Abstract
Metastatic renal cell carcinoma (mRCC) is nearly incurable and accounts for most of the mortality associated with RCC. Von Hippel Lindau (VHL) is a tumour suppressor that is lost in the majority of clear cell RCC (ccRCC) cases. Its role in regulating hypoxia-inducible factors-1α (HIF-1α) and -2α (HIF-2α) is well-studied. Recent work has demonstrated that VHL knock down induces an epithelial-mesenchymal transition (EMT) phenotype. In this study we showed that a CRISPR/Cas9-mediated knock out of VHL in the RENCA model leads to morphologic and molecular changes indicative of EMT, which in turn drives increased metastasis to the lungs. RENCA cells deficient in HIF-1α failed to undergo EMT changes upon VHL knockout. RNA-seq revealed several HIF-1α-regulated genes that are upregulated in our VHL knockout cells and whose overexpression signifies an aggressive form of ccRCC in the cancer genome atlas (TCGA) database. Independent validation in a new clinical dataset confirms the upregulation of these genes in ccRCC samples compared to adjacent normal tissue. Our findings indicate that loss of VHL could be driving tumour cell dissemination through stabilization of HIF-1α in RCC. A better understanding of the mechanisms involved in this phenomenon can guide the search for more effective treatments to combat mRCC.
Kidney and renal pelvis cancers accounted for an estimated 61,650 new cancer cases and 14,080 deaths in 20151. Patients with metastatic disease face a poor prognosis, with a five year survival of less than 12%. Renal cell carcinoma (RCC) makes up 90–95% of these cancers, with the majority of those the clear cell (ccRCC) histological subtype2,3. Treatment options for metastatic RCC (mRCC) are limited because this tumour shows resistance to traditional chemotherapy and radiation. The one treatment that has cured this condition is interleukin-2 (IL-2) therapy, but only in around 7% of patients4. Recent developments of targeted therapies, including those targeting immune checkpoint inhibitor programmed cell death-1 (PD-1), have shown modest efficacy5,6. The lack of enduring interventions to combat mRCC underscores the need for models that better recapitulate the disease and new insights into the mechanisms driving this condition.
Much of our understanding of ccRCC comes from studies on the tumour suppressor von Hippel Lindau (VHL). Hereditary cases of VHL syndrome show increased risk of ccRCC development7,8,9. Subsequent studies revealed that this gene is also silenced in up to 90% of sporadic ccRCC cases10. VHL’s best-described role involves its regulation of the hypoxia response through its recognition and targeting of the alpha subunits of hypoxia-inducible factor (HIF-1α, HIF-2α and HIF-3α) for ubiquitination and degradation11,12,13,14,15,16. In low oxygen conditions, VHL cannot recognize the HIF-αs and they combine with HIF-1β to translocate to the nucleus and enact the transcriptional program necessary for the hypoxic response17,18.
Researchers have attempted to derive murine models of ccRCC by targeting VHL for knockout19,20,21,22,23. Recent work has demonstrated that loss of Bap1 in addition to VHL may aid in modelling ccRCC in mice more consistently24. Though some of these studies show signs of early cystic ccRCC changes and local neoplasms, they all fail to produce an aggressive, metastatic form of this disease. For this reason, many studies depend on the RENCA model, the most widely used immunocompetent murine model of RCC25,26,27,28. This line was isolated from a spontaneously arising tumour in a BALB/c mouse in 197329. When implanted under the kidney capsule, this tumour metastasizes to sites seen in clinical ccRCC, including the lungs, liver and lymph nodes30. Despite the proven utility of this murine model, a major concern of its clinical applicability involves its expression of wild type VHL.
Previous work indicates that VHL loss may promote a more aggressive and metastatic tumour model. A number of studies have shown that targeting VHL function can lead to elements of epithelial-mesenchymal transition (EMT)31,32,33. This process has been identified as a central node through which carcinomas must pass to spread from their primary site to other parts of the body34. EMT involves the loss of cell-cell contact and a breaking away from the basement membrane of epithelial cells as they transition toward a more migratory and invasive cell type35. Concurrent with these phenotypic changes are an assortment of molecular changes, including loss of epithelial markers such as E-cadherin, a common occurrence in clinical ccRCC specimens31,36, and gain of mesenchymal markers such as N-cadherin and alpha smooth muscle actin (α-SMA)37. Notably, a number of studies demonstrate the role of HIF-1α in driving these changes32,33,38. Additionally, HIF-1α has been shown to cause metastasis in other tumour models38,39. These findings indicate that VHL deletion in the RENCA model may produce a more metastatic, clinically relevant model.
The clustered regularly interspaced short palindromic repeat (CRISPR) method of genetic manipulation has recently been harnessed for routine lab studies40. This breakthrough technique of gene disruption is notable for its ease of use and effectiveness in completely knocking out gene function. Based on the Streptococcus pyogenes adaptive immune system, this RNA-based technique for genome editing has quickly proved its utility in a number of biological studies41. Researchers have developed CRISPR methods in order to generate knockout mice, do genome-wide screens in cell lines, knock out genes in mice in vivo and screen for metastatic genes in vivo42,43,44,45,46,47. However, few studies thus far demonstrate CRISPR-mediated modification of murine cell lines in order to generate improved transplantable murine cancer models.
In this study, we utilized the new CRISPR genome editing tool to knock out VHL expression in RENCA cells. We show that this change produced increased aggressive behaviour in vitro and increased metastasis in vivo. This effect depended on HIF-1α, as knockout of this gene in VHL null cells reversed the noted phenotypic and molecular changes consistent with EMT. We went on to identify a set of four HIF-1α-regulated genes that are overexpressed in clinical RCC samples and are associated with poorer survival according to the TCGA database.
Results
Generation of VHL knockout RENCA cells using CRISPR
We obtained the lentiCRISPR plasmid developed by the Zhang lab44 and generated LCGFP, which has enhanced green fluorescent protein (eGFP) in place of the puromycin resistance gene in lentiCRISPR. Using the CRISPR design tool (crispr.mit.edu), we found candidate gRNA sequences targeting murine VHL. gRNAs against Renilla luciferase were also generated to serve as vector controls (Supplementary Fig. S1a and Table S1). RENCA FLuc cells, which stably express firefly luciferase, were transduced with either one of the VHL gRNAs or two. VHL gene sequencing provides an example confirming the specific ten base pair deletion in a region targeted by gRNA1 (g1) (Supplementary Fig. S1b). Protein levels of HIF-1α and expression of its downstream target Glut-1 indicated that g1 was more potent than gRNA2 (g2), given equal expression of eGFP as a proxy for transduction delivery load (Supplementary Fig. S1c–e). Notably, the maximum efficiency of VHL knockout was when both guides were used together. For this reason, we took a two target approach in the generation of the VHL knockout (RVN) and control (RC) cell lines (Supplementary Table S2).
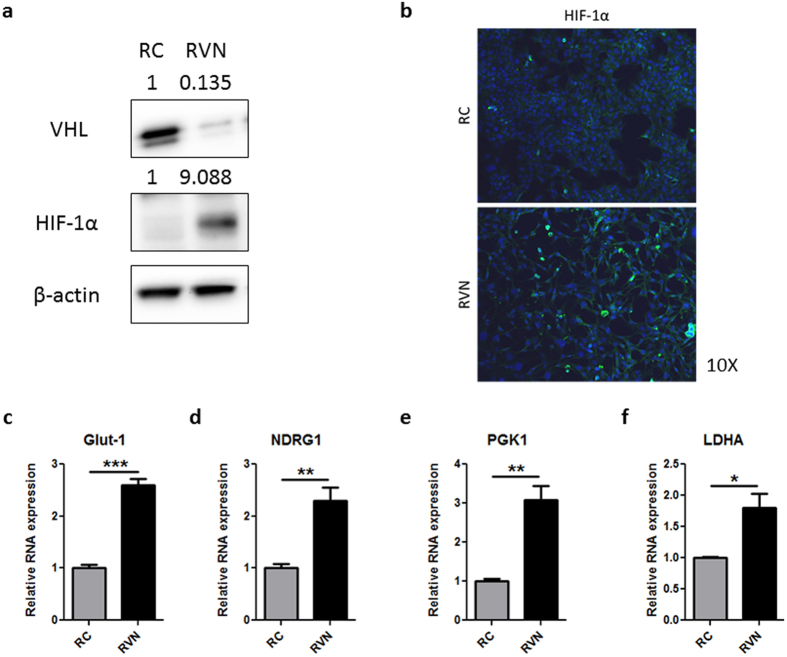
RC and RVN cells were assessed for VHL protein levels to determine the effectiveness of VHL knockout. Western blots revealed a dramatic reduction in VHL upon CRISPR-mediated gene modification (Fig. 1a). Levels of HIF-1α protein were greatly increased in the RVN cells, a change that is expected given that VHL degrades HIF-1α. Since the HIFs are transcription factors, activation of these proteins would be expected to induce nuclear localization. Immunofluorescent staining for HIF-1α in RVN cells revealed increased punctate staining, suggestive of nuclear localization, compared to RC cells (Fig. 1b). Elevation of HIF target genes such as Glut-1, N-myc downstream regulated gene 1 (NDRG1), phosphoglycerate kinase 1 (PGK1) and lactate dehydrogenase A (LDHA), were significantly increased in RVN compared to RC cells (Fig. 1c–f). Notably, Glut-1 and NDRG1 have been shown to be regulated by both HIF-1α and HIF-2α, whereas PGK1 and LDHA are HIF-1α-specific48. These findings indicate that this CRISPR approach can effectively knock out VHL, and that its well-studied role in modulating the HIF pathways is intact in RENCA cells.
Figure 1. CRISPR-mediated VHL knockout upregulates HIF pathways.
(a) Western blot for murine VHL, HIF-1α and β-actin in RC and RVN cells. Normalized quantification by densitometry shown above blot. (b) Immunofluorescent staining for HIF-1α in RC (top) and RVN (bot.) cells. Nuclei are stained blue with DAPI. Gene expression was analysed by RT-PCR for (c) Glut-1, (d) NDRG1, (e) PGK1 and (f) LDHA. n = 3 for RT-PCR studies *denotes p < 0.05, **denotes p < 0.01, ***denotes p < 0.001.
Loss of VHL induces EMT with enhanced aggressive in vitro behaviour
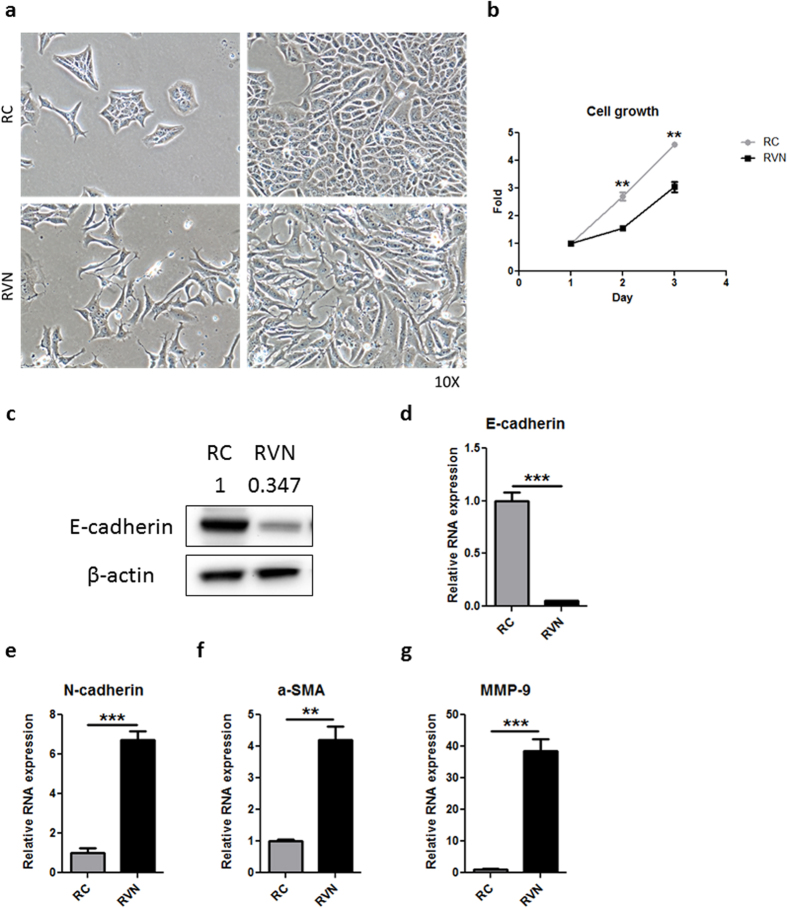
CRISPR-mediated VHL knockout induced a dramatic phenotypic change in RVN cells that was readily observable when compared to RC cells. While RC cells demonstrated the typical cobblestone epithelial morphology like wild type RENCA cells (Fig. 2a, top, Supplementary Video S1), RVN cells adopted an elongated, fibroblastic morphology with reduced cell-cell contacts (Fig. 2a, bottom, Supplementary Video S2). Interestingly, RVN cells had reduced proliferation when compared to RC cells (Fig. 2b). The morphology and proliferation changes were consistent with RVN cells undergoing EMT. For this reason, we chose to investigate whether VHL loss might also be driving EMT. Protein and RNA levels of E-cadherin, expected to be down following EMT, were reduced in RVN cells (Fig. 2c,d). Genes associated with the mesenchymal phenotype, such as N-cadherin, α-SMA and matrix metalloproteinase 9 (MMP-9), were significantly upregulated in RVN cells when compared to RC cells (Fig. 2e–g).
Figure 2. VHL loss induces morphologic and molecular changes indicative of EMT.
(a) Phase contrast 10X images of RC and RVN cells at low (left) and high (right) densities. (b) Growth chart of RC versus RVN cells. (c) Western blot for murine E-cadherin and β-actin in RC and RVN cells. Normalized quantification by densitometry shown above blot. Gene expression was analysed by RT-PCR for (d) E-cadherin, (e) N-cadherin, (f) α-SMA and g) MMP-9. n = 3 for proliferation assay and RT-PCR, **denotes p < 0.01, ***denotes p < 0.001.
Questions can be raised regarding the ability to attribute the molecular changes in our cells to loss of VHL and not a non-specific effect. For these reasons, we grew out clonal cell lines from RENCA cells transduced with RLuc g2, mVHL g1 and mVHL g3 (Supplementary Tables S1 and S2). Multiple gRNAs targeting distinct portions of VHL produced the expected VHL gene knockout and molecular phenotype consistent with RVN cells (Supplementary Fig. S2a–e). Notably, the gRNA targeting Renilla luciferase failed to produce these effects. Furthermore, rescue experiments using a doxycycline-regulated system showed that re-expression of VHL in a RENCA VHL knockout cell line leads to a reversion to an epithelial morphology (Supplementary Fig. S2f,g). Taken together, these findings demonstrate that our EMT changes are due to VHL loss and not through a non-specific effect caused by the CRISPR system or gRNAs.
Previous studies have demonstrated HIF-1α stabilization and EMT changes in human VHL wild-type RCC cell lines upon VHL knockdown33,49. We generated ACHN VHLko cells using gRNAs that targeted regions of human VHL that were orthologous to the sequences that were targeted in murine VHL to create the RVN cell lines (Supplementary Table S1). ACHN control cells were transduced with a gRNA targeting Renilla luciferase. As expected, CRISPR-mediated targeting of VHL led to a reduction in VHL protein levels and an increase in HIF-1α (Supplementary Fig. S3a). Morphologic changes in ACHN VHLko cells were consistent with those seen in RVN cells (Supplementary Fig. S3b). Molecular changes in ACHN VHLko were also as expected, with a reduction in E-cadherin and increased MMP9 expression, consistent with two separate gRNAs (Supplementary Fig. S3c,d).
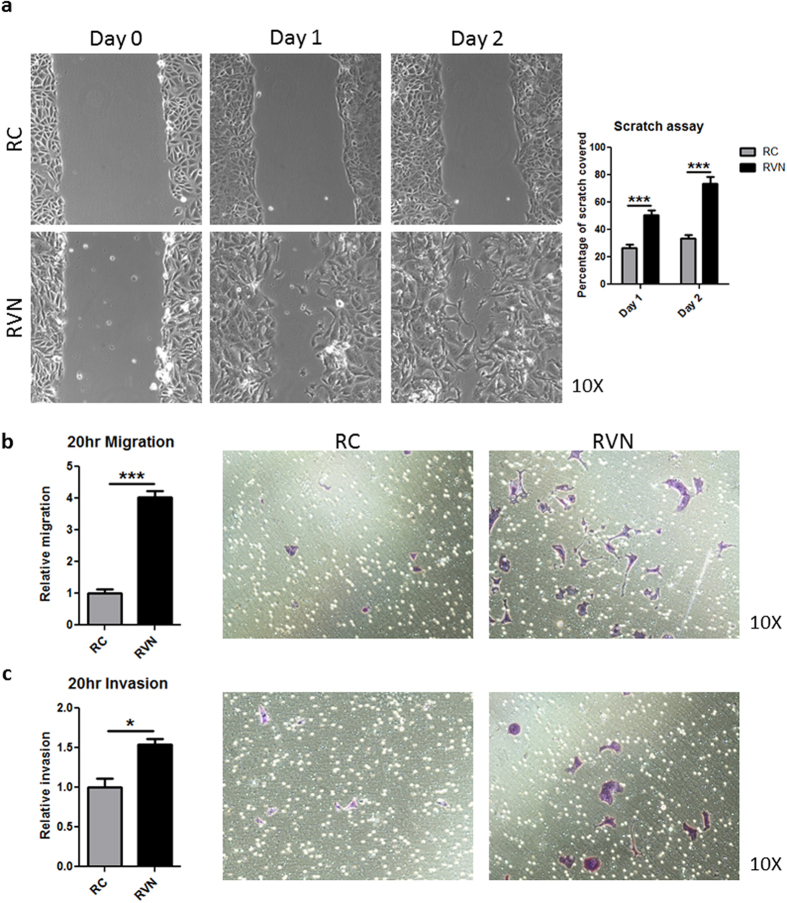
Given that the morphological and molecular changes in RVN cells were consistent with an EMT, we set out to determine whether these translated to a more aggressive phenotype. We performed a scratch assay and observed the patterns of scratch resolution for the two cell types. While RC cells resolved the scratch as a connected sheet, RVN cells moved into the open area individually (Supplementary Videos S3 and S4). We quantified the resolution of the scratch over two consecutive days (Fig. 3a). At day 1, RC cells had resolved 26.34% (+/−2.619) of the scratch compared to 50.54% (+/−3.372) for RVN cells. At day 2, RC cells had covered 33.56% (+/−2.642), while RVN cells resolved 73.57% (+/−5.227). These findings suggested a dramatically increased migratory capacity for RVN cells. Confirming this phenotype, a transwell assay showed RVN cell migration was four times greater than RC cells (Fig. 3b), and invasion of RVN cells through a matrigel transwell was more than 1.5 times that of RC cells (Fig. 3c). These findings suggest that RVN cells may have a greater metastatic potential.
Figure 3. VHL knockout induces increased RENCA cell migration and invasion in vitro.
(a) Images of a scratch assay were taken at the same location one and two days after scratching. Representative images of each cell type are shown (left) and quantification of scratch filling is shown to the right. (b) Quantification (left) and representative images (right) of RC and RVN migration are shown. (c) Quantification (left) and representative images (right) of RC and RVN invasion are shown. n = 7 for scratch assay, n = 3 for migration and invasion assays, *denotes p < 0.05, ***denotes p < 0.001.
VHL knockout drives increased metastasis
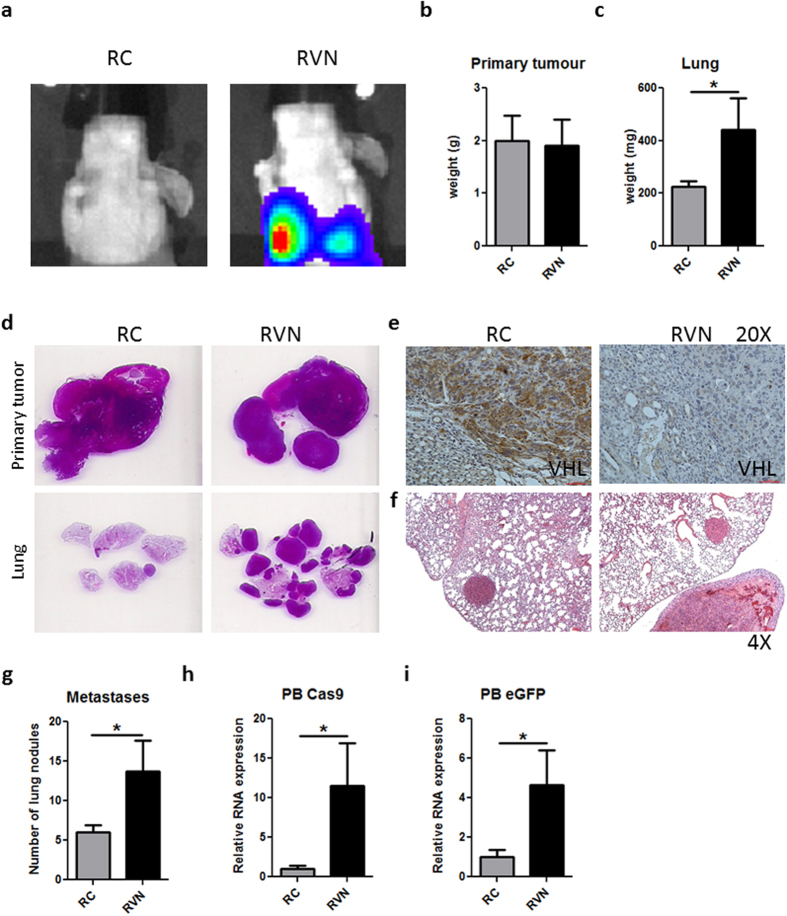
Following our demonstration of the increased aggressive phenotype of RVN cells in vitro, we sought to evaluate their ability to metastasize from a primary orthotopic tumour. Attempts to establish our RC and RVN tumours in immunocompetent BALB/c mice yielded inconsistent results characterized by failure to establish, or even rejection of, the orthotopic and subcutaneous tumours. We surmised that an immune-mediated rejection directed against CRISPR components could be at play as the use of transient expressing, non-integrating lentiCRISPR overcame this problem (data not shown). Thus, we decided to implant RC and RVN orthotopically in Nu/J mice. Animals were bioluminescently imaged four weeks post-injection and increased lung signal was revealed in RVN mice (Fig. 4a). While primary tumour size was similar in both groups, the lungs of mice implanted with RVN cells weighed significantly more (Fig. 4b,c). H&E confirmed comparable primary tumour sizes but increased lung metastasis (Fig. 4d), VHL knockout in primary tumours and increased metastatic burden in the lungs (Fig. 4e,f). H&E slides of lungs revealed significantly more lung metastases in RVN-implanted mice when compared to RC-implanted mice (Fig. 4g). Using RT-PCR for Cas9 and eGFP, circulating tumour cells were significantly increased in the peripheral blood of animals implanted with cells deficient in VHL (Fig. 4h–i).
Figure 4. Metastasis from orthotopic site is enhanced by VHL knockout.
RC and RVN cells were implanted under the kidney capsule of Nu/J mice. Animals were sacrificed at four weeks following implantation. (a) Representative bioluminescent images at day of endpoint are shown. Primary tumours (b) and lungs (c) were weighed and quantified for the two groups. (d) H&E stains are shown from primary tumour and whole lung. (e) Immunostaining for VHL in representative RC and RVN primary tumours shown at 20X. (f) 4X H&E representative images of RC and RVN lung nodules. (g) Lung nodules were counted and quantified. Gene expression of Cas9 (h) and eGFP (i) in the peripheral blood was assessed as a measure of circulating tumour cells. n = 3–4 animals per group for weights and RT-PCR, n = 6–8 per group for lung nodule counts *denotes p < 0.05.
VHL loss induces HIF-1α-mediated EMT
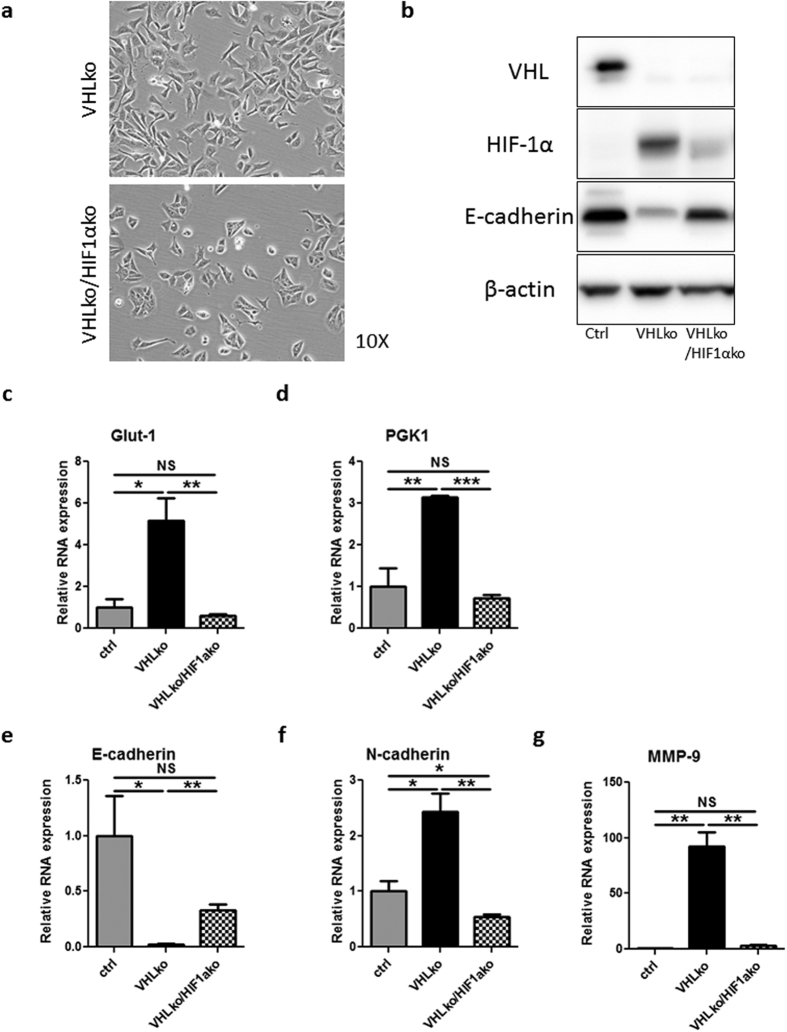
We began to interrogate the mechanism responsible for VHL-mediated EMT in RENCA cells. Previous work has implicated HIF-1α in the EMT process in RCC cells. To explore HIF-1α’s role in driving EMT in our cells, we used CRISPR to target HIF-1α or Renilla Luciferase in the V1c1 clonal VHL knockout cell line (VHLko/HIF1αko and VHLko, respectively, Supplementary Table S2). For a control, we used the clonal line targeting only Renilla luciferase (Rc1, Supplementary Table S2). The mesenchymal morphology exhibited by VHLko cells was reverted back to an epithelial appearance in VHLko/HIF1ako (Fig. 5a). HIF-1α loss in VHLko cells led to re-expression of E-cadherin (Fig. 5b,e), significantly decreased Glut-1 and PGK1 RNA expression (Fig. 5c,d), and increased expression of N-cadherin and MMP-9 (Fig. 5f,g). These morphological and molecular changes were consistent with HIF-1α mediation of VHL knockout-induced EMT. Similar findings were obtained with a VHL and HIF-1α double knockout from wildtype, non-clonal RENCA cells (Supplementary Fig. S4). It is unlikely that HIF-2α contributes to this EMT phenotype given its low level of expression compared to HIF-1α in these cells (Supplementary Fig. S5a). Furthermore, a HIF-2α small molecule inhibitor (N-(3-chloro-5-fluorophenyl)-4-nitrobenzo[c][1,2,5]oxadiazol-5-amine) was unable to reverse the phenotype or molecular changes of VHL knockout despite downregulation of HIF-2α-responsive genes erythropoietin (EPO) and vascular endothelial growth factor A (VEGF-A) (Supplementary Fig. S5b,c).
Figure 5. CRISPR-mediated knockout of HIF-1α in VHL knockout clonal cell line shows morphological and molecular reversion of EMT.
The V1c1 RENCA clonal cell line was transduced with LCGFP RLuc g1 (VHL KO) or LCGFP mHIF-1α g1 (VHLko/HIF1ako). Ctrl cells are the Rc1 clonal cell line. (a) Phase contrast 10X images of VHLko and VHLko/HIF-1αko cells. (b) Western blot for murine VHL, HIF-1α, E-cadherin and β-actin. Gene expression was analysed by RT-PCR for (c) Glut-1, (d) PGK1, (e) E-cadherin, (f) N-cadherin and (g) MMP-9. n = 3 for RT-PCR studies *denotes p < 0.05, **denotes p < 0.01, ***denotes p < 0.001, NS denotes no significance.
To further confirm the hypoxia-mediated EMT phenotype, we used Cobalt (II) chloride (CoCl2) and Dimethyloxalylglycine, N-(Methoxyoxoacetyl)-glycine methyl ester (DMOG), two hypoxia mimetic chemicals that do not affect VHL but stabilize HIFs. Treatment with either of these chemicals induced morphologic changes indicative of EMT in RENCA cells following one week of treatment (Supplementary Fig. S6a). Of interest, the VHL knockout-induced EMT appears to be independent of EMT-promoting transcription factors such Snail1/2, Twist, Zeb1 and Zeb2, as the expression levels of these genes are not elevated in RVN compared to RC cells (Supplementary Fig. S6b). Taken together with our previous findings, we can conclude that the EMT observed in VHL knockout cells can be attributed to HIF-1α.
VHL loss leads to HIF-1α-mediated upregulation of genes representative of aggressive clinical ccRCC
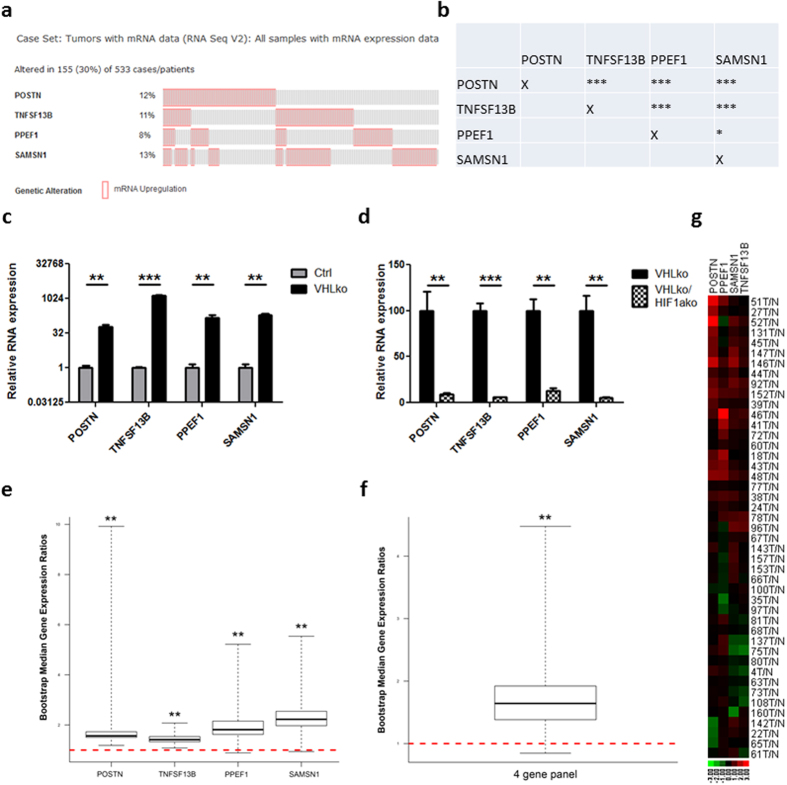
Findings from the RVN model have thus far implicated HIF-1α as a key pathway driving RCC dissemination. HIF-1α has already been associated with more aggressive ccRCC, and shown to drive metastasis in other tumour models38,39,50. However, the molecular pathways downstream of HIF-1α that modulate RCC metastatic behaviours are poorly understood. The RC and RVN cells were submitted for RNA-seq and over 8,500 genes were found to be differentially expressed between the two cell lines. Each of the top 65 upregulated genes in the RNA-Seq data set were queried with the TCGA data set, leading to a list of about ten genes that were individually indicative of a significantly poorer overall and/or progression free survival (Supplementary Table S3). Remarkably, four of these genes, of which 30% percent of patients had upregulation of one or more, seemed to be correlated in expression. These include periostin (POSTN), TNFSF13B (also known as B-cell-activating factor or BAFF), PPEF1 and SAMSN1 (Fig. 6a,b). Of note, these genes are all HIF-1α regulated, as evidenced by their induction with VHL knockout (Fig. 6c) and downregulation upon HIF-1α loss (Fig. 6d).
Figure 6. Four-gene signature shows frequent co-occurrence and upregulation in ccRCC samples.
(a) Frequency (%) of having upregulation one standard deviation higher than the mean for each of the four genes in the TCGA ccRCC database is given (left of each row). Each column represents one individual patient. Data for all patients with at least one upregulated gene is shown. (b) Table of co-occurrence analysis for the four genes. Gene expression was analysed by RT-PCR for POSTN, TNFSF13B, PPEF1 and SAMSN1 comparing (c) Ctrl and VHLko RENCA cells and (d) VHLko and VHLkoHIF1ako RENCA cells. Expression of these four genes was assessed in 45 clinical ccRCC samples from radical nephrectomies. Comparison of gene expression between tumour and normal adjacent tissue was analysed BootstRatio method for (e) each individual gene and (f) the 4 gene panel. (g) Heat map demonstrating expression of these four genes in each of the 45 clinical ccRCC samples. *denotes p < 0.05 **denotes p < 0.01 ***denotes p < 0.001.
To further verify that the upregulation of these four genes is indeed relevant in clinical ccRCC, we analysed gene expression in an independent set of clinical samples. We utilized 45 ccRCC tumour samples, recently collected from radical nephrectomies, and compared the expression of these four genes to adjacent normal tissue. POSTN, PPEF1, SAMSN1 and TNFSF13B were upregulated in a great majority of the ccRCC tumours (Supplementary Fig. S7a–d). Analysed by the BootstRatio method51, the upregulation of each gene was statistically significant (Fig. 6e, POSTN: Median.Ratio.Obs = 1.567, p < 0.01; PPEF1: Median.Ratio.Obs = 1.8139, p < 0.01; SAMSN1: Median.Ratio.Obs = 2.2257, p < 0.01; TNFSF13B: Median.Ratio.Obs = 1.4153, p < 0.01). Analysis of the geometric mean of 4 genes also showed a statistically significant upregulation in the tumours (Fig. 6f) (Median.Ratio.Obs = 1.6423, p < 0.01). In addition, we performed a log transformation with base 10 of the fold change, in which red indicates upregulation and green indicates downregulation (Fig. 6g). In this heat map, it is clear that a majority of the tumours from the 45 ccRCC patients have an upregulated pattern in this four gene set.
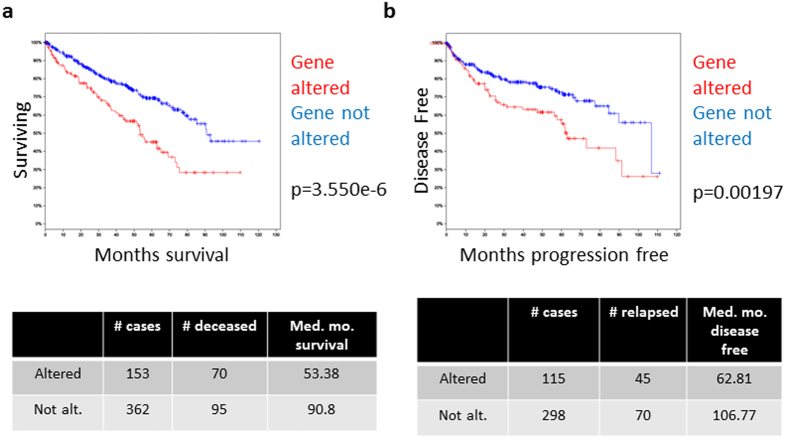
We again utilized the TCGA database to glimpse the impact of the upregulation of these 4 genes on the outcome in patients. The median overall survival for patients with upregulation of one or more of these genes was 53.38 months, compared to 90.8 months for patients without upregulation (p = 3.550e–6, Fig. 7a). Similarly, median progression-free survival for patients with gene upregulation was 62.81 months compared to 106.77 months (p = 0.00197, Fig. 7b). Collectively, our interrogation into clinical sources clearly showed that the four biomarker genes derived from our RVN model are upregulated in the ccRCC tumour tissues and that they predict a significantly poorer outcome in patients with this disease.
Figure 7. Four-gene signature predicts poor clinical outcomes in ccRCC patients.
Kaplan-Meier curves of (a) overall survival and (b) disease-free survival for the patients with upregulation of one or more of POSTN, TNFSF13B, PPEF1 and SAMSN1 is compared to patients without upregulation of any of these genes in the TCGA.
Discussion
This study demonstrates the ability to use a lentiviral CRISPR approach to manipulate a murine cell line in vitro and generate an improved mRCC tumour model in vitro and in vivo. There are a number of noteworthy aspects of the RENCA VHL null model we generated. Since VHL loss is the central hallmark of ccRCC clinically, we have now made the historically used RENCA model more genetically similar to what is seen in patients. The resulting dramatic morphologic and molecular changes, such as the E-cadherin loss that is commonly seen in the clinic36, indicates that our new ccRCC model is more similar and applicable to clinical disease. Characterizing the differences in tumour progression between RENCA VHL null cells and RENCA control cells should be the focus of future studies. We expect that these differences will reveal insights into key aspects of ccRCC.
Preliminary analysis of our cell lines indicates that many genes that correspond with a poor clinical prognosis in ccRCC patients are upregulated in the RENCA VHL null cells. We describe four genes that are HIF-1α regulated, frequently co-expressed and associated with poor survival clinically. Our independent validation of these genes showed that they are statistically significantly upregulated in ccRCC compared to adjacent normal tissue, with two or more frequently being upregulated concurrently. Interestingly, several of these genes have been tied to aggressiveness in other types of cancers. Periostin is involved in cell adhesion and motility, and some studies have shown its expression is associated with poor survival in a number of cancers, including RCC52,53. It has been recently demonstrated that periostin is associated with metastasis in head and neck cancer, and that this protein directly accelerated the growth, migration and invasion of cancer cells54. Another group found that periostin was overexpressed in a clinical case of melanoma metastasis and subsequently showed that inhibition of this protein in stromal cells leads to enhanced adhesion and reduced metastatic spread in a murine melanoma model55. TNFSF13B enhanced cell motility and invasion in a study of pancreatic cancer56. SAMSN1 was found to be expressed highly in glioblastoma multiforme, and high expression was a significant risk factor for poor survival57. Evaluating these genes and PPEF1, a serine/threonine protein phosphatase of which little is known, may produce potentially novel predictors of ccRCC prognosis. Additionally, they might provide new, clinically meaningful targets for aggressive ccRCC. Our findings warrant further validation of these markers with regards to ccRCC and an investigation into their potential mechanisms for promoting tumour aggressiveness in kidney cancer.
The role of HIF-1α in driving much of the aggressive phenotype in the RENCA VHL null cells is a noteworthy and intriguing finding of our studies. Though HIF-1α was originally thought to play a major part in ccRCC progression, recent studies have questioned this role58,59. Current thinking suggests that HIF-1α acts as a tumour suppressor, slowing growth of ccRCC cells60. Our findings showing reduced growth rate in vitro of RENCA VHL knockout cells are in line with this view. However, our results suggest that while HIF-1α may reduce proliferation, this may come with the caveat of increased migratory and invasive capacity. This agrees with the “Grow” or “Go” hypothesis, which argues that cells choose between proliferation and mobility and do not participate in both at once61. Some investigators have shown that HIF-1α inhibits primary tumour growth, whereas HIF-2α enhances it60. Combined with our findings, this suggests that VHL loss creates an environment where HIF-1α expression drives tumour metastasis and HIF-2α promotes tumour growth. There is evidence that HIF-1α expression correlates with worse prognosis for metastatic ccRCC patients50, and our study identifies some potential HIF-1α-driven genes that could contribute to a more aggressive ccRCC phenotype. Clarification of the differential roles of HIF-1α and HIF-2α with regard to metastasis promotion is critical for better understanding of this disease process and development of effective interventions.
The role of EMT in driving metastasis is a topic of great interest and pertinent in this new model for RCC metastasis. Recent work has suggested that EMT is not necessary for metastatic spread but may play a role in resistance to chemotherapy62,63. Notably, in our model, it is unclear whether the increased metastatic outgrowth we see in the VHL knockout model is through direct spread of the mesenchymal cells that have undergone the EMT. While this may be at play in our model, several other factors secondary to the mesenchymal changes in the cells could promote metastatic spread of neighbouring tumour cells. Upregulation of genes such as POSTN, TNFSF13B and SAMSN1 could provide increased metastatic potential of the tumour, as discussed. Additionally, other factors, such as CXCL1 and CXCL2 are significantly elevated with loss of VHL (data not shown) and could promote metastasis through a number of paracrine mechanisms64,65. Whether the EMT in this model leads to a direct or indirect promotion of metastasis will be the focus of future investigation.
A major question that arises from our findings involves the timing of VHL loss and how it impacts tumour behaviour. The RENCA cell line represents an already transformed tumour. We find that disrupting VHL on this background creates a more aggressive phenotype. One may wonder how well this sequence represents clinical disease since patients are thought to lose VHL early in the development of ccRCC66. It is possible that, for aggressive ccRCC, what matters is not the order in which VHL is lost, but the pathways that are disrupted in addition to its loss. The transforming mutations that drive the RENCA cell line remain elusive, and determining the molecular changes responsible for RENCA tumorigenesis should receive some attention in future studies. Once these are known, CRISPR technologies can be used to generate RCC tumours in vivo that will better model ccRCC. These models will enhance our understanding of the progression of ccRCC and could potentially provide insights into novel targeted therapies that could prove more effective in combating mRCC. This study is the first to use CRISPR-mediated technologies to explore the biology of VHL loss driving RCC aggressiveness in a new murine model and has produced intriguing clues regarding the involvement of HIF-1α in metastatic dissemination, new potential marker genes to assess poor prognosis of clinical ccRCC and possible molecular targets to combat mRCC aggressiveness.
Methods
Generation of lentiCRISPR-eGFP (LCGFP) and CRISPR cloning
LentiCRISPR (LC, Addgene #49535) and pX458 (Addgene #48138) were gifts from Dr. Feng Zhang67,68. LC was digested with NheI and MluI. A region of pX458 was amplified by PCR to produce NheI-2A-eGFP-MluI. DNA fragments ligated using Quick Ligation Kit (NE Biolabs). gRNA sequences for murine and human VHL and HIF-1α were produced by the CRISPR design tool software69 and oligonucleotides ordered from Valuegene. gRNAs were cloned into LC and LCGFP as detailed in the Zhang lab protocol70,71. gRNA sequences are listed in Supplementary Table S1. Cell lines are listed in Supplementary Table S2.
Cell culture and proliferation assay
RENCA and ACHN (ATCC) were cultured in Dulbecco’s Modification of Eagle’s Medium (DMEM) supplemented with 10% foetal bovine serum (FBS), 100 U/ml penicillin and 100 ug/ml streptomycin. RENCA cells were transduced to express firefly luciferase to generate the RENCA FLuc line as previously described72. All cell incubations were carried out at 37 °C and at 5% CO2. Puromycin selection was performed at 2 ug/ml for 5–7 days. Clonal selections were performed with Bel-Art Scienceware cloning discs according to manufacturer’s instructions. To analyse for proliferation, cells were plated in 6 well dishes at 1 × 10 ^ 5 cells per well. Each day, on three consecutive days, cells were trypsinized and counted using a Vi-CELL counter (Beckman Coulter).
Western blot, immunofluorescence and RT-PCR to analyse gene expression
Cell lysates were obtained, resolved on gels and transferred as previously described73. Blots were probed with antibodies recognizing VHL (Santa Cruz Biotechnology FL-181, 1:200), HIF-1α (Novus Biologicals H1alpha67, 1:500), E-cadherin (BD Biosciences 36/E-cadherin, 1:10,000) and β-actin (Santa Cruz Biotechnology C4, 1:5,000). Blots were imaged and densitometry performed on a ChemiDoc XRS + with associated ImageLab software (Bio-Rad).
For immunofluorescence staining, 5 × 10 ^ 5 cells per well were plated onto gelatine-coated cover glasses in a 12-well dish. Cells were fixed and stained with antibodies recognizing HIF-1α (Novus Biologicals H1alpha67, 1:100) and HIF-2α (Novus Biologicals ep190b, 1:100). Nuclei were stained with DAPI during mounting using Prolong Gold antifade mounting reagent (Life Technologies). Quantitative RT-PCR gene expression studies were conducted as previously described74. Primer sequences can be found in Supplementary Table S4.
Scratch assay, migration and invasion assays
Cells were plated in 6-well dishes at 1 × 10 ^ 5 cells per well. The following day, a scratch was applied down each well using a 10 ul pipette tip. Images were taken using an Eclipse Ti (Nikon Instruments) on the day of the scratch and for two subsequent days. Scratch resolution was analysed using Tscratch software developed by the Koumoutsakos group (CSE Lab) at ETH Zürich75.
For migration assay, 2.5 × 10 ^ 4 cells were plated in serum-free media in the upper well of an 8 um pore transwell (BD); complete media was in the bottom well. Cells were fixed and stained with crystal violet after 20 hours. For assessment of invasion, transwell inserts were coated with Corning Matrigel Growth Factor Reduced (BD). 5 random 10X images were obtained from each well and used for quantification.
Murine subcapsular model
All animal experiments were approved by the UCLA IACUC and conformed to all local and national animal care guidelines and regulations. Female 6–8 week old Nu/J mice (Jackson Laboratory) were placed in the prone position and an incision was made on the left flank30. The left kidney was partially exteriorized. A Hamilton syringe (28 gauge) was used to inject 1 × 10 ^ 5 cells in 5 ul of sterile PBS under the kidney capsule. Sectioning and H&E staining was done by the UCLA Pathology core. Two consecutive lung slides per animal were assessed for the number of visible tumour nodules. Peripheral blood was collected at endpoint through eye bleeding. Bioluminescent imaging was performed on an IVIS cooled CCD camera (Xenogen).
RNA-Seq
RC and RVN cells were cultured for two days and then processed using the RNeasy Mini Kit (Qiagen) according to manufacturer’s protocols. RNA was submitted to the UCLA Clinical Microarray Core where the sequencing was performed on the Illumina HiSeq 2000 and was single read 1 × 50. Raw data was processed by the UCLA Institute for Quantitative and Computational Biosciences. The resulting data was normalized, mapped to the genome and compared between samples. The false discovery rate cut-off was set at 0.1.
TCGA data
Data from the TCGA was queried using cBioPortal (www.cbioportal.org)76,77. All presented data utilized the Kidney Renal Clear Cell Carcinoma (TCGA, Provisional) data set.
Human samples and ethics statement
Primary tumours and corresponding adjacent normal tissues were obtained from 45 patients who received radical nephrectomy in the Department of Urology in Tongji Hospital of Huazhong University of Science and Technology in China from January 2012 to December 2015. All patients involved consented to participate in the study before surgery and signed an informed consent form, and all experiments were performed in accordance with the approved guidelines, complying with the principles for the use of human tissues in the Declaration of Helsinki. The study was approved by the Ethics Committee of Tongji Hospital, Tongji Medical School, Huazhong University of Science and Technology.
Statistics
Data is presented as mean +/− standard error mean (SEM). Comparisons between groups were analysed by student’s t-test.
Additional Information
How to cite this article: Schokrpur, S. et al. CRISPR-Mediated VHL Knockout Generates an Improved Model for Metastatic Renal Cell Carcinoma. Sci. Rep. 6, 29032; doi: 10.1038/srep29032 (2016).
Supplementary Material
Acknowledgments
The authors would like to thank Paul Pagano, Karen Jiang and Brenna Tam for technical assistance. We would also like to thank the Institute for Molecular Medicine and the Clinical and Translational Science Institute for access to common equipment that was used for these studies. We thank the Crump Institute for Molecular Imaging, the Quantitative and Computational Biosciences Collaboratory, Translational Pathology Core Laboratory and JCCC Flow Cytometry Core Laboratory for their aid in producing these findings. S. Schokrpur was supported by the UCLA/Caltech Medical Scientist Training Program (T32GM008042) and UCLA Tumour Immunology Training Grant (5T32CA009120). This project is supported by a grant provided by Cancer Research Coordinating Committee (to LW).
Footnotes
Author Contributions S.S. conceived the concept, designed and performed the experiments, collected and interpreted the data and drafted the manuscript. J.H. performed experiments and contributed to data interpretation and helped to draft the manuscript. D.L.M. and P.L. contributed greatly to the acquisition of data in animal studies and clinical samples, respectively. L.C.L. and K.H. contributed to the development of methodology. S.M. and M.P. contributed to the computational and statistical analysis and interpretation of patient clinical data. H.X. provided critical clinical materials to support the project. L.W. helped design the study, prepared and revised the manuscript. All authors read and approved the final manuscript.
References
- Howlader N. et al. SEER Cancer Statistics Review, 1975–2013, National Cancer Institute. Bethesda, MD, http://seer.cancer.gov/csr/1975_2013/, based on November 2015 SEER data submission, posted to the SEER web site, April 2016.
- Koul H. et al. Molecular aspects of renal cell carcinoma: a review. American journal of cancer research 1, 240–254 (2010). [PMC free article] [PubMed] [Google Scholar]
- Finley D. S., Pantuck A. J. & Belldegrun A. S. Tumor Biology and Prognostic Factors in Renal Cell Carcinoma. The oncologist 16, 4–13, doi: 10.1634/theoncologist.2011-S2-04 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coventry B. J. & Ashdown M. L. The 20th anniversary of interleukin-2 therapy: bimodal role explaining longstanding random induction of complete clinical responses. Cancer Management and Research 4, 215–221, doi: 10.2147/CMAR.S33979 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rini B. I. & Flaherty K. Clinical effect and future considerations for molecularly-targeted therapy in renal cell carcinoma. Urologic oncology 26, 543–549, doi: 10.1016/j.urolonc.2008.03.012 (2008). [DOI] [PubMed] [Google Scholar]
- Topalian S. L. et al. Safety, Activity, and Immune Correlates of Anti–PD-1 Antibody in Cancer. New England Journal of Medicine 366, 2443–2454, doi: 10.1056/NEJMoa1200690 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards F. M. et al. Detailed genetic mapping of the von Hippel-Lindau disease tumour suppressor gene. Journal of Medical Genetics 30, 104–107 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latif F. et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science 260, 1317–1320, doi: 10.1126/science.8493574 (1993). [DOI] [PubMed] [Google Scholar]
- Maher E. R., Neumann H. P. & Richard S. von Hippel–Lindau disease: A clinical and scientific review. European Journal of Human Genetics 19, 617–623, doi: 10.1038/ejhg.2010.175 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L. E. et al. Von Hippel-Lindau (VHL) Inactivation in Sporadic Clear Cell Renal Cancer: Associations with Germline VHL Polymorphisms and Etiologic Risk Factors. PLoS genetics 7, doi: 10.1371/journal.pgen.1002312 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berra E. et al. HIF prolyl‐hydroxylase 2 is the key oxygen sensor setting low steady‐state levels of HIF‐1α in normoxia. The EMBO journal 22, 4082–4090, doi: 10.1093/emboj/cdg392 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruick R. K. & McKnight S. L. A Conserved Family of Prolyl-4-Hydroxylases That Modify HIF. Science 294, 1337–1340, doi: 10.1126/science.1066373 (2001). [DOI] [PubMed] [Google Scholar]
- Epstein A. C. R. et al. C. elegans EGL-9 and Mammalian Homologs Define a Family of Dioxygenases that Regulate HIF by Prolyl Hydroxylation. Cell 107, 43–54, doi: 10.1016/S0092-8674(01)00507-4 (2001). [DOI] [PubMed] [Google Scholar]
- Yu F., White S. B., Zhao Q. & Lee F. S. HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proceedings of the National Academy of Sciences 98, 9630–9635, doi: 10.1073/pnas.181341498 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M. et al. HIFα Targeted for VHL-Mediated Destruction by Proline Hydroxylation: Implications for O2 Sensing. Science 292, 464–468, doi: 10.1126/science.1059817 (2001). [DOI] [PubMed] [Google Scholar]
- Jaakkola P. et al. Targeting of HIF-α to the von Hippel-Lindau Ubiquitylation Complex by O2-Regulated Prolyl Hydroxylation. Science 292, 468–472, doi: 10.1126/science.1059796 (2001). [DOI] [PubMed] [Google Scholar]
- Hu C.-J., Wang L.-Y., Chodosh L. A., Keith B. & Simon M. C. Differential Roles of Hypoxia-Inducible Factor 1α (HIF-1α) and HIF-2α in Hypoxic Gene Regulation. Molecular and cellular biology 23, 9361–9374, doi: 10.1128/MCB.23.24.9361-9374.2003 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B.-H., Rue E., Wang G. L., Roe R. & Semenza G. L. Dimerization, DNA Binding, and Transactivation Properties of Hypoxia-inducible Factor 1. Journal of Biological Chemistry 271, 17771–17778, doi: 10.1074/jbc.271.30.17771 (1996). [DOI] [PubMed] [Google Scholar]
- Albers J. et al. Combined mutation of Vhl and Trp53 causes renal cysts and tumours in mice. EMBO molecular medicine 5, 949–964, doi: 10.1002/emmm.201202231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Wang G., Shevchuk M. M., Nanus D. M. & Gudas L. J. Generation of a mouse model of Von Hippel-Lindau kidney disease leading to renal cancers by expression of a constitutively active mutant of HIF1α. Cancer research 71, 6848–6856, doi: 10.1158/0008-5472.CAN-11-1745 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnarra J. R. et al. Defective placental vasculogenesis causes embryonic lethality in VHL-deficient mice. Proceedings of the National Academy of Sciences of the United States of America 94, 9102–9107 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchett T. L., Bader H. L., Henderson J. & Hsu T. Conditional inactivation of the mouse von Hippel-Lindau tumor suppressor gene results in wide-spread hyperplastic, inflammatory and fibrotic lesions in the kidney. Oncogene 0, doi: 10.1038/onc.2014.197 (2014). [DOI] [PubMed] [Google Scholar]
- Rankin E. B., Tomaszewski J. E. & Haase V. H. Renal cyst development in mice with conditional inactivation of the von Hippel-Lindau tumor suppressor. Cancer research 66, 2576–2583, doi: 10.1158/0008-5472.CAN-05-3241 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-S. et al. Bap1 is essential for kidney function and cooperates with Vhl in renal tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America 111, 16538–16543, doi: 10.1073/pnas.1414789111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J. S. et al. Direct and differential suppression of myeloid-derived suppressor cell subsets by sunitinib is compartmentally constrained. Cancer research 70, 3526–3536, doi: 10.1158/0008-5472.CAN-09-3278 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J. S. et al. Sunitinib Mediates Reversal of Myeloid-Derived Suppressor Cell Accumulation in Renal Cell Carcinoma Patients. Clinical Cancer Research 15, 2148–2157, doi: 10.1158/1078-0432.CCR-08-1332 (2009). [DOI] [PubMed] [Google Scholar]
- Webster W. S. et al. Targeting Molecular and Cellular Inhibitory Mechanisms for Improvement of Antitumor Memory Responses Reactivated by Tumor Cell Vaccine. The Journal of Immunology 179, 2860–2869 (2007). [DOI] [PubMed] [Google Scholar]
- Xin H. et al. Sunitinib inhibition of Stat3 induces renal cell carcinoma tumor cell apoptosis and reduces immunosuppressive cells. Cancer research 69, 2506–2513, doi: 10.1158/0008-5472.CAN-08-4323 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G. P. & Hrushesky W. J. A Murine Renal Cell Carcinoma. Journal of the National Cancer Institute 50, 1013–1025, doi: 10.1093/jnci/50.4.1013 (1973). [DOI] [PubMed] [Google Scholar]
- Tracz A., Mastri M., Lee C. R., Pili R. & Ebos J. M. L. Modeling Spontaneous Metastatic Renal Cell Carcinoma (mRCC) in Mice Following Nephrectomy. Journal of Visualized Experiments, doi: 10.3791/51485 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban M. A. et al. Regulation of E-cadherin Expression by VHL and Hypoxia-Inducible Factor. Cancer research 66, 3567–3575, doi: 10.1158/0008-5472.CAN-05-2670 (2006). [DOI] [PubMed] [Google Scholar]
- Krishnamachary B. et al. Hypoxia-inducible factor-1-dependent repression of E-cadherin in von Hippel-Lindau tumor suppressor-null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer research 66, 2725–2731, doi: 10.1158/0008-5472.CAN-05-3719 (2006). [DOI] [PubMed] [Google Scholar]
- Pantuck A. J., An J., Liu H. & Rettig M. B. NF-κB–Dependent Plasticity of the Epithelial to Mesenchymal Transition Induced by Von Hippel-Lindau Inactivation in Renal Cell Carcinomas. Cancer research 70, 752–761, doi: 10.1158/0008-5472.CAN-09-2211 (2010). [DOI] [PubMed] [Google Scholar]
- Jiang J., Tang Y.-l. & Liang X.-h. EMT: a new vision of hypoxia promoting cancer progression. Cancer biology & therapy 11, 714–723 (2011). [DOI] [PubMed] [Google Scholar]
- Kalluri R. & Neilson E. G. Epithelial-mesenchymal transition and its implications for fibrosis. The Journal of clinical investigation 112, 1776–1784, doi: 10.1172/JCI20530 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langner C., Ratschek M., Rehak P., Schips L. & Zigeuner R. Expression of MUC1 (EMA) and E-cadherin in renal cell carcinoma: a systematic immunohistochemical analysis of 188 cases. Modern Pathology: An Official Journal of the United States and Canadian Academy of Pathology, Inc 17, 180–188, doi: 10.1038/modpathol.3800032 (2004). [DOI] [PubMed] [Google Scholar]
- Kalluri R. & Weinberg R. A. The basics of epithelial-mesenchymal transition. Journal of Clinical Investigation 119, 1420–1428, doi: 10.1172/JCI39104 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M.-H. et al. Direct regulation of TWIST by HIF-1α promotes metastasis. Nature cell biology 10, 295–305, doi: 10.1038/ncb1691 (2008). [DOI] [PubMed] [Google Scholar]
- Eisinger-Mathason T. S. K. et al. Hypoxia-Dependent Modification of Collagen Networks Promotes Sarcoma Metastasis. Cancer Discovery 3, 1190–1205, doi: 10.1158/2159-8290.CD-13-0118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D., Lander E. S. & Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 157, 1262–1278, doi: 10.1016/j.cell.2014.05.010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M. et al. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816–821, doi: 10.1126/science.1225829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. et al. Genome-wide CRISPR Screen in a Mouse Model of Tumor Growth and Metastasis. Cell 160, 1246–1260, doi: 10.1016/j.cell.2015.02.038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H., Li Y., Tan E.-P., Velasco-Herrera M. D. C. & Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nature Biotechnology advance online publication, doi: 10.1038/nbt.2800 (2013). [DOI] [PubMed] [Google Scholar]
- Ran F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature advance online publication, doi: 10.1038/nature14299 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell 153, 910–918, doi: 10.1016/j.cell.2013.04.025 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W. et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature, doi: 10.1038/nature13589 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. et al. One-Step Generation of Mice Carrying Reporter and Conditional Alleles by CRISPR/Cas-Mediated Genome Engineering. Cell 154, 1370–1379, doi: 10.1016/j.cell.2013.08.022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.-J., Wang L.-Y., Chodosh L. A., Keith B. & Simon M. C. Differential Roles of Hypoxia-Inducible Factor 1α (HIF-1α) and HIF-2α in Hypoxic Gene Regulation. Molecular and cellular biology 23, 9361–9374, doi: 10.1128/mcb.23.24.9361-9374.2003 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. V. et al. Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer. Nature medicine 12, 122–127, doi: 10.1038/nm1337 (2006). [DOI] [PubMed] [Google Scholar]
- Klatte T. et al. Hypoxia-inducible factor 1 alpha in clear cell renal cell carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research 13, 7388–7393, doi: 10.1158/1078-0432.CCR-07-0411 (2007). [DOI] [PubMed] [Google Scholar]
- Clèries R. et al. BootstRatio: A web-based statistical analysis of fold-change in qPCR and RT-qPCR data using resampling methods. Computers in Biology and Medicine 42, 438–445, doi: 10.1016/j.compbiomed.2011.12.012 (2012). [DOI] [PubMed] [Google Scholar]
- Morra L. & Moch H. Periostin expression and epithelial-mesenchymal transition in cancer: a review and an update. Virchows Archiv 459, 465–475, doi: 10.1007/s00428-011-1151-5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheev A. M. et al. Periostin is a novel therapeutic target that predicts and regulates glioma malignancy. Neuro Oncol 17, 372–382, doi: 10.1093/neuonc/nou161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X. et al. TGFbeta3-mediated induction of Periostin facilitates head and neck cancer growth and is associated with metastasis. Scientific reports 6, 20587, doi: 10.1038/srep20587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K. et al. Periostin Is a Key Niche Component for Wound Metastasis of Melanoma. PloS one 10, e0129704, doi: 10.1371/journal.pone.0129704 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi M. et al. Increased B cell-activating factor promotes tumor invasion and metastasis in human pancreatic cancer. PloS one 8, e71367, doi: 10.1371/journal.pone.0071367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y. et al. SAMSN1 is highly expressed and associated with a poor survival in glioblastoma multiforme. PloS one 8, e81905, doi: 10.1371/journal.pone.0081905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger N. et al. Deletions of chromosomes 3p and 14q molecularly subclassify clear cell renal cell carcinoma. Cancer 119, 1547–1554, doi: 10.1002/cncr.27947 (2013). [DOI] [PubMed] [Google Scholar]
- Shen C. et al. Genetic and Functional Studies Implicate HIF1α as a 14q Kidney Cancer Suppressor Gene. Cancer Discovery 1, 222–235, doi: 10.1158/2159-8290.CD-11-0098 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raval R. R. et al. Contrasting Properties of Hypoxia-Inducible Factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-Associated Renal Cell Carcinoma. Molecular and cellular biology 25, 5675–5686, doi: 10.1128/MCB.25.13.5675-5686.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiwarski D. J. et al. To “Grow” or “Go”: TMEM16A Expression as a Switch between Tumor Growth and Metastasis in SCCHN. Clinical Cancer Research, clincanres. 0363, 2014, doi: 10.1158/1078-0432.CCR-14-0363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K. R. et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527, 472–476, doi: 10.1038/nature15748 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527, 525–530, doi: 10.1038/nature16064 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharyya S. et al. A CXCL1 Paracrine Network Links Cancer Chemoresistance and Metastasis. Cell 150, 165–178, doi: 10.1016/j.cell.2012.04.042 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestas J. et al. The Role of CXCR2/CXCR2 Ligand Biological Axis in Renal Cell Carcinoma. The Journal of Immunology 175, 5351–5357 (2005). [DOI] [PubMed] [Google Scholar]
- Gerlinger M. et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nature genetics 46, 225–233, doi: 10.1038/ng.2891 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science (New York, NY) 339, 819–823, doi: 10.1126/science.1231143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O. et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science 343, 84–87, doi: 10.1126/science.1247005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotech 31(9), 827–832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87, doi: 10.1126/science.1247005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N. E., Shalem O. & Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature methods 11, 783–784, doi: 10.1038/nmeth.3047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok S. et al. Inhibition of CSF-1 receptor improves the antitumor efficacy of adoptive cell transfer immunotherapy. Cancer research 74, 153–161, doi: 10.1158/0008-5472.CAN-13-1816 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. et al. CSF1R signaling blockade stanches tumor-infiltrating myeloid cells and improves the efficacy of radiotherapy in prostate cancer. Cancer research 73, 2782–2794, doi: 10.1158/0008-5472.CAN-12-3981 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton J. B. et al. Suppression of prostate cancer nodal and systemic metastasis by blockade of the lymphangiogenic axis. Cancer research 68, 7828–7837, doi: 10.1158/0008-5472.CAN-08-1488 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebäck T., Schulz M. M. P., Koumoutsakos P. & Detmar M. TScratch: a novel and simple software tool for automated analysis of monolayer wound healing assays. BioTechniques 46, 265–274, doi: 10.2144/000113083 (2009). [DOI] [PubMed] [Google Scholar]
- Cerami E. et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discovery 2, 401–404, doi: 10.1158/2159-8290.CD-12-0095 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J. et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Science Signaling 6, pl1–pl1, doi: 10.1126/scisignal.2004088 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.