ZBTB16, MX1, and FHL1 in OB. MSC were treated with negative control siRNA, EZH2 siRNA #1, and EZH2 siRNA #2. MSC were cultured for 14 days with osteogenic differentiation media. (A) Knockdown of EZH2 by siRNA was confirmed by RT-PCR. (B) RT-PCR reveals a significant increase in RUNX2 expression compared with negative control-treated OB. (C) ZBTB16, MX1, and FHL1 gene expression was upregulated in EZH2 siRNA-treated OB compared with negative control-treated OB. Mean values ± SEM, n = 3 independent MSC donors (*P < 0.05, 1-way ANOVA with Dunnett's multiple comparison test).
Collectively, these studies showed that ZBTB16, MX1, and FHL1 are direct targets of EZH2 binding and activity in undifferentiated MSC leading to suppression of gene expression. However, during osteogenic lineage commitment, ZBTB16, MX1, and FHL1 expression is induced due to the loss of both EZH2 binding and H3K27me3, correlating with the appearance of the H3K4me3 modification.
Targeted knockdown of MX1 and FHL1 inhibits MSC osteogenic differentiation
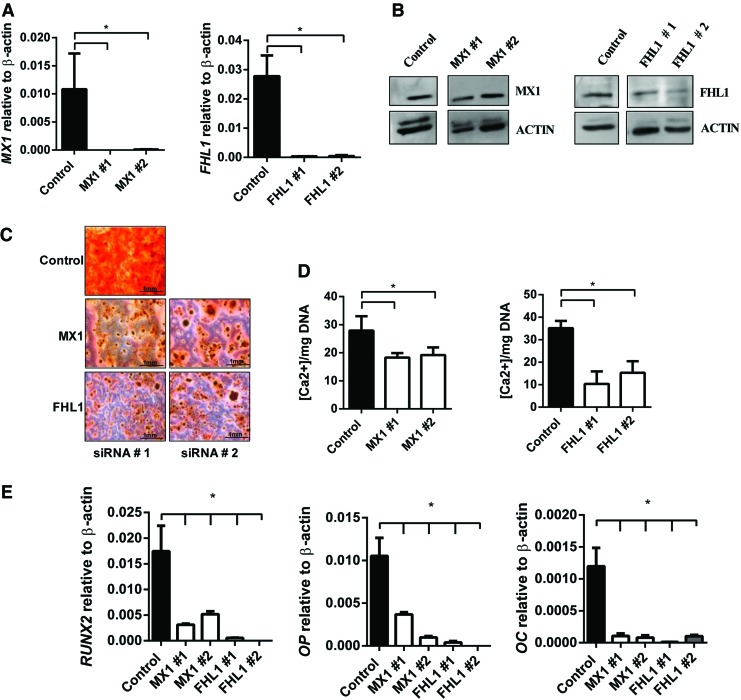
ZBTB16 has previously been implicated in regulating osteogenesis in human MSC by acting upstream of RUNX2 to promote osteogenic differentiation and the expression of key osteogenic differentiation genes [38,39]. However, the direct role of MX1 and FHL1 in regulating MSC osteogenic differentiation is not as well defined. Two independent siRNA targeting different regions of MX1 or FHL1 were used to assess the role of these factors in MSC osteogenic differentiation. Transient knockdown of MX1 and FHL1 was confirmed with real-time PCR and western blot analysis (Fig. 5A, B). Functionally, siRNA knockdown of MX1 or FHL1 reduced Alizarin red positive mineralized deposits (Fig. 5C) and reduced extracellular calcium (Fig. 5D) compared with scramble control siRNA-treated MSC. RT-PCR analysis of MX1 and FHL1 siRNA-treated MSC revealed a significant reduction in expression of RUNX2, and the mature bone-associated markers, OP and osteocalcin (OC) compared with control MSC, following osteogenic induction (Fig. 5E).
FIG. 5.
siRNA knockdown of MX1 and FHL1 inhibits osteogenesis. MSC were treated with negative control siRNA, MX1 siRNA #1, MX1 siRNA #2, FHL1 siRNA #1, and FHL1 siRNA #2 and were cultured for 14 days under osteogenic differentiation media. (A) Knockdown of MX1 and FHL1 by siRNA was confirmed by RT-PCR at 14 days of induction. (B) siRNA targeted knockdown of MX1 and FHL1 protein was confirmed by western blot analysis. (C) Representative images of Alizarin red stained mineral (x40) showing a decrease in hydroxyl appetite mineral MX1 and FHL1 siRNA-treated OB. (D) Extracellular calcium quantitated and normalized to DNA per triplicate well revealed significant decrease in calcium in siRNA-treated OB (n = 3 donors). (E) RT-PCR reveals a significant decrease in osteogenic marker RUNX2, OP, and OC in MX1 and FHL1 siRNA knockdown OB. Mean values ± SEM, n = 3 independent MSC donors (*P < 0.05, 1-way ANOVA with Dunnett's multiple comparisons test). OP, osteopontin. Color images available online at www.liebertpub.com/scd
We next assessed the proliferation rates of MX1 and FHL1 siRNA-treated MSC by BrdU incorporation. After 6 days of culture, there were no significant differences in the proliferation rates between siRNA scramble control-treated MSC compared with MX1 or FHL1 siRNA-treated MSC (Supplementary Fig. S3A). The data demonstrate that MX1 and FHL1 play an important role in promoting MSC osteogenic differentiation, similar to that described for ZBTB16.
Targeted knockdown of EZH2 and MX1 or FHL1 rescues aberrant osteogenic differentiation
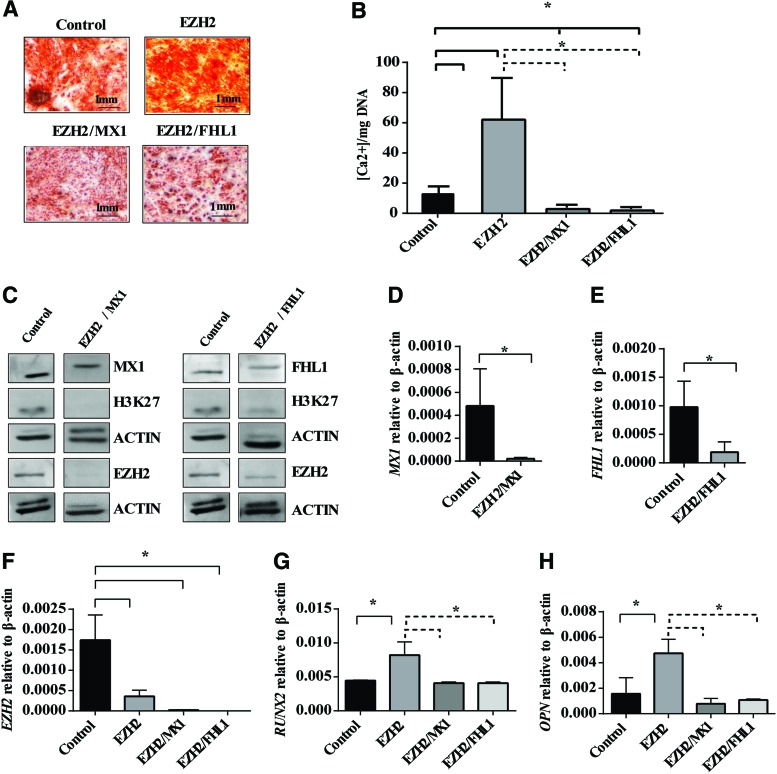
MX1 and FHL1 appear to play a functional role in promoting osteogenic differentiation in MSC, where siRNA knockdown of MX1 or FHL1 reduced Alizarin red positive mineralized deposit formation and expression of key osteogenic genes. We next assessed whether siRNA transfection of EZH2 and either FHL1 or MX1 could rescue the increased osteogenic differentiation potential observed in MSC treated with siRNA targeting EZH2 as we have previously described [25]. In the present study, MSC were treated with a combination of siRNA targeting EZH2 and MX1 (EZH2/MX1) or EZH2 and FHL1 (EZH2/FHL1) during osteogenic differentiation. Alizarin red staining of mineralized deposits and extracellular calcium quantitation revealed EZH2/MX1, and EZH2/FHL1 siRNA-treated MSC formed significantly less mineral deposits and extracellular calcium when compared to MSC treated with EZH2 siRNA or scramble control siRNA (Fig. 6A, B).
FIG. 6.
siRNA knockdown of EZH2 and MX1 or FHL1 rescued aberrant osteogenic differentiation. MSC were treated with 12 pmol of negative control siRNA (Control), EZH2 siRNA #1 (EZH2), EZH2 siRNA # 1/MX1 siRNA #1 (EZH2/MX1), and EZH2 siRNA # 1/FHL1 siRNA #2 (EZH2/FHL1) culture under osteogenic differentiation conditions for 14 days. (A) Alizarin red staining of mineral deposits in EZH2/MX1 and EZH2/FHL1 compared to control and EZH2 siRNA-treated MSC. (B) Extracellular calcium quantitated and normalized to DNA per triplicate well (n = 3 donors) for EZH2/MX1 and EZH2/FHL1 compared to control and EZH2 siRNA-treated MSC. (C) Western blot analysis of EZH2, H3K27me3, MX1, or FHL1 protein levels in EZH2/MX1 or EZH2/FHL1 siRNA-treated MSC compared to control-treated MSC under osteogenic differentiation conditions. (D) RT-PCR analysis of MX1 gen expression levels in EZH2/MX1 siRNA-treated OB compared with control siRNA OB. (E) RT-PCR analysis of FHL1 gene expression levels in EZH2/FHL1 siRNA-treated OB compared with control siRNA OB. (F) EZH2 knockdown was confirmed by RT-PCR in EZH2, EZH2/MX1, EZH2/FHL1 siRNA-treated OB compared with control OB. (G, H) RT-PCR analysis of gene expression levels for osteogenic markers RUNX2 and OP in EZH2/MX1 and EZH2/FHL1 siRNA OB when compared to EZH2 siRNA OB. Mean values ± SEM, n = 3 independent MSC donors (*P < 0.05, B, F–H, 1-way ANOVA with Dunnett's multiple comparisons test; *P < 0.05, D and E unpaired Student's t-test). Color images available online at www.liebertpub.com/scd
Western blot analysis confirmed a decrease in MX1 or FHL1, EZH2, and H3K27 methylation in EZH2/MX1 or EZH2/FHL1 siRNA-treated MSC compared to scramble control siRNA, following osteogenic induction (Fig. 6C). Real-time PCR analysis confirmed knockdown of EZH2 and MX1 or FHL1 in the EZH2/MX1 or EZH2/FHL1 siRNA-treated MSC, respectively (Fig. 6D–F). Knockdown of EZH2 in MSC increased RUNX2 and OP gene expression during osteogenic differentiation (Fig. 6G, H), as we have previously described [25]. However, knockdown of both EZH2 and MX1 or FHL1 in MSC resulted in repression of RUNX2 and OP gene expression at similar levels to scramble control-treated MSC (Fig. 6G, H). These findings demonstrate that MX1 and FHL1 are important downstream regulators of osteogenic differentiation of MSC. However, osteogenic differentiation is not solely reliant on these factors. Therefore, we suggest EZH2 functions to repress osteogenic differentiation through direct methylation of RUNX2, ZBTB16, MX1, FHL1, and many other genes, which are critical to driving MSC osteogenic differentiation.
Discussion
Our previous work has shown that the histone H3K27 methyltransferase EZH2 is an essential epigenetic modifying enzyme involved in inhibiting osteogenesis [25] and is recruited to the p16/Ink4A locus in response to the bHLH transcription factor Twist-1 to repress senescence [40]. Although we have illustrated that EZH2 can bind to some key osteogenic gene promoters to repress their expression, we have a limited knowledge in regard to what the targets of EZH2 are and how they are involved in inhibiting osteogenesis. To identify novel EZH2 targets that are involved in human MSC osteogenic lineage specification, we conducted a bioinformatics survey of publically available databases to identify putative EZH2 target genes suppressed in undifferentiated human bone marrow-derived MSC and subsequently activated following osteogenic differentiation. The present study revealed that EZH2 directly regulated the pro-osteogenic genes ZBTB16, MX1, and FHL-1 in human bone marrow-derived MSC through H3K27me3.
Upon MSC osteogenic differentiation, EZH2 and H3K27me3 were removed from the promoters of these genes, correlating with the appearance of the epigenetic activation mark H3K4me3. This is in agreement with our recent studies that identified EZH2 as a suppressor of MSC osteogenic differentiation, through the regulation of key osteogenic genes, RUNX2, OP, and OC [25]. Supportive studies demonstrated that inhibition of EZH2 methyltransferase activity following treatment with DZNep, or deactivation of EZH2 through phosphorylation of Thr 487 by CDK1, resulted in the promotion of osteogenesis [25,26]. Furthermore, EZH2 has been reported to regulate the β-catenin signaling pathway by targeting Wnt genes Wnt1, -6, -10a, and -10b in mouse peripheral preadipocytes and MSC to stimulate adipogenesis over osteogenesis [24]. As these Wnt genes are involved in the induction of RUNX2 and downstream osteogenic genes, their inhibition by recruitment of EZH2 to the promoter inhibits osteogenesis. Therefore, EZH2 acts to repress osteogenesis at multiple levels by directly inhibiting WNT genes, key osteogenic transcription factor RUNX2, and its downstream targets such as OP and OC while allowing adipogenic differentiation to occur. Collectively, these findings support the notion that EZH2 is a key regulator of MSC fate determination by selective repression of osteogenic inductive pathways.
The validity of the bioinformatics approach used here was shown by the identification of the putative EZH2 target ZBTB16, a promyelotic leukemia zinc finger transcription factor. ZBTB16 is a negative regulator of cell division in embryogenesis and controls skeletal patterning through repression of HOX, BMP, and Sonic Hedge Hog (Shh) expression. Deletion of ZBTB16 in mice leads to severe skeletal deformities due to impaired limb and axial skeleton patterning leading to transformation of anterior skeletal elements into posterior structures [41–43]. ZBTB16 is required for stylopod and zeugopod formation during early stages of limb development, before the initiation of cartilage condensation [44].
ZBTB16 has been found to either be deleted or mutated in patients exhibiting bone deformities. A 12-year-old male patient with skeletal defects, including hypoplasia of radius and ulna, short stature, mental retardation, delayed bone age, and mild facial dysmorphism, was identified with a ∼8 Mbp interstitial deletion in the 11q23 chromosomal region, which includes the ZBTB16 gene. This patient also revealed a recessive missense mutation (c. 1849ARG) within the remaining ZBTB16 allele [45,46]. This study indicated that ZBTB16 may play a critical role in the development of the skeleton and bone aging. Furthermore, Ikeda et al. (2005) found ZBTB16 was one of 24 genes upregulated during osteoblastic differentiation of OPLL cells [39].
ZBTB16 has been reported to regulate mesenchymal osteogenic and chondrogenic differentiation, acting upstream of the master transcription factors RUNX2 and SOX9, respectively, in mesenchymal progenitor cell line C3H10T1/2 [47]. Overexpression of ZBTB16 in C3H10T1/2 MSC line promoted osteogenic and chondrogenic differentiation while inhibiting adipogenic differentiation, Furthermore, these ZBTB16 overexpressing C3H10T1/2 MSC readily formed bone and cartilage when implanted into an intratibial osteochondral defect model [47]. Small interfering RNA knockdown of ZBTB16 in human and mouse MSC reduced osteogenic differentiation and expression of collagen 1A1 (COL1A1), alkaline phosphatase, RUNX2, and OC. Furthermore, overexpression of Zbtb16 in a murine MSC line increased Runx2 and Col1A expression during osteogenic differentiation [39]. Collectively, these studies and this current study have identified ZBTB16 as a regulator of osteogenic differentiation.
Mechanistically, ZBTB16 can act through its localization within nuclear bodies and plays a role in transcriptional activation or repression by regulating chromatin modifications and remodeling [48]. ZBTB16 can act as a transcriptional repressor through the recruitment of nuclear compressors SMRT, N-COR, SIN-3, and class 1 and class 2 histone deacetylases to the transcriptional complex [48–54]. ZBTB16 mediates transcriptional repression of HoxD gene expression through chromatin remodeling through long range DNA looping and interactions with polycomb protein Bmi-1 on DNA [48]. This study identified EZH2 actively binds ZBTB16 transcriptional start site and this coincides with the presence of H3K27me3 and low ZBTB16 gene expression. However, during osteogenic differentiation, EZH2 and its H3K27me3 modification is removed from the TSS followed by the addition of H3K4me3 and significant activation of ZBTB16. In the current study, we have, for the first time, demonstrated that overexpression of EZH2 in MSC, and thereby inhibiting osteogenic differentiation [25], was associated with reduced expression of ZBTB16. Conversely, siRNA-mediated knockdown of EZH2 promoted osteogenic differentiation [25] and promoted expression of ZBTB16. However, this study is the first to identify ZBTB16 as an EZH2 target in human mesenchymal stem cells.
In the present study, MX1, another known regulator of osteogenesis, was identified as a putative EZH2 target. The MX1 gene encodes a guanosine triphosphate-metabolizing protein that participates in the cellular antiviral response, where it is induced by type I and type II interferons and antagonizes the replication process of several different RNA and DNA viruses [55,56]. In vivo lineage tracing studies, using Mx1-Cre mice crossed with Rosa-YFP reporter mice, revealed that MX1-positive cells were predominantly osteolineage restricted with a small number of adipocytes found to have originated from MX1-positive cells during normal development and or bone fracture healing [57]. While the MX1-positive cell population exhibited trilineage potential, differentiating into adipocytes, osteoblast, and chondrocytes in vitro, MX1-positive cells did not contribute to chondrogenesis in vivo [57], where MX1 is expressed by less than half of the multipotent MSC and seems to be restricted to the osteogenic lineage in vivo. These findings suggest a distinct role of MX1 in murine osteogenic differentiation; however, the role of MX1 in human osteogenic differentiation is not as well defined.
Global wide gene expression analysis and ChIP analysis conducted by multiple groups indicate that human MSC express low levels of MX1, which is targeted by Ezh2 and is significantly upregulated during osteogenic differentiation. The present study demonstrated that siRNA-mediated suppression of MX1 expression decreased osteogenic differentiation of human MSC in vitro. These findings are supported by microarray studies that have identified MX1 as being differentially upregulated in transformed bone-forming human stromal cell lines versus nonbone-forming stromal cell lines [58]. Furthermore, our data suggest that EZH2-mediated repression of MX1 expression in MSC may result in an inhibition of osteogenic differentiation, where removal of EZH2 and subsequent H3K27me3 at the TSS of MX1 is critical for promotion of osteogenic lineage differentiation.
Another gene that was identified as a potential EZH2 target was four-and-a-half LIM domains 1 (FHL1), which encodes a member of the four-and-a-half-LIM-only protein family. FHL1 contains two conserved zinc finger domains with four highly conserved cytosine binding zinc atoms in each zinc finger. FHL-1 contains a TCF/LEF binding site, which can be stimulated by β-catenin or Wnt pathway agonist lithium chloride inducing muscle hypertrophy and repressing chondrogenesis. [59,60]. In support of our results, a recent study [61] identified that overexpression of FHL1 in murine MC3T3-E1 cells promotes osteogenesis. Transfection of Wnt3a or treatment with estrogen in MC3T3-E1 cells significantly increases FHL1 expression and osteogenic differentiation.
Overexpression of FHL1 increased alizarin red staining and extracellular calcium, which was associated with increased expression of osteogenic genes RUNX2, OP, and OC. Treatment with shRNAs targeting FHLl inhibited osteogenic differentiation of MC3T3-E1 cells compared to scramble control. These studies identify the role of FHL-1 in regulating muscle, chondrogenic, and osteogenic lineage specification. Our study identified that FHL1 is upregulated in human OB (Fig. 1B) and that knockdown of FHL1 by siRNA repressed osteogenic differentiation (Fig. 3B) as similar previously described in murine MC3T3-E1 cells. Furthermore, this is the first study to identify FHL1 as a novel EZH2 target during osteogenesis. EZH2 may act to repress FHL1 in MSC, preventing osteogenic lineage specification, suggesting that removal of EZH2 and hence H3K27me3 is critical for osteogenic differentiation.
Through bioinformatics analysis of publically available ChIP-on-chip, ChIP-Seq, and microarray data sets, we identified three novel EZH2 targets that may play a role in the suppression of osteogenesis by EZH2. With the use of overexpression studies, we verified that EZH2 directly inhibits ZBTB16, MX1, and FHL1 expression in MSC. Furthermore, siRNA knockdown of EZH2 in OB promoted the expression of ZBTB16, MX1, and FHL1. Knockdown studies using siRNA targeting MX1 and FHL-1 identified that MX1 and FHL-1 are critical for human MSC osteogenic differentiation and for the expression of key osteogenic transcription factor RUNX2 and its downstream targets. These findings suggest that EZH2-mediated H3K27me3 of ZBTB16, MX1, and FHL1 in MSC prevents osteogenic differentiation, keeping the MSC in a more undifferentiated state. Furthermore, EZH2 does not only act on these genes identified in this study but regulation of RUNX2 and WNT genes is also critical for the lineage specification [24–26].
Recent studies from our laboratory, and others, have identified the role of the histone H3 lysine 27 demethylase KDM6A and KDM6B in regulating MSC osteogenic differentiation [25,62]. We predict that these demethylases together or individually may be responsible for the removal of H3K27me3 from the TSS of ZBTB16, MX1, and FHL-1 during MSC to osteogenic lineage commitment. Furthermore, as we know KDM6A and KDM6B often associate with the H3K4 methyltransferases, we suggest that the recruitment of these demethylases to the TSS of these genes is critical for the removal of H3K27me3 and the subsequent addition of H3K4me3 modification. Further studies are needed to fully identify the mechanism H3K27me3 removal and the addition of H3K4me3 on these key osteogenic genes. Furthermore it is still unclear how and why is EZH2 recruited to the promoters ZBTB16, MX1, and MX1 and what factors and or signaling pathways are critical for the specific removal of EZH2 from these genes. Studies have identified EZH2 as an important regulator of the HOX genes during skeletal development and neural crest-derived cartilage and bone in mice [22,23]. Therefore, we predict that EZH2 may directly regulate ZBTB16, MX1, and FHL1 during murine and human skeletal development.
Supplementary Material
Acknowledgments
Funding National Health and Medical Research project grant APP1046053 and Fellowship APP1042677. S.H. Australian Postgraduate Award (APA) and Dawes Top-up scholarship. K.V. was supported by a Mary Overton Early Career Research Fellowship (Royal Adelaide Hospital).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S. and Marshak DR. (1999). Multilineage potential of adult human mesenchymal stem cells. Science 284:143–147 [DOI] [PubMed] [Google Scholar]
- 2.Gronthos S, Zannettino ACW, Hay SJ, Shi ST, Graves SE, Kortesidis A. and Simmons PJ. (2003). Molecular and cellular characterisation of highly purified stromal stem cells derived from human bone marrow. J Cell Sci 116:1827–1835 [DOI] [PubMed] [Google Scholar]
- 3.Chou RH, Yu YL. and Hung MC. (2011). The roles of EZH2 in cell lineage commitment. Am J Transl Res 3:243–250 [PMC free article] [PubMed] [Google Scholar]
- 4.James AW, Pang S, Askarinam A, Corselli M, Zara JN, Goyal R, Chang L, Pan A, Shen J, et al. (2012). Additive effects of sonic hedgehog and Nell-1 signaling in osteogenic versus adipogenic differentiation of human adipose-derived stromal cells. Stem Cells Dev 21:2170–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pei L. and Tontonoz P. (2004). Fat's loss is bone's gain. J Clin Invest 113:805–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng SL, Shao JS, Charlton-Kachigian N, Loewy AP. and Towler DA. (2003). MSX2 promotes osteogenesis and suppresses adipogenic differentiation of multipotent mesenchymal progenitors. J Biol Chem 278:45969–45977 [DOI] [PubMed] [Google Scholar]
- 7.Barnes GL, Javed A, Waller SM, Kamal MH, Hebert KE, Hassan MQ, Bellahcene A, Van Wijnen AJ, Young MF, et al. (2003). Osteoblast-related transcription factors Runx2 (Cbfa1/AML3) and MSX2 mediate the expression of bone sialoprotein in human metastatic breast cancer cells. Cancer Res 63:2631–2637 [PubMed] [Google Scholar]
- 8.Yang DC, Tsay HJ, Lin SY, Chiou SH, Li MJ, Chang TJ. and Hung SC. (2008). cAMP/PKA regulates osteogenesis, adipogenesis and ratio of RANKL/OPG mRNA expression in mesenchymal stem cells by suppressing leptin. PLoS One 3:e1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Alimonte I, Lannutti A, Pipino C, Di Tomo P, Pierdomenico L, Cianci E, Antonucci I, Marchisio M, Romano M, et al. (2013). Wnt signaling behaves as a “master regulator” in the osteogenic and adipogenic commitment of human amniotic fluid mesenchymal stem cells. Stem Cell Rev 9:642–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taipaleenmaki H, Abdallah BM, AlDahmash A, Saamanen AM. and Kassem M. (2011). Wnt signalling mediates the cross-talk between bone marrow derived pre-adipocytic and pre-osteoblastic cell populations. Exp Cell Res 317:745–756 [DOI] [PubMed] [Google Scholar]
- 11.Jung RE, Windisch SI, Eggenschwiler AM, Thoma DS, Weber FE. and Hammerle CH. (2009). A randomized-controlled clinical trial evaluating clinical and radiological outcomes after 3 and 5 years of dental implants placed in bone regenerated by means of GBR techniques with or without the addition of BMP-2. Clin Oral Implants Res 20:660–666 [DOI] [PubMed] [Google Scholar]
- 12.Bessa PC, Casal M. and Reis RL. (2008). Bone morphogenetic proteins in tissue engineering: the road from laboratory to clinic, part II (BMP delivery). J Tissue Eng Regen Med 2:81–96 [DOI] [PubMed] [Google Scholar]
- 13.Kugimiya F, Kawaguchi H, Kamekura S, Chikuda H, Ohba S, Yano F, Ogata N, Katagiri T, Harada Y, et al. (2005). Involvement of endogenous bone morphogenetic protein (BMP)2 and BMP6 in bone formation. J Biol Chem 280:35704–35712 [DOI] [PubMed] [Google Scholar]
- 14.Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T. and Schreiber SL. (2002). Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A 99:8695–8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS. and Zhang Y. (2002). Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039–1043 [DOI] [PubMed] [Google Scholar]
- 16.Taverna SD, Li H, Ruthenburg AJ, Allis CD. and Patel DJ. (2007). How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 14:1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sparmann A. and van Lohuizen M. (2006). Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 6:846–856 [DOI] [PubMed] [Google Scholar]
- 18.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, et al. (2006). The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439:871–874 [DOI] [PubMed] [Google Scholar]
- 19.Francis NJ, Kingston RE. and Woodcock CL. (2004). Chromatin compaction by a polycomb group protein complex. Science 306:1574–1577 [DOI] [PubMed] [Google Scholar]
- 20.Ringrose L, Ehret H. and Paro R. (2004). Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol Cell 16:641–653 [DOI] [PubMed] [Google Scholar]
- 21.Ringrose L. and Paro R. (2004). Epigenetic regulation of cellular memory by the polycomb and trithorax group proteins. Annu Rev Genet 38:413–443 [DOI] [PubMed] [Google Scholar]
- 22.Schwarz D, Varum S, Zemke M, Scholer A, Baggiolini A, Draganova K, Koseki H, Schubeler D. and Sommer L. (2014). Ezh2 is required for neural crest-derived cartilage and bone formation. Development 141:867–877 [DOI] [PubMed] [Google Scholar]
- 23.Wyngaarden LA, Delgado-Olguin P, Su IH, Bruneau BG. and Hopyan S. (2011). Ezh2 regulates anteroposterior axis specification and proximodistal axis elongation in the developing limb. Development 138:3759–3767 [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Jin Q, Lee JE, Su Ih. and Ge K. (2010). Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc Natl Acad Sci 107:7317–7322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemming S, Cakouros D, Isenmann S, Cooper L, Menicanin D, Zannettino A. and Gronthos S. (2014). EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells 32:802–815 [DOI] [PubMed] [Google Scholar]
- 26.Wei Y, Chen Y-H, Li L-Y, Lang J, Yeh S-P, Shi B, Yang C-C, Yang J-Y, Lin C-Y, Lai C-C. and Hung M-C. (2010). CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol 13:87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubo H, Shimizu M, Taya Y, Kawamoto T, Michida M, Kaneko E, Igarashi A, Nishimura M, Segoshi K, et al. (2009). Identification of mesenchymal stem cell (MSC)-transcription factors by microarray and knockdown analyses, and signature molecule-marked MSC in bone marrow by immunohistochemistry. Genes Cells 14:407–424 [DOI] [PubMed] [Google Scholar]
- 28.Easwaran H, Johnstone SE, Van Neste L, Ohm J, Mosbruger T, Wang Q, Aryee MJ, Joyce P, Ahuja N, et al. (2012). A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res 22:837–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y. and Gentry J. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smyth GK. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article3. [DOI] [PubMed] [Google Scholar]
- 31.Smyth GK. (2005). Limma: linear models for microarray data. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, eds. Springer, New York: pp 397–420 [Google Scholar]
- 32.Gentleman R, Carey V, Huber W, Irizarry R. and Dudoit S. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. (2006). Springer, Science & Business Media [Google Scholar]
- 33.Saeed A, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T. and Thiagarajan M. (2003). TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374. [DOI] [PubMed] [Google Scholar]
- 34.Isenmann S, Arthur A, Zannettino ACW, Turner JL, Shi ST, Glackin CA. and Gronthos S. (2009). TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment. Stem Cells 27:2457–2468 [DOI] [PubMed] [Google Scholar]
- 35.Nguyen TM, Arthur A, Hayball JD. and Gronthos S. (2013). EphB and Ephrin-B interactions mediate human mesenchymal stem cell suppression of activated T-cells. Stem Cells Dev 22:2751–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cakouros D, Isenmann S, Cooper L, Zannettino A, Anderson P, Glackin C. and Gronthos S. (2012). Twist-1 Induces Ezh2 recruitment regulating histone methylation along the Ink4A/Arf locus in mesenchymal stem cells. Mol Cell Biol 32:1433–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cakouros D, Isenmann S, Hemming SE, Menicanin D, Camp E, Zannetinno AC. and Gronthos S. (2015). Novel basic helix-loop-helix transcription factor hes4 antagonizes the function of twist-1 to regulate lineage commitment of bone marrow stromal/stem cells. Stem Cells Dev 24:1297–1308 [DOI] [PubMed] [Google Scholar]
- 38.Inoue I, Ikeda R. and Tsukahara S. (2006). Current topics in pharmacological research on bone metabolism: promyelotic leukemia zinc finger (PLZF) and tumor necrosis factor-alpha-stimulated gene 6 (TSG-6) identified by gene expression analysis play roles in the pathogenesis of ossification of the posterior longitudinal ligament. J Pharmacol Sci 100:205–210 [DOI] [PubMed] [Google Scholar]
- 39.Ikeda R, Yoshida K, Tsukahara S, Sakamoto Y, Tanaka H, Furukawa K. and Inoue I. (2005). The promyelotic leukemia zinc finger promotes osteoblastic differentiation of human mesenchymal stem cells as an upstream regulator of CBFA1. J Biol Chem 280:8523–8530 [DOI] [PubMed] [Google Scholar]
- 40.Cakouros D, Isenmann S, Hemming SE, Menicanin D, Camp E, Zannettino AC. and Gronthos S. (2015). Novel basic helix loop helix transcription factor hes4 antagonizes the function of twist-1 to regulate lineage commitment of bone marrow stromal/stem cells. Stem Cells Dev 24:1297–1308 [DOI] [PubMed] [Google Scholar]
- 41.Ivins S, Pemberton K, Guidez F, Howell L, Krumlauf R. and Zelent A. (2003). Regulation of Hoxb2 by APL-associated PLZF protein. Oncogene 22:3685–3697 [DOI] [PubMed] [Google Scholar]
- 42.Barna M, Hawe N, Niswander L. and Pandolfi PP. (2000). Plzf regulates limb and axial skeletal patterning. Nat Genet 25:166–172 [DOI] [PubMed] [Google Scholar]
- 43.Xu B, Hrycaj SM, McIntyre DC, Baker NC, Takeuchi JK, Jeannotte L, Gaber ZB, Novitch BG. and Wellik DM. (2013). Hox5 interacts with Plzf to restrict Shh expression in the developing forelimb. Proc Natl Acad Sci U S A 110:19438–19443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barna M, Pandolfi PP. and Niswander L. (2005). Gli3 and Plzf cooperate in proximal limb patterning at early stages of limb development. Nature 436:277–281 [DOI] [PubMed] [Google Scholar]
- 45.Wieczorek D, Koster B. and Gillessen-Kaesbach G. (2002). Absence of thumbs, A/hypoplasia of radius, hypoplasia of ulnae, retarded bone age, short stature, microcephaly, hypoplastic genitalia, and mental retardation. Am J Med Genet 108:209–213 [DOI] [PubMed] [Google Scholar]
- 46.Fischer S, Kohlhase J, Bohm D, Schweiger B, Hoffmann D, Heitmann M, Horsthemke B. and Wieczorek D. (2008). Biallelic loss of function of the promyelocytic leukaemia zinc finger (PLZF) gene causes severe skeletal defects and genital hypoplasia. J Med Genet 45:731–737 [DOI] [PubMed] [Google Scholar]
- 47.Djouad F, Tejedor G, Toupet K, Maumus M, Bony C, Blangy A, Chuchana P, Jorgensen C. and Noel D. (2014). Promyelocytic leukemia zinc-finger induction signs mesenchymal stem cell commitment: identification of a key marker for stemness maintenance? Stem Cell Res Ther 5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barna M, Merghoub T, Costoya JA, Ruggero D, Branford M, Bergia A, Samori B. and Pandolfi PP. (2002). Plzf mediates transcriptional repression of HoxD gene expression through chromatin remodeling. Dev Cell 3:499–510 [DOI] [PubMed] [Google Scholar]
- 49.Hong SH, David G, Wong CW, Dejean A. and Privalsky ML. (1997). SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARalpha) and PLZF-RARalpha oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci U S A 94:9028–9033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He LZ, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A. and Pandolfi PP. (1998). Distinct interactions of PML-RARalpha and PLZF-RARalpha with co-repressors determine differential responses to RA in APL. Nat Genet 18:126–135 [DOI] [PubMed] [Google Scholar]
- 51.Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara FF, et al. (1998). Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature 391:815–818 [DOI] [PubMed] [Google Scholar]
- 52.Rosato RR, Wang Z, Gopalkrishnan RV, Fisher PB. and Grant S. (2001). Evidence of a functional role for the cyclin-dependent kinase-inhibitor p21WAF1/CIP1/MDA6 in promoting differentiation and preventing mitochondrial dysfunction and apoptosis induced by sodium butyrate in human myelomonocytic leukemia cells (U937). Int J Oncol 19:181–191 [DOI] [PubMed] [Google Scholar]
- 53.Lemercier C, Brocard MP, Puvion-Dutilleul F, Kao HY, Albagli O. and Khochbin S. (2002). Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor. J Biol Chem 277:22045–22052 [DOI] [PubMed] [Google Scholar]
- 54.David G, Alland L, Hong SH, Wong CW, DePinho RA. and Dejean A. (1998). Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene 16:2549–2556 [DOI] [PubMed] [Google Scholar]
- 55.Watanabe S, Imaizumi T, Tsuruga K, Aizawa T, Ito T, Matsumiya T, Yoshida H, Joh K, Ito E. and Tanaka H. (2013). Glomerular expression of myxovirus resistance protein 1 in human mesangial cells: possible activation of innate immunity in the pathogenesis of lupus nephritis. Nephrology (Carlton) 18:833–837 [DOI] [PubMed] [Google Scholar]
- 56.Patzina C, Haller O. and Kochs G. (2014). Structural requirements for the antiviral activity of the human MxA protein against Thogoto and influenza A virus. J Biol Chem 289:6020–6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park D, Spencer JA, Koh BI, Kobayashi T, Fujisaki J, Clemens TL, Lin CP, Kronenberg HM. and Scadden DT. (2012). Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell 10:259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Larsen KH, Frederiksen CM, Burns JS, Abdallah BM. and Kassem M. (2010). Identifying a molecular phenotype for bone marrow stromal cells with in vivo bone-forming capacity. J Bone Miner Res 25:796–808 [DOI] [PubMed] [Google Scholar]
- 59.Lee JY, Chien IC, Lin WY, Wu SM, Wei BH, Lee YE. and Lee HH. (2012). Fhl1 as a downstream target of Wnt signaling to promote myogenesis of C2C12 cells. Mol Cell Biochem 365:251–262 [DOI] [PubMed] [Google Scholar]
- 60.Lee H-H, Lee J-Y. and Shih L-H. (2013). Proper Fhl1 expression as Wnt signaling is required for chondrogenesis of ATDC5 cells. Anim Cells Syst 17:413–420 [Google Scholar]
- 61.Wu SM, Shih LH, Lee JY, Shen YJ. and Lee HH. (2015). Estrogen enhances activity of wnt signaling during osteogenesis by inducing fhl1 expression. J Cell Biochem 116:1419–1430 [DOI] [PubMed] [Google Scholar]
- 62.Ye L, Fan Z, Yu B, Chang J, Al Hezaimi K, Zhou X, Park N-H. and Wang C-Y. (2012). Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell 11:50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.