Abstract
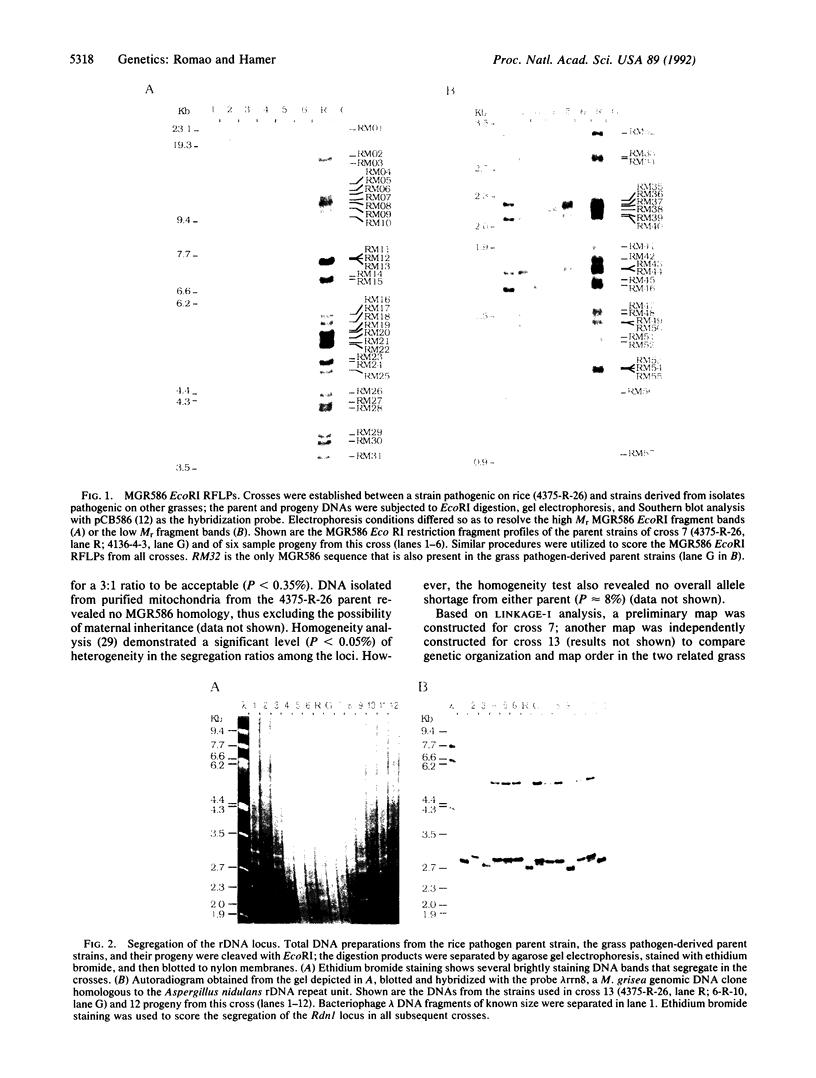
The fungal rice pathogen Magnaporthe grisea contains repetitive DNA sequences called MGR. We have used a DNA probe, MGR586, derived from these sequences and crosses between rice-pathogenic and non-rice-pathogenic laboratory strains of M. grisea to rapidly map genes in this organism. The rice-pathogenic strain contained 57 EcoRI restriction fragments that hybridize to the MGR586 probe; the other five non-rice-pathogenic parent strains contained a single MGR586 sequence. Genetic analysis of MGR segregation detected eight linkage groups and allowed the mapping of three pigmentation genes (Alb1, Rsy1, and Buf1), the mating type locus (Mat1), the nucleolar organizer (Rdn1), the Smol gene, and two restriction fragment length polymorphisms linked to Smol. Our results indicate that the MGR586 loci are randomly distributed about the M. grisea genome and permit the construction of a well-marked linkage map useful for future studies on genome organization and genetic analysis in M. grisea.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brody H., Griffith J., Cuticchia A. J., Arnold J., Timberlake W. E. Chromosome-specific recombinant DNA libraries from the fungus Aspergillus nidulans. Nucleic Acids Res. 1991 Jun 11;19(11):3105–3109. doi: 10.1093/nar/19.11.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford M. S., Chumley F. G., Weaver C. G., Valent B. Characterization of the Heterokaryotic and Vegetative Diploid Phases of MAGNAPORTHE GRISEA. Genetics. 1986 Dec;114(4):1111–1129. doi: 10.1093/genetics/114.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer J. E., Farrall L., Orbach M. J., Valent B., Chumley F. G. Host species-specific conservation of a family of repeated DNA sequences in the genome of a fungal plant pathogen. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9981–9985. doi: 10.1073/pnas.86.24.9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer J. E., Givan S. Genetic mapping with dispersed repeated sequences in the rice blast fungus: mapping the SMO locus. Mol Gen Genet. 1990 Sep;223(3):487–495. doi: 10.1007/BF00264458. [DOI] [PubMed] [Google Scholar]
- Hamer J. E. Molecular probes for rice blast disease. Science. 1991 May 3;252(5006):632–633. doi: 10.1126/science.252.5006.632. [DOI] [PubMed] [Google Scholar]
- Hamer J. E., Valent B., Chumley F. G. Mutations at the smo genetic locus affect the shape of diverse cell types in the rice blast fungus. Genetics. 1989 Jun;122(2):351–361. doi: 10.1093/genetics/122.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Leung H., Lehtinen U., Karjalainen R., Skinner D., Tooley P., Leong S., Ellingboe A. Transformation of the rice blast fungus Magnaporthe grisea to hygromycin B resistance. Curr Genet. 1990 May;17(5):409–411. doi: 10.1007/BF00334519. [DOI] [PubMed] [Google Scholar]
- Levy M., Romao J., Marchetti M. A., Hamer J. E. DNA Fingerprinting with a Dispersed Repeated Sequence Resolves Pathotype Diversity in the Rice Blast Fungus. Plant Cell. 1991 Jan;3(1):95–102. doi: 10.1105/tpc.3.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbach M. J., Vollrath D., Davis R. W., Yanofsky C. An electrophoretic karyotype of Neurospora crassa. Mol Cell Biol. 1988 Apr;8(4):1469–1473. doi: 10.1128/mcb.8.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons K. A., Chumley F. G., Valent B. Genetic transformation of the fungal pathogen responsible for rice blast disease. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4161–4165. doi: 10.1073/pnas.84.12.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. D., Barry E. G. The cytogenetics of Neurospora. Adv Genet. 1977;19:133–285. doi: 10.1016/s0065-2660(08)60246-1. [DOI] [PubMed] [Google Scholar]
- Ruvkun G., Ambros V., Coulson A., Waterston R., Sulston J., Horvitz H. R. Molecular genetics of the Caenorhabditis elegans heterochronic gene lin-14. Genetics. 1989 Mar;121(3):501–516. doi: 10.1093/genetics/121.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suiter K. A., Wendel J. F., Case J. S. LINKAGE-1: a PASCAL computer program for the detection and analysis of genetic linkage. J Hered. 1983 May-Jun;74(3):203–204. doi: 10.1093/oxfordjournals.jhered.a109766. [DOI] [PubMed] [Google Scholar]
- Valent B., Farrall L., Chumley F. G. Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics. 1991 Jan;127(1):87–101. doi: 10.1093/genetics/127.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent B., Farrall L., Chumley F. G. Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics. 1991 Jan;127(1):87–101. doi: 10.1093/genetics/127.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990 Dec 25;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]