Abstract
Recent initiatives to develop more effective and affordable drugs, controlling mosquitoes and development of a preventative vaccine have been launched with the goal of completely eradicating malaria. To this end, Novartis (Surrey, UK) and GlaxoSmithKline (Middlesex, UK) screened their chemical libraries of approximately two million small molecules for antimalarial properties, which resulted in a set of over 20,000 ‘highly druggable’ initial hits. Efforts in academia are centered on specific pathway targets. One such high-throughput screening effort has been focused on hemozoin formation, a unique heme detoxification pathway found in the malaria parasite. This review discusses the current approaches and limitations of high-throughput screening discovery of hemozoin inhibitors. In the future, new methods must be developed to validate the mechanism of action of these hit compounds within the parasite.
Malaria burden
Malaria is preventable and curable, yet it remains one of the world’s deadliest diseases. Approximately half of the world’s population is at risk from contracting malaria, especially young children and pregnant women in Sub-Saharan Africa [1]. In addition, there is a strong correlation between malaria endemic regions and world poverty. A vast majority of malaria victims live in the poorest regions of the world, where the cost of prevention and treatment can consume 30% of a family’s income [2]. The protozoan, Plasmodium falciparum, is the primary causative agent of this infectious disease in humans, which resulted in 200–300 million cases and 655,000 deaths in 2010 [1]. While this disease has been eradicated from the USA, it still remains a major public health concern throughout more tropical regions.
In the past decade, there has been a re-energized effort to develop antimalarial treatments. These include strategies to replenish the drug pipeline with new small-molecule therapeutics, develop combination drug therapies against resistance and realize an effective vaccine. These efforts have been largely driven by new public– private partnerships, such as the Medicines for Malaria Venture, the Roll Back Malaria Campaign, the President’s Global Health Initiative and the Bill and Melinda Gates Foundation. These public–private partnerships have registered a number of new antimalarials based on combination therapies of older mainline treatments (e.g., Eurartesim® and Pyramax®) [3]. Given the relative maturity of the malaria drug portfolio, there has been a shortage of new chemical entities entering the malaria drug pipeline. There are, however, signs that new collaborative approaches to discovery may soon pay significant dividends.
Parasite life cycle
The malaria parasite is transmitted between hosts through the female Anopheles mosquito [1]. Following a blood meal, the mosquitoes deposit sporozoites from their salivary glands. These sporozoites invade host hepatocytes where they begin the next parasitic stage [4]. Here, the parasite develops into hepatic schizonts and eventually merozoites, which are released into the bloodstream, initiating the intraerythrocytic stage [5]. It is during this stage of infection that erythrocytes are destroyed and parasitic toxic waste is discharged, causing the characteristic symptoms of fever and chills in the victims. Once the merozoite enters the red blood cell it begins to mature into a ring form, continues to grow to a trophozoite, and finally differentiates into an erythrocytic schizont before the red blood cell ruptures. For each originally infected erythrocyte, approximately 20 new merozoites are then expelled into the bloodstream and remain there until they encounter another erythrocyte to invade, allowing infection to continue [6]. Throughout this process, a few of the merozoites enter into the sexual parasite stage where gametocytes are formed. The gametocytes continue to circulate within the bloodstream until they are taken up by a mosquito through a blood meal and can then undergo fertilization and maturation within the parasite before being transmitted to another human host. A majority of the antimalarials currently on the market target the parasite specifically in the intraerythrocytic stage [5]. However, if malaria is to be completely eradicated, novel drugs must be developed that target all three (hepatic, erythrocytic and gametocyte) stages.
Heme detoxification pathway
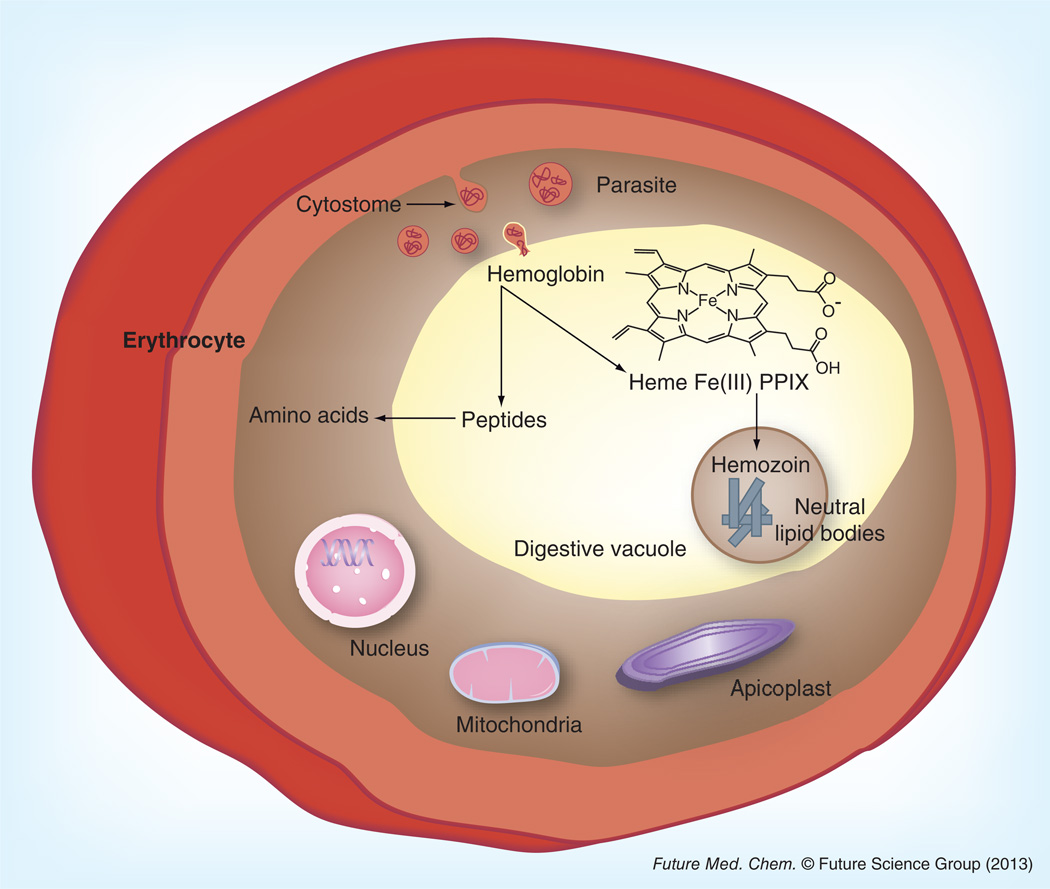
During the trophozoite stage of the intraerythrocytic life cycle, P. falciparum ingests up to 80% of the host hemoglobin through a protozoan, phagocytic organelle known as the cytostome (Figure 1) [7]. The cytostome then transports hemoglobin into an acidic digestive vacuole. Here, it is broken down by proteolytic enzymes in an ordered catabolic process into small peptides to be used as nutrients by the parasite [8]. Consequently, for every molecule of hemoglobin that is consumed, four molecules of heme (ferriprotoporphyrin IX, [Fe(III)PPIX]) are released. Due to the high toxicity of free heme, organisms must rapidly convert this molecule into an inert form, many through the enzyme heme oxygenase [9,10]. Hematophagous organisms, such as the Plasmodium, Schistosoma and Boophilus species, do not contain any functional heme oxygenase activity [10]. Instead, they must utilize a unique pathway to crystalize heme into a nontoxic biomineral, known as hemozoin [10]. Similar to the tight regulation of intracellular heme levels in vertebrates, these parasites do not tolerate high levels of free heme without harmful effects [11]. Thus, it is likely that both the catabolism and crystallization of heme must occur at comparable rates to rid the cell from any harm. Egan et al. found the kinetics for the conversion of heme to hemozoin to be so fast that the fraction of toxic Fe(III)PPIX would never exceed 1% of the total heme found in the parasite, even if hemoglobin degradation occurred at a constant rate [12].
Figure 1. Proposed representation of hemozoin formation within the intraerythrocytic life cycle of Plasmodium falciparum.
Host hemoglobin is taken up by the parasite and transported to the digestive vacuole through the cytostome. In this acidic organelle the hemoglobin is digested into small peptides and four toxic heme units (ferriprotoporphyrin IX). Neutral lipid bodies mediate the detoxification of the heme byproduct through the formation of hemozoin.
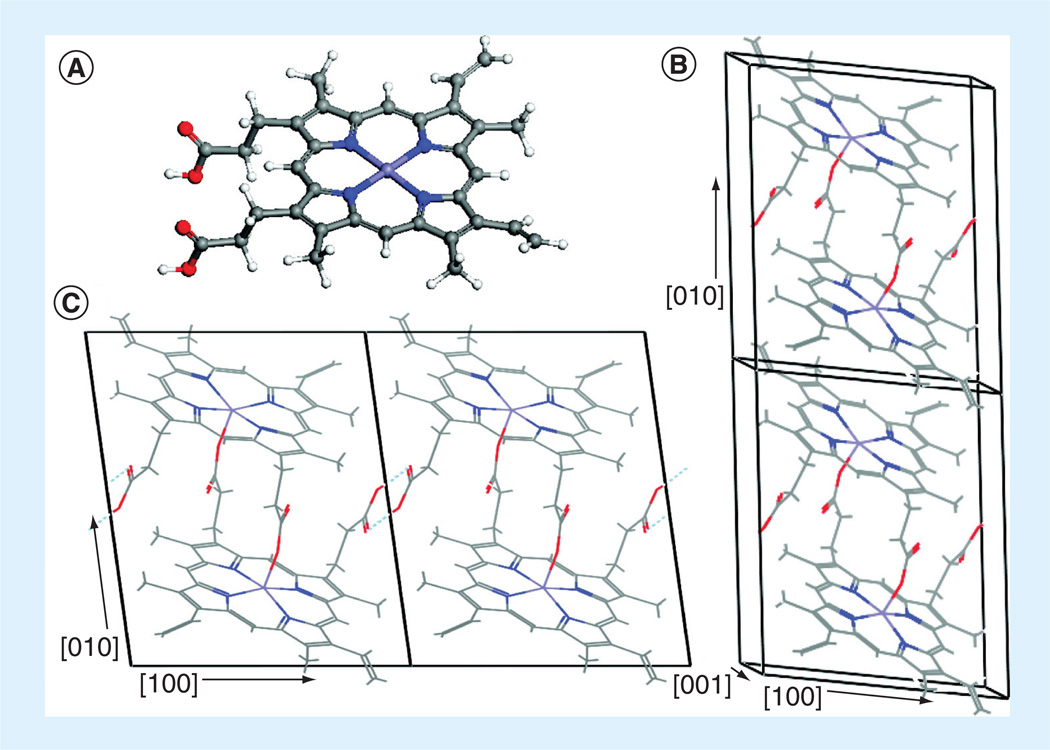
Hemozoin is a biologically unique dimer of five coordinate Fe(III)PPIX linked by reciprocating monodentate carboxylate linkages from one of the protoporphyrin IX’s propionate moieties. The biomineral is composed of an extended network of these dimeric units hydrogen bonded together via the second propionic acid group of protoporphyrin IX (Figure 2) [13]. More recently, the crystal structure of purified Plasmodium hemozoin was solved using powder x-ray diffraction, revealing that these cross-linked dimers form a network of sheets with 11.0 Å thickness [14]. The sheets are held together through π–π interactions, causing iron atoms to be partially exposed to the solvent, which allows for small molecule and ligand interaction with the metal. The formation of such an insoluble biomineral sequesters the bulk of the reactive iron, preventing any deleterious reactions. Disruption of this process has been demonstrated to be a prime target for antimalarial drugs, primarily since hemozoin is unique to the parasite [8].
Figure 2. β-hematin molecular crystal (only the unit cell of the major phase is illustrated).
(A) The Fe(III)PPIX (heme) monomer, (B) stacking of the Fe(III)PPIX dimers along the [010] direction, and (C) the hydrogen bond between the free propionic acid groups.
In some of the illustrations, hydrogen atoms are not illustrated for clarity [13]. Blue: Nitrogen; Gray: Carbon; Red: Oxygen; Violet: Iron; White: Hydrogen. This figure can be viewed in full color at: www.future-science.com/doi/full/10.4155/FMC.13.113
For the past 20 years, investigators have pondered the mechanism of formation of the unique heme crystallite, hemozoin. Theories include enzyme-catalyzed heme polymerases [15], proteins (specifically histidine-rich protein and heme detoxification protein) [16,17] and lipid mediation [18], or a combination of the two [19]. However, the most recent data suggests that neutral lipids are sufficient to effectively mediate the formation of hemozoin. One piece of evidence to support this finding is the discovery of neutral lipids present in the digestive vacuoles of early trophozoite stage parasites [20]. Analysis of transmission electron micrographs illustrated these lipid nanospheres surrounding hemozoin crystals, providing strong evidence that hemozoin formation is a lipid-mediated process [20]. Through ESI–MS/ MS, the composition of the lipid nanospheres was identified as a specific blend of mono- and diglycerols: monostearic, monopalmitic, dipalmitic, dioleic and dilinoleic glycerol in a specific 4:2:1:1:1 ratio [20]. In the presence of the lipid blend under physiologically relevant conditions, β-hematin (abiological hemozoin) formation was found to have a half-life of approximately 2 mins [21]. Hoang et al. demonstrated the partitioning of Fe(III)PPIX into neutral lipid droplets through the quenching of a lipid-specific fluorescent probe (nile red) following the addition of heme [22]. The in vitro kinetics of β-hematin formation mediated by neutral lipids is kinetically competent to handle the necessary flux of monomeric Fe(III) PPIX to prevent it from reaching toxic levels physiologically. While heme begins to localize and accumulate within this parasitic organelle, the acidic environment containing neutral lipid bodies allows for the rapid detoxification of this lethal molecule. The requirement for lipids in this process is exemplified through favored formation of the iron (III)–carboxylate bond in a hydrophobic environment [23]. Lipid mediation of hemozoin formation was also found to be more efficient than autocatalysis, as the digestive vacuole membrane could serve as a scaffold or nucleation site for the growing crystal [24]. Furthermore, Kapishnikov et al. observed crystals lying on their long axis along the digestive vacuole membrane, an unlikely orientation unless the lipid membrane was involved in crystal growth nucleation [25]. These results not only provide the criteria for lipid-mediated formation of hemozoin, but also critical information for the development of approaches to prevent this detoxification from occurring.
In the 1940s, chloroquine became the most widely used antimalarial drug and continued to be effective against P. falciparum for the next 40 years, until resistance spread throughout all endemic regions [26]. Despite this resistance and diminished efficacy of quinine and its derivatives, hemozoin remains a suitable target for antimalarial drugs. Resistance to these drugs arises from mutations or changes in expression levels in two membrane proteins found in the digestive vacuole, namely the P. falciparum chloroquine-resistant transporter protein (PfCRT) [27] and a homolog of P-glycoprotein (Pf PGH1) in the case of mefloquine [28]. These proteins are believed to reduce drug concentrations in the digestive vacuole, as PfCRT appears to actively or passively move chloroquine out of this compartment [29,30]. The putative target of these drugs (Fe[III]PPIX) arises from the host and is not under genetic control of the parasite, rather it is a chemically fixed structure and is invariantly present in large amounts in the environment in which the parasite lives. Thus, the parasite would appear to have little choice but to produce hemozoin, making it a unique drug target pathway.
Antimalarial hemozoin inhibitors
In the 1600s, an extract of the Cinchona bark was found to contain antimalarial properties [31]. This natural product extract, known as quinine, inspired numerous antimalarials, including chloroquine and halofantrine, which have been demonstrated to act as hemozoin inhibitors. Chloroquine, once the most successful antimalarials, is a neutral, weak base that is distributed evenly throughout the cytoplasm, but accumulates within the acidic digestive vacuole through an ion-trapping mechanism [26]. Upon entering the acidic environment, chloroquine is protonated and becomes membrane impermeable, causing it to reach millimolar concentrations compared with nanomolar concentrations in the plasma [32]. This property allowed for low doses of chloroquine to be administered to malaria victims, as it is concentrated in the location of biological activity.
In vitro studies have reported quinoline antimalarial compounds to decrease the rate of β-hematin formation [33]. However, it was only within the last 5 years that crystallographic information of the interaction between antimalarials and heme was obtained. Prior to this, the only analysis available was solution proton NMR (1H NMR) spectra that provided evidence of a π–π complex between Fe(III)PPIX and chloroquine [34]. This was supported through isothermal titration calorimetry experiments, which suggested a cofacial π–π complex of chloroquine with two heme µ-oxo dimers [35]. Furthermore, using solid-state 13C and 15N NMR, de Dios et al. determined that chloroquine coprecipitates with Fe(III)PPIX under acidic conditions and the aggregates consisted of covalent chloroquine-Fe(III)PPIX complexes [36]. This covalent complex would prevent coordination of the carboxylate–iron covalent bond in the dimeric units and ultimately, the formation of hemozoin.
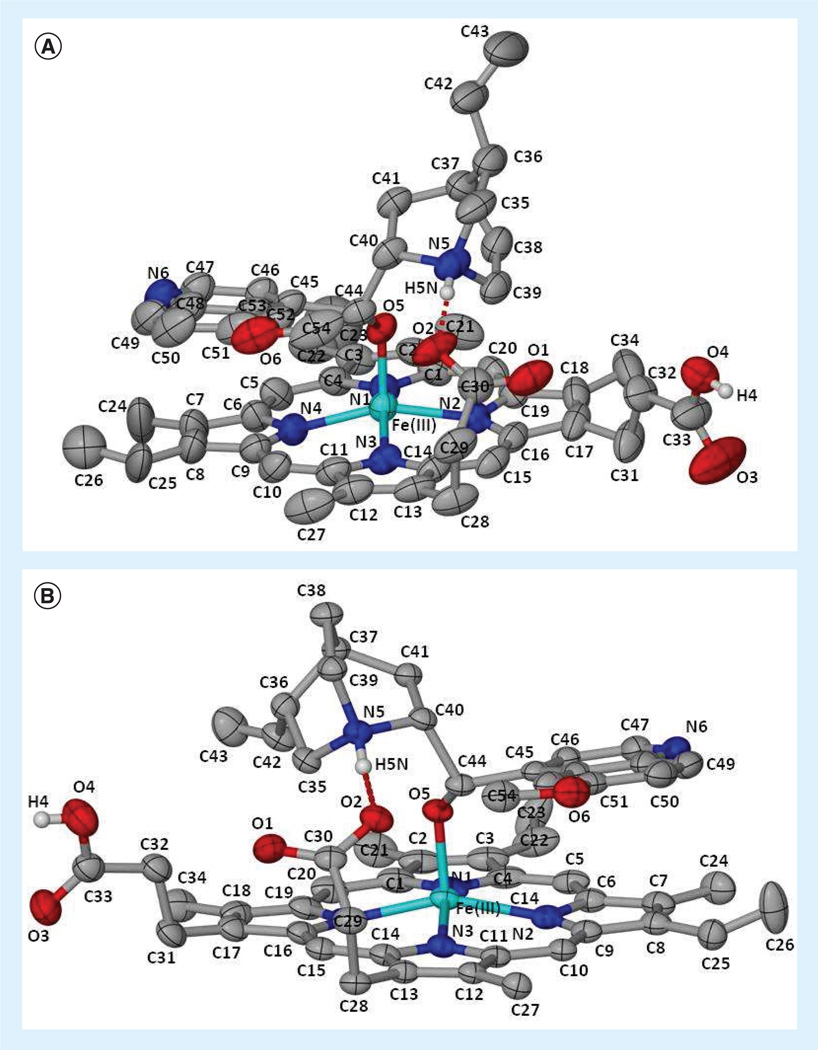
In 2008, de Villiers et al. provided the first example of a crystal structure of Fe(III)PPIX with an antimalarial drug. The crystal structure revealed that the hydroxyl group of halofantrine coordinated to the iron (III) center of the heme monomer with a bond length of 1.840 Å, while the phenanthrene group interacted with the porphyrin rings through π-stacking in a 1:1 ratio. Hydrogen bonding between a propionate group of Fe(III)PPIX and the protonated nitrogen of the compound was also observed [37]. In 2012, de Villiers et al. solved two additional crystal structures of heme–antimalarial complexes (quinine and quinidine) (Figure 3). The benzylic alcohol of the compounds forms a five-coordinate complex with the iron center of the porphyrin ring, containing Fe–O bond lengths of 1.866 and 1.862 Å for quinine and quinidine, respectively [38]. Furthermore, π-stacking between the quinoline aromatic components and the porphyrin pyrrole rings was observed. Together, these crystal structures provide important structural paradigms for the future rational drug design of possible hemozoin inhibitors.
Figure 3. Solved crystal structures of heme-antimalarial complexes.
(A) Quinine–Fe(III)PPIX and (B) quinidine–Fe(III)PPIX illustrating the atomic numbering scheme. Hydrogen bonds are illustrated as broad dashed lines (red). Displacement ellipsoids are drawn at the 50% probability level and solvent molecules and non-relevant hydrogen atoms have been removed for clarity [38]. Blue: Nitrogen; Cyan: Iron; Gray: Carbon; Red: Oxygen; White: Hydrogen. This figure can be viewed in full color at: www.future-science.com/doi/full/10.4155/FMC.13.113
While it is established that quinoline molecules inhibit hemozoin formation, the mechanism for how this occurs is not fully understood; however, there are many hypotheses throughout the literature. One possible mode of action is through the formation of a complex with the Fe(III)PPIX monomer prior to its biomineraliziation. Chloroquine binds to Fe(III)PPIX with a dissociation constant between 10−7 and 10−6 M [39]. This indicates high affinity for and specific tight association between chloroquine and heme. One hypothesis is that this noncovalent interaction could occur immediately following hemoglobin digestion, yet before the heme byproduct partitions into the neutral lipids. If Fe(III)PPIX is bound between two chloroquine molecules, then nucleation of the hemozoin crystal cannot occur. While direct evidence of this is absent, quinoline molecules have been found to significantly increase the time for crystals to form, indicating they may prevent nucleation [40]. Alternatively, chloroquine may be interacting with heme inside of the neutral lipid bodies, after hemozoin formation has already been initiated. In this case, chloroquine could cap the end of the hemozoin crystal, preventing additional heme units from extending growth. A theoretical model of the interaction of hemozoin and quinolines demonstrates the ability of the protonated drug to noncovalently bind to the fast-growing face of the crystal, but has yet to be confirmed [41]. Even if a mechanism is established for one hemozoin inhibitor, it may not coincide with every compound known to target this pathway. Consequently, further studies are required to provide more insight into the mechanisms of action of the quinoline drugs.
The xanthone family of compounds was seren-dipitously discovered to have antimalarial activity after being extracted from the sea urchin, Strongylocentrotus purpuratus [42]. Approximately 17 xanthone derivatives were subsequently screened for activity against P. falciparum, with the two most active having IC50 values of approximately 40 nM [43]. Not only were these compounds found to be potent in the parasite, but their mechanism of action was also determined to be similar to that of chloroquine. When xanthones were incubated with hemin under physiological conditions, soluble heme–xanthone complexes were observed spectroscopically. The formation of these complexes suggests inhibition of the biomineralization of β-hematin [44]. Kelly et al. also investigated the interactions of the xanthone family with heme using 1H NMR and π – π stacking between the aromatic portion of the drug and the pyrrole rings of heme was observed, similar to that of the quinoline family [45]. Their results show that xanthones interact with heme dimeric units, preventing large aggregates from forming. Further evidence of xanthones acting as hemozoin inhibitors was found in their accumulation within the digestive food vacuole. This was illustrated through laser scanning confocal microscopy, as the aromatic core contains intrinsic fluorescent properties allowing the colocalization with Lyso-tracker® Red to be observed [43]. Despite the correlation observed between inhibitory activity in the in vitro heme biomineralization assay and the parasite growth assay, the mode of action for xanthones must still be confirmed within the parasite [46]. Applying this information on the mechanism of interaction and structural properties between xanthones and Fe(III)PPIX, additional derivatives of this family can be synthetically designed for optimal activity against this target.
Another example of an antimalarial derived from a natural product extract is artemisinin. In the 1970s, artemisinin was isolated from a Chinese medicinal drug, known as qinghao, and since then other endoperoxide molecules have also been found to have antimalarial properties [47]. While the exact mechanism of action of the artemisinin family of compounds is highly debated, it is generally agreed that the endoperoxide bridge of these molecules is activated by iron, causing free radicals to form inside the parasite [48]. The free radicals can then continue to alkylate heme and possibly even proteins, inducing parasite death either by creating redox-active heme adducts [49] or by damaging DNA and disrupting cell division [50]. Further evidence of this was observed through the addition of free radical scavengers and iron chelators, which resulted in antagonistic properties [48]. The endoperoxide moiety is essential for biological activity, as derivatives of artemisinin without this bridge do not possess antimalarial activity. Another hypothesis of artemisinin activation is the requirement of hemoglobin uptake [51]. Klonis et al. determined that ring-stage parasites, which have low amounts of digested hemoglobin, were less sensitive to artemisinin activity compared with later stage parasites. While these results may suggest that hemoglobin itself may be an activator for artemisinin, it could also support the findings of other studies that indicate metal-dependent activation as iron is released during the hemoglobin digestion process. Paradoxically, even though hemoglobin was found to be an activator for artemisinin, this compound also inhibited its uptake by the digestive vacuole, causing hemoglobin to accumulate in the parasite [51,52]. Following oxygen activation, it is proposed that artemisinin can form adducts with monomeric heme units to prevent hemozoin formation from occurring. Upon mixing hemin and radioactive-labeled artemisinin in an aqueous solution, adducts were observed using HPLC and TLC, which possessed unique retention times from the individual reactants [50]. Once the hemin–artemisinin adduct is incorporated into the growing crystal structure, further heme unit additions can be prevented [53].
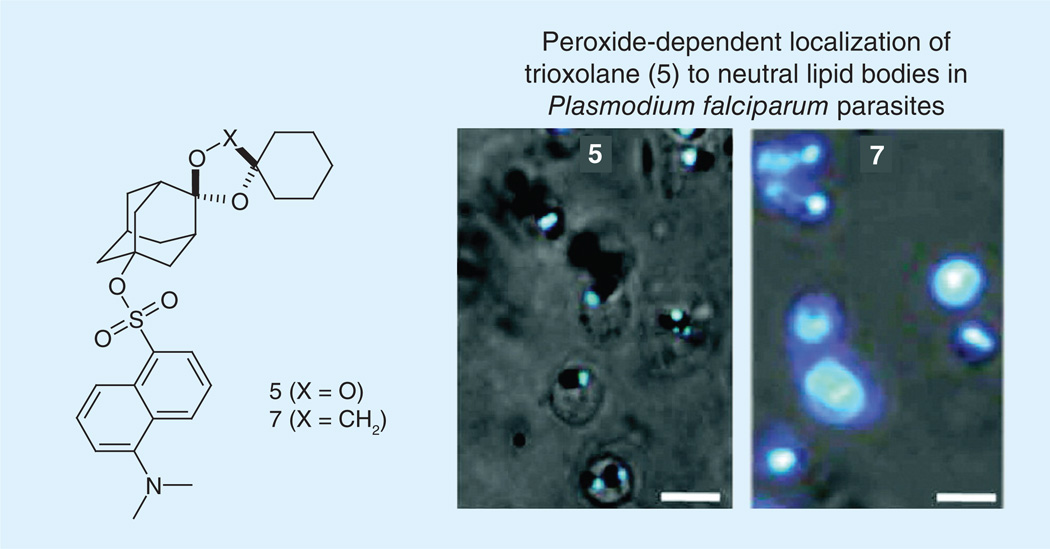
Hartwig et al. co-localized artemisinin compounds to the digestive food vacuole through the use of fluorescently labeled synthetic peroxides. Trophozoites were treated with adamantane-linked fluorescent 1,2,4-trioxolanes and allowed to incubate prior to confocal microscopy. Fluorescence was observed solely within spherical neutral lipid bodies that were closely associated with the digestive vacuole [54]. However, cyclohexane-linked fluorescent trioxolanes were observed uniformly throughout the cytoplasm, indicating the importance of the endoperoxide moiety for specific localization (Figure 4). One potential target for these endoperoxide compounds is the inhibition of hemozoin formation since they are found to accumulate within these lipid droplets, the location of this process [55].
Figure 4. Parasites were incubated with 500 nM of (5) trioxolane or (7) inactive trioxolane.
Fluorescence from the adamantyl-tagged trioxolane (5) is seen only within spherical neutral lipid droplets. By contrast, the inactive trioxolane derivative (7) distributes indiscriminately in the parasite, suggesting specific neutral lipid droplet localization requires endoperoxide activation. Images illustrate merged fluorescence with phase contrast view. Scale bars represent 5 µm.
Data taken from [54].
While hemozoin inhibition is one hypothesis for the mechanism of action, Haynes et al. provides evidence that artemisinin derivatives are not correlated with this process. Dihydroartemisinin was found to be inactive in a β-hematin inhibitory activity assay (which identifies ligands that bind hematin via π–π interactions), but retained activity in a heme biomineralization assay (confirming axial binding with the porphyrin iron) [56]. According to these screens, endoperoxide compounds do not interact with heme in a similar fashion to quinolines and are not involved in hemozoin inhibition. Crystallographic studies with Fe(III)PPIX may be one way to help distinguish these conflicting hypotheses and can then be applied to the development of high-throughput assays for determining mechanisms of interaction.
While a great deal of knowledge regarding current antimalarial drugs has been elucidated, definitive evidence concerning the exact modes of action has yet to be determined. Discovering the precise location within the digestive vacuole of hemozoin inhibition could indicate whether the drug binds to monomeric heme or an already nucleated crystal. Even still, it is vague as to whether each compound within the class of hemozoin inhibitors function through identical mechanisms. Once such necessary studies have been performed, scaffolds can be altered to possess preferential properties, such as aromatic rings, weak bases and hydrogen bonding to improve efficacy.
High-throughput screening
High-throughput screening (HTS) allows for the rapid screening of large libraries of compounds to measure their ability to affect a specific process or pathway [57]. Once the exclusive domain of pharmaceutical companies, HTS is increasingly employed by academia not only for drug screens, but also to help understand fundamental biological processes [58]. There are two main approaches for HTS: phenotypic and target-based screens. A phenotypic screen simply identifies if a compound affects the phenotype of a cell or animal, but the relevant target must subsequently be identified. The alternative approach is to directly test for activity against a known biological target of interest. It is debated as to which strategy is more advantageous. Starting with a well-validated target that accurately represents the physiology allows for the rapid reduction of hit compounds, but may require a longer assay development period. Phenotypic screens result in higher variability in the types of compounds discovered, potentially leading to greater success; however, it can be more challenging to elucidate the target pathway of a specific compound [59].
Phenotypic screening for antimalarial activity
Several in vitro antimalarial drug susceptibility high-throughput assays have been published, including fluorescence-based and ELISAs. Fluorescent nucleic acid intercalating dyes, such as SYBR® Green I and PicoGreen®, have been used to measure parasite growth in erythrocytes [60]. This method is reliant on the fact that mature erythrocytes do not possess nuclei and thus, the only DNA present in culture is that from the parasite. This provides a simple, robust and inexpensive approach to monitoring parasite growth following treatment with small molecules. An alternate ELISA-based assay detects the Plasmodium enzyme pLDH [61]. Since glycolysis is the main energy source of the parasite, pLDH is present in great quantities, providing a standard by which to measure parasite growth [62]. In earlier studies, this enzyme was found to be structurally distinct between parasite and host, allowing for specificity [63].
In 2009, GlaxoSmithKline (Middlesex, UK) screened their library of approximately two million compounds against the Plasmodium falciparum 3D7 strain by measuring the amount of pLDH present. This screen resulted in 13,533 compounds identified to inhibit parasite growth above an 80% threshold [63]. Novartis (Surrey, UK) conducted a similar screening of approximately 1.7 million compounds in the same strain using the SYBR Green I fluorescence-based assay [64]. They identified 6549 compounds with over 50% inhibitory activity and potent enough to be a possible drug lead [64]. Since the malaria disease is not a priority for commercial business, the structures of these hit compounds and their pharmacological data were released to the public. This ‘open innovation strategy’ is recently gaining popularity allowing for collaborations among large corporations and academia [65]. In order to help facilitate drug-lead identification, researchers were encouraged to test these hits further using target-based assays [66].
In vitro β-hematin inhibition
Following the initial primary screen for phenotypic activity, the hit compounds can then be tested in secondary screens developed for specific target pathways, such as heme detoxification. Garavito et al. report about 70% of the free heme found within the digestive vacuole to be catabolized by the enzyme reduced glutathione, with the remaining heme being converted into hemozoin. As a result, they developed an in vitro micro assay to detect molecules that inhibit this heme-reduced glutathione-dependent degradation [67]. Therefore, hit compounds from this study combined with those of hemozoin inhibition screens will result in a more effective drug molecule than simply testing for a single detoxification method. Comparing the results of this assay with those of hemozoin inhibition screens may also provide more specific insights into the mechanisms of various antimalarials. For instance, quinine testing reported similar values of remaining reduced glutathione to chloroquine and amodiaquine, but was significantly less active at inhibiting hemozoin formation [68]. This indicates that quinine activity is more tailored towards preventing interactions between heme and reduced glutathione than hemozoin crystal growth.
In addition, there are high-throughput assays developed to test for in vitro β-hematin inhibition. β-hematin has been demonstrated to be the synthetic analog to hemozoin chemically, spectroscopically and crystallographically, and can be grown in the laboratory under conditions mimicking the physiology of the parasite [8]. Prior to screening of compound libraries, the assays were first validated using antimalarials with well-determined target pathways. Quinoline derivatives were most commonly used as positive controls for hemozoin inhibition, while antifolates or mitochondrial inhibitors were chosen as negative controls [68]. Originally, assays developed to screen for β-hematin inhibition relied on the incorporation of radioactive [14C] hemin into the growing crystal, followed by quantitation using a scintillation counter [69]. While this is a simple and sensitive assay, it is more expensive and requires the use of radioactive materials. Other in vitro assays were later developed to resolve the complications of working with radioactive material through using the innate absorption properties of hemin or through colorimetric quantification. While these methods do not require radiolabeled hematin, other limitations include centrifugation and transfer steps and lengthy (24 h) incubation times, which decrease the throughput of the assay [70]. Tween® 20 was found to be a good initiator for β-hematin formation, allowing for an inexpensive, simple and quick (4-h incubation) assay without any purification or transfer steps [71]. However, the IC50 dose–response values of known antimalarials, such as chloroquine, were tenfold greater with Tween 20 than reported with the blend of physiological neutral lipids [71]. The increased value may indicate that Tween 20 does not accurately mimic the biological nature found within the parasite and may instead initiate hemozoin formation through an alternative method.
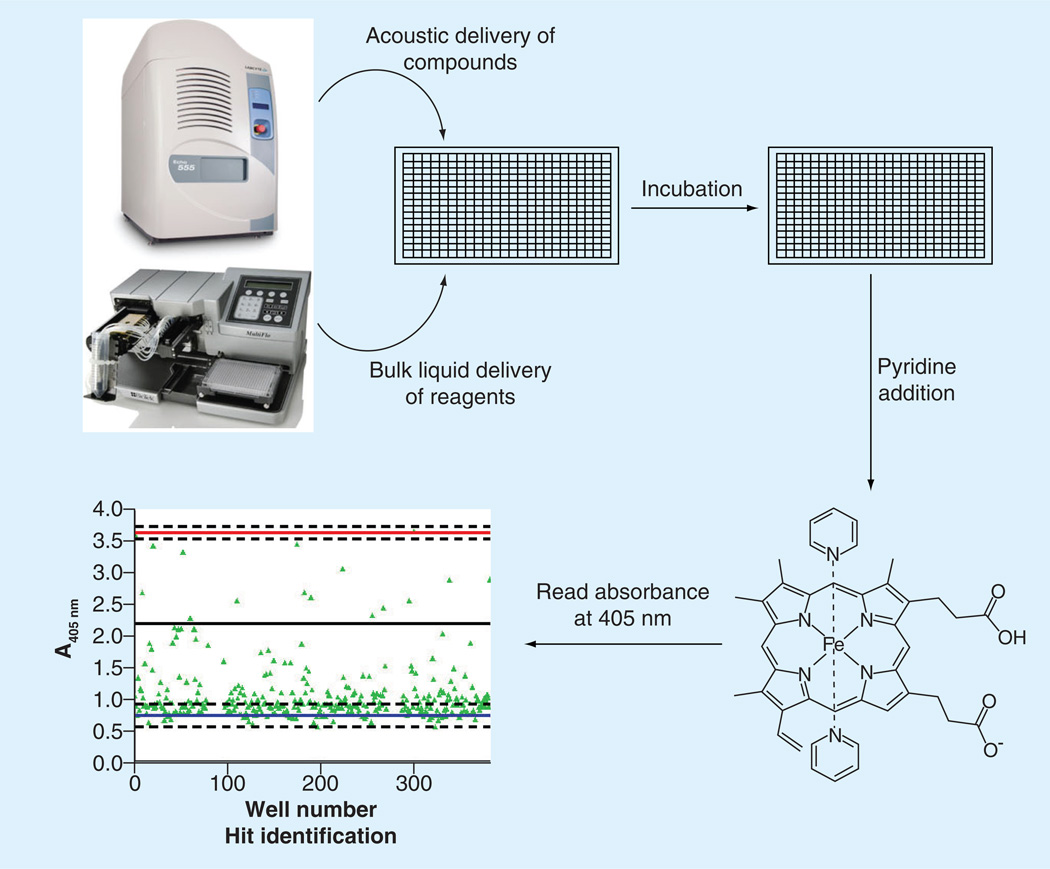
More recently, chemical colorimetric high-throughput assays have been developed and used to screen compound libraries. Results from these screens were subsequently examined for parasite efficacy, which previously mentioned assays failed to test. Due to differential solubility characteristics of heme and β-hematin through the addition of pyridine, the amount of heme that was not converted into the crystal can be quantified spectrophotometrically [72]. One spectrophotometric assay was developed for 384-well microtiter plates, which incubated a hemin solution at 60°C for 2 h before quantifying with pyridine [73]. While rapid, this assay did not effectively recapitulate the biology that is occurring in the digestive food vacuole. In fact, the mechanism of formation for β-hematin in this assay is a complex phase-transfer catalysis mechanism that is most likely not occurring in the parasite. Consequently, of the 644 pathway hits that they identified from their pilot screen of 16,000 compounds, only 17 (2.6%) were active against the parasite. Carter et al. created a high-throughput in vitro detergent based assay that more closely represented the biological environment in which hemozoin formation occurs. This method eliminated the use of radioactive materials and transfer steps, and additionally only required a short (4 h) incubation time, increasing the throughput of the assay. Not only did the positive and negative controls of known antimalarials hold true using the NP-40 detergent, but also chloroquine (53.0 µM) and amodiaquine (21.0 µM) testing resulted in similar dose–response concentrations to the values reported of the native lipid environment (85.3 and 23.1 µM, respectively) [74]. This detergent-mediated assay was subsequently used in a pilot screen of 38,400 compounds to test for β-hematin activity (Figure 5) [74]. A total of 161 inhibitors of β-hematin were identified from this screen conducted in 384-well microtiter plates, which correlated to a 0.42% hit rate and 113 compounds exhibiting IC50 values less than the positive control drug, amodiaquine. The compounds identified as β-hematin inhibitors were then screened for in vitro parasite activity and 48 (30%) were found to inhibit more than 90% of parasite growth in a chloroquine-sensitive strain (D6), while 40 compounds retained activity in a multidrug-resistant strain (C235). The fact that this screen resulted in a high percentage of parasite active hits, with a low false-positive hit rate (0.016%), demonstrates the importance for development of an assay that accurately replicates the physiological environment present in vivo [74].
Figure 5. Workflow of the high-throughput β-hematin assay developed by Sandlin et al.
Test compounds were delivered using a noncontact liquid handler, followed by the addition of buffer, hemin and detergent. The 384-well plates incubated at 37°C for 4 h before the addition of 5% (v/v) pyridine. The absorbance values of test compounds were compared with positive and negative controls to establish hits [74].
Validation of target pathway
Hits from compound library screens can begin to be prioritized based on their potencies found from both the drug susceptibility and in vitro target-based screens. However, even if a compound is able to inhibit β-hematin in vitro and reduce parasitic growth, it is not definitive that hemozoin inhibition is the sole cause of parasite death. In order to establish this target in the parasite, the levels of intracellular heme and hemozoin, following drug treatment, must be compared. An increase in the ratio of free heme to hemozoin with increasing drug concentration, corresponding with death of the parasite, would suggest the compound is inhibiting hemozoin formation. On the other hand, if elevated drug concentration does not alter the heme to hemozoin ratio, but the parasite still dies, then another biological mechanism is likely the in vivo target.
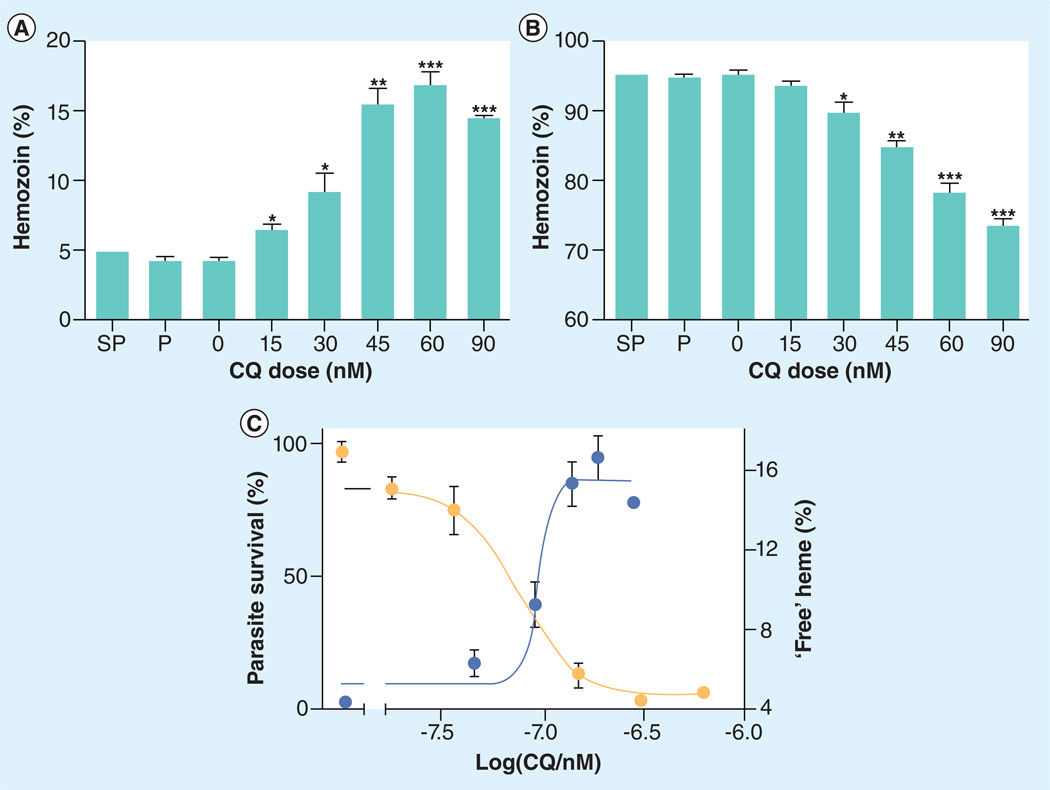
Combrinck et al. provided the first direct evidence that chloroquine prevents hemozoin formation within the malaria parasite. Following chloroquine treatment, free heme levels rose within a chloroquine-sensitive P. falciparum strain (D10) in a dose-dependent fashion, correlating strongly with parasite survival (Figure 6) [75]. Other suspected hemozoin inhibitors (amodiaquine and mefloquine) similarly demonstrated a rise in heme and decrease in hemozoin levels compared with the control culture (without drug treatment). As expected, antifolate compounds such as pyrimethamine, and the combination therapy sulfanilamide–pyrimethamine, provided no significant perturbation on the levels of free heme or hemozoin in the parasite. While this is the first assay to directly correlate a rise in free heme levels with parasite survival following a drug treatment, it remains a laborious and lengthy one. There is a need to adapt this process to an HTS format to examine the multitude of identified β-hematin inhibitors in the primary screens aforementioned. Once compounds have been determined to inhibit β-hematin, possess antimalarial activity and decrease hemozoin levels within the parasite, then pharmacokinetic studies can be prioritized.
Figure 6. Iron and heme species in untreated and drug-treated trophozoites exposed to varying concentrations of chloroquine or other antimalarials at 2.5-times their respective IC50 values.
Parasites were synchronized with sorbitol and cultured for 32 h with or without drug. Fraction of total heme present as (A) free heme and (B) hemozoin. Asterisks indicate statistical significance relative to control (two-tailed t-test): *p < 0.05; **p < 0.01; ***p < 0.001, n = 3 except for P where n = 6. (C) Parasite survival curve (yellow circles, left axis) determined using the lactate dehydrogenase assay.
CQ: Chloroquine; P: Pyrimethamine; SP: Sulfanilamide–pyrimethamine.
This figure can be viewed in full color at: www.future-science.com/doi/full/10.4155/FMC.13.113
Reproduced with permission from [75] © American Chemical Society.
High-content screening as a tool
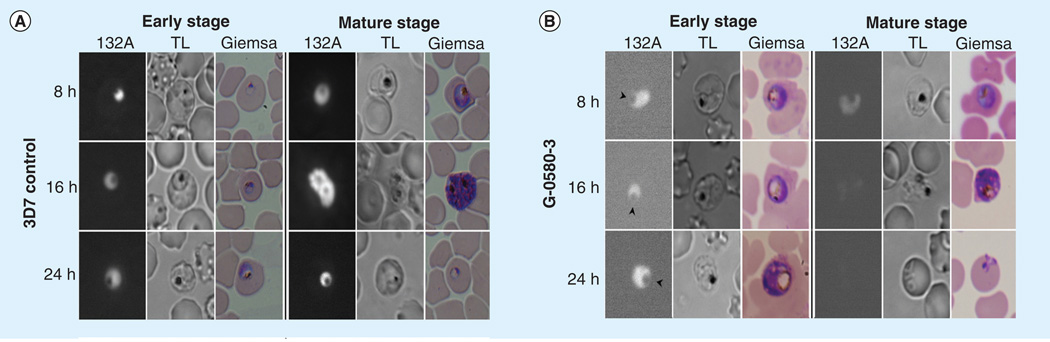
Another approach to overcome this bottleneck in drug discovery is through the use of high-content screening (HCS) [76]. HCS can be utilized in the antimalarial drug-discovery process as a secondary screen in observing the morphology and phenotypic analysis of the parasite following treatment. Cervantes et al. demonstrated the potential of HCS of P. falciparum through the use of RNA probes. Similar to fluorescence-based drug susceptibility assays, they found a relationship between parasitemia and fluorescence intensity from the probes. However, in addition to determining IC50 values, the HCS allowed the observation of cell cycle arrest, enlargement of the digestive vacuole and a decline in hemozoin formation following treatment, information that cannot be obtained with a simple fluorescence or radioactive-based assay (Figure 7) [77]. High-content imaging can also provide further analysis of P. falciparum under drug pressure during various stages of the intraerythrocytic life cycle and even cytotoxic effects on erythrocytes [78].
Figure 7. Morphological analysis of Plasmodium falciparum with IC80 concentrations of extracts.
Infected erythrocytes were synchronized and live parasites were stained with RNA probe 132A. (A) Images of the Plasmodium falciparum 3D7 strain without any drug treatment at the ring and mature stage (control). (B) Displays the phenotype of extract G-0580-3, and the arrowheads indicate the enlarged food vacuoles at the early stages of the cell cycle.
Reproduced from [78].
By designing an appropriate high-content screen, it will be possible to more directly observe the target and activity of a potential lead compound. The unique autofluorescence properties of hemozoin (excitation wavelength: 632.5 nm; emission wavelength: 655–700 nm) provide a valuable handle on which to base such a screen, particularly given the lack of fluorescence for Fe(III)PPIX [79]. In fact, previous reports have utilized this intrinsic fluorescence property to image the fate of hemozoin in host cells [80,81]. Thus, one could imagine a high-content screen tailored to observe hemozoin autofluorescence in the parasite following drug treatment. A dose– responsive decrease in autofluorescence would provide direct evidence of a compound’s ability to inhibit hemozoin formation in the parasite. Coupling this with a DNA-intercalating dye, such as SYBR Green I, parasite survival could be determined simultaneously. Furthermore, through high-content imaging the morphology of the parasite can be assessed, as previously described, to further analyze the effects of drug compounds on organelle and overall parasite morphologies. In the future, such a high-content screen will facilitate the determination of the target pathway in comprehensive phenotypic assays.
Future perspective
With the rising increase of resistance to current antimalarial drugs, especially along the Thai– Cambodian border, there is a great need for novel, inexpensive and more effective compounds to be discovered. The Medicines for Malaria Venture has several new initiatives to help spawn research on this disease and has made open collaboration easier through releasing the screening results from GlaxoSmithKline, Novartis, and St Jude Children’s Research Hospital (TN, USA) to the public [82]. Using the information from these screens, research groups can focus on target pathway validation of the hit compounds in hopes of finding antimalarial compounds that are potent against the resistant strains of P. falciparum. Through confocal microscopy, MS and various other methods of analysis, we will be able to gain further knowledge as to how drugs are interrupting the biological pathways of the parasite. Advances in x-ray crystallography may allow for additional structures to be solved of hemozoin inhibitors bound to heme. This structural information will provide evidence of the molecular interactions occurring and will allow for a rational drug design.
Similarly, further R&D is required for a simple and more high-throughput assay for target validation within the parasite. Not only validation of the hemozoin formation pathway, but also for determining the mode of action for compounds not inhibiting the formation of hemozoin. Tools such as MS and NMR can begin to look at the metabolome of P. falciparum as it develops throughout the intraerythrocytic life cycle [83,84]. Eventually, the regulation of metabolites following treatment may even be able to provide evidence as to which target pathway that a drug may inhibit. By determining the mechanism of action of lead compounds, they can be optimized against resistant strains in addition to creating combination therapies to effectively eliminate the parasite through multiple biological pathways [85].
Executive summary.
Heme detoxification pathway
-
▪
In the quest for a more effective antimalarial, one of the main target pathways being researched is hemozoin formation due to its uniqueness to the Plasmodium species.
-
▪
The hemozoin crystal is formed through a biomineralization of ferriprotoporphryin IX within the acidic digestive food vacuole as a way to combat the toxicity of this hemoglobin degradation byproduct.
Antimalarial hemozoin inhibitors
-
▪
Many of the current antimalarial drugs target this biological pathway, including the quinoline and xanthone families of compounds. Their mechanism of action has been proposed through spectroscopic as well as structural (NMR and x-ray crystallography) information.
Phenotypic screening for antimalarial activity
-
▪
GlaxoSmithKline (Middlesex, UK), Novartis (Surrey, UK) and St Jude Children’s Research Hospital (TN, USA) screened their chemical libraries for activity against the malaria parasite (Plasmodium falciparum), resulting in over 30,000 potential antimalarial compounds.
-
▪
Following the initial phenotypic screens, secondary target-based screens were conducted to test the hit compounds for their effectiveness against inhibiting the formation of β-hematin in vitro.
Validation of target pathway
-
▪
Even with the combination of these two screens, one cannot definitively conclude that the hit compounds are in fact acting against this biological pathway in the parasite. Recently, a heme speciation assay was developed to provide further evidence that the hemozoin formation pathway is being targeted in the parasite.
High-content screening as a tool
-
▪
In the past year, high-content screening has been utilized in observing the morphology of the parasite following the addition of RNA probes. This concept can be adapted in secondary screens of lead compounds to give additional information on the mechanism of action.
-
▪
Understanding the biological pathway that lead compounds target is essential to discovering a more potent antimalarial. In addition, more high-throughput methods must be developed in order to more efficiently determine the mode of action targeted within the malaria parasite.
Acknowledgements
The authors would like to thank MF Richards for critical reading of this manuscript.
The authors would like to acknowledge support from the NIH (1R01AI083145–01), Medicines for Malaria Venture, and the Waite Philip Fishel Endowment fund.
Key Terms
- Trophozoite
The stage of the intraerythrocytic life cycle where the greatest amount of hemozoin is present in the parasite.
- Hemozoin
A nontoxic crystal formed by the Plasmodium species in response to ferriprotoporphryin IX accumulation within the acidic digestive vacuole.
- β-hematin
The synthetic analog of hemozoin, found to have identical structural and spectrophotometric properties.
- Chloroquine
A 4-aminoquinoline antimalarial drug that has been found to target hemozoin formation. Strains of Plasmodium falciparum acquired resistance to this drug by decreasing its local concentration in the digestive vacuole, causing this drug to no longer be effective.
- Plasmodium falciparum chloroquine-resistant transporter
Transporter protein found in the membrane of the digestive vacuole that is the primary determinant of chloroquine resistance.
- High-content screening
An automated imaging approach that measures the phenotypic cellular processes that occur as a result of compound activity.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
References
- 1.World Malaria Report. Geneva, Switzerland: WHO press; 2012. [Google Scholar]
- 2.Teklehaimanot A, Mejia P. Malaria and poverty. Ann. NY Acad. Sci. 2008;1136:32–37. doi: 10.1196/annals.1425.037. [DOI] [PubMed] [Google Scholar]
- 3.Sigma-Tau Group. ACT to combat malaria receives marketing authorization from EMA Venture. Geneva, Switzerland: 2011. [Google Scholar]
- 4.Matuschewski K, Nunes AC, Nussenzweig V, Menard R. Plasmodium sporozoite invasion into insect and mammalian cells is directed by the same dual binding system. EMBO J. 2002;21(7):1597–1606. doi: 10.1093/emboj/21.7.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prudencio M, Rodriguez A, Mota MM. The silent path to thousands of merozoites: the Plasmodium liver stage. Nat. Rev. Microbiol. 2006;4(11):849–856. doi: 10.1038/nrmicro1529. [DOI] [PubMed] [Google Scholar]
- 6.Tilley L, Dixon MWA, Kirk K. The Plasmodium falciparum-infected red blood cell. Int. J. Biochem. Cell Biol. 2011;43(6):839–842. doi: 10.1016/j.biocel.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Goldberg DE, Slater AFG, Cerami A, Henderson GB. Hemoglobin degradation in the malaria parasite plasmodium-falciparum-an ordered process in a unique organelle. Proc. Natl Acad. Sci. USA. 1990;87(8):2931–2935. doi: 10.1073/pnas.87.8.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egan TJ. Haemozoin formation. Mol. Biochem. Parasitol. 2008;157(2):127–136. doi: 10.1016/j.molbiopara.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Choi AMK, Alam J. Heme oxygenase-1: function, regulation, and implication of a novel stress-inducible protein in oxidant-induced lung injury. Am. J. Respir. Cell Mol. Biol. 1996;15(1):9–19. doi: 10.1165/ajrcmb.15.1.8679227. [DOI] [PubMed] [Google Scholar]
- 10.Sigala PA, Crowley JR, Hsieh S, Henderson JP, Goldberg DE. Direct tests of enzymatic heme degradation by the malaria parasite Plasmodium falciparum . J. Biol. Chem. 2012;287(45) doi: 10.1074/jbc.M112.414078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ponka P. Cell biology of heme. Am. J. Med. Sci. 1999;318(4):241–256. doi: 10.1097/00000441-199910000-00004. [DOI] [PubMed] [Google Scholar]
- 12.Egan TJ, Chen JYJ, de Villiers KA, et al. Haemozoin (beta-haematin) biomineralization occurs by self-assembly near the lipid/water interface. FEBS Lett. 2006;580(21):5105–5110. doi: 10.1016/j.febslet.2006.08.043. [DOI] [PubMed] [Google Scholar]
- 13.Marom N, Tkatchenko A, Kapishnikov S, Kronik L, Leiserowitz L. Structure and formation of synthetic hemozoin: insights from first-principles calculations. Cryst. Growth Des. 2011;11(8):3332–3341. [Google Scholar]
- 14.Klonis N, Dilanian R, Hanssen E, et al. Hematin-hematin self-association states involved in the formation and reactivity of the malaria parasite pigment, hemozoin. Biochemistry. 2010;49(31):6804–6811. doi: 10.1021/bi100567j. [DOI] [PubMed] [Google Scholar]
- 15.Slater AFG, Cerami A. Inhibition by chloroquine of a novel heme polymerase enzyme-activity in malaria trophozoites. Nature. 1992;355(6356):167–169. doi: 10.1038/355167a0. [DOI] [PubMed] [Google Scholar]
- 16.Sullivan DJ, Gluzman IY, Goldberg DE. Plasmodium hemozoin formation mediated by histidine-rich proteins. Science. 1996;271(5246):219–222. doi: 10.1126/science.271.5246.219. [DOI] [PubMed] [Google Scholar]
- 17.Jani D, Nagarkatti R, Beatty W, et al. HDP – a novel heme detoxification protein from the malaria parasite. PLoS Pathogens. 2008;4(4) doi: 10.1371/journal.ppat.1000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitch CD, Cai GZ, Chen YF, Shoemaker JD. Involvement of lipids in ferriprotoporphyrin IX polymerization in malaria. Biochim. Biophys. Acta. 1999;1454(1):31–37. doi: 10.1016/s0925-4439(99)00017-4. [DOI] [PubMed] [Google Scholar]
- 19.Pandey AV, Babbarwal VK, Okoyeh JN, et al. Hemozoin formation in malaria: a two-step process involving histidine-rich proteins and lipids. Biochem. Biophys. Res. Commun. 2003;308(4):736–743. doi: 10.1016/s0006-291x(03)01465-7. [DOI] [PubMed] [Google Scholar]
- 20.Pisciotta JM, Coppens I, Tripathi AK, et al. The role of neutral lipid nanospheres in Plasmodium falciparum haem crystallization. Biochem. J. 2007;402:197–204. doi: 10.1042/BJ20060986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoang AN, Ncokazi KK, de Villiers KA, Wright DW, Egan TJ. Crystallization of synthetic haemozoin (beta-haematin) nucleated at the surface of lipid particles. Dalton Trans. 2010;39(5):1235–1244. doi: 10.1039/b914359a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoang AN, Sandlin RD, Omar A, Egan TJ, Wright DW. The neutral lipid composition present in the digestive vacuole of Plasmodium falciparum concentrates heme and mediates beta-hematin formation with an unusually low activation energy. Biochemistry. 2010;49(47):10107–10116. doi: 10.1021/bi101397u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hempelmann E, Motta C, Hughes R, Ward SA, Bray PG. Plasmodium falciparum: sacrificing membrane to grow crystals? Trends Parasitol. 2003;19(1):23–26. doi: 10.1016/s1471-4922(02)00011-9. [DOI] [PubMed] [Google Scholar]
- 24.Stiebler R, Correa Soares JBR, Timm BL, et al. On the mechanisms involved in biological heme crystallization. J. Bioenerg. Biomemb. 2011;43(1):93–99. doi: 10.1007/s10863-011-9335-x. [DOI] [PubMed] [Google Scholar]
- 25.Kapishnikov S, Weiner A, Shimoni E, et al. Oriented nucleation of hemozoin at the digestive vacuole membrane in Plasmodium falciparum . Proc. Natl Acad. Sci. USA. 2012;109(28):11188–11193. doi: 10.1073/pnas.1118120109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR. Epidemiology of drug-resistant malaria. Lancet Infect. Dis. 2002;2(4):209–218. doi: 10.1016/s1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- 27.Martin RE, Marchetti RV, Cowan AI, Howitt SM, Broeer S, Kirk K. Chloroquine transport via the malaria parasite’s chloroquine resistance transporter. Science. 2009;325(5948):1680–1682. doi: 10.1126/science.1175667. [DOI] [PubMed] [Google Scholar]
- 28.Raj DK, Mu J, Jiang H, et al. Disruption of a Plasmodium falciparum multidrug resistance-associated protein (Pf MRP) alters its fitness and transport of antimalarial drugs and glutathione. J. Biol. Chem. 2009;284(12):7687–7696. doi: 10.1074/jbc.M806944200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ecker A, Lehane AM, Clain J, Fidock DA. PfCRT and its role in antimalarial drug resistance. Trends Parasitol. 2012;28(11):504–514. doi: 10.1016/j.pt.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez CP, Rohrbach P, McLean JE, Fidock DA, Stein WD, Lanzer M. Differences in trans-stimulated chloroquine efflux kinetics are linked to PfCRT in Plasmodium falciparum . Mol. Microbiol. 2007;64(2):407–420. doi: 10.1111/j.1365-2958.2007.05664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Achan J, Talisuna AO, Erhart A, et al. Quinine, an old anti-malarial drug in a modern world: role in the treatment of malaria. Malar. J. 2011;10(144) doi: 10.1186/1475-2875-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sullivan DJ, Gluzman IY, Russell DG, Goldberg DE. On the molecular mechanism of chloroquine’s antimalarial action. Proc. Natl Acad. Sci. USA. 1996;93(21):11865–11870. doi: 10.1073/pnas.93.21.11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Egan TJ, Ncokazi KK. Quinoline antimalarials decrease the rate of beta-hematin formation. J. Inorg. Biochem. 2005;99(7):1532–1539. doi: 10.1016/j.jinorgbio.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 34.de Dios AC, Casabianca LB, Kosar A, Roepe PD. Structure of the amodiaquine-FPIX mu oxo dimer solution complex at atomic resolution. Inorg. Chem. 2004;43(25):8078–8084. doi: 10.1021/ic0489948. [DOI] [PubMed] [Google Scholar]
- 35.Vippagunta SR, Dorn A, Matile H, et al. Structural specificity of chloroquine-hematin binding related to inhibition of hematin polymerization and parasite growth. J. Med. Chem. 1999;42(22):4630–4639. doi: 10.1021/jm9902180. [DOI] [PubMed] [Google Scholar]
- 36.de Dios AC, Tycko R, Ursos LMB, Roepe PD. NMR studies of chloroquine-ferriprotoporphyrin IX complex. J. Phys. Chem. 2003;107(30):5821–5825. [Google Scholar]
- 37.de Villiers KA, Marques HM, Egan TJ. The crystal structure of halofantrine-ferriprotoporphyrin IX and the mechanism of action of arylmethanol antimalarials. J. Inorg. Biochem. 2008;102(8):1660–1667. doi: 10.1016/j.jinorgbio.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 38.de Villiers KA, Gildenhuys J, le Roex T. Iron(III) protoporphyrin IX complexes of the antimalarial cinchona alkaloids quinine and quinidine. ACS Chem. Biol. 2012;7(4):666–671. doi: 10.1021/cb200528z. [DOI] [PubMed] [Google Scholar]
- 39.Fitch CD. Antimalarial schizontocides - ferriprotoporphyrin-IX interaction hypothesis. Parasitol. Today. 1986;2(12):330–331. doi: 10.1016/0169-4758(86)90051-7. [DOI] [PubMed] [Google Scholar]
- 40.Solomonov I, Osipova M, Feldman Y, et al. Crystal nucleation, growth, and morphology of the synthetic malaria pigment beta-hematin and the effect thereon by quinoline additives: the malaria pigment as a target of various antimalarial drugs. J. Am. Chem. Soc. 2007;129(9):2615–2627. doi: 10.1021/ja0674183. [DOI] [PubMed] [Google Scholar]
- 41.Buller R, Peterson ML, Almarsson O, Leiserowitz L. Quinoline binding site on malaria pigment crystal: a rational pathway for antimalaria drug design. Cryst. Growth Des. 2002;2(6):553–562. [Google Scholar]
- 42.Riscoe M, Kelly JX, Winter R. Xanthones as antimalarial agents: discovery, mode of action, and optimization. Curr. Med. Chem. 2005;12(21) doi: 10.2174/092986705774370709. [DOI] [PubMed] [Google Scholar]
- 43.Kelly JX, Winter R, Peyton DH, Hinrichs DJ, Riscoe M. Optimization of xanthones for antimalarial activity: the 3,6-bis-omega-diethylaminoalkoxyxanthone series. Antimicrob. Agents Chemother. 2002;46(1):144–150. doi: 10.1128/AAC.46.1.144-150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ignatushchenko MV, Winter RW, Bachinger HP, Hinrichs DJ, Riscoe MK. Xanthones as antimalarial agents; studies of a possible mode of action. FEBS Lett. 1997;409(1) doi: 10.1016/s0014-5793(97)00405-5. [DOI] [PubMed] [Google Scholar]
- 45.Kelly JX, Winter R, Riscoe M, Peyton DH. A spectroscopic investigation of the binding interactions between 4,5-dihydroxyxanthone and heme. J. Inorg. Biochem. 2001;86(2–3):617–625. doi: 10.1016/s0162-0134(01)00217-3. [DOI] [PubMed] [Google Scholar]
- 46.Ignatushchenko MV, Winter RW, Riscoe M. Xanthones as antimalarial agents: stage specificity. Am. J. Trop Med Hyg. 2000;62(1):77–81. doi: 10.4269/ajtmh.2000.62.77. [DOI] [PubMed] [Google Scholar]
- 47.Hsu E. Reflections on the ‘discovery’ of the antimalarial qinghao. Br. J. Clin.Pharmacol. 2006;61(6):666–670. doi: 10.1111/j.1365-2125.2006.02673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meshnick SR, Thomas A, Ranz A, Xu CM, Pan HZ. Artemisinin (qinghaosu) - the role of intracellular hemin in its mechanism of antimalarial action. Mol. Biochem.Parasitol. 1991;49(2):181–190. doi: 10.1016/0166-6851(91)90062-b. [DOI] [PubMed] [Google Scholar]
- 49.Cazelles J, Robert A, Meunier B. Alkylation of heme by artemisinin, an antimalarial drug. C. R. Acad. Sci. Paris, Chimie. 2001;4(2):85–89. [Google Scholar]
- 50.Hong YL, Yang YZ, Meshnick SR. The interaction of artemisinin with malarial hemozoin. Mol. Biochem. Parasitol. 1994;63(1):121–128. doi: 10.1016/0166-6851(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 51.Klonis N, Crespo-Ortiz MP, Bottova I, et al. Artemisinin activity against Plasmodium falciparum requires hemoglobin uptake and digestion. Proc. Natl Acad. Sci. USA. 2011;108(28):11405–11410. doi: 10.1073/pnas.1104063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoppe HC, van Schalkwyk DA, Wiehart UIM, Meredith SA, Egan J, Weber BW. Antimalarial quinolines and artemisinin inhibit endocytosis in Plasmodium falciparum . Antimicrob. Agents Chemother. 2004;48(7):2370–2378. doi: 10.1128/AAC.48.7.2370-2378.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weissbuch I, Leiserowitz L. Interplay between malaria, crystalline hemozoin formation, and antimalarial drug action and design. Chem. Rev. 2008;108(11):4899–4914. doi: 10.1021/cr078274t. [DOI] [PubMed] [Google Scholar]
- 54.Hartwig CL, Lauterwasser EMW, Mahajan SS, Hoke JM, Cooper RA, Renslo AR. Investigating the antimalarial action of 1,2,4-trioxolanes with fluorescent chemical probes. J. Med. Chem. 2011;54(23):8207–8213. doi: 10.1021/jm2012003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hartwig CL, Rosenthal AS, D’Angelo J, Griffin CE, Posner GH, Cooper RA. Accumulation of artemisinin trioxane derivatives within neutral lipids of Plasmodium falciparum malaria parasites is endoperoxide-dependent. Biochem. Pharmacol. 2009;77(3):322–336. doi: 10.1016/j.bcp.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haynes RK, Monti D, Taramelli D, Basilico N, Parapini S, Olliaro P. Artemisinin antimalarials do not inhibit hemozoin formation. Antimicrob. Agents Chemother. 2003;47(3):1175–1175. doi: 10.1128/AAC.47.3.1175.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Janzen WP. High throughput screening as a discovery tool in the pharmaceutical industry. Lab. Robotics Autom. 1996;8(5):261–265. [Google Scholar]
- 58.Dove A. High-throughput screening goes to school. Nat. Methods. 2007;4(6):523–529. [Google Scholar]
- 59.Sams-Dodd F. Target-based drug discovery: is something wrong? Drug Discov. Today. 2005;10(2):139–147. doi: 10.1016/S1359-6446(04)03316-1. [DOI] [PubMed] [Google Scholar]
- 60.Johnson JD, Dennull RA, Gerena L, Lopez-Sanchez M, Roncal NE, Waters NC. Assessment and continued validation of the malaria SYBR green I-based fluorescence assay for use in malaria drug screening. Antimicrob. Agents Chemother. 2007;51(6):1926–1933. doi: 10.1128/AAC.01607-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Orjuela-Sanchez P, Duggan E, Nolan J, Frangos JA, Carvalho LJM. A lactate dehydrogenase ELISA-based assay for the in vitro determination of Plasmodium berghei sensitivity to anti-malarial drugs. Malaria J. 11(2012) doi: 10.1186/1475-2875-11-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vivas L, Easton A, Kendrick H, et al. Plasmodium falciparum: stage specific effects of a selective inhibitor of lactate dehydrogenase. Exp. Parasitol. 2005;111(2):105–114. doi: 10.1016/j.exppara.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 63.Gamo FJ, Sanz LM, Vidal J, et al. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- 64.Plouffe D, Brinker A, McNamara C, et al. In silico activity profiling reveals the mechanism of action of antimalarials discovered in a high-throughput screen. Proc. Natl Acad. Sci. USA. 2008;105(26):9059–9064. doi: 10.1073/pnas.0802982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Butler D. GlaxoSmithKline goes public with malaria data. Nature. 2010 Jan 26 [Google Scholar]
- 66.Cressey D. Data sharing aids the fight against malaria. Nature News. 2012 Feb 14 [Google Scholar]
- 67.Garavito G, Monje MC, Maurel S, Valentin A, Nepveu F, Deharo E. A non-radiolabeled heme-GSH interaction test for the screening of antimalarial compounds. Exp. Parasitol. 2007;116(3):311–313. doi: 10.1016/j.exppara.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 68.Chong CR, Sullivan DJ. Inhibition of heme crystal growth by antimalarials and other compounds: implications for drug discovery. Biochem. Pharmacol. 2003;66(11):2201–2212. doi: 10.1016/j.bcp.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 69.Kurosawa Y, Dorn A, Kitsuji-Shirane M, et al. Hematin polymerization assay as a high-throughput screen for identification of new antimalarial pharmacophores. Antimicrob. Agents Chemother. 2000;44(10):2638–2644. doi: 10.1128/aac.44.10.2638-2644.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Basilico N, Pagani E, Monti D, Olliaro P, Taramelli D. A microtitre-based method for measuring the haem polymerization inhibitory activity (HPIA) of antimalarial drugs. J. Antimicrob. Chemother. 1998;42(1):55–60. doi: 10.1093/jac/42.1.55. [DOI] [PubMed] [Google Scholar]
- 71.Huy NT, Uyen DT, Maeda A, et al. Simple colorimetric inhibition assay of heme crystallization for high-throughput screening of antimalarial compounds. Antimicrob. Agents Chemother. 2007;51(1):350–353. doi: 10.1128/AAC.00985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carter MD, Phelan VV, Sandlin RD, Bachmann BO, Wright DW. Lipophilic mediated assays for beta-hematin inhibitors. Comb. Chem. High Throughput Screen. 2010;13(3):285–292. doi: 10.2174/138620710790980496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rush MA, Baniecki ML, Mazitschek R, et al. Colorimetric high-throughput screen for detection of heme crystallization inhibitors. Antimicrob. Agents Chemother. 2009;53(6):2564–2568. doi: 10.1128/AAC.01466-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sandlin RD, Carter MD, Lee PJ, et al. Use of the NP-40 detergent-mediated assay in discovery of inhibitors of beta-hematin crystallization. Antimicrob. Agents Chemother. 2011;55(7):3363–3369. doi: 10.1128/AAC.00121-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Combrinck JM, Mabotha TE, Ncokazi KK, et al. Insights into the role of heme in the mechanism of action of antimalarials. ACS Chem. Bio. 2013;8(1):133–137. doi: 10.1021/cb300454t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haney SA, editor. High Content Screening: Science, Techniques and Applications. Hoboken, NJ, USA: John Wiley and Sons, Inc.; 2008. [Google Scholar]
- 77.Cervantes S, Prudhomme J, Carter D, et al. High-content live cell imaging with RNA probes: advancements in high-throughput antimalarial drug discovery. BMC Cell Biol. 2009;10(45):1–10. doi: 10.1186/1471-2121-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cervantes S, Stout PE, Prudhomme J, et al. High content live cell imaging for the discovery of new antimalarial marine natural products. BMC Infect. Dis. 2012;12(1):1–9. doi: 10.1186/1471-2334-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bellemare MJ, Bohle DS, Brosseau CN, et al. Autofluorescence of condensed heme aggregates in malaria pigment and its synthetic equivalent hematin anhydride (beta-hematin) J. Phys. Chem. B. 2009;113(24):8391–8401. doi: 10.1021/jp8104375. [DOI] [PubMed] [Google Scholar]
- 80.Schwarzer E, Bellomo G, Giribaldi G, Ulliers D, Arese P. Phagocytosis of malarial pigment haemozoin by human monocytes: a confocal microscopy study. Parasitology. 2001;123:125–131. doi: 10.1017/s0031182001008216. [DOI] [PubMed] [Google Scholar]
- 81.Carney CK, Schrimpe AC, Halfpenny K, et al. The basis of the immunomodulatory activity of malaria pigment (hemozoin) J. Biol. Inorg. Chem. 2006;11(7):917–929. doi: 10.1007/s00775-006-0147-0. [DOI] [PubMed] [Google Scholar]
- 82.Openhealth News. Medicines for Malaria Venture and EMBL-EBI establish one-stop shop for malaria drug data. Hinxton, UK: European Molecular Biology Laboratory – European Bioinformatics Institute; 2012. [Google Scholar]
- 83.Olszewski KL, Morrisey JM, Wilinski D, et al. Host-parasite interactions revealed by Plasmodium falciparum metabolomics. Cell Host Microbe. 2009;5(2):191–199. doi: 10.1016/j.chom.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gebregiworgis T, Powers R. Application of NMR metabolomics to search for human disease biomarkers. Comb. Chem. High Throughput Screen. 2012;15(8):595–610. doi: 10.2174/138620712802650522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Touze J, Fourcade L, Pradines B, Hovette P, Paule P, Heno P. Mechanism of action of antimalarials. Value combined atovaquone/ proguanil. Med. Trop. (Mars) 2002;62(3):219–224. [PubMed] [Google Scholar]