Abstract
Because of the role DNA damage and depletion play in human disease, it is important to develop and improve tools to assess these endpoints. This unit describes PCR-based methods to measure nuclear and mitochondrial DNA damage and copy number. Long amplicon quantitative polymerase chain reaction (LA-QPCR) is used to detect DNA damage by measuring the number of polymerase-inhibiting lesions present based on the amount of PCR amplification; real-time PCR (RT-PCR) is used to calculate genome content. In this unit we provide step-by-step instructions to perform these assays in Homo sapiens, Mus musculus, Rattus norvegicus, Caenorhabditis elegans, Drosophila melanogaster, Danio rerio, Oryzias latipes, Fundulus grandis, and Fundulus heteroclitus, and discuss the advantages and disadvantages of these assays.
Keywords: QPCR, DNA damage, mitochondrial DNA copy number, mitochondrial DNA, C. elegans
INTRODUCTION
Certain types of DNA lesions (e.g. bulky adducts and single-strand breaks) have the ability to inhibit or block the action of DNA polymerases (Ponti et al., 1991). Some of these types of damage are repaired via nucleotide excision repair (NER), a repair mechanism that mitochondria lack (LeDoux et al., 1992; Scheibye-Knudsen et al., 2015). As a result, mitochondrial DNA (mtDNA) accumulates these lesions at higher rates than nuclear DNA (nucDNA), decreasing levels of DNA replication and transcription and potentially causing deleterious health effects (Meyer et al., 2013; Niranjan et al., 1982; Stairs et al., 1983). Despite active NER and other repair mechanisms, polymerase-stalling lesions in nuclear DNA can also accumulate with aging as repair capacity decreases (Meyer et al., 2007; Sedelnikova et al., 2004).
Mitochondrial genome depletion occurs in many mitochondrial diseases (Copeland, 2012; Suomalainen and Isohanni, 2010), and has also been implicated in the disease pathogenesis of common diseases like cancer and Parkinson’s disease (Coskun et al., 2012; Yu, 2011).
Given the relevance of DNA damage and depletion in the context of organismal health, it is important to develop and improve tools to assess these endpoints. This unit provides updated protocols to measure nuclear and mitochondrial DNA damage and genome copy number using PCR-based assays initially developed in 1992 (Kalinowski et al., 1992). The DNA damage assay is able to quantitatively measure the number of polymerase-stalling lesions based on the amount of amplification obtained from a long amplicon quantitative PCR (LA-QPCR) (Furda et al., 2014; Furda et al., 2012; Hunter et al., 2010; Kalinowski et al., 1992). To calculate DNA copy number we utilize a real-time PCR (RT-PCR) assay in which by using a standard curve or comparing average cycle threshold (Ct) values we can calculate actual or relative DNA content (Rooney et al., 2015; Venegas and Halberg, 2012). This unit also includes updated support protocols with instructions for sample preparation, quantification, and generation of plasmids for copy number calculation using standard curves. Strengths and limitations of the assay are discussed in the Commentary.
BASIC PROTOCOL 1
ANALYSIS OF MITOCHONDRIAL AND NUCLEAR DNA DAMAGE
The purpose of this assay is to measure the number of DNA lesions capable of blocking or inhibiting polymerase activity. An LA-QPCR is run for the sample of interest, and the amount of resulting amplification will be inversely correlated with the number of polymerase-blocking lesions present in the DNA template. Assuming a Poisson distribution of lesions (Ayala-Torres et al., 2000), the amplification for treated samples can be compared with the amplification for control samples and a relative lesion frequency can be calculated (Furda et al., 2014; Furda et al., 2012). Control or reference samples are defined as having no damage for the purposes of this calculation.
Materials
Nuclease-free H2O
LongAmp Hot Start Taq 2× Master Mix (New England Biolabs)
10 µM primers, diluted with 0.1× TE buffer
Template DNA
Sterile, aerosol filter tips and pipettes dedicated to LA-QPCR
0.2 ml PCR tubes
96-well format thermocycler
Dedicated workstation (e.g. PCR hood equipped with UV lamp for sterilization)
Protocol steps
Long-Amplicon Quantitative Polymerase Chain Reaction (LA-QPCR)
-
1
UV-sterilize the work area.
-
2If running several samples, prepare a fresh master mix immediately before using by adding its components in the following order: nuclease-free water (16 µl per reaction, for a final volume of 50 µl), LongAmp Master Mix (25 µl per reaction), and primers (2 µl of each 10 µM primer working solution per reaction). Gently mix and spin.We set up reactions at room temperature. Avoid high-speed vortexing of the master mix, as the LongAmp Master Mix contains detergents. Always prepare enough master mix for two more reactions than needed (more than two if running a large number of reactions). We have successfully set up 25 instead of 50 µl reactions to decrease costs, if desired, by proportionally reducing all component volumes by 50%.
-
3Add 15 ng of purified template DNA (5 µl of 3ng/µl DNA for a 50 µl reaction) or 5 µl of C. elegans lysate to each 0.2 ml PCR tube. Also include no template and 50 % control reactions; they will be used for background subtraction and cycle number optimization (described in more detail below).No template and 50 % controls must be included for quality control purposes. Nuclease-free water or 0.1× TE buffer must be used in place of the DNA template in the case of the no template control. Control template DNA must be diluted 1:1 with nuclease-free water or 0.1× TE buffer and then used as template for the 50 % control reactions. To reduce pipetting error, prepare a larger volume (at least 20 µL) of 50% template than needed.
-
4
Carefully dispense 45 µl of the master mix prepared in step 2 to each PCR tube, avoiding introduction of bubbles or residual liquid on tube walls. Spin down using a PCR tube minicentrifuge.
-
5Set up thermocycler with reaction conditions. Our standard conditions are: an initial denaturation step of 2 min at 94 °C, followed by an optimized number of cycles of a denaturation step of 15 sec at 94 °C, and a combined annealing/extension step of 12 minutes at 62–68 °C (11 min 30 sec works for C. elegans). Also include a final extension step of 10 minutes at 72 °C. Refer to Table 1 for the target-specific PCR parameters we have optimized in the laboratory.The recommended annealing/extension temperature range is based on our optimized primers from Table 1; this temperature will vary based on the primers used (see Table 2). The optimal cycle number is the one at which the PCR is in the exponential phase i.e. a 50 % dilution of a template results in approximately 50 % the amplification observed for the 100 % template.
-
6
Place your PCR reactions into the thermocycler and start the program. Make sure the thermocycler lid is heated to 99 °C throughout the entire PCR reaction time. UV-sterilize the workstation once finished.
-
7
Keep the reaction products at 4–8 °C until ready to analyze.
Table 1.
LA-QPCR primers, targets and conditions optimized for LongAmp PCR Kit
| Species | Genome | Forward Primer Seq | Reverse Primer Seq | Annealing Temperature (°C) |
Cycle Number |
Reference |
|---|---|---|---|---|---|---|
| C. elegans | mt | 10.9 kb mito fragment | 64 | 26 | This unit, Hunter et al. (2010) | |
| 5’-CCA TCA ATT GCC CAA AGG GGA GT-3’ |
5’-TGT CCT CAA GGC TAC CAC CTT CTT CA-3’ |
|||||
| nuc | 9.3 kb nuc fragment from the unc-2 gene | 64 | 29 | This unit, Hunter et al. (2010) | ||
| 5’-TGG CTG GAA CGA ACC GAA CCA T-3’ |
5’-GGC GGT TGT GGA GTG TGG GAA G-3’ |
|||||
|
M. musculus |
mt | 10.9 kb mito fragment | 64 | 17 | This unit, Furda et al. (2012) | |
| 5’-GCC AGC CTG ACC CAT AGC CAT AAT AT-3’ |
5’-GAG AGA TTT TAT GGG TGT AAT GCG G-3’ |
|||||
| nuc | 8.7 kb nuc fragment of the β-globin gene, accession number X14061 |
62 | 27 | This unit, Ayala-Torres et al. (2000) | ||
| 5’-TTG AGA CTG TGA TTG GCA ATG CCT-3’ |
5’-CCT TTA ATG CCC ATC CCG GAC T-3’ |
|||||
| H. sapiens | mt | 8.9 kb mito fragment, accession number J01415 | 64 | 17 | This unit, Furda et al. (2012) | |
| 5’-TCT AAG CCT CCT TAT TCG AGC CGA-3’ |
5’-TTT CAT CAT GCG GAG ATG TTG GAT GG-3’ |
|||||
| nuc | 13.5 kb nuc fragment from the 5’ flanking region near the β-globin gene, accession number J00179 |
64 | 28 | This unit, Furda et al. (2012) | ||
| 5’-CGA GTA AGA GAC CAT TGT GGC AG-3’ |
5’-GCA CTG GCT TAG GAG TTG GAC T-3’ |
|||||
| D. rerio | mt | 10.3 kb mito fragment | 66 | 29 | This unit, Hunter et al. (2010) | |
| 5’-TTA AAG CCC CGA ATC CAG GTG AGC-3’ |
5’-GAG ATG TTC TCG GGT GTG GGA TGG-3’ |
|||||
| nuc | 10.7 kb nuc fragment of the AHR2 gene | 68 | 30 | |||
| 5’-AGA GCG CGA TTG CTG GAT TCA C-3’ |
5’-GTC CTT GCA GGT TGG CAA ATG G-3’ |
|||||
|
F. heteroclitus |
mt | 13.1 kb mitochondria fragment, accession number NC_012312 |
63 | 22 | This unit, Hunter et al. (2010), Jung et al. (2009) |
|
| 5’-AAG GAA ACA AGG AGC CGG TA-3’ |
5’-ACG TAG CGA GAA GGG TTA GG-3’ |
|||||
| nuc | 11.5 kb fragment of the CFTR gene, accession number AY028263 |
65 | 29 | |||
| 5’-CAG CCG CCC GCA AAT TCT CA-3’ |
5’-CAG AAT GCG GGC CTT GCT GA-3’ |
|||||
|
R. norvegicus |
mt | 12.1 kb mito fragment | 66 | 19 | This unit | |
| 5’- TCG CCC CAA CCC TCT CCC TT -3’ |
5’- TCG CCC CAA CCC TCT CCC TT -3’ |
|||||
| nuc | 12.9 kb nuc fragment from chromosome 12 genomic scaffold 5380 |
66 | 23 | This unit | ||
| 5’- CCT GCT GGG CTT GCC TTG GT -3’ |
5’- AGC AGG GGA GGT GGA TGG GA -3’ |
|||||
Table 2.
Long- and short-amplicon QPCR primers and targets (not yet optimized for LongAmp PCR kit)
| Species | Genome | Forward Primer Seq | Reverse Primer Seq | Annealing Temperature (°C) |
Reference |
|---|---|---|---|---|---|
| M. musculus | mt | 117 bp mito fragment | 60 | Ayala-Torres et al. (2000), Furda et al. (2012) | |
| 5’-CCC AGC TAC TAC CAT CAT TCA AGT-3’ |
5’-GAT GGT TTG GGA GAT TGG TTG ATG T-3’ |
||||
| nuc | 6.6 kb nuc fragment of the DNA polymerase gene β, accession number AA79582 |
64 | Furda et al. (2012) | ||
| 5’-TAT CTC TCT TCC TCT TCA CTT CTC CCC tgg-3’ |
5’-CGT GAT GCC GCC GTT GAG GGT CTC CTG-3’ |
||||
| H. sapiens | mt | 221 bp mito fragment | 62 | Furda et al. (2012) | |
| 5’-CCC CAC AAA CCC CAT TAC TAA ACC CA-3’ |
5’-TTT CAT CAT GCG GAG ATG TTG GAT GG-3’ |
||||
| nuc | 12.2 kb nuc fragment from region of the DNA polymerase gene β, accession number L11607 |
64 | Furda et al. (2012) | ||
| 5’-CAT GTC ACC ACT GGA CTC TGC AC-3’ |
5’-CCT GGA GTA GGA ACA AAA ATT GCT G-3’ |
||||
| 10.4 kb nuc fragment encompassing exons 2–5 of HPRT gene, accession number J00205 |
64 | Furda et al. (2012) | |||
| 5’-TGG GAT TAC ACG TGT GAA CCA ACC-3’ |
5’-GCT CTA CCC TGT CCT CTA CCG TCC-3’ |
||||
|
D. melanogaster |
mt | 151 bp mito fragment | 61 | Hunter et al. (2010) | |
| 5’-GCT CCT GAT ATA GCA TTC CCA CGA-3’ |
5’-CAT GAG CAA TTC CAG CGG ATA AA-3’ |
||||
| 14.2 kb mito fragment | 66 | ||||
| 5’-GCC GCT CCT TTC CAT TTT TGA TTT CC-3’ |
5’-TGC CAG CAG TCG CGG TTA TAC CA-3’ |
||||
| nuc | 152 bp nuc fragment | 65 | |||
| 5’-CGA GGG ATA CCT GTG AGC AGC TT-3’ |
5’-GTC ACT TCT TGT GCT GCC ATC GT-3’ |
||||
| 11.5 kb nuc fragment of the β-tubulin gene | 67 | ||||
| 5’-GTA TTC CTG CGC CAG GAG GAT CG-3’ |
5’-CAG ATG CTG GAG CTG CCT TTG GA-3’ |
||||
| 10.3 kb nuc fragment of the β-tubulin gene | 67 | ||||
| 5’-GAG GAG CCT TGC GAA CAA CAG CA-3’ |
5’-CAA TGA CAG CTG CGC CTC GAG AT-3’ |
||||
| D. rerio | mt | 198 bp mito fragment | 62 | Hunter et al. (2010) | |
| 5’-CAA ACA CAA GCC TCG CCT GTT TAC-3’ |
5’-CAC TGA CTT GAT GGG GGA GAC AGT-3’ |
||||
| nuc | 233 bp nuc fragment | 60 | |||
| 5’-ATG GGC TGG GCG ATA AAA TTG G-3’ |
5’-ACA TGT GCA TGT CGC TCC CAA A-3’ |
||||
| C. elegans | mt | 195 bp mito fragment | 63 |
Hunter et al. (2010) Meyer et al. (2007) |
|
| 5’-CAC ACC GGT GAG GTC TTT GGT TC-3’ |
5’-TGT CCT CAA GGC TAC CAC CTT CTT CA-3’ |
||||
| nuc | 225 bp nuc fragment | 63 |
Meyer et al. (2007) Boyd et al. (2010) |
||
| 5’-TCC CGT CTA TTG CAG GTC TTT CCA-3’ |
5’-GAC GCG CAC GAT ATC TCG ATT TTC-3’ |
||||
| 13.7 kb nuc fragment from the polymerase epsilon gene | 68 | Hunter et al. (2010) | |||
| 5’-AGT CGT TGA ACG CAG TGG TGT CAT-3’ |
5’-CAG TCT TTC TTC GAC GCA TTC AAC G-3’ |
||||
| O. latipes | mt | 184 bp mito fragment | 59 | Rooney et al. (2015) | |
| 5’-AAC TCC AAG TAG CAG CTA TGC AC-3’ |
5’-GAG GGG TAG AAG GCT TAC AAA AA-3’ |
||||
| nuc | 140 bp nuc fragment | 57 | |||
| 5’-CTC ACA AAC ATC TTT GCA CTC AG-3’ |
5’-AGA ACC TCT CTC CAA AAC ATT CC-3’ |
||||
| F. grandis | mt | 206 bp mito fragment | 55 | ||
| 5’-TTT ACA CAT GCA AGT ATC CG-3’ |
5’-CCG AAG GCT ATC AAC TTG AG-3’ |
||||
| nuc | 234 bp nuc fragment | 62 | |||
| 5’-GCC GCT GCC TTC ATT GCT GT-3’ |
5’-ATG AGC TGG GTG TGC GCT GA-3’ |
||||
| F. heteroclitus | nuc | 234 bp nuc fragment | 62 |
Hunter et al. (2010) Jung et al. (2009) |
|
| 5’-GCC GCT GCC TTC ATT GCT GT-3’ |
5’-ATG AGC TGG GTG TGC GCT GA-3’ |
||||
Product Quantification and Quality Check
-
8Quantify PCR products following the steps listed in Support Protocol 3. In this case 10 µl of PCR product are used per well instead of DNA extract (add 90 µl of 1× TE instead of 95 µl to account for the volume difference). Also, the PCR product does not need to be diluted. Refer to Supplemental File 1 for an example of this procedure.Be careful not to open tubes with PCR product in the same room used for setting up the PCR. This prevents cross-contamination of new PCR reactions, which is critical because the same reaction, amplifying the same target, is repeated over and over and the product can volatilize and contaminate an entire room. When running LA-QPCR with new parameters or for the first time, run the PCR products on an agarose electrophoresis gel first to make sure a unique product of the correct size is present (no extra bands). After only one product is observed, perform the product quantification.
-
9Average the duplicate fluorescence values for all samples. Subtract the average value of the “no template control” (blank) sample from all other samples. Make sure that the value of the blank-corrected 50 % control signal is 40 – 60 % of the blank-corrected control sample signal.The fluorescence value for the “no template control” (blank) should be between 5000–7000 (this value will vary based on the plate reader used, but once established will be consistent over time). Higher values are indicative of cross-contamination. If the 50 % control is between 40 and 60 % of the control sample, this indicates that the PCR reaction is in the exponential phase; this means that all amplification is directly proportional to the starting amount of template. If this is not the case, the cycle number needs to be increased or decreased in order to achieve exponential amplification.
-
10
As a quality control check, LA-QPCR for each sample needs to be run twice, and the values obtained should to be compared using correlation analysis. Plot the blank-corrected values from one run against the other and calculate the correlation coefficient. If the correlation is good (we recommend r2>0.9), the values for replicate PCRs are averaged; if it is poor, a third LA-QPCR run is necessary in order to eliminate outliers. Comparing the values obtained from all three runs using correlation analysis helps identify and remove outliers.
Calculating DNA lesion frequencies
-
11Average all copy number values (obtained by following Basic Protocol 2), and divide each sample’s copy number value by this average.Skip this step if using the logarithmic curve method described in Basic Protocol 2.
-
12
Divide each sample’s blank-corrected fluorescence value by its corresponding ratio obtained in step 11. If using the logarithmic curve method described in Basic Protocol 2, divide by the calculated normalization factors here, instead of the ratios from step 11. This is done to normalize the amount of PCR product to the amount of DNA copies in each template.
-
13Average all normalized fluorescence values for the control samples, and divide each normalized sample value by this average. The resulting number is the amplification relative to control.Each normalized control value is also divided the by the average of all normalized control values.
-
14Take the negative natural logarithm (-ln) of each relative amplification value. The resulting number is the lesion frequency for each sample. We usually represent this value as lesions per 10 kb. In order to do this, multiply the lesion frequency by 10 and divide by the size (in kb) of the LA-QPCR DNA target. Refer to Supplemental Files 1 and 3 to see lesion number calculations.Lesion frequencies need to be calculated for at least two separate LA-QPCR runs of the same sample (technical replicates). We usually collect three different samples from each treated and control group (biological replicates), and perform the experiment at least twice (total n = 6 at minimum).
Statistical Analysis
-
15
Take the average of the lesions/10 kb values of biological replicates; this is the level of DNA damage for the treatment or control group.
-
16
Graph the averaged lesions/10kb values as mean ± standard error (use standard deviation if interested in representing the distribution of damage levels within a population).
-
17Perform statistical analysis with parametric tests. If comparing two samples, perform a t-test or one-way analysis of variance (ANOVA). If comparing more than two samples, always perform an ANOVA. If there is more than one independent variable perform a multifactor ANOVA first; if the result is significant then compare desired subsets of the data with post-hoc tests.In our experience, DNA damage data has always been normally distributed. In the event that your dataset is not, data transformation techniques or non-parametric alternatives to the tests recommended in this step should be used.
BASIC PROTOCOL 2
ANALYSIS OF MITCHONDRIAL AND NUCLEAR GENOME COPY NUMBER
Although the number of genome copies can be measured by amplifying a short target sequence using the quantitative PCR assay described in steps 1–10 of Basic Protocol 1 (primers for short-amplicon QPCR are in Table 2)(Furda et al., 2012; Rooney et al., 2015), we currently perform this analysis using RT-PCR instead. This method is particularly advantageous if a standard curve is run along with the samples of interest, because the actual number of copies can be calculated (Bratic et al., 2009; Leung et al., 2013). Mitochondrial DNA content from purified DNA samples can be calculated without a standard curve by using the comparative Ct method, resulting in a measure of mtDNA content relative to nucDNA copy number (Rooney et al., 2015; Venegas and Halberg, 2012) or, if the goal is to normalize the results from LA-QPCR (Basic Protocol 1, step 12), by creating a logarithmic curve based on the average genome Ct value (see below).
Materials
Nuclease-free water
100,000 copies/µl aliquots of pCR 2.1 plasmid containing cloned species-specific nuclear or mitochondrial gene – if calculating with standard curve; Refer to Support Protocol 5
40 µl young adult (24 hr post-L4) glp-1 worm lysate (20 worms; 1567 copies/µl) – alternative to C. elegans nuclear plasmid for standard curve calculations; refer to Support Protocol 1
Template DNA
10 µM primers
Power SYBR Green PCR Master Mix (Life Technologies)
0.2 ml PCR tubes
Optical 96-well PCR plate and optical adhesive film
Real-Time PCR System
Plate vortexer
Centrifuge
Protocol steps
Real-time PCR (RT-PCR)
-
1Prepare a serial dilution with which to calculate a standard curve. Using0.2 ml PCR tubes proceed as follows: dilute a 100,000 copies/µl aliquot of the plasmid down to 32,000 copies/µl and 24,000 copies/µl. Serially dilute each preparation 1:1 until getting a 2,000 copies/µl dilution (32,000 copies/µl preparation) and a 3,000 copies/µl dilution (24,000 copies/µl preparation). If calculating nuclear copy number for worms, and using worm glp-1 lysate instead of a plasmid, add 40 µl of nuclease-free water to lysate; concentration will now be 784 copies/µl. Serially dilute this preparation 1:1 until getting a 24.5 copies/µl dilution.Skip this step if not calculating copy number using a standard curve. Single use aliquots of plasmids and glp-1 lysates are used to prevent freeze/thaw cycles. Glp-1 worms are used because when grown at 25 °C they do not develop a germline, and therefore have a constant number of nuclear DNA copies, as described in (Leung et al., 2013). Plotting the Ct values obtained against the known number of copies per standard dilution, and analyzing by logarithmic regression allows us to calculate the exact number of copies in our sample.
-
2If running several samples, prepare a fresh master mix immediately before using by adding its components in the following order: nuclease-free water (8.5 µl per reaction, for a final volume of 25 µl), Power SYBR Green Master Mix (12.5 µl per reaction), and primers (1 µl of each 10 µM primer working solution per reaction). Gently mix and spin.We set up reactions at room temperature. Always prepare enough master mix for two more reactions than needed (more than two if running a large number of reactions).
-
3
Aliquot 23 µl of the master mix prepared in step 2 to each well.
-
4Add 2 µl of each plasmid dilution to triplicate wells in the PCR plate, including a 0 copies control (nuclease-free water or 0.1× TE). The number of plasmid copies per well for the standard curve would be as follows: 64,000, 48,000, 32,000, 24,000, 16,000, 12,000, 8,000, 6,000, 4,000. If using glp-1 worm lysate, the number of nuclear DNA copies per well for the standard curve would be as follows: 1,568, 784, 392, 196, 98 and 49.Skip this step if not calculating copy number with a standard curve.
-
5Add 6 ng of purified template DNA (2 µl of 3ng/µl DNA for a 25 µl reaction) or 2 µl of worm lysate to wells in triplicate. Also include a no template control reaction.A no template control must be included for quality control purposes. If running plasmid serial dilutions for a standard curve, then the 0 copies/µl reaction is the no template control. Nuclease-free water or 0.1× TE buffer must be used in place of the DNA template.
-
6
Cover plate with optical film and spin, vortex and spin again (vortex at around 1650 rpm for 30 seconds using a plate vortexer; centrifuge at 400 rcf for 10 seconds).
-
7Set up a program in the RT-PCR system with the following conditions: 2 min at 50 °C, 10 min at 95 °C, 40 cycles of 15 sec at 95 °C and 60 sec at primer-specific annealing temperature. A dissociation curve is also calculated to ensure presence of only one PCR product.Refer to Table 3 for target-specific PCR parameters we have optimized in the laboratory.
-
8Place your PCR reactions on the real-time system and start the program.We usually perform one RT-PCR run per sample, with triplicate technical and biological replicates, and perform the experiment at least twice (total n = 6 at minimum).
Table 3.
Real-time PCR primers and targets
| Species | Genome | Forward Primer Seq | Reverse Primer Seq | Annealing Temperature (°C) |
Reference |
|---|---|---|---|---|---|
| C. elegans | mt | 75 bp mito fragment of nd1 mito | 60 | Bratic et al. (2009) | |
| 5’-AGC GTC ATT TAT TGG GAA GAA GAC-3’ |
5’- AAG CTT GTG CTA ATC CCA TAA ATG T -3’ |
||||
| nuc | 164 bp nuc fragment of cox4 | 60 | Rooney et al. (2015) | ||
| 5’-GCC GAC TGG AAG AAC TTG TC-3’ |
5’-GCG GAG ATC ACC TTC CAG TA-3’ |
||||
| D. rerio | mt | 195 bp nd1 mito fragment | 60 | This unit | |
| 5’-CGT TTA CCC CAG ATG CAC CT-3’ |
5’-GTG CGA TTG GTA GGG CGA TA-3’ |
||||
| nuc | 90 bp nuc fragment of vtg2 | 60 | |||
| 5’-TGG ATA CCT GAC CGA GAG CT-3’ |
5’-AGA CAA CTC TTA CGG CTG GC-3’ |
||||
| H. sapiens | mt | 107 bp mito fragment of tRNA-Leu(UUR) gene | 62 | Venegas and Halberg (2012) | |
| 5’-CAC CCA AGA ACA GGG TTT GT-3’ |
5’-TGG CCA TGG GTA TGT TGT TA-3’ |
||||
| nuc | 86 bp nuc fragment of β2-microglobulin gene | 62 | |||
| 5’- TGC TGT CTC CAT GTT TGA TGT ATC T -3’ |
5’- TCT CTG CTC CCC ACC TCT AAG T -3’ |
||||
|
R. norvegicus |
mt | 181 bp mito fragment | 60 | This unit | |
| 5’- CAA ACC TTT CCT GCA CCT CC -3’ |
5’- AGG CGT TCT GAT GAT GGG AA -3’ |
||||
| nuc | 144 bp nuc fragment from 3',5'-cyclic AMP phosphodiesterase (PDE4-1, PDE4-2) gene |
62 | |||
| 5’-GTT CCC GCC TTC TTC CTC TG-3’ |
5’-GTT TGC TTG CCG ACT CCT TG-3’ |
||||
|
F. heteroclitus |
mt | 131 bp mito fragment of 16S rRNA gene | 60 | This unit | |
| 5’-AAA ATT AAC GGC CCC AAC CC-3’ |
5’-CCG AGT TCC TTC TTC CCC TT-3’ |
||||
| nuc | 234 bp nuc fragment of CFTR gene | 60 |
Hunter et al. (2010) Jung et al. (2009) |
||
| 5’-GCC GCT GCC TTC ATT GCT GT-3’ |
5’-ATG AGC TGG GTG TGC GCT GA-3’ |
||||
Data analysis
-
9
Obtain the Ct information from the PCR results. Compare the triplicate values to each other; if any given one varies by more than 0.5 Ct from the others it can be thrown out from the calculations. Average technical replicates.
-
10Plot standard curve Ct values against the known number of copies in each well, and perform a logarithmic regression. The resulting equation should be as follows: Ct = m*ln(x) + b, where m is the slope, b is the y-intercept and x is the sample’s genome copy number. Another way to look at this equation is by isolating the x variable as follows: x = e(Ct − b)/mSkip this step if not calculating copy number with a standard curve.
-
11Use the equation from step 10 and calculate the copy number for each sample. If working with worm lysates as DNA template, obtain the copy number per worm by multiplying the total copy number per sample by the template volume equivalent to one worm (e.g. if 6 worms were picked into 90 µl, then you would multiply by 15) and then divide by the volume of template added to the well (2 µl). Refer to Supplemental File 2 for example calculations.Skip this step if not calculating copy number with a standard curve.
-
12To calculate the mitochondrial genome content of purified DNA samples the comparative Ct method can be used instead of a standard curve (Venegas and Halberg, 2012). Subtract the mtDNA averaged Ct values from the nucDNA averaged Ct values (both from step 9); this is the ΔCt. Calculate the relative mitochondrial DNA content by raising 2 to the power of ΔCt and then multiplying by 2 (calculate fold difference). Expressed as equations this would be:Skip this step if calculating copy number with a standard curve. Note that this method only provides mtDNA content relative to nucDNA; if a calculation of actual genome copy number is desired, utilize standard curves.
-
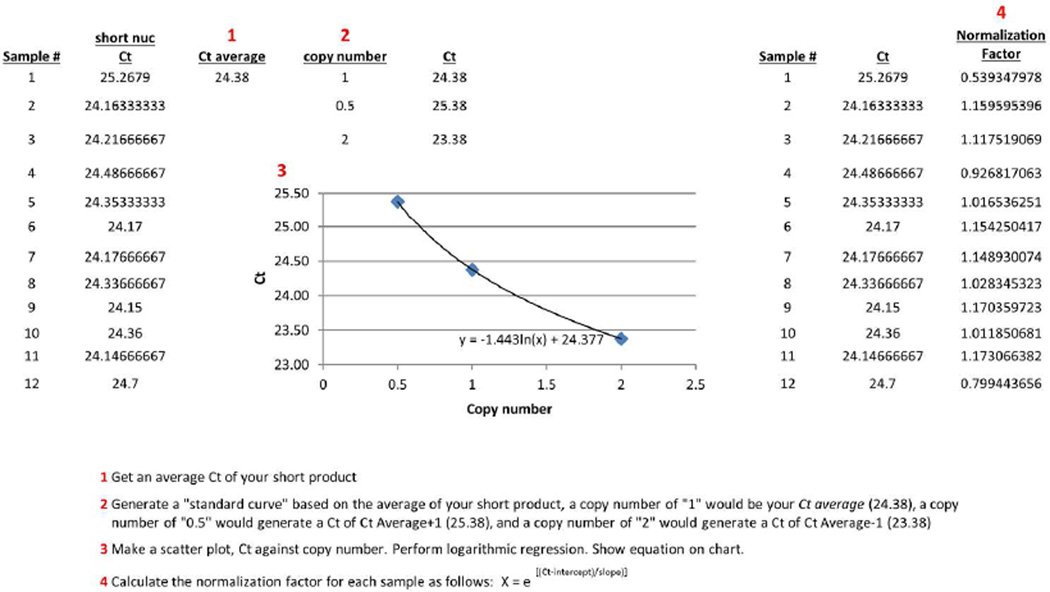
13If copy number is being calculated solely for normalization of LA-QPCR, a normalization ratio can be obtained without utilizing standard curves or the comparative Ct method. This is accomplished by creating a logarithmic curve using the Ct values for all samples. To do this, average all Ct values for the experiment (after averaging all technical replicates per sample in step 9). Generate a "standard curve" based on the experiment Ct average; designate a copy number of "1" to the Ct average. Calculate the Ct for a copy number of "0.5" with the equation Ct = Ct Average+1, and calculate it for a copy number of "2" with the equation Ct = Ct Average−1. Make a scatter plot with these hypothetical values, Ct against copy number. Perform logarithmic regression; the equation will be Ct = m*ln(x) + b, where x is the sample’s hypothetical copy number. Another way to look at this equation is by isolating the x variable as follows: x = e(Ct − b)/m. Calculate x for each sample using the equation from the exponential regression; this is the normalization factor (see Figure 1 for a visual representation of this method; refer to Supplemental File 3 for example calculations).Skip this step if calculating copy number with a standard curve.
-
14
Perform statistical analysis with parametric tests. If comparing two samples, perform a t-test or one-way analysis of variance (ANOVA). If comparing more than two samples, always perform an ANOVA. If there is more than one independent variable perform a multifactor ANOVA first; if the result is significant then compare desired subsets of the data with post-hoc tests.
-
15Data obtained from C. elegans lysates is typically best represented graphically as normalized to nucDNA copy number in order to indicate copy number per cell. Normalize mtDNA copy number from each sample by dividing the number of mtDNA copies per worm by the number of nucDNA copies per worm. Graph this data as a mtDNA:nucDNA ratio.Skip this step if not working with C. elegans lysates.
Figure 1. Calculation of normalization factors for DNA damage assay using logarithmic regression.
A standard curve is generated based on the calculated Ct values and corresponding hypothetical copy numbers. Logarithmic regression is performed and the resulting equation is utilized to calculate normalization factors.
SUPPORT PROTOCOL 1
DNA TEMPLATE EXTRACTION FROM C. elegans (SMALL NUMBER OF WORMS)
Worms of interest are lysed to obtain their DNA and use it as a template for PCR reactions. This method is much faster and less labor-intensive than the traditional batch DNA extraction (Furda et al., 2012; Hunter et al., 2010; Rooney et al., 2015).
Materials
Worm lysis buffer (see recipe)
Worms of interest
Platinum wire worm pick
0.2 ml PCR tubes
Ice or cryogenic 96-well plate (PCR cooler)
−80 °C freezer
96-well format thermocycler or heat block
Protocol steps
Aliquot 90 µl of lysis buffer into each PCR tube. Place PCR tubes with buffer on ice or on a cryogenic 96-well plate.
- Pick 6 worms (L4 stage or later) into each PCR tube. Immediately place tubes with worms in the −80 °C freezer. Do not leave worms on buffer unfrozen for more than 5 min.If using worms younger than L4 stage, pick 9 worms per 90 µl of lysis buffer instead.
Place tubes with worms in the −80 °C freezer for at least 10 minutes in order to disrupt the nematode’s cuticle.
Set up a program in the thermocycler to lyse the worms as follows: 65 °C for 1 hr, then 95°C for 15 min, and once finished hold at 4–8 °C.
Remove PCR tubes from the −80 °C freezer and place in thermocycler. Start program. Make sure lid is set to be heated to 99 °C.
- Use worm lysate as template for LA-QPCR immediately or store at −80°C until further use.In our experience, worm lysate cannot be stored at 4 °C if it is intended for use in LA-QPCR. Storing at −80 °C, even if this exposes the lysate to multiple freeze/thaw cycles, preserves the integrity of the DNA better than storing it at 4 °C. This is not an issue if the lysate is going to be used solely for short amplicon PCR or RT-PCR.
SUPPORT PROTOCOL 2
DNA TEMPLATE EXTRACTION FROM C. elegans (LARGE NUMBER OF WORMS) OR ANIMAL TISSUE
Traditionally, DNA is extracted from the cells or tissue of interest and purified before use in PCR reactions. Extracting DNA from cells can be done easily following standard procedures and commercial kits that result in very high molecular weight DNA that is not oxidized during extraction; we recommend QIAGEN Genomic-tip 20/G kit, using a 1 × 106 cell pellet and following the kit’s tissue protocol (the cell culture protocol isolates the nuclei and discards the mtDNA). However, extracting DNA from other samples like animal tissue or large numbers of C. elegans requires special care to preserve adequate DNA integrity for LA-QPCR. Before using the extraction kits these samples must also be snap frozen first and manually ground (or homogenized if using fresh soft tissue) (Furda et al., 2012; Hunter et al., 2010).
Materials
K medium (for C. elegans, see recipe)
Worms or tissue of interest
20 % glycerol solution (for animal tissues)
Liquid nitrogen
Dry ice
Genomic-tip 20/G kit (QIAGEN)
Reagents needed for Genomic-tip kit: Buffers G2, QBT, QC and QF, RNase A and Proteinase K (QIAGEN)
Isopropanol
70 % ethanol
15 ml screw-cap conical tubes
Orbital shaker (C. elegans)
Glass Pasteur pipettes (C. elegans)
Liquid nitrogen-cooled mortar and pestle (C. elegans and tough animal tissue)
1–2 ml cryotubes
−80 °C freezer
Handheld homogenizer (softer animal tissue)
2.2 ml centrifuge tubes
Water bath (50 °C)
Refrigerated microcentrifuge
Tabletop centrifuge with 15 ml tube buckets (C. elegans)
Protocol steps
- Wash off 3000–5000 worms from a culture dish with K medium into a 15 ml conical and centrifuge at 2200 × g for 2 min. Carefully remove supernatant and refill with 10 ml of K medium. Place tube on shaker for 20 min in order to allow worms to clear their guts.Skip this step if extracting DNA from animal tissue.
- Centrifuge tube for 2 min at 2200 × g. Perform a wash by carefully removing supernatant, refilling tube with K medium and then centrifuging again using the same parameters. Repeat washing step, this time leaving a small volume of the supernatant behind.Skip this step if extracting DNA from animal tissue.
- Using the Pasteur pipette, resuspend worms in leftover supernatant and freeze them by dripping the suspension directly into a mortar with liquid nitrogen.Skip this step if extracting DNA from animal tissue.
Frozen worm pellets can be placed in cryotubes and stored at −80 °C until further processing. Snap freeze tissue samples (up to 20 mg per sample; optional for soft tissues) by placing the tissue in a cryotube with 20 % glycerol solution and storing it at −80 °C.
Cool mortar and pestle by packing dry ice underneath the mortar, and pouring liquid nitrogen on the mortar and pestle and letting it evaporate.
Place about 6 frozen worm pellets or the frozen tissue sample on the chilled mortar and carefully grind with the pestle until a squeaking sound is heard. If using fresh soft tissue, homogenize it directly in buffer G2 and RNAse A following the Genomic Tips protocol.
Using a sterilized spatula, scoop up powder from mortar into a 15 ml conical tube with buffer G2 and RNAse A as described by the Genomic Tips protocol.
- Continue following the genomic tips tissue protocol in order to isolate DNA.It is possible to extract DNA from cultured cells or tissue with an automated protocol, but in this case mtDNA must be digested prior to amplification. Refer to Furda et al. (2012) and see Support Protocol 4.
SUPPORT PROTOCOL 3
DNA QUANTIFICATION
An important step in ensuring a successful PCR is accurately measuring the amount of template DNA. The same amount of template must be used for all samples and all runs. This quantification is not necessary if using worm lysates, as a precise number of age-synchronized worms were picked into the lysis buffer (in principle, it would similarly be possible to base LA-QPCR on small nucDNA quantification, although we have not optimized this approach). PicoGreen dye is used as it fluoresces >1,000-fold brighter when bound to DNA, and has a 25 pg/ml limit of detection.
Materials
1× TE buffer
λ DNA/Hind III Fragments (Invitrogen)
Extracted DNA samples
50 µl aliquots (stored at −20 °C) of Quant-iT PicoGreen dsDNA reagent (Molecular Probes)
0.1× TE buffer
96-well white- or black-bottomed plates
Fluorescence plate reader capable of measuring 485 nm excitation and 528 nm emission
Protocol steps
- Prepare a DNA serial dilution to calculate a standard curve by diluting λ DNA/Hind III fragments to 15, 10, 5, 2.5, 1.25 and 0 ng/µl in 1× TE buffer.We recommend preparing large volumes (e.g. 2 ml) of the standard curve and using them repeatedly in order to reduce variability among measurements. Store at 4 °C.
- Dilute the DNA extracts 1:10 in 1× TE in order to get the DNA concentration in the range of the standard curve.Depending on the amount of worms, cells or tissue used for extraction, it might be necessary to further dilute the DNA or use the undiluted extract.
Add 95 µl of 1× TE to each well to be used in the 96-well plate (add 90 µl to standard wells). The number of wells needed is the number of samples and standards in duplicate.
Add 5 µl of each sample and 10 µl of each standard to the wells containing 1× TE in duplicate.
- Turn the overhead lab lights off to prepare PicoGreen solution. Remove a PicoGreen aliquot from the −20 °C freezer and allow it to thaw at room temperature. Once thawed, prepare a solution of 5 µl PicoGreen per 1 ml 1× TE. You need 100 µl of this solution per well undergoing quantification.Make excess PicoGreen solution to account for pipetting loss.
- Add 100 µl of PicoGreen solution to each well to be analyzed. Cover plate with aluminum foil and incubate at room temperature for 10 minutes.PicoGreen solution is best added with a multichannel pipette.
Set up the plate reader. Set filters to 485 nm excitation and 528 nm emission and add a 20 sec shaking step before the reading step. Measure fluorescence.
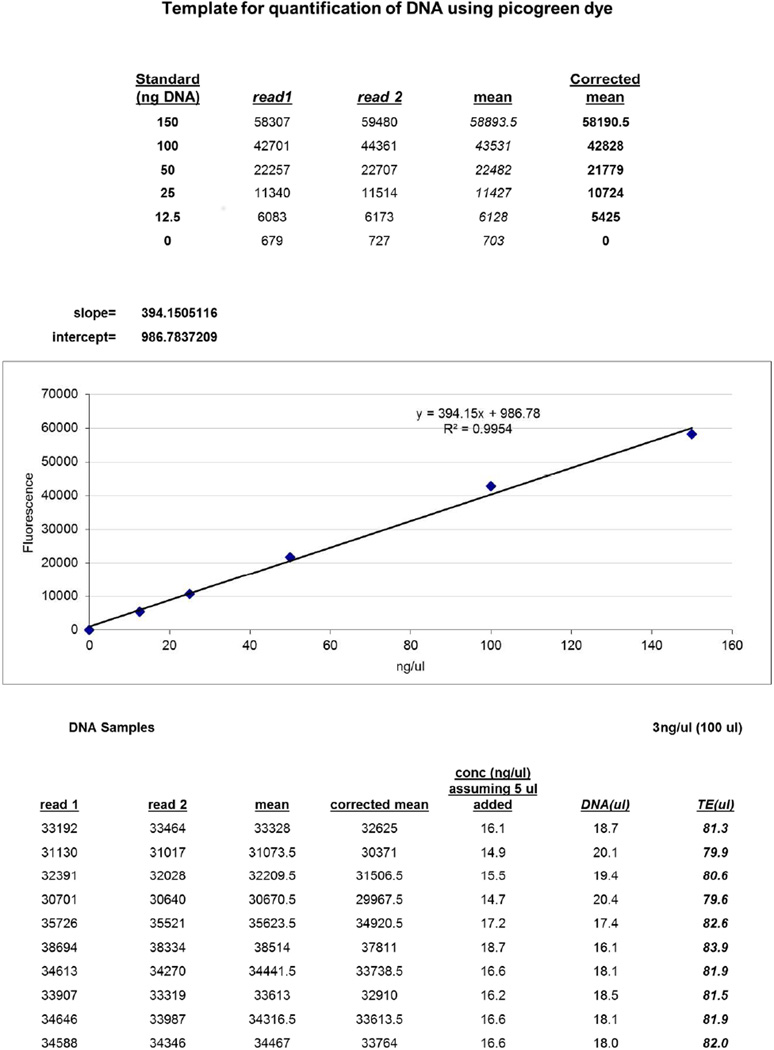
Calculate the DNA concentration of the samples by comparing sample fluorescence values to the standard curve values (Figure 2; refer to Supplemental File 4 for example calculations). Plot the fluorescence readings for the standard curve against the standard curve concentrations, and perform correlation analysis. Use the resulting equation to calculate the DNA concentration in the samples of interest. If the fluorescence reading or concentration for a sample is higher than the highest standard, dilute sample further and measure again. If sample signal is too low (close to the 0 or 1.25 ng/µl standard readings) use undiluted samples and measure again.
- Dilute desired amount of DNA extract in 0.1× TE buffer so that the concentration is 3ng/µl. DNA is now ready to use for PCR assays.The final dilution that results in a concentration of 3 ng/µL should be carried out using a preceding concentration of no more than 15 ng/µL to ensure accurate measurement and dilution. If the preceding dilution is more concentrated, carry out as many additional dilutions to obtain a penultimate concentration of no more than 15 ng/µL.To save time and reagents, and especially if some samples have very high concentrations of DNA, it is also possible to carry out the initial (rough) quantification using a nanodrop or similar spectrophotometer, and then carry out the first dilution and proceed using PicoGreen as described above. Nanodrop and PicoGreen methods do not give identical values, however, so this initial quantification should be considered an estimate.
Figure 2. Example of DNA quantification template.
The fluorescence values for the standard curve are plotted against the standard concentrations and correlation analysis is performed. Using the equation, the actual DNA concentration of the samples can be calculated. Based on this concentration, the DNA can be diluted with 0.1× TE to bring the concentration down to 3 ng/ul.
SUPPORT PROTOCOL 4
LINEARIZATION OF MITOCHONDRIAL DNA FOLLOWING AUTOMATED DNA EXTRACTION
As mentioned earlier, it is possible to extract DNA from samples in a fully automated manner by using a QIAcube from QIAGEN. However, after QIAcube extraction the mtDNA appears to be mostly in the supercoiled conformation, making primer access difficult (Furda et al., 2012). Utilizing restriction enzymes to linearize the mtDNA in a region outside the amplification target can alleviate this.
Materials
225 ng purified DNA
Restriction enzyme (HaeII for human, PvuII for mouse and XhoI for rat, and associated CutSmart or NEBuffer buffer; New England Biolabs)
Nuclease-free water
Ice (optional)
0.2 ml PCR tubes
96-well format thermocycler
Protocol steps
Calculate the volume of the purified DNA needed to perform the digest (need 225 ng, refer to Support Protocol 3). Final digestion volume will be 50 µl per sample.
Calculate the amount of nuclease-free water needed in order to bring the volume up to 44 µl (subtract the value obtained in step 1 from 44 µl)
Pipette the calculated amount of purified DNA and nuclease-free water into a PCR tube. Also add 5 µl of the appropriate buffer.
Add 1 µl of the enzyme and incubate in a thermocycler following the time and temperature parameters described in the enzyme supplier protocol.
Place samples in ice or store at 4 °C immediately following incubation. No further purification is needed for PCR assays.
SUPPORT PROTOCOL 5
GENERATING STANDARD CURVE PLASMIDS FOR MITCHONDRIAL AND NUCLEAR GENOME COPY NUMBER ANALYSIS
In order to measure the absolute mitochondrial and nuclear genome copy numbers by RT-PCR, we have generated a standard curve by cloning the respective short (100–250 bp) target sequences into the pCR2.1 plasmid. We adopted the restriction enzymes cloning method for our work. Bacterial glycerol stocks are available from us upon request.
Materials
Nuclease-free water
Taq DNA polymerase with standard Taq buffer
Respective primers
QIAquick PCR Purification Kit (QIAGEN)
Restriction enzymes (HindIII, and Xho-1 and associated NEBuffer buffer; New England Biolabs)
Agarose
1× TE buffer
Gel Extraction Kit
T4 DNA ligase and associated buffer; New England Biolabs
Heat shock competent E. coli (Any recA− cloning strain)
SOC medium (Sambrook and Russell, 2001)
Ampicillin
LB Agar ampicillin plates (100 µM/ml) (Sambrook and Russell, 2001)
LB broth (Sambrook and Russell, 2001)
Plasmid purification kit
Sterile, aerosol filter tips and pipettes dedicated to PCR
0.2 ml PCR tubes
NanoDrop2000 UV-Vis Spectrophotometer
96-well format thermocycler
Dedicated workstation (e.g. PCR hood equipped with UV lamp for sterilization)
Nucleic acid electrophoresis unit
37 °C shaker incubator
Heat block
1.7 ml microcentrifuge tubes
Sterile round bottom polystyrene test tube with snap cap
Protocol steps
Obtain genomic DNA from animal tissue by following the steps listed in support protocol 2. Quantify the extracted DNA using a NanoDrop2000 UV-Vis Spectrophotometer at 260nm.
- Design primers with modifications made to the 5’-end of the forward and reverse primers: restriction site addition and 5'-extension to the restriction site. Refer to Table 4 for some species-specific cloning primers used in our work.The restriction site should be the same or provide the same sticky end to the choice of restriction enzymes used to digest the multiple cloning site of the cloning vector. For our cloning strategy we selected two sticky end cutters that create different 5'-overhangs: HindIII which recognizes the hexamer AAGCTT and XhoI which recognizes the hexamer CTCGAG. To increase the efficiency of the restriction enzyme - cleaving the end fragment of the primer DNA, the 5’ end of the restriction site is extended with 4 –6 random nucleotides.
- Perform a PCR to obtain sufficient amounts of template DNA by amplifying the insert region from genomic DNA. First prepare the reaction mix by adding the components in the following order: nuclease-free water (for a final volume of 50 µl), Taq Polymerase buffer (5×) (10 µl per reaction), dNTPs (5 µl per reaction), MgCl2 (50mM) (3 µl per reaction), forward and reverse primers (10 µM) (1.5 µl per reaction), Taq polymerase (2.5 U/µl) (0.5 µl per reaction), template DNA (5 µl per reaction or 200 to 500 ng per reaction). The above reagents are mixed in a 0.2 ml PCR tube and briefly spun down to remove any air bubbles.Reactions are set-up at room temperature. Since the amplified products are subjected to several downstream applications, it is advisable to scale-up the reaction numbers or reaction volume.
Perform the PCR reaction in a thermocycler with a heated lid using the conditions: an initial denaturation step at 94 °C for 2 min, followed by 30–35 cycles of a denaturation step at 94 °C of 15 sec, and a combined annealing/extension step at 62–65°C for 30 sec and a final extension step at 72 °C for 10 min. Pause the reaction at 8 °C until removed from the thermocycler.
PCR reaction clean-up: The amplified DNA fragment(s) should be purified from the reaction mixture using a commercial PCR purification kit such as QIAGEN’s QIAquick PCR purification kit prior to setting up the restriction enzyme digestion reaction.
- Double digest the plasmid and insert DNA by mixing the restriction digestion reaction in separate 1.7 ml microcentrifuge tubes in the following order: nuclease-free water (to a final volume of 50 µl per reaction), restriction enzyme buffer (10×) (5 µl per reaction), vector DNA (2 µg total) (10 µl per reaction); insert DNA (up to 40 µl of the PCR product can be used for each 50 µl reaction), restriction enzymes 1 and −2 (10–20 units/µl) (2 µl each per reaction).Both enzyme digestion reactions can be carried out simultaneously if the enzymes work equally in the commercially available buffers. For reference, check the New England Biolabs double digest finder to decide on the most compatible buffer. The Restriction enzyme is added last and mixed gently by pipetting the solution up and down.
Incubate the reaction mix at 37 °C in a heat block, filled with water, for up to 2 hours.
Purify the digested vector and insert DNA from the small DNA fragment (product of digestion) by running the reaction mix in a 2% agarose preparative gel at 100 volts for 45 minutes in 1× TE buffer.
Extract the respective products (vector and insert DNA) from the agarose gel using a commercial gel extraction kit.
- Ligate the digested and purified insert and linear vector DNA using the bacteriophage T4 DNA ligase enzyme. A standard sticky-end ligation reaction is set up by adding the ingredients in the following order: nuclease-free water (to a final volume of 20 µl per reaction), ligase buffer 10× (2 µl per reaction), digested vector DNA (~50 ng/µl) (2 µl per reaction), digested insert DNA (in molar concentrations as per the ratio; appropriate volume for respective ratios), T4 DNA ligase 20–40 NEB U/µl (1 µl per reaction). The reaction is set-up in a 0.2 ml PCR tube. Mix the contents gently by pipetting the solution up and down followed by a brief spin and incubate at 16°C overnight in a thermocycler.Reactions are set up with vector to insert DNA in a molar ratio of 1:1, 1:2 and 1:4 along with a control reaction containing the digested vector DNA only to determine the self-ligated non-recombinant background.
- Transform the ligation mix into chemically competent E. coli cells by heat shock. Briefly, thaw E.coli cells (50 µl per reaction) on ice-water. Add 5 to 10 µl of the ligation mix and mix gently. Incubate the mixture for 30 minutes on ice-water. Heat shock the cells for 45 sec at 42 °C and place them immediately on ice. Add 1ml of SOC medium and incubate at 37 °C for 1 hour. Pellet the culture by centrifuging at 5000 rpm. Remove 800 µl of the supernatant and plate out the re-suspended cultures on LB agar ampicillin plates (100 µM/ml). Incubate the plates overnight at 37 °C.We use commercially available competent E.coli cells to enable uptake of the circular vector DNA. As to the E. coli strain, use any recA- cloning strain such as TOP10 or DH5a. The negative control i.e. the vector only ligation mixture and a positive control i.e. an uncut known vector is also used.
- Pick and culture the E. coli transformants in 2 ml of LB broth containing either ampicillin (50 µg/ml) or kanamycin (50 µg/ml) antibiotics for a maximum of 16 h in a 37°C incubator shaker.We use a sterile aerosol filter tip to pick the transformed colonies from the LB plate. We also use a sterile round bottom polystyrene test tube with snap cap to culture bacteria
- Isolate the cloned plasmid from E. coli using a commercially available plasmid purification kit following the manufacturer’s protocol.We use QIAGEN QIAprep Spin Miniprep Kit (50); however, for this purpose any commercially available kit can be used.
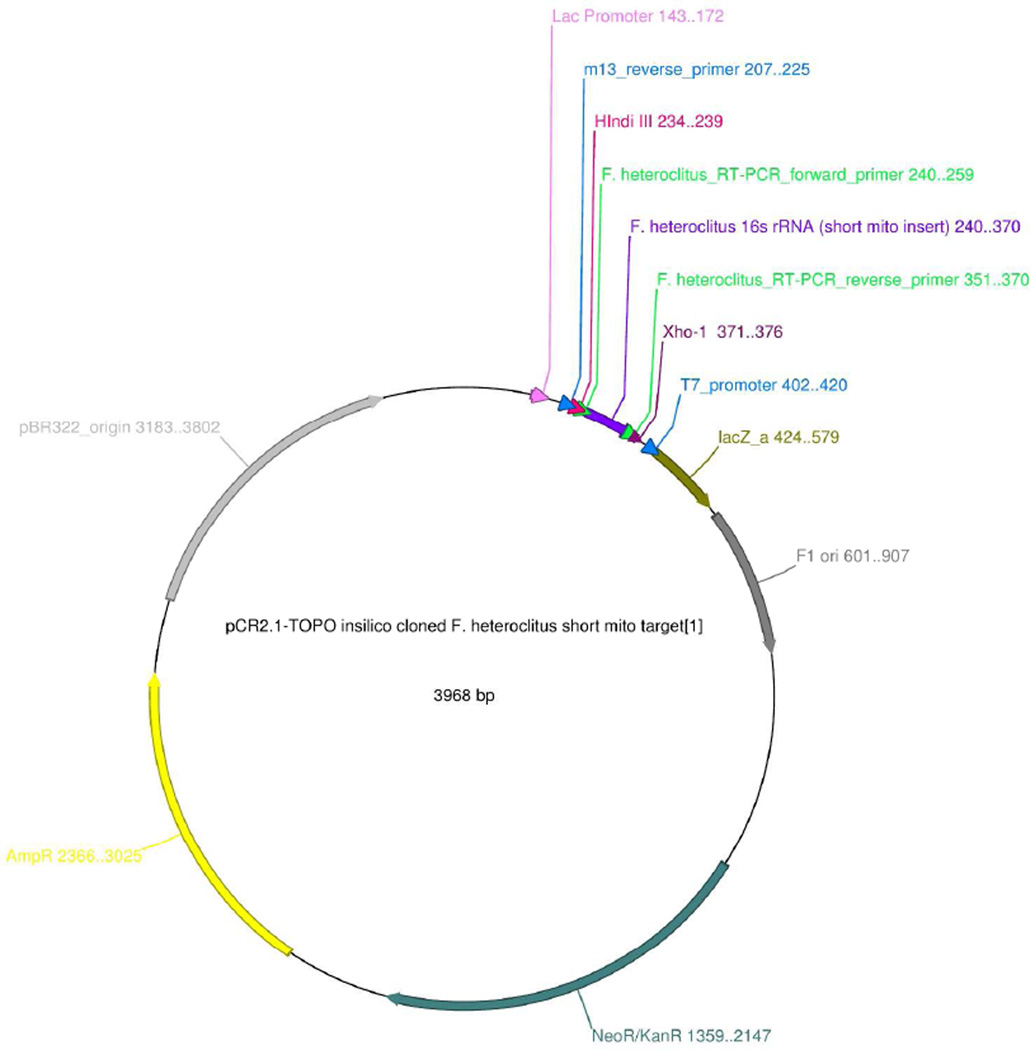
Analyze the transformants, after obtaining the plasmid, for the presence of the right insert either by restriction analysis or sequencing using M13 and or T7 sequencing primers (see Figure 3).
Table 4.
Cloning PCR primers and conditions
| Species | Genome | Target gene |
Forward primer Seq | Reverse primer Seq | Amplicon (bp) |
Annealing temperature (oC) |
Reference |
|---|---|---|---|---|---|---|---|
| C. elegans | mt | nd-1 | Not Reported | Not Reported | NR | NR | Bratic et al. (2009) |
| nuc | cox-4 | 5’- ATCT AAGCTT GCC GAC TGG AAG AAC TTG TC -3’ |
5’- ATAG CTCGAG GCG GAG ATC ACC TTC CAG TA -3’ |
184 | 60 | This unit | |
| H. sapiens | mt | tRNA- Leu(UUR) |
5’- ATCTAAGCTT GCC TTC CCC CGT AAA TGA TA -3’ |
5’- ATAG CTCGAG AGG AAT GCC ATT GCG ATT AG -3’ |
215 | 55 | This unit |
| nuc | B2M | 5’- ATCT AAGCTT TGC TGT CTC CAT GTT TGA TGT ATC T -3’ |
5’- ATAG CTCGAG TCT CTG CTC CCC ACC TCT AAG T -3’ |
106 | 62 | This unit | |
| F. heteroclitus | mt | 16S rRNA | 5’- ATAG AAGCTT AAA ATT AAC GGC CCC AAC CC -3’ |
5’- ACTT CTCGAG CCG AGT TCC TTC TTC CCC TT -3’ |
131 | 64 | This unit |
| nuc | CFTR | 5’- ATAG AAGCTT GCC GCT GCC TTC ATT GCT GT -3’ |
5’- AATT CTCGAG ATG AGC TGG GTG TGC GCT GA -3’ |
234 | 65 | Hunter et al. (2010) | |
| D. rerio | mt | nd-1 | 5’- ATCT AAGCTT CGT TTA CCC CAG ATG CAC CT -3’ |
5’- ATAG CTCGAG GTG CGA TTG GTA GGG CGA TA -3’ |
215 | 60 | This unit |
| nuc | vtg2 | 5’- ATCT AAGCTT TGG ATA CCT GAC CGA GAG CT -3’ |
5’- ATAG CTCGAG AGA CAA CTC TTA CGG CTG GC -3’ |
110 | 60 | This unit |
Underlined regions denote restriction sites.
Figure 3. Example of an in silico cloned short product target in pCR2.1 plasmid.
Figure shows cloned pCR2.1 containing the F. heteroclitus short mito target (131 bp, 16s rRNA) inserted at the multiple cloning site (MCS) using unique restriction enzymes HindIII and XhoI. The target was PCR amplified using primers that contained these restriction elements at the 5’ end of the forward and reverse primers. Sequencing primers, M13_reverse and T7_Promoter, are highlighted in blue.
REAGENTS AND SOLUTIONS
K-medium
2.36g Potassium chloride (KCl)
3.0g Sodium chloride (NaCl)
1.0L ddH2O
Autoclave to sterilize
Store at room temperature, indefinitely under sterile conditions
3.3X Worm Lysis Buffer (40 ml) – Adapted from (Cheng, 2001)
82.5 mM Tricine (pH 8)
264 mM Potassium acetate
36.2% w/v glycerol
7.425% v/v DMSO
16.95 ml nuclease-free water
Store at −20 °C indefinitely
COMMENTARY
Background Information
The LA-QPCR assay has been used to detect DNA damage, traditionally from cultured cells and model organisms, in the laboratory for more than 20 years (Kalinowski et al., 1992; Meyer, 2010). To date, it has been optimized for a variety of species, including humans, rats, mice, yeast, fruit fly, nematodes, plant and fish species, and others (Castro et al., 2012; Colton et al., 2014; Furda et al., 2014; Hunter et al., 2010; Jung et al., 2009; Kumar et al., 2014; Meyer et al., 2007). It consists of amplifying a long target in the genome of interest (~10 kb) and quantifying the PCR product. If polymerase-stalling lesions are present, the amplification product will be less than expected; comparing the level of amplification in the treated samples to the control samples allows us to estimate the lesion frequency. For this to be possible the amplification must be exponential; the amount of PCR product has to be directly proportional to the amount of starting template (Furda et al., 2012; Meyer, 2010).
There are several advantages to using this assay. First, little template and therefore sample is needed (we have used as little as 50–100 pg per sample, using individual nematodes). Second, the assay is highly specific, because it is primer-based, allowing analysis of the species, genome, and even genomic regions of interest. The DNA extraction methods here described isolate both mitochondrial and nuclear DNA; this allows us to measure damage in both genomes for each sample without separation of genomes that can lead to artifactual differences (Furda et al., 2012; Meyer, 2010). Different regions of nuclear genome can be analyzed, in order to compare region-specific damage or repair (e.g., Van Houten et al. (2000) and (Meyer et al., 2007); but note that not all of the targets described in those papers have been re-optimized for the current polymerase). Third, the assay detects many kinds of damage, providing an integrative although not complete measure of damage present. However, there are several limitations to this assay as well. Only lesions that stall the DNA polymerase can be detected, and it cannot discern among those lesions. For nucDNA damage measurements, there is the possibility that the region amplified is not representative of the entire genome. The lesion frequencies are calculated by using the control lesion levels as a damage-free baseline; however, of course, in fact all templates have some level of damage. Finally, if the template is obtained by batch extraction, lesion measurements cannot be made at the individual cell level (Meyer, 2010).
Traditionally, quantitative PCR is also used to measure genome copy number, by amplifying a small region (~200 bp) of the genome. This small target is unlikely to have DNA lesions and therefore is a good representation of the genome content present (Furda et al., 2012; Santos et al., 2006). These values are used to normalize the amplification from LA-QPCR to the actual amount of starting copies, which can vary dramatically for mtDNA from cell to cell. It is also helpful when amplifying C. elegans lysates, as it can account for errors in picking (wrong number of worms per lysate), and worm size in the case of growth-variable populations (Hunter et al., 2010). The method presented in this protocol is based on RT-PCR instead and, if standard curves are run alongside samples, it has the advantage of allowing us to calculate the actual number of mitochondrial and nuclear genome copies. If standard curves are not available, we use the comparative Ct method and as a result the measure of mtDNA content is relative to nucDNA content (Bratic et al., 2009; Rooney et al., 2015; Venegas and Halberg, 2012). If the goal is to normalize results from LA-QPCR, and standard curves are not available, we employ a method that utilizes logarithmic regression to calculate normalization factors per sample.
Critical Parameters
The biggest concern when performing these PCR-based assays is obtaining high-quality DNA that has not been sheared, oxidized or otherwise damaged. Standard DNA extraction techniques do not yield high molecular weight DNA that is suitable for the LA-QPCR assay, although such DNA can be used for copy number analysis. Similarly, phenol-chloroform-based extraction methods cause DNA oxidation that may obscure the damage of interest. Further discussion of these concerns, and examples of how to analyze DNA integrity, are presented in (Meyer, 2010).
Another major concern is cross-contamination of newly isolated template DNA with PCR products generated in previous PCR reactions. To avoid this, high-quality reagents must be used and gloves worn at all times. UV-irradiate the work area before and after sample preparation. Use dedicated pipettes and tips and designate a PCR-only workstation. Never open the completed reactions in the same room where the dedicated PCR workstation is located. Always include a no template control to monitor for contamination.
Another parameter to keep in mind is pipetting consistency and accuracy. Small changes in volume can greatly increase variability in the PCR results, as can introduction of air bubbles and creation of microenvironments in residual drops on tube walls or caps.
The normality of the data obtained must be verified, and parametric or non-parametric statistical tests should be run based on this analysis.
Troubleshooting
The most common problem encountered is contamination. If despite taking precautions the results from the no template controls suggest the presence of contamination, the first thing to do is replace the reagents. Particularly, economical reagents that are aliquoted and kept in the workstation like nuclease-free water and TE buffers need to be exchanged for fresh replacements. It is also important to decontaminate all work areas and instruments (e.g., PCR hood, pipettors, racks, etc.).
If designing primers for a new species, or if the optimization of cycle number or other PCR parameters is problematic, please refer to Furda et al. (2012), Hunter et al. (2010) and Meyer (2010) for suggestions. Follow our PCR parameter recommendations keeping in mind that all PCR conditions may require some re-optimization in the context of different thermocyclers and other equipment.
Using the hot start master mix version of the PCR kits greatly reduces the chances of problems arising and reduces noise and variability. We have not yet optimized our PCR parameters for the regular, non-master mix kit.
Anticipated Results
These assays yield genome copy number values and DNA damage values for nuclear and mitochondrial genomes. Examples of results obtained with these assays are available as supplemental files. For the DNA damage assay, we expect the data to be normally distributed. However, this needs to be corroborated for every new experiment. In the event that the data is not normally distributed, non-parametric statistical tests should be used to analyze the data. It is important to keep in mind that the limit of detection for the DNA damage assay is ~1 lesion per 105 bases (Meyer, 2010), although this value depends on the number of replicates and variability between them. Lesion frequencies below this value cannot be considered DNA damage. The assay can be used to measure DNA repair by examining damage levels over time, as long as there is not significant cell division that results in production of new, undamaged nucDNA, or mtDNA replication. Either event will dilute the number of lesions/10 kb by increasing the number of total bases, confounding measurement of damage removal or repair.
Time Considerations
Manual DNA purification from tissue takes one to two days, depending on the number of samples and how quickly they elute, with some down time during incubations and elutions. Worm lysis can take up to 2 hours, depending on the number of samples. DNA quantification (pre- or post-PCR) takes approximately 45 minutes. Setting up the LA-QPCR or RT-PCR can take an hour or more depending on the number of samples. Running the PCR program varies depending on the number of cycles, but is frequently 6–8 hours for the LA-QPCR assay.
Supplementary Material
Acknowledgments
We thank Aleksandra Trifunovic for sharing the mtDNA copy number plasmid for C. elegans with us. This work was supported by NIEHS R01-ES017540-01A2 and P42ES010356.
Footnotes
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
LITERATURE CITED
- Boyd WA, Crocker TL, Rodriguez AM, Leung MCK, Wade Lehmann D, Freedman JH, Van Houten B, Meyer JN. Nucleotide excision repair genes are expressed at low levels and are not detectably inducible in Caenorhabditis elegans somatic tissues, but their function is required for normal adult life after UVC exposure. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2010;683:57–67. doi: 10.1016/j.mrfmmm.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratic I, Hench J, Henriksson J, Antebi A, Burglin TR, Trifunovic A. Mitochondrial DNA level, but not active replicase, is essential for Caenorhabditis elegans development. Nucleic acids research. 2009;37:1817–1828. doi: 10.1093/nar/gkp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro MdR, Suarez E, Kraiselburd E, Isidro A, Paz J, Ferder L, Ayala-Torres S. Aging increases mitochondrial DNA damage and oxidative stress in liver of rhesus monkeys. Experimental gerontology. 2012;47:29–37. doi: 10.1016/j.exger.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. Amplification of long nucleic acid sequences by PCR. Google Patents. 2001 [Google Scholar]
- Colton MD, Kwok KWH, Brandon JA, Warren IH, Ryde IT, Cooper EM, Hinton DE, Rittschof D, Meyer JN. Developmental toxicity and DNA damage from exposure to parking lot runoff retention pond samples in the Japanese medaka (Oryzias latipes) Marine Environmental Research. 2014;99:117–124. doi: 10.1016/j.marenvres.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland WC. Defects in mitochondrial DNA replication and human disease. Critical Reviews in Biochemistry and Molecular Biology. 2012;47:64–74. doi: 10.3109/10409238.2011.632763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun P, Wyrembak J, Schriner SE, Chen H-W, Marciniack C, LaFerla F, Wallace DC. A mitochondrial etiology of Alzheimer and Parkinson disease. Biochimica et Biophysica Acta (BBA) - General Subjects. 2012;1820:553–564. doi: 10.1016/j.bbagen.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furda A, Santos J, Meyer J, Van Houten B. Quantitative PCR-Based Measurement of Nuclear and Mitochondrial DNA Damage and Repair in Mammalian Cells. In: Keohavong P, Grant SG, editors. Molecular Toxicology Protocols. Vol. 1105. Humana Press; 2014. pp. 419–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furda AM, Bess AS, Meyer JN, Van Houten B. Analysis of DNA damage and repair in nuclear and mitochondrial DNA of animal cells using quantitative PCR. Methods Mol Biol. 2012;920:111–132. doi: 10.1007/978-1-61779-998-3_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter SE, Jung D, Di Giulio RT, Meyer JN. The QPCR assay for analysis of mitochondrial DNA damage, repair, and relative copy number. Methods. 2010;51:444–451. doi: 10.1016/j.ymeth.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Cho Y, Meyer JN, Di Giulio RT. The long amplicon quantitative PCR for DNA damage assay as a sensitive method of assessing DNA damage in the environmental model, Atlantic killifish (Fundulus heteroclitus) Comparative biochemistry and physiology. Toxicology & pharmacology : CBP. 2009;149:182–186. doi: 10.1016/j.cbpc.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski DP, Illenye S, Van Houten B. Analysis of DNA damage and repair in murine leukemia L1210 cells using a quantitative polymerase chain reaction assay. Nucleic acids research. 1992;20:3485–3494. doi: 10.1093/nar/20.13.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RA, Oldenburg DJ, Bendich AJ. Changes in DNA damage, molecular integrity, and copy number for plastid DNA and mitochondrial DNA during maize development. Journal of Experimental Botany. 2014;65:6425–6439. doi: 10.1093/jxb/eru359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDoux SP, Wilson GL, Beecham EJ, Stevnsner T, Wassermann K, Bohr VA. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–1973. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- Leung M, Rooney J, Ryde I, Bernal A, Bess A, Crocker T, Ji A, Meyer J. Effects of early life exposure to ultraviolet C radiation on mitochondrial DNA content, transcription, ATP production, and oxygen consumption in developing Caenorhabditis elegans. BMC Pharmacology and Toxicology. 2013;14:9. doi: 10.1186/2050-6511-14-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JN. QPCR: a tool for analysis of mitochondrial and nuclear DNA damage in ecotoxicology. Ecotoxicology. 2010;19:804–811. doi: 10.1007/s10646-009-0457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JN, Boyd WA, Azzam GA, Haugen AC, Freedman JH, Van Houten B. Decline of nucleotide excision repair capacity in aging Caenorhabditis elegans. Genome biology. 2007;8:R70. doi: 10.1186/gb-2007-8-5-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JN, Leung MCK, Rooney JP, Sendoel A, Hengartner MO, Kisby GE, Bess AS. Mitochondria as a Target of Environmental Toxicants. Toxicological Sciences. 2013;134:1–17. doi: 10.1093/toxsci/kft102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjan B, Bhat N, Avadhani N. Preferential attack of mitochondrial DNA by aflatoxin B1 during hepatocarcinogenesis. Science. 1982;215:73–75. doi: 10.1126/science.6797067. [DOI] [PubMed] [Google Scholar]
- Ponti M, Forrow SM, Souhami RL, D'Incalci M, Hartley JA. Measurement of the sequence specificity of covalent DNA modification by antineoplastic agents using Taq DNA polymerase. Nucleic acids research. 1991;19:2929–2933. doi: 10.1093/nar/19.11.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney J, Ryde I, Sanders L, Howlett EH, Colton M, Germ K, Mayer G, Greenamyre JT, Meyer J. PCR Based Determination of Mitochondrial DNA Copy Number in Multiple Species. In: Palmeira CM, Rolo AP, editors. Mitochondrial Regulation. Vol. 1241. New York: Springer; 2015. pp. 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning : a laboratory manual. 3rd. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol. 2006;314:183–199. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- Scheibye-Knudsen M, Fang EF, Croteau DL, Wilson DM, Iii, Bohr VA. Protecting the mitochondrial powerhouse. Trends in Cell Biology. 2015;25:158–170. doi: 10.1016/j.tcb.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedelnikova OA, Horikawa I, Zimonjic DB, Popescu NC, Bonner WM, Barrett JC. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nature cell biology. 2004;6:168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- Stairs PW, Guzelian PS, Van Tuyle GC. Benzo[a]pyrene differentially alters mitochondrial and nuclear DNA synthesis in primary hepatocyte cultures. Res Commun Chem Pathol Pharmacol. 1983;42:95–106. [PubMed] [Google Scholar]
- Suomalainen A, Isohanni P. Mitochondrial DNA depletion syndromes – Many genes, common mechanisms. Neuromuscular Disorders. 2010;20:429–437. doi: 10.1016/j.nmd.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Van Houten B, Cheng S, Chen Y. Measuring gene-specific nucleotide excision repair in human cells using quantitative amplification of long targets from nanogram quantities of DNA. Mutation Research/DNA Repair. 2000;460:81–94. doi: 10.1016/s0921-8777(00)00018-5. [DOI] [PubMed] [Google Scholar]
- Venegas V, Halberg M. Measurement of Mitochondrial DNA Copy Number. In: Wong PDL-JC, editor. Mitochondrial Disorders. Vol. 837. Humana Press; 2012. pp. 327–335. [DOI] [PubMed] [Google Scholar]
- Yu M. Generation, function and diagnostic value of mitochondrial DNA copy number alterations in human cancers. Life Sciences. 2011;89:65–71. doi: 10.1016/j.lfs.2011.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.