Abstract
With ~ 40 years of research completed after the development of self-contained underwater breathing apparatus, drug discovery opportunities in the sea are still too numerous to count. Since the FDA approval of the direct-from-the-sea calcium channel blocker ziconotide, marine natural products have been validated as a source for new medicines. However, the demand for natural products is extremely high due to the development of high-throughput assays and this bottleneck has created the need for an intense focus on increasing the rate of isolating and elucidating the structures of new bioactive secondary metabolites. In addition to highlighting the drug discovery potential of the marine environment, this review discusses several of the pressing needs to increase the rate of drug discovery in marine natural products, and describes some of the work and new technologies that are contributing in this regard.
Keywords: analytical techniques, antineoplastic agents, drug discovery, infectious disease agents, marine microorganisms, marine natural products, synthetic methods
1. Introduction
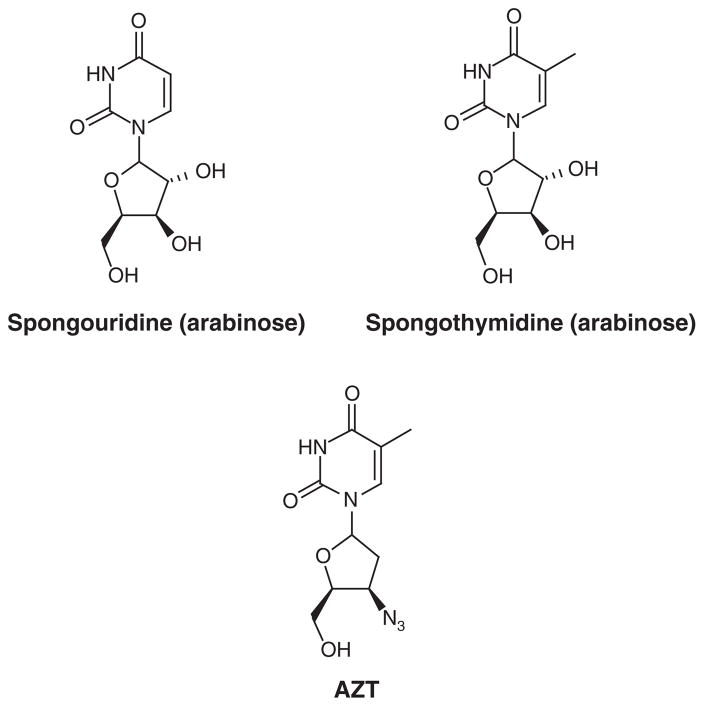
For many outside the field of natural products chemistry, traditional medicines have generally been associated with preparations from terrestrial organisms. In fact, some records in Chinese herbal pharmacopeia detail the use of marine seaweeds as early as 2000 years ago [1]. However, it can be assumed that access to the world’s oceans was severely limited and, consequently, the development of medicines from this environment was slow. The generation of global interest in the marine environment increased after the discovery of the atypical nucleosides spongouridine and spongothymidine (Figure 1)[2]. These arabinose-containing nucleosides were credited with inspiring later development of synthetic reverse-transcriptase inhibitors such as the well-known drug zidovudine (AZT, Figure 1)[3]. In the 1970s, the development of self-contained underwater breathing apparatus (SCUBA) technology and diving physiology by the military provided greater access to the marine environment and, with the help of such late pioneers as Paul J Scheuer, Luigi Minale, D John Faulkner and Kenneth J Rinehart, the field of marine natural products experienced rapid development. However, after over half a century, the enormity of the work ahead is evident by the estimations of marine species that represent an unexplored biosphere [4]. It was estimated by Pettit et al. in 1989 that only 0.5% of the marine organisms had been evaluated for antineoplastic constituents [5]; certainly less than this number were evaluated for other metabolic, infectious, neurological, cardiovascular and inflammation targets as funding during that time focused heavily on finding cancer treatments [6]. Other disease research has since benefited from the support of federal agencies and several non-profit organizations, effectively expanding the range of disease screening opportunities for new and previously reported marine natural products.
Figure 1.
Spongothymidine and spongouridine inspire the development of synthetic antiviral drugs such as AZT.
FDA approval of the conesnail peptide toxin ziconotide (Figure 2) in December 2004 has created a considerable buzz about marine natural products and their potential. Marketed under the trade name Prialt® (Elan Pharmaceuticals) and used for the treatment of chronic pain, the toxin was the first approved drug sourced directly from a marine organism. The clinical development of any drug is a long road, but the approval of ziconotide highlights a number of similar drug candidates moving synchronously through the pipeline. For example, the recent decision [201] by the European Medicines Agency to give a positive opinion of the drug trabectedin (ET-743, Figure 3) moves it even closer to full approval for use by patients with soft tissue sarcoma in the EU and, consequently, would represent a second structurally unmodified drug to have originated from the sea.
Figure 2.
Conesnail toxin ziconotide utilized for chronic pain.
Figure 3.
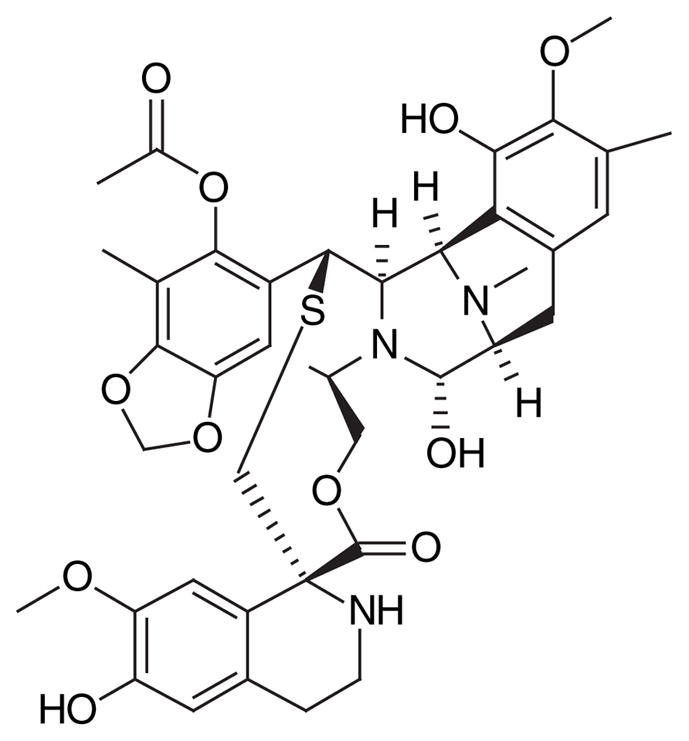
Anticancer drug ET-743 originally isolated from a marine tunicate.
Several excellent reviews published within the last 3 years detail the expanding collection of marine derived natural products, including their associated biological activities [7,8], status in clinical trials [6] and their synthesis [9] and biosynthesis [10,11]. The pharmacopeia described in these reviews certainly establishes the fact that the seas have been an extremely productive source in a relatively short period of time. A number of isolated natural products of marine origin are in clinical trials, approved or approaching full approval pending Phase III results (Table 1).
Table 1.
The status of isolated marine natural products in clinical trials.

| |||||
|---|---|---|---|---|---|
| Compound (class) | Kahalalide F (depsipeptide) | Aplidine (depsipeptide) | Bryostatin 1 (macrocycle) | ET-743 (isoquinoline) | Ziconotide (polypeptide) |
| Origin | Elysia rufescens | Aplidium albicans | Bugula neritina | Ecteinascidia turbinata | Conus magnus |
| Sourcing | Synthesis | Synthesis | Recollection, aquaculture | Aquaculture, synthesis | Synthesis |
| Activity | Cytotoxicity | Cytotoxicity | Cytotoxicity | Cytotoxicity | Analgesic |
| Target | Cell lysosomes [12] | JNK activation [13] | PKC-δ [14] | Minor groove DNA [15] | Neuronal Ca 2+ channels [16] |
| Status and references | Phase II | Phase II | Phase II | Phase II/III | 2004 |
| Various cancer [17] | Various cancer [18] | Cancer combination therapies [19] | Orphan drug approval [20], positive opinion EMEA | Approval [21] | |
JNK: Janus kinase; PKC: Protein kinase C.
The purpose of this brief review is to highlight how modern tools and emerging technologies in natural products chemistry are expanding the promise of drug discovery in the marine environment, while utilizing some examples of their applications towards the development of marine derived drugs. Many relevant compound structures are included throughout the text. However, due to this review’s limited space and scope, some compound structures have been omitted and readers are invited to consult the aforementioned reviews and associated references for structure information.
2. Expanded diversity among marine derived structures
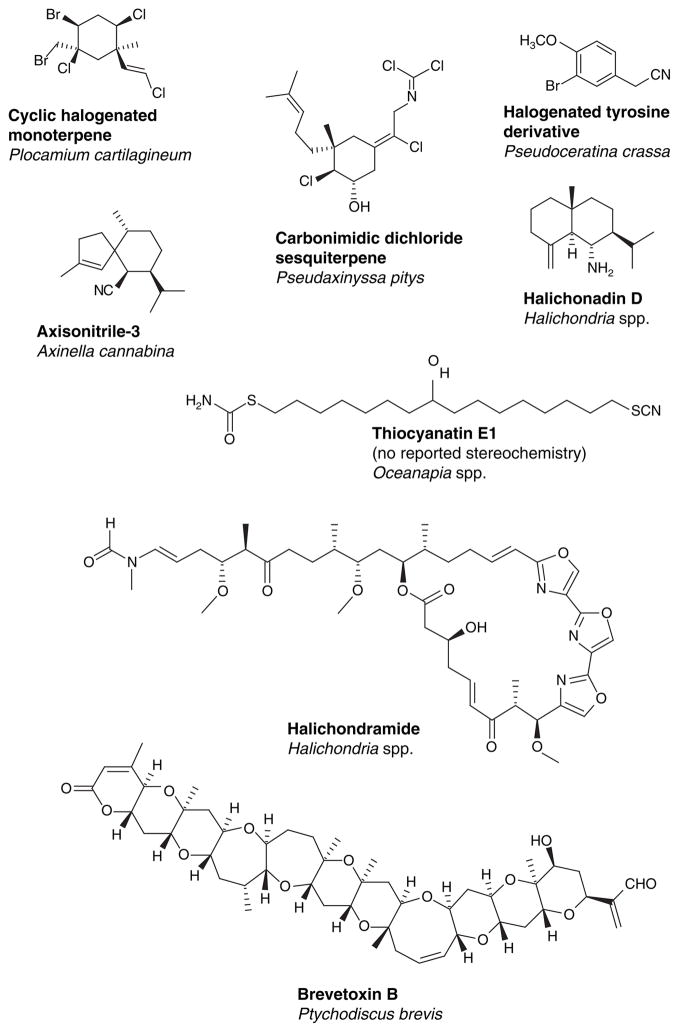
Due to the vast size and limited accessibility, the marine environment will continue to produce chemistry and biology that expand the current boundaries of basic and applied sciences. Previous research clearly indicates that the biosynthetic pathways and secondary metabolism of ocean species differs significantly from their terrestrial counterparts [22]. Environmental pressures are the primary driving force for this diversity and as an example, communication between terrestrial organisms requires small primarily volatile molecules, whereas in water, signaling molecules can be larger and more complex, as long as they are water soluble [23]. Functional groups rarely found in terrestrial species, such as isonitrile, isothiocyanate and halogenated moieties occur often in marine natural products [24]. Marine natural products possess both unique functionalities and unusual carbon skeletons. Some examples of diversity and unique structures among marine natural products are illustrated in Figure 4.
Figure 4.
Examples of the diversity and unique structural features of marine-derived compounds.
2.1 Isolation and characterization
High-throughput screening in the 1990s enabled the biological testing of thousands of samples within a short period of time [25] and this created a bottleneck effect between the assays and the natural products chemists struggling to keep up with the demand for new compounds. This situation negatively impacted on the pharmaceutical industry’s opinion of natural products at the time, but provided inspiration for improvements in the technologies used for isolation and structure elucidation of natural products, primarily focusing on increased throughput. High-performance liquid chromatography (HPLC), coupled with techniques including detection with diode array (HPLC-DAD), circular dichroism (HPLC-CD), nuclear magnetic resonance (HPLC-NMR) and mass spectrometry (HPLC-MS), has made it possible to speed the purification and characterization of secondary metabolites from natural sources. Structure elucidation has become possible even with submilligram quantities of sample thanks to recent developments in NMR technology that include more sensitive probes and higher magnetic fields. Cryoprobes have been an important advance in the sensitivity of NMR although they are relatively expensive to maintain in comparison with standard probes. By cooling the probe hardware with helium, sensitivity can be increased approximately four fold while insulation maintains the desired temperature of the sample. Acquiring NMR spectra on extremely limited amounts of samples is becoming routine with either the tube-based 1 mm microprobes or tubeless capillary NMR (capNMR) probes. With smaller receiver coil diameters, microprobes use a non-conventional sample tube that only requires 10 μl sample volume, but have been logistically challenging to use. CapNMR probes are equipped with a receiver coil wrapped around a capillary-scale flow cell that can be tuned for different nuclei. The extremely small solenoid flow cells in such capNMR probes can be oriented perpendicular to the magnetic field, thus providing higher intrinsic sensitivity [26]. Automation of the injection of samples into capNMR probes has also reduced logistical issues associated with traditional tube-based NMR probes and provided a route to a relatively high-throughput routine NMR [27].
Limitations of HPLC (speed, efficiency, high pressure limit) have recently been overcome by the introduction of ultra-high-performance liquid chromatography (UHPLC) [28]. UHPLC instrumentation and small particle size allow analysis at extremely high pressure of ≥ 1034 bar [29].
The coupling of HPLC with highly sensitive spectroscopy methods provides structural information without time-consuming and laborious purification processes. Techniques such as preparative HPLC-MS have become widely used in the pharmaceutical industry as a purification method because of its more universal applicability.
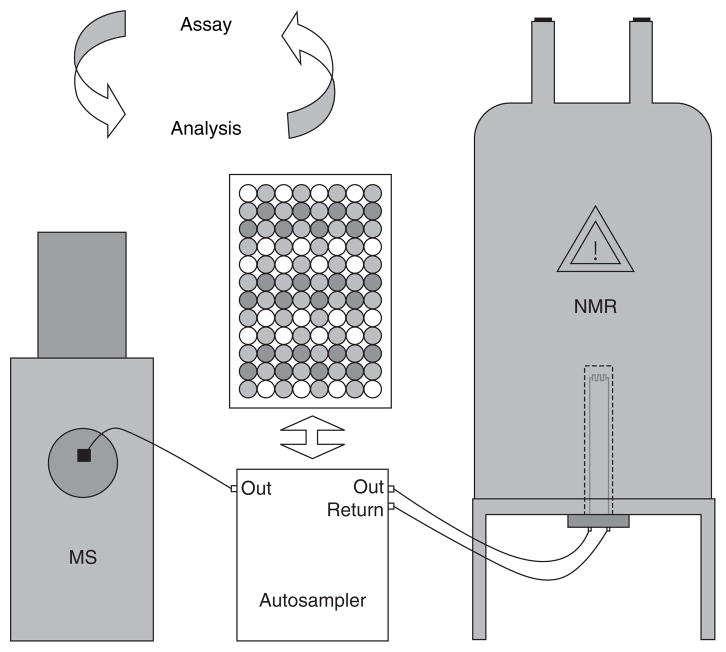
Tracking bioactivity in complex mixtures is now more efficient with the assistance of microtiter-based bioassays. After separation by HPLC, a crude extract is fractionated into a 96-well plate and duplicated. While one plate is characterized using mass spectrometry, the other plate is sent to the assay. Autosampler-integrated capNMR probes can likewise be used for the high-throughput analysis of large compound libraries or HPLC fractions (Figure 5). The activity profile obtained from this rapid screening is then matched with the HPLC chromatogram and the structural information obtained from the spectroscopy of the fractions.
Figure 5.
Using automated systems with standard mass and NMR spectrometers having capillary flow probes in combination with high-throughput assays significantly increases the rate of isolation and identification of bioactive natural products.
NMR: Nuclear magnetic resonance; MS: Mass spectrometry.
Hyphenated NMR methods offer some significant advances in natural product research. The combination of HPLC and NMR enables structure elucidation using small amounts of extract and can help avoid long and laborious isolation processes [30]. NMR requires little sample preparation and, with the latest technology, good quality data can be obtained with low millimolar concentrations of sample [27,30]. The combination of capillary NMR probes with liquid chromatography results in an exceedingly powerful tool – capillary HPLC-NMR. New developments in the miniaturization of solid-phase extraction interfaces and cryoprobes will continue to improve the sensitivity of such HPLC-NMR hyphenated techniques [30].
High resolution mass spectrometry (HRMS) is widely used today with sensitive analyzers such as time-of-flight (TOF), that enable the rapid dereplication of bioactive compounds derived from marine natural sources. A reliable and simple dereplication method is essential in order to avoid purification of many natural products that have already been published [31]. The introduction of HPLC/HRMS techniques has made it possible to thoroughly screen crude extracts without extensive purification. Molecular weight information can be compared with an existing marine natural product database and, after a relatively short time and using as little as 40 μg of crude extract [32], one can decide whether the sample should be purified further using more comprehensive methods. HPLC/HRMS spectrometry can also be used for the purpose of bioactivity screening. As reported by Davis et al.[33], a mixture of compounds is incubated with a target protein and the components that bind to the protein are selected by size exclusion chromatography. The unbound components are retained on the column while the bound ones are eluted and identified with HPLC/HRMS.
2.2 Sample collection
Analytical methods for isolation and purification of active metabolites are important, but the development of sample collection methods directly contributes to the available species diversity for research. SCUBA technology improvements have led into the development of closed circuit underwater breathing apparatus (CCUBA), which allows the exploration of underwater environments as deep as 150 m with extended bottom time. This technology led to a deep-water collection of Myrmekioderma styx and isolation of > 30 new sesquiterpenes and diterpenes from this individual sponge [34]. In addition to CCUBA, exploration of deep sea environments is now possible using remotely operating vehicles (ROVs). An estimated 50% of the Earth’s oceans are deeper than 3000 m and ROVs provide researchers access to these unexplored environments. Examples of deep sea organisms that were collected using ROVs include jellyfish Bumpy (Stellamedusa ventana) and the eel-like halosaurs [35]. The fact that the collected jellyfish represents a new species of large scyphomedusae shows how much there is still left to be explored and discovered in the deep sea environment.
3. Addressing the supply issues of marine natural products
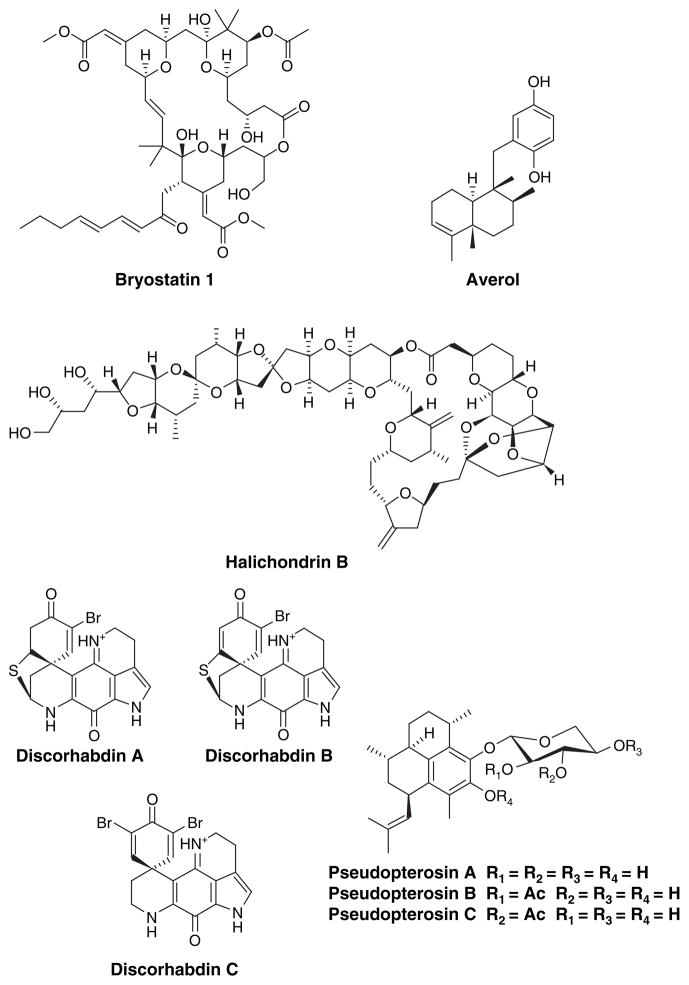
Despite their high potency and unique structural features, marine natural products have been facing major difficulties fitting into the conventional drug discovery process. One of the most significant issues is supply of materials. The reported yield of natural products from marine organisms can be as low as 1 mg from 3 kg of organism [6] and is certain to be much less based on existing detection limits. Most marine natural products under clinical investigations are supplied from chemical synthesis. Isolation procedures can be lengthy and natural supplies frequently cannot sustain gram scale production. In general, the natural abundance of source organisms will not support the development of marine natural products based on wild harvest, and massive collections of marine invertebrates have serious environmental consequences. However, some of the compounds are supplied by large scale isolation from aquaculture organisms and this method has been applied to the production of ET743, bryostatin 1, avarol, halichondrin B, discorhabdins and pseudopterosins (Figure 6)[36].
Figure 6.
Marine natural products produced through large-scale aquaculture.
Isolation of multi-gram quantities of GMP-bryostatin 1 was reported in 1991 [37]. The yield of this large-scale isolation was 10 g from 10,000 gallons of marine bryozoan. Purification of the crude extract was guided by HPLC with photodiode array detection and a 3 H-phorbol dibutyrate receptor binding assay and took six steps from crude extract to pure compound to be ready for clinical studies. A total of 1 g of ET743 was obtained from 1 metric ton of E. turbinata[38], and the same weight of sponge Lissodendoryx spp. yielded only 300 mg of halichondrin B [39].
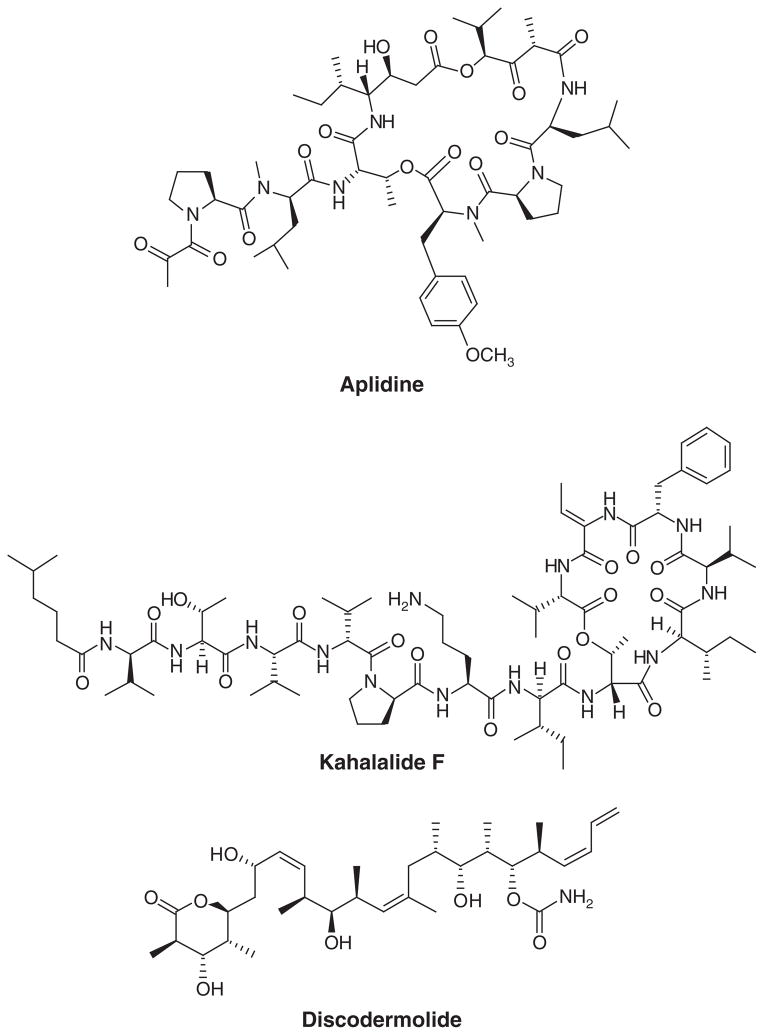
The complexity of some chemical structures of marine natural products represents a major roadblock in the design of economic synthetic routes. Large-scale synthesis of nonribosomal peptides is an exception as they can be assembled from amino acid building blocks using strategies of conventional peptide synthesis. However, peptides can still face higher costs due to the presence of unusual amino acids or peptide conformations that are commonplace in marine natural products. The anticancer candidates aplidine [17], kahalalide F [18] (Figure 7) and the clinically available ziconotide [40] are at present produced using synthesis and have individual synthetic hurdles. Other non-peptide related compounds have been produced through this method, sometimes at more inflated costs. The total synthesis of discodermolide (Figure 7) was achieved in 39 steps and yielded 60 g for a Phase I clinical trial [41]. The supply of ET743 for clinical trials was produced using a semisynthetic approach by converting the structurally related cyanosafracin B in 18 steps [42].
Figure 7.
Marine natural products produced through large-scale synthesis.
Another method to overcome the supply issue comes from the discovery of sponge- and algae-associated bacteria producing previously reported bioactive natural products [43]. This recent development supports culture and growth in fermentation systems as a viable method to provide a sustainable source of active metabolites of pharmacological interest. Sponges are well known to harbor diverse microbes and represent a significant source of bioactive natural compounds. Intimate associations between microbes and eukaryotic hosts are common in nature and marine animals and plants are well known to have developed highly specific relationships with numerous microbes [43]. The association between microorganisms and macroorganisms may involve a symbiotic microorganism, a specific and permanently associated non-symbiotic microorganism or a commensally present microorganism. Marine sponges are a particularly good reservoir and microorganisms can account for > 60% of the sponge’s wet weight [44]. Sponges can contain large and diverse bacterial populations, as reported on Aplysina aerophobia[45] and Rhopaloeides odorabile[46].
Through true sponge–microbe symbiosis, sponges may benefit from the provision of nutrients, transportation of waste products or active metabolites, chemical defense or contribution to mechanical structure. Recent studies of microbial communities associated with sponges have resulted in the identification of new marine microbes, novel natural compounds and host specificity [43,47,48].
3.1 Accessing sponge symbionts
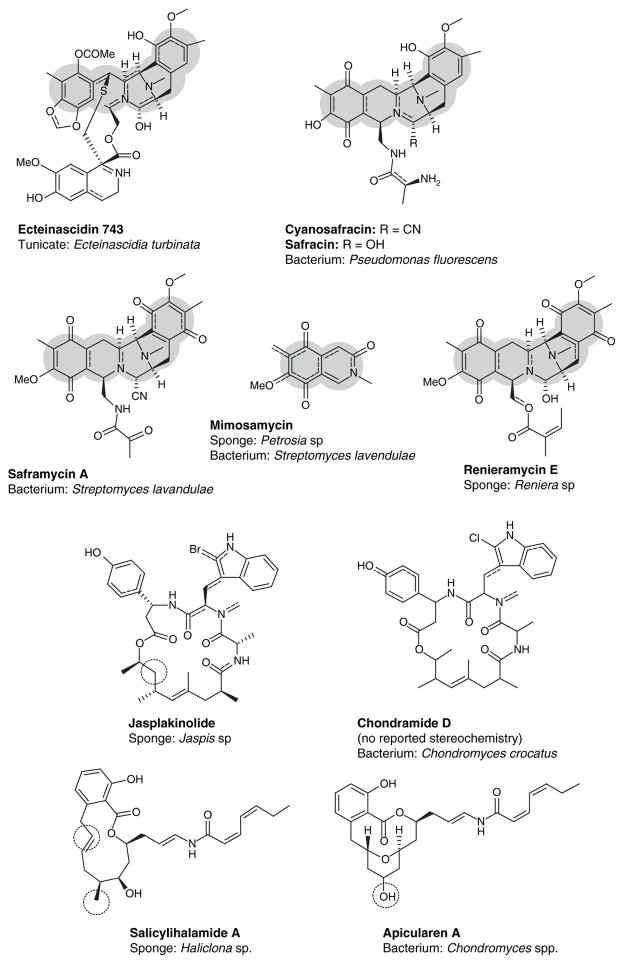
According to Hildebrand et al.[49], there are a number of criteria that should be met to prove the symbiotic origin of bioactive metabolites: i) the similarity of chemical structure between metabolites from microbes and marine animals (Figure 8); ii) the compounds should be localized in the host; iii) the associated microbes should be present persistently and be correlated with production of the bioactive compound; iv) the reduction or elimination of symbionts influences the production of metabolites; and v) the biosynthetic genes of the symbionts are present and can be detected.
Figure 8. The structures of marine natural products sharing a range of similarity to metabolites isolated from microorganisms.
Areas of gray highlight some structural similarities; dashed circles highlight areas of minor differences.
The long-term association and co-evolution of sponges and bacteria has resulted in a close linkage of the metabolic pathways in both symbiotic partners. The development of these relationships has been a major reason why many sponge-associated bacteria have not been cultured. In addition, even if the symbiont can be cultured outside the host, there can be additional problems in attempting to produce desired metabolites due to suboptimal conditions that inhibit their biosynthesis [47].
Studies on the production of bioactive compounds from sponge-associated microorganisms has used two general approaches: optimization of the in vitro culture conditions by extensive study of the microorganism’s environmental requirements and the identification and cloning of the genes that are used by the microorganism for biosynthesis [50].
The former can be especially complex due to the numerous factors that can influence the microorganism’s behavior, and in some cases the requirements may be beyond what can be achieved in a large scale setting. The more promising alternative is to clone the biosynthetic genes and transfer them from the natural host to a more productive host. This metagenomic approach provides genetic insights into the biology of bacteria that are not accessible with standard microbiological methods and at the same time makes the genetic information available, which is required to produce the compounds [43,47].
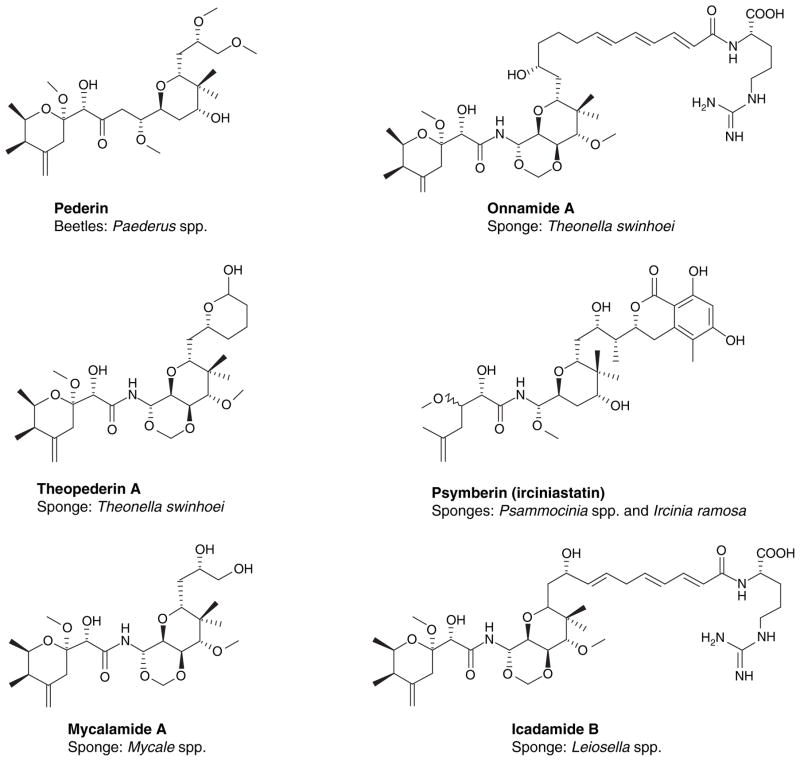
Metagenomics offers the possibility to directly access the genome of the complex marine ecosystem. The first isolation of a biosynthetic gene cluster from an invertebrate metagenome was reported for pederins (Figure 9) in the beetle Paederus fuscipes. The genes belong to uncultivatable symbiont related to Pseudomonas aeruginosa, but coincidentally, sequence homology was found in the sponge Theonella swinhoei[51]. Later, the biosynthetic gene cluster of onnamide A (Figure 9) was identified from metagenome of sponge Theonella swinhoei based on a homology approach [52]. Metagenomic analysis of the sponge Discodermolide dissoluta revealed a great diversity and a novel group of sponge-specific polyketide synthase (PKS) ketosynthase domains. The metagenomic libraries of bacterial genomes associated with this sponge were screened for PKS type I. This project was hampered by the fact that no closely related biosynthetic gene cluster had been reported; as a consequence, the homology approach was inapplicable. The isolated multimodular PKS gene cluster responsible for the biosynthesis of discodermolide was not detected. The diversity of the sponge–microbe community was eubacterial and the majority was γ-proteobacteria that were closely related to Entotheonella[53]. This report showed that isolation of microbial gene clusters from sponges is typically not a trivial task and that it is common for a single bacterium to contain more than one pathway for a particular secondary metabolite.
Figure 9.
The pederin class is isolated from diverse sources.
Haygood’s group provided evidence that bryostatin 1 and others are produced by an uncultured symbiont, a novel species of γ-proteobacteria named Candidatus Endobugula sertula[54–56]. The putative biosynthetic genes from Candidatus Endobugula sertula were identified and contained five modular polyketide synthase genes, a discrete acyltransferase, a β-ketosynthase, a hydroxyl-methyl glutaryl CoA synthase and a methyl transferase. Interestingly, the abovementioned bry genes share features with the pederin/onnamide clusters [57].
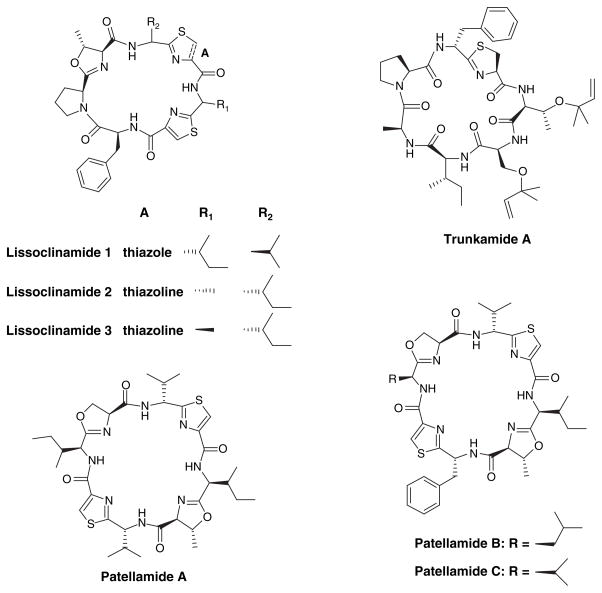
The ascidian genus Lissoclinum harbors both natural products and symbiotic cyanobacteria [58]. They produce cyclic peptides, such as lissoclinamides, trunkamide A and patellamides (Figure 10). Genome sequencing of Prochloron didemni isolated from Lissoclinum patella has shown that only a single non-ribosomal peptide synthase gene was identified and did not correlate with patellamide production. Surprisingly, patellamides are coded by a ribosomal peptide synthase [59].
Figure 10.
Cyclic peptides produced by microorganisms living in association with ascidians of the genus Lissoclinum.
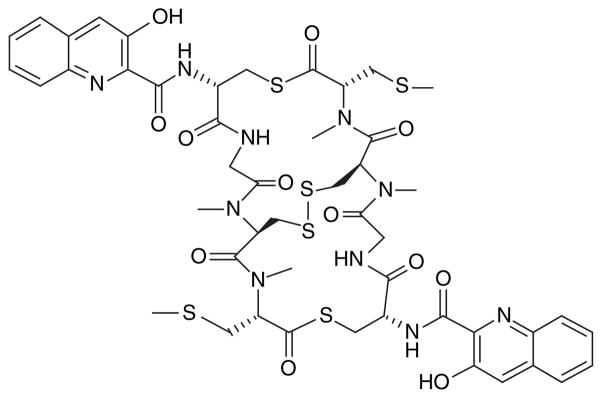
The successful isolation of the biosynthetic gene cluster must be followed by expression in a heterologous host. A number of microorganisms can be potential hosts for heterologous expression, including Escherichia coli, Streptomyces spp. and Pseudomonas putida. Although some suggest E. coli as a robust system [60], the organism may not be a perfect host in the expression of metagenome products from antibiotic gene clusters because the promoters of some high GC-content microbes fail to initiate transcription [61]. The gene cluster for production of patellamides A and C has been expressed in E. coli although the yield still needs improvement [59]. Due to their capacity as a prolific source of secondary metabolites, Streptomyces spp. can be an ideal heterologous host for expression of marine natural products. They are naturally equipped with antibiotic production machinery and, therefore, fulfill the task as a bioreactor [60]. The biosynthetic pathway of the antitumor compound thiocoraline (Figure 11) was expressed successfully in the heterologous hosts Streptomyces lividans and Streptomyces albus. Pseudomonas spp. have advantages in that they are widely distributed in the environment, P. putida is also well known as a producer of secondary metabolites [62].
Figure 11.
Antitumor compound thiocoraline has been expressed in heterologous hosts.
4. Creating clinical leads from bioactive natural products
As nature designs organic molecules for applications not involving humans, a bioactive natural product frequently has some therapeutic hurdles (i.e., toxicity, delivery) to overcome before the compound can be used for medicinal purposes. This could be more prevalent among compounds isolated from the marine environment because of the unique environmental factors involved in their biosynthesis [22,63]. Regardless, the discovery and development of marine natural products offers an excellent opportunity as many of these compounds have non-specific affinities for proteins, DNA, RNA or processes in which one or all of these targets are present. Such natural product scaffolds are now often described as ‘privileged structures’ from which libraries may be produced through semi- or total synthesis. Since Evans and others [64] coined this terminology, the perception of natural products chemistry has moved well beyond typical bioassay guided isolation. With the aid of synthetic and in silico technologies in development since the 1980s, bioactive natural products and their fully synthetic counterparts have collectively acted as a divining rod for biologically active molecular fragments [65].
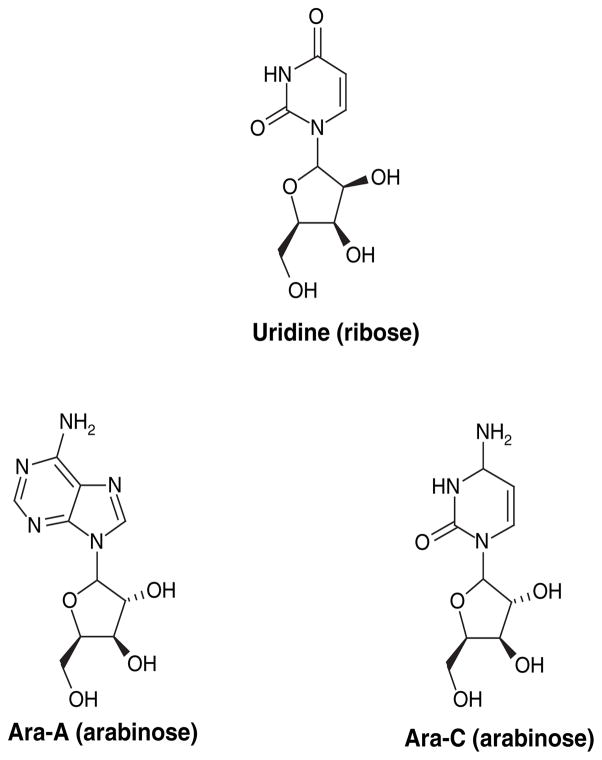
It is well established that synthetic modifications can play a big role in the improvement of pharmacokinetic and toxicity profiles for natural-product drug candidates. As a general example, a well written review by Rivkin, Chou and Danishefsky [66] provides a detailed account of the rational development of a second generation E-9,10-dehydroepothilone derivative with a significantly improved preclinical therapeutic index compared with its predecessors. One well-known class of marine natural product-inspired drugs is the non-traditional nucleosides, which incorporate sugar moieties other than ribose or deoxyribose. For several years, this class of compounds have been modified to target retro-viruses on the heels of the successful drug zidovudine [101]. According to Newman and Cragg’s review [6] and other sources [67,68], this breakthrough in AIDS treatment and other antileukemia and antiviral drugs, such as cytosine ara-binoside (Ara-C, Figure 12)[102] and adenine arabinoside (Ara-A, Figure 12)[103], may not have been discovered unless the then present dogma was changed by the isolation of the marine natural products spongothymidine and spongouridine.
Figure 12.
Spongothymidine and spongouridine inspire the development of synthetic antiviral and antileukemia drugs Ara-A and Ara-C based on non-traditional arabinonucleoside chemistry.
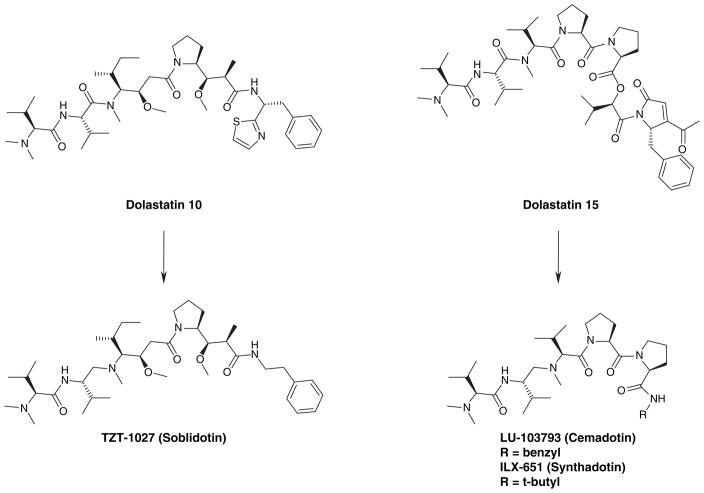
Several successful cases of FDA-approved, clinically-optimized analogs of terrestrial natural products exist. As the field of marine natural products matures and the ongoing clinical trials of marine-derived leads progress, further work will certainly accomplish similar success. At the moment, there are quite a few examples of synthetic marine natural product analogs that have produced leads at present in clinical trials [69]. Dolastatin 10 was originally isolated from a seahare, but later found in an associated cyanobacterium [70] and was highly promising as an antitumor lead until reported toxicity issues in 2004 reduced its clinical potential [71]. Dolastatin 15 was isolated from the same mollusc, but stalled in preclinical trials due to an inefficient total synthesis and poor water solubility [69]. Since those initial setbacks, an intensive structure–activity relationship study was undertaken and three synthetic analogs, TZT-1027 (soblidotin) [104], LU-103793 (cemadotin) [105] and ILX-651 (synthadotin) [106], emerged as clinical leads with improved potency and efficacy [72]. Halichondrin B was first isolated from a marine sponge of the genus Halichondria[73] and showed promising antimitotic activity, but its structural complexity hampered development of the natural product beyond the preclinical stage. During total synthetic studies [74], E7389 (norhalichondrin B) [107] was identified as a structurally simpler and equally potent analog of halichondrin B that is now in Phase II clinical trials [202]. Figures 13 and 14 show dolastatin 10 and 15, halichondrin B and their corresponding synthetic analogs highlighting their structural modifications. Other noteworthy examples of marine natural product synthetic analogs with improved potential are included in Table 2 with associated references made available for further details.
Figure 13.
Dolastatins 10 and 15 and improved analogs at present in clinical trials.
Figure 14.
E7389, a structurally simpler analog of Halichondrin B is now in Phase II clinical trials.
Table 2.
Marine natural product synthetic analogs with improved therapeutic potential over the original natural product.
| Compound | Derived from | Activity | Clinical status |
|---|---|---|---|
| GTS-21 [108] | Anabaseine [75] | Alzheimer nicotinic receptor antagonist | Phase II [76] |
| IPL-576-092 [109] | Contignasterol [77] | Antiasthmatic | Phase II (stalled) [78] |
| KRN-7000 [110] | Agelasphin-9b [79] | Immunomodulator (antitumor, hepatitis C) | Phase I [80] |
| PT-650 [111] | ET-743 [81] | Cytotoxicity | Pre-clinical (stalled) [82] |
4.1 Rationally guided modification of marine natural products
In some cases, the analogs produced from a particular core structure are numerous and structure–activity relationships are based solely on the chemist’s intuition. Thus, the design of synthetic analogs is dependent on the capacity of the chemist to account for this mass of information, but in silico rational design can now play a significant role in answering questions about the structural requirements for biological activity [83,84]. For marine natural product analogs, using computational studies to predict the most effective structural modifications for activity is especially important in cases where total synthesis is not efficient and natural supplies are low.
The design of product-based libraries typically rely on virtual semisynthetic approaches in combination with both liquid or solid-phase library synthesis and have assisted in the development of successful natural-product prodrugs [85]. Computationally demanding hybrid approaches attempt to merge the developing area of computational chemistry and traditional synthetic studies and have been used to generate compound libraries based on smaller building blocks or intermediate-size fragments [83]. In cases where bioactive marine natural products cannot be used clinically, the same compounds can be used as molecular probes of their target. A recent study of the marine-derived latrunculins used modeling of the actin binding site of latrunculin A (Figure 15) to examine requirements for its affinity. Information from this model was used in the design of analogs that were submitted for in vitro comparison with the parent; one analog produced through fewer synthetic steps had a relatively identical potency compared to latrunculin A [86].
Figure 15.
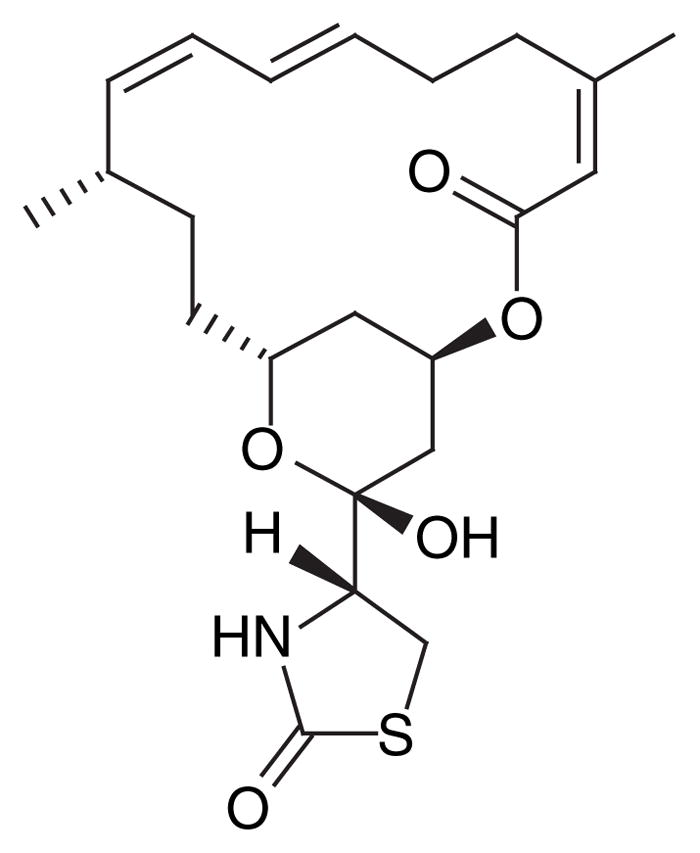
Actin-binding marine-derived latrunculin A is cytotoxic, but offers the opportunity for target optimization using in silico modeling.
4.2 Biosynthetic engineering of marine natural product analogs
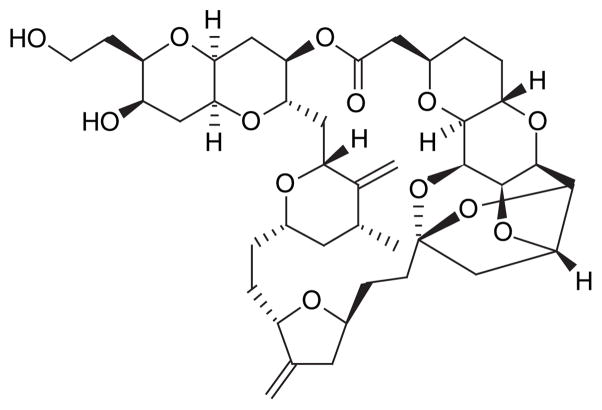
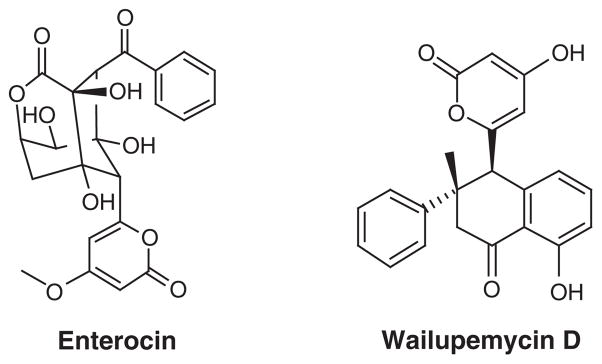
Marine microorganisms have been exploited for their in situ transformations of natural product scaffolds, where, in turn, isolation of the small quantities of metabolic products from the fermentation media proposes to produce either more bioactive or metabolically stable compounds [87]. Marine microorganisms that are responsible for the production of a particular compound can also be used for the production of analogs through engineering of the associated biosynthetic gene cluster. As mentioned earlier, the identification and isolation of the biosynthetic gene cluster for biosynthesis is particularly elusive, but the technology for genetic manipulation of those gene clusters is now available, allowing for the creation of what has been described as unnatural natural products [88] or more often referred to as the method of combinatorial biosynthesis [89]. Following the identification of the gene cluster responsible for enterocin (Figure 16) biosynthesis in the marine microorganism Streptomyces maritimus[90], Piel et al. engineered and expressed the gene cluster in S. lividans, producing enterocin, wailupemycin D (Figure 16) and several other known and new compounds. This group later went on to block benzoyl-CoA production in the same engineered strain and created a series of enterocin and wailupemycin analogs with precursor mimics [91]. Essentially, the method of combinatorial biosynthesis can be applied to an indefinite number of marine organisms and provides an alternative route to otherwise synthetically challenging derivatives of marine-derived drug leads.
Figure 16. Streptomyces maritimus.
is the marine bacteria responsible for producing enterocin and wailupemycin D.
5. Conclusion
In the recent data presented by Newman and Cragg [92], there has been a subtle decrease in the number of small-molecule new chemical entities from the mid-1980s to 2005, which could be correlated directly to the development of higher-throughput assays and the use of previously developed synthetic and natural product-based libraries. In the years following, combinatorial chemistry became a mainstream tool for building compound diversity and natural products took a metaphorical backseat, which may have negatively impacted the number of compounds that made it beyond clinical trials [93]. It is undeniable that nature has provided a bounty of chemistry in the world’s oceans; and only the present technology limits the development of this resource for new drugs.
As the previously mentioned new improvements in technology associated with collection, isolation and identification of new chemistry become commonly accustomed to scientists, the frequency of new leads identified is expected to increase [94]. Marine microorganisms [47] will certainly be of particular interest in the future, not only for their potential association with previously isolated natural products, but for their ability to produce new biologically active compounds. The promise of worms, bryozoans, molluscs, tunicates and smaller encrusting or slow growing sponges, which were considered either too small or too few in number to study thoroughly, will become accessible thanks to the reduction of the critical mass required for a compound’s structure and bioactivity determination.
6. Expert opinion
Although the approval of ziconotide is a major historical event in the eyes of marine natural-product chemists, it is yet to be determined if this instance of a truly marine-derived drug will cause a ripple effect for the entire pharmaceutical industry. Questions concerning sourcing still linger and until these issues are more consistently resolved through either a synthetic or a biotechnology-based approach, the throughput of a traditional natural products isolation approach will cause industry leaders to hesitate. On the other hand, the poor return for the two decades of combinatorial chemistry has highlighted the need to learn more from bioactive natural products. This emerging impetus to find novel bioactive compounds, coupled with the development of new technology to increase the throughput of isolation and identification of natural products, should further bolster the interest in the marine environment as well as future stories of success, such as that of ET-743. Based on the present knowledge of the potential diversity of marine macro- and microorganisms, the reported diversity of chemical structures represents a keyhole view of what remains to be discovered from this environment. Significant advances in the area of analytical chemistry, diving technology, molecular biology and marine microbiology are helping to improve the rate of drug discovery and development. Ideally, future work in natural products chemistry should aim to match the demands of high-throughput assays. The numbers may never match the power of purely synthetic techniques, but the effort will undoubtedly be fruitful in terms of chemical diversity.
Two technologies in particular will have dramatic impacts in this regard, capNMR probes and metagenomic analysis. CapNMR probes using either automated sample injectors or HPLC will dramatically reduce the handling and time required for identification of components in bioactive extracts from marine organisms. Essentially, HPLC fractions will be routinely screened for bioactivity and analyzed by NMR simultaneously. Although the technique of metagenomic analysis will likely encounter early difficulties associated with research on marine microorganisms, it will eventually provide access to biosynthetic chemistry that is relatively difficult to mimic with total synthesis and concurrently help address some drug supply issues.
Footnotes
Declaration of interest
The authors state no conflict of interest and have received no payment in preparation of this manuscript.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.CHENGKUI Z, TSENG CK, JUNFU Z, CHANG CF. Chinese seaweeds in herbal medicine. Hydrobiologia. 1984;116/117(1):152–154. [Google Scholar]
- 2.BERGMANN W, BURKE DC. Contributions to the study of marine products. XXXIX. The nucleosides of sponges. III. Spongothymidine and spongouridine. J Org Chem. 1955;20(11):1501–1507. [Google Scholar]
- 3.HORWITZ JP, CHUA J, NOEL M. Nucleosides. V. The monomesylates of 1-(2′-Deoxy-β-D-luxofuranosyl)thymine. J Org Chem. 1964;29(7):2076–2078. [Google Scholar]
- 4•.SOGIN ML, MORRISON HG, HUBER JA, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA. 2006;103(32):12115–12120. doi: 10.1073/pnas.0605127103. Updated estimation of the diversity of deep waters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5•.PETTIT GR, HERALD CL, SMITH CR. Biosynthetic Products for Cancer Chemotherapy. Vol. 6. Elseveir Scientific; Amsterdam: 1989. Early review describing the discovery of natural-product anticancer agents. [Google Scholar]
- 6••.NEWMAN DJ, CRAGG GM. Marine natural products and related compounds in clinical and advanced preclinical trials. J Nat Prod. 2004;67(8):1216–1238. doi: 10.1021/np040031y. Review of marine natural products in preclinical and clinical trials until 2003. [DOI] [PubMed] [Google Scholar]
- 7••.BLUNT JW, COPP BR, HU WP, et al. Marine natural products. Nat Prod Rep. 2007;24(1):31–86. doi: 10.1039/b603047p. Yearly review of isolated marine natural product structures in the literature. [DOI] [PubMed] [Google Scholar]
- 8•.HILL RA. Marine natural products. Ann Rep Prog Chem B Org Chem. 2007;103:125–139. Similar yearly review of marine derived structures in the literature. [Google Scholar]
- 9••.MORRIS JC, NICHOLAS GM, PHILLIPS AJ. Marine natural products: synthetic aspects. Nat Prod Rep. 2007;24(1):87–108. doi: 10.1039/b602832m. Review describing the total synthesis of marine natural products. [DOI] [PubMed] [Google Scholar]
- 10•.MOORE BS. Biosynthesis of marine natural products: microorganisms (Part A) Nat Prod Rep. 2005;22(5):580–593. doi: 10.1039/b404737k. Review series describing work with marine natural product biosynthesis. [DOI] [PubMed] [Google Scholar]
- 11•.MOORE BS. Biosynthesis of marine natural products: macroorganisms (Part B) Nat Prod Rep. 2006;23(4):615–629. doi: 10.1039/b508781n. Review series describing work with marine natural product biosynthesis. [DOI] [PubMed] [Google Scholar]
- 12.GARCIA-ROCHA M, BONAY P, AVILA J. The antitumoral compound kahalalide facts on cell lysosomes. Cancer Lett. 1996;99(1):43–50. doi: 10.1016/0304-3835(95)04036-6. [DOI] [PubMed] [Google Scholar]
- 13.CUADRADO A, GONZALEZ L, SUAREZ Y, MARTINEZ T, MUNOZ A. JNK activation is critical for aplidin-induced apoptosis. Oncogene. 2004;23(27):4673–4680. doi: 10.1038/sj.onc.1207636. [DOI] [PubMed] [Google Scholar]
- 14.KOS FJ, BEAR HD. Involvement of protein kinase C-δ in CD28-triggered cytotoxicity mediated by a human leukaemic cell line YT. Immunology. 1998;94(4):575–579. doi: 10.1046/j.1365-2567.1998.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MINUZZO M, CERIBELLI M, PITARQUE-MARTI M, et al. Selective effects of the anticancer drug Yondelis (ET-743) on cell-cycle promoters. Mol Pharmacol. 2005;68(5):1496–1503. doi: 10.1124/mol.105.013615. [DOI] [PubMed] [Google Scholar]
- 16.OLIVERA BM, CRUZ LJ, DE SANTOS V, et al. Neuronal calcium channel antagonists. Discrimination between calcium channel subtypes using w-conotoxin from Conus magus venom. Biochemistry. 1987;26(8):2086–2090. doi: 10.1021/bi00382a004. [DOI] [PubMed] [Google Scholar]
- 17.MAROUN JA, BELANGER K, SEYMOUR L, et al. Phase I study of aplidine in a daily × 5 1-h infusion every 3 weeks in patients with solid tumors refractory to standard therapy. A National cancer institute of Canada clinical trials group study: NCIC CTG IND 115. Ann Oncol. 2006;17(9):1371–1378. doi: 10.1093/annonc/mdl165. [DOI] [PubMed] [Google Scholar]
- 18.HAMANN MT. Technology evaluation: Kahalalide F, PharmaMar. Curr Opin Mol Ther. 2004;6(6):657–665. [PMC free article] [PubMed] [Google Scholar]
- 19.NEWMAN DJ. The Bryostatins. In: Cragg GM, et al., editors. Anticancer Agents from Natural Products. Taylor and Francis; Boca Raton, Florida, USA: 2005. pp. 137–150. [Google Scholar]
- 20.GROSSO F, JONES RL, DEMETRI GD, et al. Efficacy of trabectedin (ecteinascidin-743) in advanced pretreated myxoid liposarcomas: a retrospective study. Lancet Oncol. 2007;8(7):595–602. doi: 10.1016/S1470-2045(07)70175-4. [DOI] [PubMed] [Google Scholar]
- 21.ALONSO D, KHALIL Z, SATKUNANATHAN N, LIVETT BG. Drugs from the sea: conotoxins as drug leads for neuropathic pain and other neurological conditions. Mini Rev Med Chem. 2003;3(7):785–787. doi: 10.2174/1389557033487746. [DOI] [PubMed] [Google Scholar]
- 22.COSTANTINO V, FATTORUSSO E, MENNA M, TAGLIALATELA-SCAFATI O. Chemical diversity of bioactive marine natural products: an illustrative case study. Curr Med Chem. 2004;11(13):1671–1692. doi: 10.2174/0929867043364973. [DOI] [PubMed] [Google Scholar]
- 23.BHAKUNI DS, RAWAT DS. Bioactive Marine Natural Products. Springer; New York: 2005. p. xv.p. 382. [Google Scholar]
- 24.GROLS H, KONIG GM. Terpenoids from marine organisms: unique structures and their pharmacological potential. Phytochem Rev. 2006;5:115–141. [Google Scholar]
- 25.THIERICKE R. High throughput screening technologies. In: Hillisch A, et al., editors. Modern Methods of Drug Discovery. Birkhäuser Verlag; Basel, Boston, USA: 2003. pp. 71–85. [DOI] [PubMed] [Google Scholar]
- 26•.SCHROEDER HC, GRONQUIST M. Extending the scope of NMR spectroscopy with microcoil probes. Angew Chem Int Ed. 2006;45(43):7122–7131. doi: 10.1002/anie.200601789. Review of mass-sensitive NMR probe technology. [DOI] [PubMed] [Google Scholar]
- 27•.JANSMA A, CHUAN T, ALBRECHT RW, et al. Automated microflow NMR: routine analysis of five-microliter samples. Anal Chem. 2005;77(19):6509–6515. doi: 10.1021/ac050936w. Description of methods using capillary probes for high-throughput NMR analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DONG MW. Modern HPLC for Practicing Scientists. Wiley-Interscience; Hoboken, NJ, USA: 2006. p. 286. [Google Scholar]
- 29•.DE VILLIERS A, LESTREMAU F, SZUCS R, et al. Evaluation of ultra high performance liquid chromatography, part 1. possibilities and limitations. J Chromatogr A. 2006;1127:60–69. doi: 10.1016/j.chroma.2006.05.071. Review of the ultra-high-performance liquid chromatography system. [DOI] [PubMed] [Google Scholar]
- 30•.JAROSZEWSKI JW. Hyphenated NMR methods in natural products research, part 2: HPLC-SPE-NMR and other new trends in NMR hyphenation. Planta Med. 2005;71(9):795–802. doi: 10.1055/s-2005-873114. Description of a popular hyphenated technique using NMR. [DOI] [PubMed] [Google Scholar]
- 31.CORCORAN O, SPRAUL M. LC-NMR-MS in drug discovery. Drug Discov Today. 2003;8(14):624–631. doi: 10.1016/s1359-6446(03)02749-1. [DOI] [PubMed] [Google Scholar]
- 32.LEE MS, KERNS EH. LC/MS applications in drug development. Mass Spectr Rev. 1999;18(3–4):187–279. doi: 10.1002/(SICI)1098-2787(1999)18:3/4<187::AID-MAS2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 33.DAVIS RG, ANDEREGG RJ, BLANCHARD SG. Iterative size-exclusion chromatography coupled with liquid chromatographic mass spectrometry to enrich and identify tight-binding ligands from complex mixtures. Tetrahedron. 1999;55(39):11653–11667. [Google Scholar]
- 34•.PENG J, WALSH K, WEEDMAN V, et al. The new bioactive diterpenes cyanthiwigins E-AA from the Jamaican sponge Myrmekioderma styx. Tetrahedron. 2002;58(39):7809–7819. This study demonstrates chemical productivity in sponges collected from deep water. [Google Scholar]
- 35.RASKOFF KA, MATSUMOTO GI. Stellamedusa ventana, a new mesopelagic scyphomedusa from the eastern Pacific representing a new subfamily, the Stellamedusinae. J Mar Biol Assoc UK. 2004;84(1):37–42. [Google Scholar]
- 36.MENDOLA D, LOZANO SAN, DUCKWORTH AR, OSINGA R. The promise of aquaculture for delivering sustainable supplies of new drugs from the sea: examples from in sea and tank-based invertebrate culture projects from around the world. In: Proksch P, Müller WEG, editors. Frontiers in Marine Biotechnology. Horizon Bioscience; Norfolk, UK: 2006. pp. 72–105. [Google Scholar]
- 37•.SCHAUFELBERGER DE, KOLECK MP, BEUTLER JA. The large-scale isolation of bryostatin 1 from Bugula neritina following current good manufacturing practices. J Nat Prod. 1991;54(5):1265–1270. doi: 10.1021/np50077a004. Overview of early effort to produce a highly pure bioactive marine natural product in large scale. [DOI] [PubMed] [Google Scholar]
- 38•.SIPKEMA D, OSINGA R, SCHATTON W, et al. Large scale production of pharmaceutical by marine sponge: sea, cell or synthesis? Biotech Bioeng. 2005;90(2):201–222. doi: 10.1002/bit.20404. This paper discusses the issue of drug supply in marine natural products. [DOI] [PubMed] [Google Scholar]
- 39.HART JB, LILL RE, HICKFORD SJH, BLUNT JW, MUNRO MHG. The Halicondrins: chemistry, biology, supply and delivery. In: Fusetani N, editor. Drugs from the Sea. Karger; Basel, Switzerland: 2000. pp. 134–153. [Google Scholar]
- 40•.MILJANICH GP. Ziconotide: neuronal calcium channel blocker for treating severe chronic pain. Curr Med Chem. 2004;11(23):3029–3040. doi: 10.2174/0929867043363884. This paper discusses the discovery and synthesis of ziconotide. [DOI] [PubMed] [Google Scholar]
- 41.MICKEL SJ. Total synthesis of the marine natural product (+)-discodermolide in multigram quantities. Pure Appl Chem. 2007;79(4):685–700. [Google Scholar]
- 42.CUEVAS C, PEREZ M, MARTIN MJ, et al. Synthesis of ecteinascidin ET-743 and phthalascidin Pt-650 from cyanosafracin B. Org Lett. 2000;2(16):2545–2548. doi: 10.1021/ol0062502. [DOI] [PubMed] [Google Scholar]
- 43.TAYLOR MW, RADAX R, STEGER D, WAGNER M. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Bio Rev. 2007;71(2):295–347. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.WILKINSON CR. Microbial association in sponges. II. Numerical analysis of sponge and water bacterial population. Mar Biol. 1978;49(2):169–176. [Google Scholar]
- 45.HENTSCHEL U, HOPKE J, HORN M, et al. Molecular evidence for uniform microbial community in sponges from different oceans. Appl Environ Microbiol. 2002;68(9):4431–4440. doi: 10.1128/AEM.68.9.4431-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.WEBSTER NS, WILSON KJ, BLACKALL LL, HILL RT. Phylogenetic diversity of bacteria associated with the marine spongeRhopaloides odorabile. Appl Environ Microbiol. 2001;67(1):434–444. doi: 10.1128/AEM.67.1.434-444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.SALOMON CE, MAGARVEY NA, SHERMAN DA. Merging the potential of microbial genetics with biological and chemical diversity: an even brighter future for marine natural product drug discovery. Nat Prod Rep. 2004;21(1):105–121. doi: 10.1039/b301384g. [DOI] [PubMed] [Google Scholar]
- 48••.NEWMAN DJ, HILL RT. New drugs from marine microbes: the tide is turning. J Ind Microbiol Biotech. 2006;33(7):539–544. doi: 10.1007/s10295-006-0115-2. Review of bioactive compounds from marine microorganisms. [DOI] [PubMed] [Google Scholar]
- 49••.HILDEBRAND M, WAGGONER LE, LIM GE, et al. Approaches to identify clone and express symbiont bioactive metabolite genes. Nat Prod Rep. 2004;21(1):122–142. doi: 10.1039/b302336m. Review of the metagenomic approach to marine natural products. [DOI] [PubMed] [Google Scholar]
- 50.MÜLLER WEG, GREBENJUK VA, PENNEC GL, et al. Sustainable production of bioactive compounds by sponge-cell culture and gene cluster approach. Mar Biotech. 2004;6(2):105–117. doi: 10.1007/s10126-002-0098-6. [DOI] [PubMed] [Google Scholar]
- 51••.PIEL J, BUTZKE D, FUSETANI N, et al. Exploring the chemistry of uncultivated bacterial symbionts: antitumor polyketides of pederin family. J Nat Prod. 2005;68(3):472–479. doi: 10.1021/np049612d. This article describes methods to explore previously uncultivatible marine microorganisms. [DOI] [PubMed] [Google Scholar]
- 52••.PIEL J, HUI D, WEN G, et al. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of marine sponge Theonella swinhoei. Proc Natl Acad Sci USA. 2004;101(46):16222–16227. doi: 10.1073/pnas.0405976101. This article describes work towards isolation of the gene cluster producing onnamide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.SCHIRMER A, GADKARI R, REEVES CD, et al. Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissolute. Appl Environ Microbiol. 2005;71(8):4840–4849. doi: 10.1128/AEM.71.8.4840-4849.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.HAYGOOD MG, DAVIDSON SK. Small-subunit rRNA genes and in situ hybridization with oligonucleotides specific for bacterial symbionts in the larvae of the bryozoan Bugula neritina and proposal of “Candidatus Endobugula sertula”. Appl Environ Microbiol. 1997;63(11):4612–4616. doi: 10.1128/aem.63.11.4612-4616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DAVIDSON SK, ALLEN SW, LIM GE, ANDERSON C, HAYGOOD MG. Evidence for the biosynthesis of bryostatins by the bacterial symbiont Candidatus Endobugula sertula of the bryozoan Bugula neritina. Appl Environ Microbiol. 2001;67(10):4531–4537. doi: 10.1128/AEM.67.10.4531-4537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.HILDEBRAND M, WAGGONER LE, LIU H, et al. bryA: an usual modular polyketide synthase gene from the uncultivated bacterial symbiont of the marine bryozoan Bugula neritina. Chem Biol. 2004;11(11):1543–1552. doi: 10.1016/j.chembiol.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 57.SUDEK S, LOPANIK NB, WAGGONER LE, et al. Identification of the putative bryostatin polyketide synthase gene cluster from Candidatus Endobugula sertula, the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J Nat Prod. 2007;60(1):67–74. doi: 10.1021/np060361d. [DOI] [PubMed] [Google Scholar]
- 58.SCHMIDT EW, SUDEK S, HAYGOOD MG. Genetic evidence support secondary metabolic diversity in Prochloron spp., the cyanobacterial symbiont of tropical ascidian. J Nat Prod. 2004;67(8):1341–1345. doi: 10.1021/np049948n. [DOI] [PubMed] [Google Scholar]
- 59••.SCHMIDT E, NELSON JT, RASKO DA, et al. Patellamide A and C biosynthesis by microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci USA. 2005;102(20):7315–7320. doi: 10.1073/pnas.0501424102. Isolation of a ribosomal polyketide synthase biosynthetic gene cluster. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60•.WATANABE K, OIKAWA H. Robust platform for de novo production of heterologous polyketides and non-ribosomal peptides in Escherichia coli. Org Biomol Chem. 2007;5(4):593–602. doi: 10.1039/b615589h. Justification to use E. coli for metagenomic approaches. [DOI] [PubMed] [Google Scholar]
- 61.GUSTAFSSON C, GOVINDARAJAN S, MINSHULL J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22(7):346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 62.LI X, QIN L. Metagenomics-based drug discovery and marine microbial diversity. Trend Microbiol. 2005;23:539–543. doi: 10.1016/j.tibtech.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 63.SENNETT SH. Marine chemical ecology: applications in marine biomedical prospecting. In: McClintock JB, Baker BJ, editors. Marine Chemical Ecology. CRC Press; Boca Raton, Florida, USA: 2001. pp. 523–542. [Google Scholar]
- 64•.EVANS BE, RITTLE KE, BOCK MG, et al. Methods for drug discovery: development of potent, selective, orally effective cholecystokinin antagonists. J Med Chem. 1988;31(12):2235–2246. doi: 10.1021/jm00120a002. Original paper defining the term privileged structure. [DOI] [PubMed] [Google Scholar]
- 65.COSTANTINO L, BARLOCCO D. Privileged structures as leads in medicinal chemistry. Curr Med Chem. 2006;13(1):65–85. [PubMed] [Google Scholar]
- 66••.RIVKIN A, CHOU T-C, DANISHEFSKY SJ. On the remarkable antitumor properties of fludelone: how we got there. Angew Chem Int Ed. 2005;44(19):2838–2850. doi: 10.1002/anie.200461751. Review of a successful example of rational natural product drug design. [DOI] [PubMed] [Google Scholar]
- 67.NEWMAN DJ, CRAGG GM, SNADER KM. The influence of natural products upon drug discovery. Nat Prod Rep. 2000;17(3):215–234. doi: 10.1039/a902202c. [DOI] [PubMed] [Google Scholar]
- 68.SUCKLING CJ. Chemical approaches to the discovery of new drugs. Sci Prog. 1991;75(298 Part 3–4):323–359. [PubMed] [Google Scholar]
- 69.RAWAT DS, JOSHI MC, JOSHI P, ATHEAYA H. Marine peptides and related compounds in clinical trial. Anticancer Agents Med Chem. 2006;6:33–40. doi: 10.2174/187152006774755519. [DOI] [PubMed] [Google Scholar]
- 70.LUESCH H, MOORE RE, PAUL VJ, et al. Isolation of dolastatin 10 from the marine cyanobacterium Symploca species VP642 and total stereochemistry and biological evaluation of its analogue symplostatin 1. J Nat Prod. 2001;64(7):907–910. doi: 10.1021/np010049y. [DOI] [PubMed] [Google Scholar]
- 71.VON MEHREN M, BALCERZAK SP, KRAFT AS, et al. Phase II trial of dolastatin-10, a novel anti-tubulin agent, in metastatic soft tissue sarcomas. Sarcoma. 2004;8(4):107–111. doi: 10.1155/2004/924913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.FLAHIVE E, SRIRANGAM J. The dolastatins: novel antitumor agents from Dolabella aricularia. In: Cragg GM, et al., editors. Anticancer Agents from Natural Products. Taylor and Francis; Boca Raton, Florida, USA: 2005. pp. 191–213. [Google Scholar]
- 73.HIRATA Y, UEMURA D. Halichondrins – antitumor polyether macrolides from a marine sponge. Pure Appl Chem. 1986;58(5):701–710. [Google Scholar]
- 74.TOWLE MJ, SALVATO KA, BUDROW J, et al. In vitro and in vivo anticancer activities of synthetic macrocyclic ketone analogues of halichondrin B. Cancer Res. 2001;61(3):1013–1021. [PubMed] [Google Scholar]
- 75.KEM WR. Alzheimer’s drug design based upon an invertebrate toxin (anabaseine) which is a potent nicotinic receptor agonist. Invert Neurosci. 1997;3(2/3):251–259. doi: 10.1007/BF02480382. [DOI] [PubMed] [Google Scholar]
- 76.CHEN L, YAMADA K, NABESHIMA T, SOKABE M. α7 Nicotinic acetylcholine receptor as a target to rescue deficit in hippocampal LTP induction in β-amyloid infused rats. Neuropharmacology. 2006;50(2):254–268. doi: 10.1016/j.neuropharm.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 77.BURGOYNE DL, ANDERSEN RJ, ALLEN TM. Contignasterol, a highly oxygenated steroid with the unnatural 14 β configuration from the marine sponge Petrosia contignata. Thiele, 1899. J Org Chem. 1992;57(2):525–528. [Google Scholar]
- 78.KEYZERS RA, DAVIES-COLEMAN MT. Anti-inflammatory metabolites from marine sponges. Chem Soc Rev. 2005;34(4):355–365. doi: 10.1039/b408600g. [DOI] [PubMed] [Google Scholar]
- 79.NATORI T, MORITA M, AKIMOTO K, KOEZUKA Y. Agelasphins, novel antitumor and immunostimulatory cerebrosides from the marine sponge Agelas mauritianus. Tetrahedron. 1994;50(9):2771–2784. [Google Scholar]
- 80.ISHIKAWA A, MOTOHASHI S, ISHIKAWA E, et al. A Phase I study of α-galactosylceramide (KRN7000)-pulsed dendritic cells in patients with advanced and recurrent non-small cell lung cancer. Clin Cancer Res. 2005;11(5):1910–1917. doi: 10.1158/1078-0432.CCR-04-1453. [DOI] [PubMed] [Google Scholar]
- 81.RINEHART KL. Antitumor compounds from tunicates. Med Res Rev. 2000;20(1):1–27. doi: 10.1002/(sici)1098-1128(200001)20:1<1::aid-med1>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 82.GONZALEZ JF, SALAZAR L, DE LA CUESTA E, AVENDANO C. Synthesis of phthalascidin analogs. Tetrahedron. 2005;61(31):7447–7455. [Google Scholar]
- 83.STAHURA F, BAJORATH J. Computational analysis of natural molecules and strategies for the design of natural product-based compound libraries. In: Boldi AM, editor. Combinatorial Synthesis of Natural Product Based Libraries. Taylor & Francis; New York, USA: 2006. pp. 53–64. [Google Scholar]
- 84.WEI DQ, AL E. Theoretical studies of alzheimer’s disease drug candidate 3-[(2,4-dimethoxy)benzylidene]-anabaseine (GTS-21) and its derivatives. Biochem Biophys Res Commun. 2005;338(2):1059–1064. doi: 10.1016/j.bbrc.2005.10.047. [DOI] [PubMed] [Google Scholar]
- 85••.BERTELS S, FRORMANN S, JAS G, BINDSEIL KU. Synergistic use of combinatorial and natural product chemistry. In: Grabley S, et al., editors. Drug Discovery from Nature. Springer; New York, USA: 1999. pp. 72–105. This paper describes the use of natural products for developing synthetic libraries. [Google Scholar]
- 86.FURSTNER A, KIRK D, FENSTER MDB, et al. Diverted total synthesis: preparation of a focused library of latrunculin analogues and evaluation of their actin–binding properties. Proc Natl Acad Sci USA. 2005;102(23):8103–8108. doi: 10.1073/pnas.0501441102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.HAMANN MT. Enhancing marine natural product structural diversity and bioactivity through semisynthesis and biocatalysis. Curr Pharm Des. 2003;9(11):879–889. doi: 10.2174/1381612033455297. [DOI] [PubMed] [Google Scholar]
- 88.MOORE BS, KALAITZIS JA, XIANG L. Exploiting marine actinomycete biosynthetic pathways for drug discovery. Antonie Van Leeuwenhoek. 2005;87(1):49–57. doi: 10.1007/s10482-004-6541-0. [DOI] [PubMed] [Google Scholar]
- 89•.GALM U, SHEN B. Expression of biosynthetic gene clusters in heterologous hosts for natural product production and combinatorial biosynthesis. Expert Opin Drug Discov. 2006;1(5):409–437. doi: 10.1517/17460441.1.5.409. The article summarizes the effect of metagenomics technology on marine natural products research. [DOI] [PubMed] [Google Scholar]
- 90•.PIEL J, HOANG K, MOORE BS. Natural metabolic diversity encoded by the enterocin biosynthesis gene cluster. J Am Chem Soc. 2000;122(22):5415–5416. Early example of combinatorial biosynthesis of marine natural products. [Google Scholar]
- 91•.KALAITZIS JA, IZUMIKAWA M, XIANG L, et al. Mutasynthesis of enterocin and wailupemycin analogues. J Am Chem Soc. 2003;125(31):9290–9291. doi: 10.1021/ja035973o. Another related example of combinatorial biosynthesis of marine natural products. [DOI] [PubMed] [Google Scholar]
- 92••.NEWMAN DJ, CRAGG GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70(3):461–477. doi: 10.1021/np068054v. Updated review of clinically approved natural products drugs. [DOI] [PubMed] [Google Scholar]
- 93••.LAHANA R. How many leads from HTS? Drug Discov Today. 1999;4(10):447–448. doi: 10.1016/s1359-6446(99)01393-8. The article describes reasons for the poor performance of high-throughput screening. [DOI] [PubMed] [Google Scholar]
- 94.KOEHN FE, CARTER GT. The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 2005;4(3):206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
Patents
- 101.WELLCOME FOUNDATION LTD. 199451. EP. 1986
- 102.UPJOHN CO. 3512. FRM. 1965
- 103.ICN PHARMACEUTICALS, INC. 2413226. DE. 1974
- 104.TEIKOKU HORMONE MFG. CO., LTD. 9303054. WO. 1993
- 105.BASF AKTIENGESELLSCHAFT. 5831002. US. 1998
- 106.AEGERA THERAPEUTICS, INC. 2005042030. WO. 2005
- 107.HARVARD COLLEGE. 9317690. WO. 1993
- 108.UNIVERSITY OF FLORIDA. 9215306. WO. 1992
- 109.INFLAZYME PHARMACEUTICALS LTD. 9802450. WO. 1998
- 110.BREWERY KIRIN. 9402168. WO. 1994
- 111.HARVARD COLLEGE. 2000018233. WO. 1999
Websites
- 201.http://www.pharmamar.com/en/press Pharmamar website press release (2007).
- 202.http://www.eisai.com/pipeline.asp?ID=173 Eisai website pipeline (2007).