Abstract
Background
Previous research has demonstrated that ectopic expression of Ran-binding protein (RanBP) in Arabidopsis results in more axillary buds and reduced apical dominance compared to WT plants. However, the function of RanBP in poplar, which has very typical secondary growth, remains unclear. Here, the Populus deltoides (Marsh.) RanBP gene (PdRanBP) was isolated and functionally characterized by ectopic expression in a hybrid poplar (P. davidiana Dode × P. bolleana Lauche).
Results
PdRanBP was predominantly expressed in leaf buds and tissues undergoing secondary wall expansion, including immature xylem and immature phloem in the stem. Overexpression of PdRanBP in poplar increased the number of sylleptic branches and the proportion of cells in the G2 phase of the cell cycle, retarded plant growth, consistently decreased the size of the secondary xylem and secondary phloem zones, and reduced the expression levels of cell wall biosynthesis genes. The downregulation of PdRanBP facilitated secondary wall expansion and increased stem height, the sizes of the xylem and phloem zones, and the expression levels of cell wall biosynthesis genes.
Conclusions
These results suggest that PdRanBP influences the apical and radial growth of poplar trees and that PdRanBP may regulate cell division during cell cycle progression. Taken together, our results demonstrated that PdRanBP is a nuclear, vascular tissue development-associated protein in P. deltoides.
Electronic supplementary material
The online version of this article (doi:10.1186/s12863-016-0403-4) contains supplementary material, which is available to authorized users.
Keywords: Molecular cloning, Functional analysis, PdRanBP, Populus deltoides, Vascular tissue
Background
Forests provide the raw materials for a very large amount of wood products. The process of wood formation and development is mediated by the activity of the vascular cambium, which is a meristematic cell population that facilitates vascular tissues development in tree stems [1]. The development of vascular tissue (secondary xylem and secondary phloem) includes the emergence of new tissues through regular cell division, horizontal and radial extension and, ultimately, cell maturation [2–4]. However, the mechanisms that regulate secondary wall thickening and subsequent expansion of the stems remain largely unknown. Genetic engineering could be used to improve specific traits in plants without the need for long-term breeding, and other valuable traits can be stably inherited from the parental genetic material [5–9].
Small GTP-binding genes play diverse roles in a multitude of cellular processes, such as microtubule organization, vesicle-mediated intracellular trafficking, signal transduction, and cell growth and division in plants and animals [10, 11]. The Ras-related nuclear protein (Ran or RAN) is a member of an important family of small GTP-binding proteins. Ran interacts with importin or exportin proteins to regulate a variety of biochemical processes, including nuclear envelope assembly, nucleo-cytoplasmic signal transfer, cell cycle progression, light signalling, resistance to pathogens, and the regulation of hormone sensitivities [12–17]. Ran-binding protein (RanBP) is vital for the transit of nuclear proteins between the stages of mitosis and interphase. Lee et al. [18] found that expression of the pea (Pisum sativum L., cv. Alaska) Ran gene (PsRan1) is regulated by various light sources via a phytochrome-mediated signalling pathway. Overexpression of the wheat (Triticum aestivum L.) RAN gene (TaRAN1) increased the amount of primordial tissue, reduced the number of lateral roots, and stimulated hypersensitivity to exogenous auxin in Arabidopsis thaliana (L.) and rice (Oryza sativa L.) [19]. Virus-induced gene silencing (VIGS) of the Nicotiana benthamiana (Domin.) RanBP gene (NbRanBP1) caused leaf yellowing, abnormal leaf morphology, and stunted growth in transgenic N. benthamiana plants. Defence-related genes were induced and mitochondrial membrane potential was reduced in NbRanBP1 VIGS plants [20]. Transgenic Arabidopsis expressing the antisense Arabidopsis RanBP1c gene (AtRanBP1c) displayed enhanced primary root growth but suppressed lateral root growth. Antisense AtRanBP1c transgenic plants were hypersensitive to auxin and had an increased mitotic index in both the lateral and primary roots [21]. The overexpression of the O. sativa RAN gene (OsRAN2) resulted in extreme sensitivity to abscisic acid (ABA), osmotic stress, and salinity in rice and A. thaliana [22].
Molecular and genetic studies in tree species (e.g., poplar and Eucalyptus gunnii) and Arabidopsis have uncovered a number of wood-associated transcription factors and other proteins that might be involved in secondary wall formation [23–25]. Among the identified transcription factors, the best-characterized are the NAC and MYB families. Populus trichocarpa (Torr. & Gray) wood-associated NAC domain transcription factors (PtrWNDs) are master switches that activate a suite of downstream transcription factors, such as PtrNAC150, PtrNAC156, PtrNAC157, PtrMYB90, PtrMYB18, PtrMYB74, PtrMYB75, PtrMYB121 and PtrMYB128. These proteins are involved in the coordinated regulation of secondary wall biosynthesis during wood formation [26]. P. deltoides PdMYB221 has been shown to be involved in the negative regulation of secondary wall formation through the direct and indirect suppression of gene expression related to secondary wall biosynthesis [27]. It has recently been shown that P. tomentosa PtoMYB92 activates the lignin biosynthetic pathway; specifically, this factor activates the expression of the lignin biosynthetic genes CCOAOMT1, CCR2 and C3H3 by binding to their promoters [28]. Eucalyptus gunnii (J.T. Hook) cinnamoyl coenzyme A reductase (EgCCR) is expressed in all lignifying cells (vessel elements and xylem fibres) of xylem tissues and is associated with primary and secondary xylem formation in Arabidopsis thaliana [29]. Coleman et al. [30] showed that overexpression of the Gossypium hirsutum sucrose synthase gene (GhSuSy) in hybrid poplar (Populus alba L. × Populus grandidentata Michx.) induced thicker cell walls and greater wood density. Furthermore, a recent study in Chinese white poplar (Populus tomentosa Carr.) showed that genes associated with lignin biosynthesis, including 4-coumarate:cinnamate-4-hydroxylase(C4H), cinnamyl alcohol dehydrogenase(CAD), and caffeoyl CoA 3-O-methyltransferase (CCoAOMT), were transcribed in the lignified xylem [31]. These studies have significantly improved our understanding of secondary xylem differentiation and secondary wall formation.
Populus deltoides (Marsh.), which is widely distributed between the northern latitudes of 40° to 60° in North America, was introduced into China in 1972. This tree is a black poplar tree of the Aigeiros section in the Populus genus, exhibiting good quality, high yield, disease resistance and strong adaptability. Therefore, P. deltoides is widely used as an important species for poplar breeding. However, compared with our understanding of the function of the small GTP-binding protein in Arabidopsis, N. benthamiana, O. sativa and other plants, the functions of small GTP-binding protein genes in tree species remain largely unknown.
In this study, we isolated the P. deltoides small GTP-binding protein gene (PdRanBP), and observed its expression primarily in leaf buds as well as in immature xylem and immature phloem in the stem. Additionally, the downregulation of PdRanBP promoted vegetative growth in poplar. Interestingly, the overexpression of PdRanBP induced the formation of sylleptic branches and reduced apical dominancy in hybrid poplar plantlets. This study provides new data that will help to determine the molecular mechanism of PdRanBP in P. deltoides growth and vascular tissue development.
Results
Isolation and phylogenetic analysis of PdRanBP
A 670-bp cDNA fragment of P. deltoides RanBP was amplified by reverse transcription PCR (RT-PCR) and sequenced. The gene from which the cDNA was derived was named PdRanBP (Fig. 1a). PdRanBP shares 99, 89 and 100 % sequence identity with the open reading frames (ORFs) of P. trichocarpa RanBP6 (PtRanBP6), PtRanBP18 and hpPdRanBP of hybrid poplar (P. davidiana Dode × P. bolleana Lauche) (Additional file 1), respectively. PdRanBP encodes a polypeptide that is predicted to contain 221 amino acids and to have a molecular weight of 25033.5 Da and an isoelectric point (pI) of 6.38 (Fig. 1a).
Fig. 1.
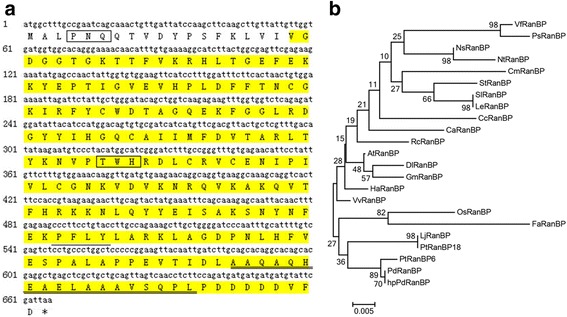
Characterization of the PdRanBP protein. a Nucleotide and deduced amino acid sequence of the coding region (cDNA) of PdRanBP from Populus deltoides. The highlighted yellow sequence indicates the conserved RAN subfamily domain. The 2Fe-2S ferredoxin-type iron-sulphur-binding domains are boxed. The EGF-like domain is underlined with a thin black line. The double-underlined region is the VWFC domain. The PdRanBP protein sequence was analysed and characterized using the SMART analysis service (http://smart.embl-heidelberg.de/). b A neighbor- joining phylogenetic tree was constructed based on an alignment of the protein sequences of AtRanBP [GenBank: AY116939.1], CaRanBP [GenBank: AJ299064.1], CcRanBP [GenBank: AB372270.1], CmRanBP [GenBank: AB015288.1], DlRanBP [GenBank: JF461291.1], FaRanBP [GenBank: FJ610236.1], GmRanBP [GenBank: AK243772.1], HaRanBP [GenBank: AF495716.1], LeRanBP [GenBank: L28714.1], LjRanBP [GenBank: Z73959.1], NtRanBP [GenBank: L16787.1], NsRanBP [GenBank: AY563049.1], OsRanBP [GenBank: AB015288.1], PsRanBP [GenBank: EF194277.1], PtRanBP6 [GenBank: XM_002308612.1], PtRanBP18 [GenBank: XM_002324810.2], RcRanBP [GenBank: XM_002515509.1], SlRanBP [GenBank: NM_001247091.1], StRanBP [GenBank: DQ222522.1], VfRanBP [GenBank: Z24678.1], VvRanBP [GenBank: FQ396597.1], hpPdRanBP [KU841447] and PdRanBP [KU841446]. MEGA 4.0 software was used for this alignment. The 23 RanBP amino acid sequences are shown in Additional file 2. The scale bar indicates the distance calculated using the multiple alignment
Nucleotide sequence analysis revealed that the PdRanBP cDNA sequence has 82–100 % similarity to the RanBP cDNA sequences from twenty-two other plant species. Phylogenetic analysis (Fig. 1b) of the RanBP amino acid sequences derived from P. deltoides and the twenty-two other plant species showed that PdRanBP clusters closely with hybrid poplar (P. davidiana Dode × P. bolleana Lauche) hpPdRanBP and P. trichocarpa PtRanBP6. In addition, the PdRanBP amino acid sequence has 100 and 99.0 % similarity (Additional file 2) to the hpPdRanBP and PtRanBP6 sequences, respectively. We found that PdRanBP contains a core domain between residues 19 and 220 that is structurally similar to the GTP-binding domains of other small GTPases. Based on these findings, PdRanBP is a conserved member of the Ras superfamily of small GTPases. In addition, three other important domains were identified in PdRanBP. Two 2Fe-2S ferredoxin-type iron-sulphur binding domains may exist between residues 4 and 6, and between residues 106 and 108; the conserved cysteine residues of these domains are important elements of various metabolic enzymes. An epidermal growth factor (EGF)-like domain signature was identified in N-terminal half of the PdRanBP protein (between residues 163 and 166). These domains bind to specific cell-surface receptors with a high affinity and induce their dimerization. This event is essential for the activation of tyrosine kinases and the initiation of a signal transduction cascade that results in DNA synthesis and cell proliferation. A von Willebrand factor type C (VWFC) domain is located at the N-terminus (between residues 195 and 212) of PdRanBP; this domain is thought to participate in oligomerization (but not the initial dimerization step) during the formation of large protein complexes (Fig. 1a and Additional file 3).
The expression pattern of PdRanBP in different organs and tissues in P. deltoides
The PdRanBP gene showed highly divergent expression patterns in the tissues tested. Compared with the floral buds, PdRanBP was expressed at 3.9- to 6.3-fold higher levels in the immature xylem, immature phloem and mature phloem. However, only weak expression of PdRanBP was detected in the mature xylem (Fig. 2a). The observed patterns of gene expression were the same when using either TUA1 (Fig. 2a) or UBQ1 (Fig. 2b) as the control gene. PdRanBP was predominantly expressed in leaf buds and in the immature xylem and immature phloem, indicating that PdRanBP expression correlates with leaf bud development and wood formation in P. deltoides.
Fig. 2.
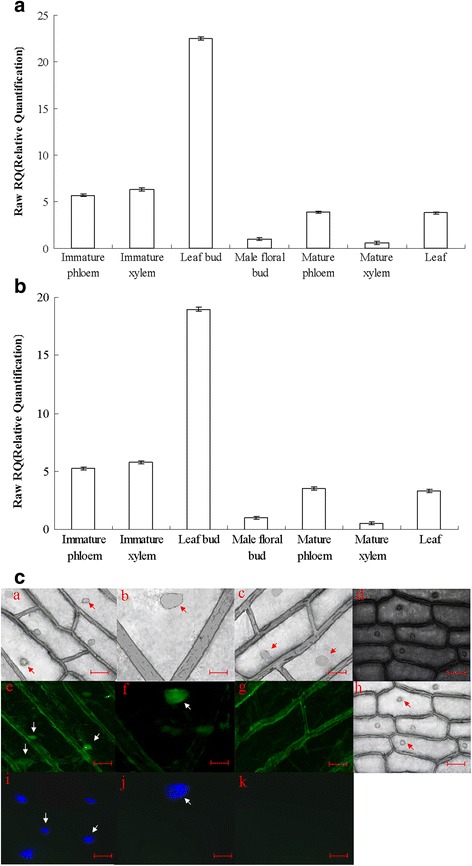
qRT-PCR analysis of the expression of PdRanBP in different vascular tissues and organs of P. deltoides, and detection of immediate and stable expression of GFP-tagged PdRanBP. a, b qRT-PCR analysis of PdRanBP expression in the vascular tissues and other organs of P. deltoides during secondary cell wall development. Aliquots of 1000 ng total RNA were reverse-transcribed into cDNA. The signals were normalized to the constitutively expressed poplar α-tubulin (TUA1) (a) and Ubiquitin (UBQ1) (b) genes. The values are the mean ± standard error (SE) of three replicates. PdRanBP was predominantly expressed in the leaf buds, immature xylem and immature phloem of P. deltoides. c Nuclear localization of EGFP-PdRanBP fusion protein in onion epidermal cells. Dark-field images were captured for green fluorescence (e and f), GFP-only control (g) and the corresponding bright-field images for e, f, g are a, b, c. Bright-field images (h) were captured for cell morphology, and the corresponding dark-field images for h is d. i and j, Nuclei counterstained with 4′, 6-diamidino-2-phenylindole (DAPI); the corresponding GFP-only control images of i and j are shown in k. The scale bars are 200 μm in a, c, d, e, g, h, i and k, and 800 μm in b, f, and j
Detection of immediate and stable expression of GFP-tagged PdRanBP
Using gene gun technology, nuclear localization of GFP-tagged PdRanBP was observed in transient expression conditions in onion cells (Fig. 2c, white arrows in panels e and f) and stable expression conditions in poplar stem cells (Fig. 2c, white arrows in panels i and j). Control cells (g and k) did not exhibit any green fluorescence (g) or 4′,6-diamidino-2-phenylindol (DAPI) staining (k) at the settings at which the images were collected.
Generation of PdRanBP-overexpressing and PdRanBP-downregulated poplar lines
PdRanBP-overexpressing (OE) and PdRanBP-downregulated (DR) poplar lines (PdRanBP antisense lines) were initially screened by PCR amplification of the NptII-specific sequence. Six lines (of the 60 independent lines subjected to PCR detection) with high or low level expression of PdRanBP were selected for further characterization. Compared with the wild-type (WT) plants, PdRanBP was upregulated by 120.82, 150.88 and 192.41 % in the PdRanBP-OE lines G9, G10 and G15, respectively (P = 0.000, Fig. 3a). PdRanBP expression in four independent PdRanBP-DR lines (GA106, GA515, GA516 and GA521) was reduced by 64.46, 50, 44.61, and 25 %, respectively, compared with the WT condition (P = 0.007, Fig. 3a). The same patterns of gene expression were observed using either TUA1 (Fig. 3a) or UBQ1 (Fig. 3b) as the control gene.. These selected transgenic and WT plants were multiplied clonally in vitro, and five plants from each poplar line were cultivated in soil in the greenhouse until a growth age of 120 days.
Fig. 3.
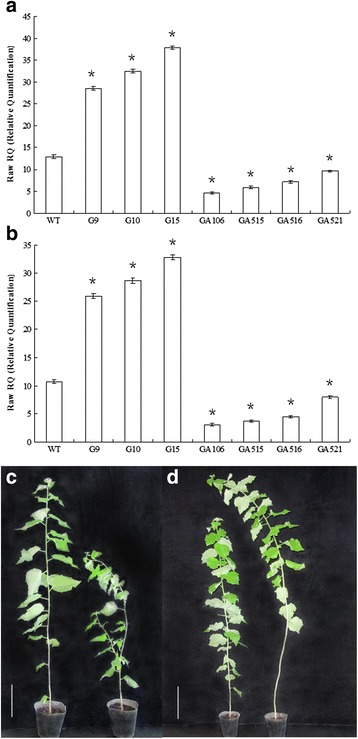
Overexpression of PdRanBP stunted growth and induced sylleptic branches; PdRanBP downregulation produced taller plants with thicker stems. a, b qRT-PCR analysis confirming PdRanBP overexpression in the transgenic poplar lines G9 and G15. TUA1 (a) and UBQ1 (b) were used as control genes. The error bars represent the standard error (SE) of three replicates. WT: wild poplar; G9, G10 and G15: transgenic poplar lines overexpressing PdRanBP (OE); GA106, GA515, GA516 and GA521: transgenic poplar lines in which PdRanBP is downregulated (DR). The asterisks indicate significant differences between the transgenic lines and WT (*P < 0.05). c Phenotypic comparison of 120-day-old WT poplar (left) and the PdRanBP-OE line G15 (right). d Phenotypic comparison of 120-day-old WT plants (left) and the PdRanBP-DR line GA106 (right). The scale bars correspond to 20.76 cm in B, and 23.8 cm in C
PdRanBP overexpression causes slow growth and induces sylleptic branches in hybrid poplar
Three PdRanBP-OE lines with high levels of transgene expression were selected for in vitro growth monitoring and phenotypic analyses. Growth was retarded and sylleptic branches were observed in PdRanBP-OE plantlets after 120 days of growth in pots; this effect was particularly pronounced in PdRanBP-OE line G15 (Fig. 3c). The stem growth of PdRanBP-OE poplars was reduced, with a mean decrease in plant height of 44.75 % compared with WT plants (Fig. 4e). A decrease in stem width and average internode length, as well as inhibition of leaf development, were observed in PdRanBP-OE lines (Figs. 3c and 4d, f, g). In addition, significant decreases in the width and number of cell layers in the xylem and phloem zones were detected in PdRanBP-OE lines compared with WT plants (Figs. 4a, b and 5a, b, d, e). The width and number of cell layers in the cambium were also decreased in PdRanBP-OE plants (Fig. 4c). Relative-quantitative real-time PCR (qRT-PCR) and phenotypic data analysis showed that the slow growth correlated positively with the expression level of PdRanBP. These experiments demonstrated that increased levels of PdRanBP blocked secondary wall synthesis and led to defects in secondary wall expansion.
Fig. 4.
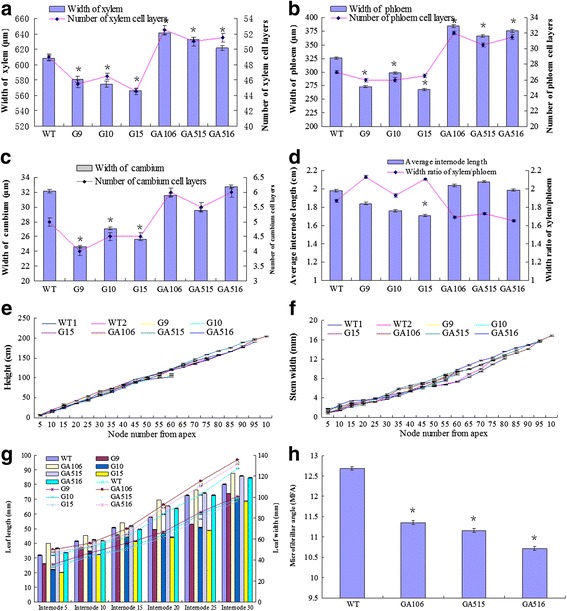
Anatomical features of the stem, plant height, stem width, leaf size and microfibril angle in 120-day-old WT and transgenic poplars expressing sense or antisense PdRanBP. For the morphological analyses, the stem base diameter was measured 5 cm above the soil surface using digital callipers. The values are the mean ± standard error (SE) of measurements from three plants. The asterisks indicate significant differences from the wild-type (*P < 0.05). WT: wild-type poplar; WT1: control of PdRanBP-OE poplar; WT2: control for the PdRanBP-DR poplar; G9, G10 and G15: PdRanBP-OE poplar lines; GA106, GA515 and GA516: PdRanBP-DR poplar lines. a The width of the xylem (left panel) and number of xylem cell layers (right panel) in PdRanBP-OE, PdRanBP-DR and control lines. The measurements were made at the 15th internode. b The width of the phloem (left panel) and number of phloem cell layers (right panel) in the PdRanBP-OE, PdRanBP-DR and control lines. c The width of the cambium (left panel) and number of cambium cell layers (right panel) in the PdRanBP-OE, PdRanBP-DR and control lines. d The average internode length (left panel) and the xylem:phloem width ratio (right panel) in PdRanBP-OE, PdRanBP-DR and control lines. The average internode length (cm) was calculated by dividing the total height by the total number of nodes. e, f The PdRanBP-DR lines were taller and had wider stems at every fifth node from the apex. g A comparison of leaf size in 120-day-old transgenic poplars. Leaf length (left panel) and width (right panel) measurements were performed at the 5th to 30th nodes to compare fully expanded leaves. The values are the mean ± standard error (SE) of 10 leaves. (H) Analysis of the microfibril angle (MFA) in PdRanBP transgenic poplar plants. The PdRanBP-DR poplar lines had lower MFAs than the control line
Fig. 5.
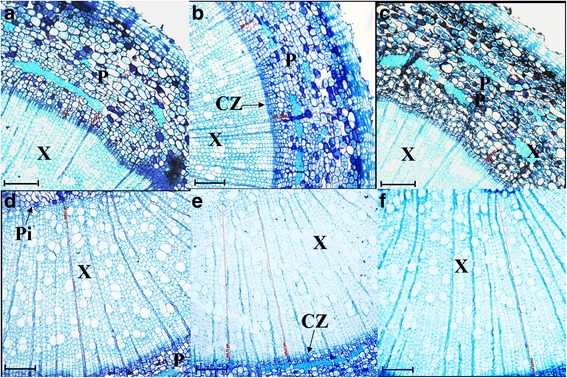
Transverse view of the anatomical features in WT and transgenic poplars expressing sense or antisense PdRanBP. The 15th internodes of 120-day-old plants were used to examine the secondary wall cell layers and width of the vascular tissues (a, b, c, d, e and f). Transverse anatomical structure of the stem of a wild-type (WT) poplar (a, d), PdRanBP-OE transgenic poplar (b, e) and PdRanBP-DR transgenic poplar (c, f). CZ, cambial zone; P, secondary phloem; Pi, pith; C, cortex cells; X, secondary xylem. Scale bars = 100 μm
PdRanBP downregulation promotes growth and facilitates secondary wall expansion
Consistent with the in vitro observations, soil-grown PdRanBP-DR GA106, GA515 and GA516 plants were larger overall and showed increased shoot growth compared with WT plants (Figs. 3d and 4e). Increases were also observed in leaf size (leaf length and leaf width), stem diameter, the number of internodes, and average internode length in PdRanBP-DR plants (Fig. 4d, f, g). Compared to WT plants, increases in the width and number of cell layers were detected in cross-sections of the primary-secondary transition zone in the stems of transgenic hybrid poplar compared with the WT (every internode from 3th to 7th joint, data not shown). However, these changes were much more pronounced in the stem sections of wood-producing stem tissues (15th internode; Figs. 4a, b, c and 5a, c, d, f). For example, the areas of xylem and phloem in the stele; the number of cell layers in the xylem and phloem; and the xylem, phloem and cambium widths were all increased by the downregulation of PdRanBP (Fig. 4a, b, c and Additional file 4).
The microfibril angle is clearly altered in PdRanBP-DR poplar
The microfibril angle (MFA) values in PdRanBP-DR poplar lines were 10.49 %–15.46 % lower than that in WT plants, and these differences were statistically significant (P = 0.000, Fig. 4h). Thus, PdRanBP is likely to be a valuable gene for improving timber strength (i.e., stiffness) in trees.
Verification of primer specificity and gene-specific PCR amplification efficiency
Ten secondary wall-associated genes encoding transcription factors and other proteins and two reference genes from P. deltoides were selected to verify primer specificity and amplification efficiency. The gene name, accession number, gene description, primer sequences, amplification efficiency and correlation coefficients are listed in Additional file 5. The melting temperatures (Tm) of all PCR products ranged from 76.32 °C for PtrFRA1 to 84.83 °C for PtrCAD10 (Additional file 6). The amplification efficiency (E) of the PCR reactions varied from 91.29 % for PtrGT8 to 100.005 % for PtrCCoAOMT1, and the correlation coefficients (R2) ranged from 0.9933 for PtrC4H1 and 0.9995 for PtrGT8 (Additional files 5 and 7).
PdRanBP overexpression and downregulation alter the expression of secondary wall-associated genes
Significant changes in the transcript abundance of ten secondary wall-associated transcription genes were observed in PdRanBP-OE and PdRanBP-DR lines, as determined using qRT-PCR. Five genes associated with cell wall biosynthesis (trans-cinnamate 4-hydroxylase 1 (PtrC4H1), cinnamyl alcohol dehydrogenase 10 (PtrCAD10), caffeoyl CoA 3-O-methyltransferase 1 (PtrCCoAOMT1), glycosyltransferase 8 (PtrGT8) and cinnamoyl coenzyme A reductase 7 (PtrCCR7)), three gene associated with MFA (sucrose synthase 1 (PtrSuS1), beta-tubulin 7 (PtrTUB7) and fragile fibre 1 (PtrFRA1)), and two myeloblastosis (MYB) genes (MYB; PtrMYB90, PtrMYB18) were selected for investigation. Compared with WT plants, the expression level of PtrCCR7 decreased by 80.10 %–82.60 % in the three PdRanBP-OE lines (P = 0.003, Fig. 6a) and upregulated by 182.5 %–209.40 % in the three PdRanBP-DR lines (P = 0.004, Fig. 6a). The expression levels of PtrCCoAOMT1 were significantly decreased by 67.80 %–75.90 %, in the three PdRanBP-OE tested lines (P = 0.002, Fig. 6a) and significantly upregulated by 109.3 %–314.20 % in the three PdRanBP-DR tested lines (P = 0.025, Fig. 6a). PtrFRA1 expression decreased by 57.3 %–78.20 % in the three tested PdRanBP-OE lines (P = 0.001, Fig. 6a) and increased by 103.9 %–314.20 % in the three tested PdRanBP-DR lines (P = 0.000, Fig. 6a). The observed patterns of gene expression were the same when using either TUA1 (Fig. 6a) or UBQ1 (Fig. 6b) as the control gene. These results indicated that PdRanBP might be involved in the regulation of cell wall-related transcription factors/genes and as well as with cell wall biogenesis.
Fig. 6.
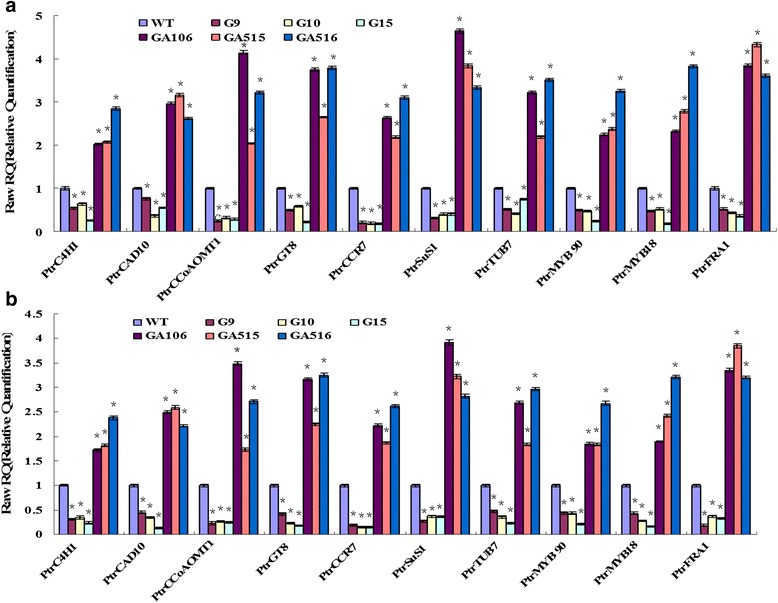
qRT-PCR analysis of secondary cell wall-associated gene expression in PdRanBP-OE, PdRanBP-DR and WT seedlings. Total RNA was isolated from three biological replicates of the wild-type (WT), PdRanBP-OE and PdRanBP-DR plants. The expression of trans-cinnamate 4-hydroxylase 1 (PtrC4H1), cinnamyl alcohol dehydrogenase 10 (PtrCAD10), caffeoyl CoA 3-O-methyltransferase 1 (PtrCCoAOMT1), glycosyltransferase 8 (PtrGT8), cinnamoyl coenzyme A reductase 7 (PtrCCR7), sucrose synthase 1 (PtrSuS1), beta-tubulin 7 (PtrTUB7), fragile fibre 1 (PtrFRA1), and two myeloblastosis (MYB) genes (PtrMYB90, PtrMYB18) were examined. TUA1 (a) and UBQ1 (b) were used as reference genes, and the expression level of each gene in the WT background was set to 1. The data are the mean ± standard errors (SE). The asterisks indicate significant differences from WT (*P < 0.05)
Transgenic poplar lines increased the proportion of cells in the G2 phase of the cell cycle in poplar
Fluorescence-activated cell sorting (FACS) was used to determine whether the cell cycle was altered in the PdRanBP transgenic poplar lines. The average number of G2 phase cells was clearly increased in the stems and leaf buds of the PdRanBP-OE lines compared with WT plant cells (1.34 % to 2.47 % in stems, P = 0.001; 1.47 % to 2.66 % in buds and P = 0.001, Fig. 7). In terms of the cell cycle, these results suggested that PdRanBP primarily increased the tendency of cells to remain in G2, thus regulating cell division.
Fig. 7.
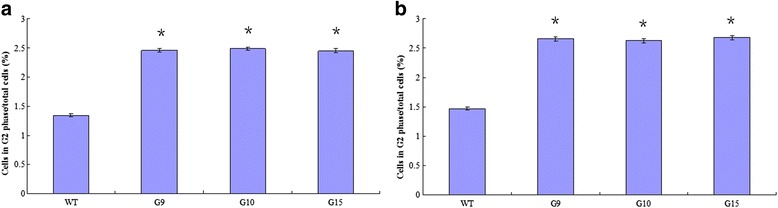
Effect of the PdRanBP transgene on the cell cycle phases of poplar cells. a The proportion of cells in G2 in the top stems of wild-type (WT) and PdRanBP-OE cutting seedlings (G9, G10 and G15). The number of cells in G2 was calculated as a proportion of the total cell population. The results are the mean ± SE from different lines analysed in three independent experiments. b The G2 population in the leaf buds of the WT and PdRanBP-OE cutting seedlings (G9, G10 and G15). The asterisks indicate significant differences compared with WT (*P < 0.05)
Discussion
The evaluation of gene expression levels of PdRanBP in different organs and tissues in P. deltoides has contributed to our understanding of the function of this gene in plant growth. Haizel et al. [32] reported that the Arabidopsis AtRanBP genes were expressed in the stems, leaves, roots and flowers, with the highest level of expression being in meristematic tissues, such as the shoot and the root apical meristem. Tian et al. [33] demonstrated that the wheat TaRanBP gene is expressed similarly in its stem, leaf and root tissues. Wang et al. [34] found that the transcript levels of the wheat RAN gene (TaRAN1) were high in young stems and flower buds but low in old leaves. The fescue (Festuca arundinacea) Ran GTPase homologous (FaRan) gene is broadly expressed in stems, inflorescence meristems, old mature leaves, young leaves and plumules [35]. In this study, high PdRanBP expression was observed in in leaf buds, immature xylem and immature phloem. The stem-specific expression pattern of PdRanBP in P. deltoides is also consistent with the pattern of AtRanBP in Arabidopsis [32], TaRanBP in wheat [33], and FaRan in Fescue [35]. This tissue-specific expression pattern indicated that PdRanBP might be involved in stem development and wood formation in P. deltoides.
The green fluorescent protein (GFP) of the jellyfish Aequorea victoria can be visualized directly through emission of green light upon excitation with blue light or long UV [36, 37]. Recently, transient or stable expression of gfp has been described in several transformed angiosperm and gymnosperm plants [38, 39]. DNA particle bombardment has been used to produce transgenic soybeans [40], beans [41], peanuts [42], cowpeas [43] and poplars [44, 45]. In this study, the fusion of GFP to the C- terminus of full-length PdRanBP resulted in exclusive nuclear labelling in onion epidermal cells and poplar stem cells. This nuclear localization of GFP-tagged PdRanBP was consistent with the expression of the RanBP gene in various other plants, such as Nicotiana benthamiana, Oryza sativa and Triticum aestivum [19, 20, 34].
Two types of cell division occur during secondary xylem development: periclinal and anticlinal. Periclinal division determines the number of secondary xylem cells in each radial file, while anticlinal division occurs in the initial cambial cells and determines the number of radial files in the secondary xylem cells [46–49]. To investigate the role of PdRanBP in secondary tissues development, we examined transverse sections of the xylem, phloem and cambium regions, which represent the different anatomical features of vascular tissue. The number of cell layers, widths of the vascular tissues (xylem, phloem and cambium), average internode length, and stem height and width were significantly increased in PdRanBP-DR plants compared with WT plants. This result is similar to those observed for the P. deltoides remorin gene PdREM in aspen. The average internode length and the widths of the secondary xylem and secondary phloem and were also increased in PdREM-DR lines [50]. Based on these experiments, it appears that PdRanBP suppresses cell enlargement directly or indirectly by blocking secondary cell wall synthesis and expansion.
Yeast two-hybrid and co-immunoprecipitation analyses demonstrated the specific interaction of basic helix-loop-helix (bHLH) transcription factors with human RanBP17 [51]. In plants, a bHLH transcription factor was identified as a secondary cell wall regulator that can bind to the promoters of secondary cell wall biosynthesis genes and play an important role in the secondary cell wall regulatory network [52]. bHLH proteins can interact with MYBs [53], and the MYB–bHLH interaction is necessary to control secondary cell wall synthesis in the xylem [54]. In this study, the expression levels of PtrMYB90 and PtrMYB18 were significantly decreased (by 67.80 %–75.90 %) in the three tested PdRanBP-OE lines (P = 0.004 and P = 0.003, respectively), In addition, these genes were significantly upregulated (by 109.3 %–314.20 %) in the three tested PdRanBP-DR lines (P = 0.007 and P = 0.011, respectively). We speculated that PdRanBP interacted with MYB, then with MYB–bHLH transcription factors, and ultimately formed protein complexes that induced changes in the expression of secondary cell wall formation-associated genes in poplar.
The cambium is derived from the shoot apical meristem (SAM). Apical regions have common roles in promoting primary growth and accelerate the differentiation of functional cell types. Lu et al. [35] reported that overexpression of the tall fescue FaRan gene reduced apical dominance and induced over-proliferation of axillary buds in the rosette leaf axils of transgenic Arabidopsis. Wang et al. [19] found that overexpression of wheat (T. aestivum) TaRAN1 increased primordia, delayed flowering, and reduced apical dominance in Arabidopsis. In the present study, PdRanBP was enriched in the shoot apices (i.e., the stem tip of the 5-cm collected branches) of PdRanBP-DR poplar lines (increased secondary wall growth) but not in control and PdRanBP-OE lines (Figs. 3 and 4). This result indicated that PdRanBP regulates secondary growth via differences in gene expression in stems. PdRanBP overexpression induced sylleptic branches and reduced apical dominance, whereas PdRanBP downregulation promoted seedling height and shoot growth (Figs. 3 and 4). The apical and radial growth (e.g., stem height and width, and average internode length) of PdRanBP-DR lines were greater than in PdRanBP-OE lines (Figs. 3 and 4), indicating that PdRanBP affects the apical and radial growth of poplar trees.
The MFA is an important property of wood tissues. The angle at which microfibrils are arranged with respect to the longitudinal axis of the cell determines the stiffness of the wood. A high MFAs results in increased longitudinal shrinkage and low wood stiffness. The stiffness of the cell wall increases fivefold as the MFA decreases from approximately 40° to 10° [55]. Thus, a low MFA of wood is a highly undesirable property for the genetic improvement of poplar. MFA is under genetic control [56–58] and can be directly measured in immature trees, providing an attractive option for early selection and trait improvement in poplar. The P. deltoides gene PdCYTOB, which encodes a cytokinin-binding protein, is related to the wood properties of P. deltoides. The MFA of antisense-PdCYTOB transformed hybrid poplar (P. davidiana × P. bolleana) decreased by 4.9 %–24.4 % compared with WT plants in the greenhouse [59]. In a previous study, a 10.0–17.5 % reduction in MFA was observed in PdREM antisense-expressing transgenic poplar lines compared with control lines [50]. In the present research, the MFA values of the PdRanBP-DR poplar lines GA106, GA515 and GA516 ranged from 10.72° to 11.35°, with a mean of 11.08° and an average SD of 0.045; these differences were statistically significant (P = 0.001). All of the transgenic poplar hybrids expressing antisense PdRanBP constructs had lower MFAs than the untransformed lines, suggesting that PdRanBP gene might play an important role in improving microfibril angles.
Schulze et al. carried out yeast two-hybrid assays, finding that mouse β1-tubulin or β5-tubulin can interact with RanBP10. RanBP10 also interacted with the β5-tubulin isoform in yeast cells, thereby exhibiting nonselective for association with β-tubulins [60]. In plants, Spokevicius showed that a Eucalyptus grandis β-tubulin gene (EgrTUB1) is involved in determining the orientation of cellulose microfibrils in plant secondary fibre cell walls and that the cellulose microfibril angle (MFA) correlates with EgrTUB1 expression [61]. In PdRanBP transgenic poplar lines, the downregulation of PdRanBP significantly increased the expression of PtrTUB7, and was associated with a lower MFA. The molecular mechanism by which PdRanBP decreases the MFA in transgenic plants is unclear. We hypothesize that poplar PdRanBP may interact with tubulin proteins, such as PtrTUB7, and thereby direct microfibril orientation and determine the MFA in secondary fibre cell walls. Another hypothesis is that PdRanBP regulates MFA-associated genes (e.g., PtrFRA1 and PtrSuS1) (Fig. 6); in this way,, downregulation of PdRanBP expression would alter the MFA.
The overexpression of PdRanBP increased the proportion of cells in the G2 phase. This finding echoes the results of other studies in yeast and rice [19, 34]. Wang et al. [19, 34] found that the average number of cells in G2 increased significantly in TaRAN1-transformed yeast or rice cells compared with WT cells. We propose that PdRanBP, like many other Ran/RanBPs, regulates cell division during cell cycle progression.
Conclusions
In conclusion, the cloning and detailed characterization of PdRanBP from the developing xylem of poplar trees support the notion that this gene is associated with tree growth and vascular tissue development. PdRanBP is predominantly expressed in leaf buds and particular cell types (e.g., immature xylem and immature phloem) within the vascular system. These results indicate that PdRanBP is potentially involved in vascular tissue development and wood formation. Full-length PdRanBP-GFP fusion proteins were exclusively observed in the nucleus of onion epidermal cells and poplar stem cells. Using a transgenic approach, we showed that PdRanBP might function as a negative regulator in P. deltoides to enhance secondary cell wall synthesis and promote cell wall expansion. Further characterization of the wood-associated PdRanBP gene will open up new avenues of research that may lead to the optimization of molecular breeding and genetic engineering strategies for improved wood quality.
Endnotes
All the References within the text were designated using the Endnotes X6 software.
Methods
Plant growth conditions and sampling
A 15-year-old P. deltoides specimen was used to isolate the PdRanBP gene and to analyse its tissue-specific expression pattern. Leaves, leaf buds, stems (immature xylem, mature phloem, immature phloem, mature xylem), and male flower buds were harvested three times from different areas of the plant for the analysis (Additional file 8). The hybrid poplar (P. davidiana × P. bolleana) is a breed that was developed in China by crossing P. davidiana and P. bolleana and was used for genetic transformation experiments to characterize PdRanBP function.
Isolation, plant expression vector construction and genetic transformation of PdRanBP
Total RNA was prepared using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) and cDNA was synthesized using M-MLV reverse transcriptase (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The coding region of PdRanBP was amplified from cDNA by reverse transcription PCR (RT-PCR), using the primer pair P1 (Additional file 9), which was designed according to the PtRanBP6 sequence (accession no. XM_002308612.1). The PCR product was ligated into the pGEM-T Easy vector (Promega) and sequenced; the vector was termed pGEM-T-PdRanBP.
The ORF of PdRanBP was amplified from pGEM-T-PdRanBP using the primers P2 and P3 (Additional file 9), yielding DNA fragments with different restriction sites at the 5′ end and 3′ ends. The amplified DNA constructs were inserted into the XbaI and SalI sites of the intermediate vector pGEM-T to yield pGEM-T-sense PdRanBP and pGEM-T-antisense PdRanBP, respectively. XbaI and SalI were used to digest pGEM-T-sense PdRanBP, pGEM-T-antisense PdRanBP, and the plant expression vector pBI121 (Clontech Labs, Inc., Palo Alto, CA, USA). Lastly, sense and antisense PdRanBP constructs were cloned into the pBI121 vector to generate pBI121-sense PdRanBP and pBI121-antisense PdRanBP, respectively (Additional file 10). The vectors were confirmed by sequencing, separately transformed into Agrobacterium tumefaciens (strain GV3101), and subsequently transformed into hybrid poplar (P. davidiana × P. bolleana) using the leaf disk transformation method [62].
Characterization of transformed poplars
The transgenic poplar were grown in a greenhouse at the Chinese Academy of Forestry under natural light conditions, with an 18 h light/6 h dark photoperiod at a temperature of 22 °C/15 °C (day/night). Transgenic poplars were identified by PCR using P6 primers (Additional file 9) to amplify the NptII-sensitive selective marker gene.
Analysis of the expression of PdRanBP and secondary wall-associated transcription factors/genes by qRT-PCR
qRT-PCR was used to analyse the expression patterns and levels of PdRanBP in different tissues of the P. deltoides tree and transgenic poplar plants. The expression patterns and levels of secondary cell wall-related genes in 120-day-old PdRanBP-OE and PdRanBP-DR transgenic plants were also assessed (Additional files 9 and 5). The qRT-PCR analysis was performed using the α-tubulin (TUA1) and Ubiquitin (UBQ1) gene as internal controls [63], according to the instructions of the SYBR® Premix Ex TaqTM Kit (Takara, Tokyo, Japan). The reactions were run on an ABI Prism 7500 sequence detector (Applied Biosystems, Foster City, CA, USA) using SYBR Green PCR Master Mix (Applied Biosystems). Each PCR reaction (final volume 20 μl) contained 1 μl of first-strand cDNA, 200 nM of primers and 1× SYBR Green PCR Master Mix. Three replicates were conducted in parallel, and statistical analysis of the data was performed following the ABI Prism 7500 Sequence Detection System Users Guide. Gene-specific primer pairs (Additional files 9 and 5) were designed using the software Primer premier 5.0 (Premier Biosoft Int., Palo Alto, CA, USA).
Standard curves were constructed to calculate the gene-specific PCR efficiency from 10-fold series dilutions of the mixed cDNA templates for each primer pair. The correlation coefficients (R2) and slope values could be obtained from the standard curve, and the corresponding PCR amplification efficiencies (E) were calculated according to the following equation: E = (10-1/slope-1) × 100 [64].
Construction of the expression vector EGFP-PdRanBP and plant cell transformation
The enhanced green fluorescent protein (EGFP) gene was amplified by PCR from the EGFP vector (Clontech, Palo Alto, CA, USA) using the P5 primer pair (Additional file 9). After digestion of the amplified DNA fragment with XbaI, the 715-bp fragment was inserted into the XbaI site of pBI121, downstream of the CaMV 35S promoter, yielding the EGFP-PdRanBP vector (Additional file 10). The sequence of the EGFP-PdRanBP plasmid was confirmed by DNA sequencing, and the vector was transformed into onion cells and poplar (P. davidiana × P. bolleana) cells using DNA particle bombardment [43, 65].
EGFP fluorescence analysis
To detect fluorescent signals in onion cells transformed with the EGFP-PdRanBP vector, at least 10 independently transformed lines were observed using an inverted fluorescence microscope (Carl Zeiss, Oberkochen, Germany) with a blue high-sensitivity filter block. The images were captured using a computationally controlled digital camera (AP-1; Apogee Instruments Inc., Tucson, AZ, USA). The images were processed using AxioVision software (Carl Zeiss Inc., Thornwood, NY, USA). The selected sections were processed further using Photoshop 5.0 (Adobe Systems, Mountain View, CA, USA).
Toluidine blue O staining, DAPI staining and microscopy
Cross sections (approximately 5–10 mm thick) of the internodes of PdRanBP transgenic hybrid poplars and WT stems, as well as the tips of EGFP-PdRanBP transgenic hybrid poplars and WT stems, were fixed overnight at room temperature (RT, 22 °C) in a formalin–alcohol–acetic acid (FAA). The samples were then embedded in paraffin wax, cut into 8-μm sections using a microtome (Leitz, Wetzlar, Germany), and dehydrated through an alcohol series. WT stems and cross sections of the internodes of PdRanBP transgenic hybrid poplars were stained with toluidine blue O (TBO), as described by Abbott et al. [66]. WT stems and cross sections of the tips of EGFP-PdRanBP transgenic hybrid poplars were briefly stained with DAPI (1 mg/mL in mounting medium [Vectashield; Vector Labs, Burlingame, CA, USA]), as described by Jasencakova et al. [67].
The number of radial cell layers and the overall widths of the xylem, phloem and cambium region of PdRanBP transgenic hybrid poplar were measured using an inverted fluorescence microscope. The number of nuclei was determined by counterstaining with DAPI (Carl Zeiss). The images were obtained using a digital camera system (AP-1; Apogee Instruments Inc., Tucson, AZ, USA).
MFA measurement
Blocks of stems were excised 5 cm above ground level from each transgenic and control poplar line. The MFA was determined by the method described by Franklin et al. [68–70]. Briefly, macerated fibres were acquired from the samples by incubation in glacial acetic acid/hydrogen peroxide solution (1:1 v/v) at 60 °C overnight. Individual fibres were identified on microscope slides, and the MFA was measured by polarized microscopy using an Olympus BX51 microscope (Melville, NY, USA).
Flow cytometric analysis
The stems and leaf buds of PdRanBP-OE transgenic and WT plants were cut with a razor blade. The cells were treated with nuclear isolation buffer [71] and prepared for FACS by staining with propidium iodide (PI) (Annexin-V-FLUOS staining kit, Roche) [72, 73]. Briefly, the cells were fixed in ethanol overnight at 4 °C, washed, and resuspended in 0.4 mL of 30 mM sodium citrate, pH 7.0, containing 0.1 mg/mL RNase A for 2 h at 37 °C. These steps were followed by incubation in 4 mg/mL PI (final concentration). Each sample was analysed using a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA,USA).
Phylogenetic analysis and statistical analyses
The Populus trichocarpa RanBP sequence (PtRanBP6, NCBI accession no. XM_002308612.1) and other RanBP protein sequences were obtained from GenBank. These sequences were then aligned to generate a phylogenetic tree using the MEGA 4.0 software program [74, 75], using the neighbour-joining method.
The growth, wood properties, and all qRT-PCR results were analysed using one-way analysis of variance. Asterisks and/or ‘sig’ indicate significant differences (P < 0.05; ANOVA, Fisher test) between the transgenic lines and WT. The statistical analyses were performed using the statistical program SPSS 11.0 (SPSS Inc., Chicago, IL, USA).
Abbreviations
ABA, Abscisic acid; ANOVA, One way analysis of variance; AtRanBP1c, Arabidopsis RanBP1c gene; bHLH, basic helix-loop-helix; C4H, 4-coumarate:cinnamate-4-hydroxylase; CAD, Cinnamyl alcohol dehydrogenase; CCoAOMT, Caffeoyl CoA 3-O-methyltransferase; EgCCR, Eucalyptus gunnii (J.T. Hook) cinnamoyl coenzyme A reductase; EGF, Epidermal growth factor; EGFP, The enhanced green fluorescent protein; EgrTUB1, Eucalyptus grandis b-tubulin gene; FAA, Formalin–alcohol–acetic acid; FACS, Fluorescence-activated cell sorter; FaRan, Fescue (Festuca arundinacea) Ran GTPase homologous gene; GFP, Green fluorescent protein; GhSuSy, Gossypium hirsutum sucrose synthase gene; MFA, Microfibril angle; MYB, Myeloblastoma; NbRanBP1, Nicotiana benthamiana (Domin.) RanBP gene; OsRAN2, Oryza sativa RAN gene; PdRanBP, P. deltoides small GTP-binding protein gene; PdRanBP-DR, PdRanBP-downregulated; PdRanBP-OE, PdRanBP-overexpressing; PI, Propidium iodide; PsRan1, Pea (Pisum sativum L., cv. Alaska) Ran gene; PtRanBP6, P. trichocarpa RanBP 6; PtrCCR7, P. trichocarpa cinnamoyl coenzyme A reductase 7; PtrFRA1, P. trichocarpa fragile fibre 1; PtrGT8, P. trichocarpa glycosyltransferase 8; PtrSuS1, P. trichocarpa sucrose synthase 1; PtrTUB7, P. trichocarpa beta-tubulin 7; RanBP, Ran-binding protein; SAM, Shoot apical meristem; SND1, Secondary wall-associated NAC domain protein 1; TaRAN1, Wheat (Triticum aestivum L.) RAN gene; TBO, Toluidine blue O; TUA1, α-tubulin; UBQ1, Ubiquitin; VIGS, Virus-induced gene silencing; VWFC, Von Willebrand factor type C; WT, Wild-type
Acknowledgements
We are grateful to Dr Jinxing Lin (Institute of Botany, Chinese Academy of Sciences), Dr Fan Liu (Beijing Vegetable Research Center, Beijing) and Dr Yanguang Chu (Research Institute of Forestry, Chinese Academy of Forestry) for helpful advice and technical assistance.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31400570) and Fundamental Research Funds for the Central Non-profit Research Institution of the Chinese Academy of Forestry (CAFYBB2014QB051).
Availability of data and materials
The open reading frame (ORF) and amino acid sequences of P. deltoides PdRanBP and those for hybrid poplar (P. davidiana Dode × P. bolleana Lauche) hpPdRanBP were deposited into the National Center for Biotechnology Information (NCBI) under nucleotide numbers KU841446 and KU841447. We deposited our phylogenetic data in TreeBase, with the following URL: http://purl.org/phylo/treebase/phylows/study/TB2:S18967.
Authors’ contributions
SL and QH contributed equally to this work and should be considered co-first authors. Conceived and designed the experiments: SL, XS and QH. Performed the experiments: SL, XL and CD. Analysed the data: SL, ML and ZH. Contributed reagents/materials/analysis tools: SL, BZ and JZ. Wrote the paper: SL. All of the authors read and approved the final manuscript.
Authors’ information
Mr SL is a research assistant of Chinese Academy of Forestry;
Mr QH is a associate researcher of Chinese Academy of Forestry;
Mrs BZ is a researcher of Chinese Academy of Forestry;
Mr JZ is a postdoctor of molecular biology and bionformatics in north carolina state university of USA;
Mr XL is a research assistant of Chinese Academy of Forestry;
Mr ML is a researcher of Chinese Academy of Forestry, Secretary of National Poplar Committee of China;
Mr ZH is a professor of Chinese Academy of Sciences;
Mr CD is a research assistant of Chinese Academy of Forestry;
Mrs XS is a professor of Chinese Academy of Forestry.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional files
The open reading frames (ORFs) of P. trichocarpa RanBP6 and both PtRanBP18 and hpPdRanBP of hybrid poplar (P. davidiana Dode × P. bolleana Lauche). (DOC 23 kb)
The amino acid sequences of P. deltoides PdRanBP, and the RanBP sequences from other plants. (TXT 6 kb)
Alignment of the deduced amino acid sequences of P. deltoides PdRanBP, and the RanBP sequences from other plants. (DOC 2829 kb)
The anatomical features of the stem of the WT and transgenic poplar plants in transverse view. (DOC 36 kb)
Primers used to detect the expression of secondary wall-associated genes by qRT-PCR. (DOC 38 kb)
Melting curves of two reference genes and ten secondary wall-associated genes. (DOC 1689 kb)
Standard curves of two reference genes and ten secondary wall-associated genes. (DOC 152 kb)
Details of the different vascular tissues and other organs sampled from Populus deltoides. (DOC 28 kb)
Primers used for gene isolation, plant vector construction and qRT-PCR gene expression analysis in poplar. (DOC 137 kb)
Diagrams of the vectors used for transgenic analysis. (A) Construction of the pBI121-sense PdRanBP vector overexpressing the poplar PdRanBP gene; (B) Construction of the pBI121-antisense PdRanBP vector expressing the antisense poplar PdRanBP gene; (C) Construction of the EGFP-PdRanBP vector expressing the EGFP-PdRanBP fusion protein. (DOC 137 kb)
Contributor Information
Shaofeng Li, Email: lishaof2009@163.com.
Qinjun Huang, Email: huangqj@caf.ac.cn.
Bingyu Zhang, Email: cafzhang@126.com.
Jianhui Zhang, Email: jianhuizhang@163.com.
Xue Liu, Email: 2396777010@qq.com.
Mengzhu Lu, Email: lumz@caf.ac.cn.
Zanmin Hu, Email: zmhu@genetics.ac.cn.
Changjun Ding, Email: changjunding@126.com.
Xiaohua Su, Phone: 86-10-62889627, Email: suxh@caf.ac.cn.
References
- 1.Larson PR. The Vascular Cambium. Berlin: Springer-Verlag; 1994. pp. 594–600. [Google Scholar]
- 2.Schuetz M, Smith R, Ellis B. Xylem tissue specification, patterning, and differentiation mechanisms. J Exp Bot. 2013;64(1):11–31. doi: 10.1093/jxb/ers287. [DOI] [PubMed] [Google Scholar]
- 3.Gorshkova T, Brutch N, Chabbert B, Deyholos M, Hayashi T, Lev-Yadun S, Mellerowicz EJ, Morvan C, Neutelings G, Pilate G. Plant fiber formation: state of the art, recent and expected progress, and open questions. Crit Rev Plant Sci. 2012;31(3):201–228. doi: 10.1080/07352689.2011.616096. [DOI] [Google Scholar]
- 4.Schrader J, Nilsson J, Mellerowicz E, Berglund A, Nilsson P, Hertzberg M, Sandberg G. A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell. 2004;16(9):2278–2292. doi: 10.1105/tpc.104.024190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whetten R, Sun YH, Zhang Y, Sederoff R. Functional genomics and cell wall biosynthesis in loblolly pine. Plant Mol Biol. 2001;47(1–2):275–291. doi: 10.1023/A:1010652003395. [DOI] [PubMed] [Google Scholar]
- 6.Hertzberg M, Aspeborg H, Schrader J, Andersson A, Erlandsson R, Blomqvist K, Bhalerao R, Uhlen M, Teeri TT, Lundeberg J, Sundberg B, Nilsson P, Sandberg G. A transcriptional roadmap to wood formation. Proc Natl Acad Sci U S A. 2001;98(25):14732–14737. doi: 10.1073/pnas.261293398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, Park S, Kamdem DP, Keathley DE, Retzel E, Paule C, Kapur V, Han KH. Novel gene expression profiles define the metabolic and physiological processes characteristic of wood and its extractive formation in a hardwood tree species. Robinia pseudoacacia Plant Molecular Biology. 2003;52(5):935–956. doi: 10.1023/A:1025445427284. [DOI] [PubMed] [Google Scholar]
- 8.Sterky F, Bhalerao RR, Unneberg P, Segerman B, Nilsson P, Brunner AM, Charbonnel-Campaa L, Lindvall JJ, Tandre K, Strauss SH, Sundberg B, Gustafsson P, Uhlen M, Bhalerao RP, Nilsson O. A Populus EST resource for plant functional genomics. Proc Natl Acad Sci U S A. 2004;101(38):13951–13956. doi: 10.1073/pnas.0401641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dungey HS, Matheson AC, Kain D, Evans R. Genetics of wood stiffness and its component traits in Pinus radiata. Can J For Res. 2006;36(5):1165–1178. doi: 10.1139/x06-014. [DOI] [Google Scholar]
- 10.Vernoud V, Horton AC, Yang Z, Nielsen E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 2003;131(3):1191–1208. doi: 10.1104/pp.013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuksel B, Memon AR. Comparative phylogenetic analysis of small GTP-binding genes of model legume plants and assessment of their roles in root nodules. J Exp Bot. 2008;59(14):3831–3844. doi: 10.1093/jxb/ern223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke PR, Zhang C. Ran GTPase: a master regulator of nuclear structure and function during the eukaryotic cell division cycle? Trends Cell Biol. 2001;11(9):366–371. doi: 10.1016/S0962-8924(01)02071-2. [DOI] [PubMed] [Google Scholar]
- 13.Dasso M. The Ran GTPase: theme and variations. Curr Biol. 2002;12(14):R502–508. doi: 10.1016/S0960-9822(02)00970-3. [DOI] [PubMed] [Google Scholar]
- 14.Hetzer M, Gruss OJ, Mattaj IW. The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat Cell Biol. 2002;4(7):E177–184. doi: 10.1038/ncb0702-e177. [DOI] [PubMed] [Google Scholar]
- 15.Ciciarello M, Mangiacasale R, Lavia P. Spatial control of mitosis by the GTPase Ran. Cell Mol Life Sci. 2007;64(15):1891–1914. doi: 10.1007/s00018-007-6568-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Fiore B, Ciciarello M, Lavia P. Mitotic functions of the Ran GTPase network: the importance of being in the right place at the right time. Cell Cycle. 2004;3(3):305–313. doi: 10.4161/cc.3.3.641. [DOI] [PubMed] [Google Scholar]
- 17.Quimby BB, Dasso M. The small GTPase Ran: interpreting the signs. Curr Opin Cell Biol. 2003;15(3):338–344. doi: 10.1016/S0955-0674(03)00046-2. [DOI] [PubMed] [Google Scholar]
- 18.Lee Y, Kim MH, Kim SK, Kim SH. Phytochrome-mediated differential gene expression of plant Ran/TC4 small G-proteins. Planta. 2008;228(1):215–224. doi: 10.1007/s00425-008-0745-x. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Xu Y, Han Y, Bao S, Du J, Yuan M, Xu Z, Chong K. Overexpression of RAN1 in rice and Arabidopsis alters primordial meristem, mitotic progress, and sensitivity to auxin. Plant Physiol. 2006;140(1):91–101. doi: 10.1104/pp.105.071670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho HK, Park JA, Pai HS. Physiological function of NbRanBP1 in Nicotiana benthamiana. Molecules and Cells. 2008;26(3):270–277. [PubMed] [Google Scholar]
- 21.Kim SH, Arnold D, Lloyd A, Roux SJ. Antisense expression of an Arabidopsis ran binding protein renders transgenic roots hypersensitive to auxin and alters auxin-induced root growth and development by arresting mitotic progress. Plant Cell. 2001;13(12):2619–2630. doi: 10.1105/tpc.13.12.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zang A, Xu X, Neill S, Cai W. Overexpression of OsRAN2 in rice and Arabidopsis renders transgenic plants hypersensitive to salinity and osmotic stress. J Exp Bot. 2010;61(3):777–789. doi: 10.1093/jxb/erp341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L, Ramsay T, Zinkgraf M, Sundell D, Street NR, Filkov V, Groover A. A resource for characterizing genome-wide binding and putative target genes of transcription factors expressed during secondary growth and wood formation in Populus. Plant J. 2015;82(5):887–898. doi: 10.1111/tpj.12850. [DOI] [PubMed] [Google Scholar]
- 24.Liu L, Zinkgraf M, Petzold HE, Beers EP, Filkov V, Groover A. The Populus ARBORKNOX1 homeodomain transcription factor regulates woody growth through binding to evolutionarily conserved target genes of diverse function. The New phytologist. 2015;205(2):682–694. doi: 10.1111/nph.13151. [DOI] [PubMed] [Google Scholar]
- 25.Zhong R, Ye ZH. MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant & Cell Physiology. 2012;53(2):368–380. doi: 10.1093/pcp/pcr185. [DOI] [PubMed] [Google Scholar]
- 26.Zhong R, McCarthy RL, Lee C, Ye ZH. Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol. 2011;157(3):1452–1468. doi: 10.1104/pp.111.181354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang X, Zhuang Y, Qi G, Wang D, Liu H, Wang K, Chai G, Zhou G. Poplar PdMYB221 is involved in the direct and indirect regulation of secondary wall biosynthesis during wood formation. Scientific Reports. 2015;5:12240. doi: 10.1038/srep12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li C, Wang X, Ran L, Tian Q, Fan D, Luo K. PtoMYB92 is a Transcriptional Activator of the lignin biosynthetic pathway during secondary cell wall formation in Populus tomentosa. Plant & Cell Physiology. 2015;56(12):2436–2446. doi: 10.1093/pcp/pcv157. [DOI] [PubMed] [Google Scholar]
- 29.Baghdady A, Blervacq AS, Jouanin L, Grima-Pettenati J, Sivadon P, Hawkins S. Eucalyptus gunnii CCR and CAD2 promoters are active in lignifying cells during primary and secondary xylem formation in Arabidopsis thaliana. Plant Physiol Biochem. 2006;44(11–12):674–683. doi: 10.1016/j.plaphy.2006.10.027. [DOI] [PubMed] [Google Scholar]
- 30.Coleman HD, Yan J, Mansfield SD. Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc Natl Acad Sci U S A. 2009;106(31):13118–13123. doi: 10.1073/pnas.0900188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang XH, Li XG, Li BL, Zhang DQ. Genome-wide transcriptional profiling reveals molecular signatures of secondary xylem differentiation in Populus tomentosa. Genet Mol Res. 2014;13(4):9489–9504. doi: 10.4238/2014.November.11.14. [DOI] [PubMed] [Google Scholar]
- 32.Haizel T, Merkle T, Pay A, Fejes E, Nagy F. Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. Plant J. 1997;11(1):93–103. doi: 10.1046/j.1365-313X.1997.11010093.x. [DOI] [PubMed] [Google Scholar]
- 33.Tian B, Lin ZB, Ding Y, Ma QH. Cloning and characterization of a cDNA encoding Ran binding protein from wheat. DNA Seq. 2006;17(2):136–142. doi: 10.1080/10425170600609306. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Xu W, Xu Y, Chong K, Xu Z, Xia G. Wheat RAN1, a nuclear small G protein, is involved in regulation of cell division in yeast. Plant Sci. 2004;167(6):1183–1190. doi: 10.1016/j.plantsci.2004.03.011. [DOI] [Google Scholar]
- 35.Lü S, Fan Y, Jin C. Overexpression of a Ran GTPase homologous gene, FaRan from tall fescue, in transgenic Arabidopsis. Biol Plant. 2011;55(2):331–334. doi: 10.1007/s10535-011-0048-0. [DOI] [Google Scholar]
- 36.Reichel C, Mathur J, Eckes P, Langenkemper K, Koncz C, Schell J, Reiss B, Maas C. Enhanced green fluorescence by the expression of an Aequorea victoria green fluorescent protein mutant in mono- and dicotyledonous plant cells. Proc Natl Acad Sci U S A. 1996;93(12):5888–5893. doi: 10.1073/pnas.93.12.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263(5148):802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 38.Caniard A, Zerbe P, Legrand S, Cohade A, Valot N, Magnard JL, Bohlmann J, Legendre L. Discovery and functional characterization of two diterpene synthases for sclareol biosynthesis in Salvia sclarea (L.) and their relevance for perfume manufacture. BMC Plant Biol. 2012;12:119. doi: 10.1186/1471-2229-12-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weyens N, Boulet J, Adriaensen D, Timmermans J-P, Prinsen E, Van Oevelen S, D’Haen J, Smeets K, Van Der Lelie D, Taghavi S. Contrasting colonization and plant growth promoting capacity between wild type and a gfp-derative of the endophyte Pseudomonas putida W619 in hybrid poplar. Plant Soil. 2012;356(1–2):217–230. doi: 10.1007/s11104-011-0831-x. [DOI] [Google Scholar]
- 40.Christou P, McCabe DE, Swain WF. Stable transformation of soybean callus by DNA-coated gold particles. Plant Physiol. 1988;87(3):671–674. doi: 10.1104/pp.87.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell DR, Wallace KM, Bathe JH, Martinell BJ, McCabe DE. Stable transformation of Phaseolus vulgaris via electric-discharge mediated particle acceleration. Plant Cell Rep. 1993;12(3):165–169. doi: 10.1007/BF00239099. [DOI] [PubMed] [Google Scholar]
- 42.Brar GS, Cohen BA, Vick CL, Johnson GW. Recovery of transgenic peanut (Arachis hypogaea L.) plants from elite cultivars utilizing ACCELL® technology. Plant J. 1994;5(5):745–753. doi: 10.1111/j.1365-313X.1994.00745.x. [DOI] [Google Scholar]
- 43.Ikea J, Ingelbrecht I, Uwaifo A, Thottappilly G. Stable gene transformation in cowpea (Vigna unguiculata L. Walp.) using particle gun method. Afr J Biotechnol. 2003;2(8):211–218. doi: 10.5897/AJB2003.000-1044. [DOI] [Google Scholar]
- 44.Devantier YA, Moffatt B, Jones C, Charest PJ. Microprojectile-mediated DNA delivery to the Salicaceae family. Can J Bot. 1993;71(11):1458–1466. doi: 10.1139/b93-176. [DOI] [Google Scholar]
- 45.Su X, Chu Y, Li H, Hou Y, Zhang B, Huang Q, Hu Z, Huang R, Tian Y. Expression of multiple resistance genes enhances tolerance to environmental stressors in transgenic poplar (Populus × euramericana ‘Guariento’) PLoS One. 2011;6(9):e24614. doi: 10.1371/journal.pone.0024614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han X, Ma S, Kong X, Takano T, Liu S. Efficient agrobacterium-mediated transformation of hybrid Poplar Populus davidiana Dode × Populus bollena Lauche. Int J Mol Sci. 2013;14(2):2515–2528. doi: 10.3390/ijms14022515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao C, Craig JC, Petzold HE, Dickerman AW, Beers EP. The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 2005;138(2):803–818. doi: 10.1104/pp.105.060202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karpinska B, Karlsson M, Srivastava M, Stenberg A, Schrader J, Sterky F, Bhalerao R, Wingsle G. MYB transcription factors are differentially expressed and regulated during secondary vascular tissue development in hybrid aspen. Plant Mol Biol. 2004;56(2):255–270. doi: 10.1007/s11103-004-3354-5. [DOI] [PubMed] [Google Scholar]
- 49.Patzlaff A, McInnis S, Courtenay A, Surman C, Newman L, Smith C, Bevan M, Mansfield S, Whetten R, Sederoff R. Characterisation of a pine MYB that regulates lignification. Plant J. 2003;36(6):743–754. doi: 10.1046/j.1365-313X.2003.01916.x. [DOI] [PubMed] [Google Scholar]
- 50.Li S, Su X, Zhang B, Huang Q, Hu Z, Lu M. Molecular cloning and functional analysis of the Populus deltoides remorin gene PdREM. Tree Physiol. 2013;33(10):1111–1121. doi: 10.1093/treephys/tpt072. [DOI] [PubMed] [Google Scholar]
- 51.Lee JH, Zhou S, Smas CM. Identification of RANBP16 and RANBP17 as novel interaction partners for the bHLH transcription factor E12. J Cell Biochem. 2010;111(1):195–206. doi: 10.1002/jcb.22689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitney IP. Thermocycle-regulated wall regulator interacting bHLH encodes a protein that interacts with secondary-cell-wall-associated transcription factors. Masters Theses May 2014 - current. 2015. p. 174. [Google Scholar]
- 53.Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004;40(1):22–34. doi: 10.1111/j.1365-313X.2004.02183.x. [DOI] [PubMed] [Google Scholar]
- 54.Legay S, Sivadon P, Blervacq AS, Pavy N, Baghdady A, Tremblay L, Levasseur C, Ladouce N, Lapierre C, Seguin A, Hawkins S, Mackay J, Grima-Pettenati J. EgMYB1, an R2R3 MYB transcription factor from eucalyptus negatively regulates secondary cell wall formation in Arabidopsis and poplar. The New phytologist. 2010;188(3):774–786. doi: 10.1111/j.1469-8137.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 55.Andersson S, Serimaa R, Torkkeli M, Paakkari T, Saranpää P, Pesonen E. Microfibril angle of Norway spruce [Picea abies (L.) Karst.] compression wood: comparison of measuring techniques. J Wood Sci. 2000;46(5):343–349. doi: 10.1007/BF00776394. [DOI] [Google Scholar]
- 56.Donaldson L. The use of pit apertures as windows to measure microfibril angle in chemical pulp fibers. Wood Fiber Sci. 1991;23(2):290–295. [Google Scholar]
- 57.Kumar S. Genetic parameter estimates for wood stiffness, strength, internal checking, and resin bleeding for radiata pine. Can J For Res. 2004;34(12):2601–2610. doi: 10.1139/x04-128. [DOI] [Google Scholar]
- 58.Kumar S, Jayawickrama K, Lee J, Lausberg M. Direct and indirect measures of stiffness and strength show high heritability in a wind-pollinated radiata pine progeny test in New Zealand. Silvae Genetica. 2002;51(5–6):256–260. [Google Scholar]
- 59.Li S, Su X, Zhang B, Huang Q, Chu Y, Ding C. Functional identification of wood-property candidate gene PdCYTOB in Populus deltoides. Chinese Bulletin of Botany. 2011;46:642–651. doi: 10.3724/SP.J.1259.2011.00642. [DOI] [Google Scholar]
- 60.Schulze H, Dose M, Korpal M, Meyer I, Italiano JE, Shivdasani RA. RanBP10 is a cytoplasmic guanine nucleotide exchange factor that modulates noncentrosomal microtubules. J Biol Chem. 2008;283(20):14109–14119. doi: 10.1074/jbc.M709397200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spokevicius AV, Southerton SG, MacMillan CP, Qiu D, Gan S, Tibbits JF, Moran GF, Bossinger G. β‐tubulin affects cellulose microfibril orientation in plant secondary fibre cell walls. Plant J. 2007;51(4):717–726. doi: 10.1111/j.1365-313X.2007.03176.x. [DOI] [PubMed] [Google Scholar]
- 62.Zhang B, Su X, Zheng S. Establishment of a highly efficient plant regeneration system of Populus davidiana × P. bolleana and study of its genetic stability. Journal of Beijing Forestry University. 2008;30:68–73. [Google Scholar]
- 63.Brunner AM, Yakovlev IA, Strauss SH. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 2004;4:14. doi: 10.1186/1471-2229-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313(4):856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 65.Iida A, Seki M, Kamada M, Yamada Y, Morikawa H. Gene delivery into cultured plant cells by DNA-coated gold particles accelerated by a pneumatic particle gun. Theor Appl Genet. 1990;80(6):813–816. doi: 10.1007/BF00224198. [DOI] [PubMed] [Google Scholar]
- 66.Abbott JC, Barakate A, Pincon G, Legrand M, Lapierre C, Mila I, Schuch W, Halpin C. Simultaneous suppression of multiple genes by single transgenes. Down-regulation of three unrelated lignin biosynthetic genes in tobacco. Plant Physiol. 2002;128(3):844–853. doi: 10.1104/pp.010698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jasencakova Z, Meister A, Walter J, Turner BM, Schubert I. Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell. 2000;12(11):2087–2100. doi: 10.1105/tpc.12.11.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Franklin G. Preparation of thin sections of synthetic resins and wood-resin composites, and a new macerating method for wood. Nature. 1945;155(3924):51. doi: 10.1038/155051a0. [DOI] [Google Scholar]
- 69.Leney L. A technique for measuring fibril angle using polarized light. Wood Fiber Sci. 1981;13(1):13–16. [Google Scholar]
- 70.Wang H, Drummond J, Reath S, Hunt K, Watson P. An improved fibril angle measurement method for wood fibres. Wood Sci Technol. 2001;34(6):493–503. doi: 10.1007/s002260000068. [DOI] [Google Scholar]
- 71.Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science. 1983;220(4601):1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- 72.Sazer S, Sherwood SW. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J Cell Sci. 1990;97(Pt 3):509–516. doi: 10.1242/jcs.97.3.509. [DOI] [PubMed] [Google Scholar]
- 73.Gil S, Sarun S, Biete A, Prezado Y, Sabes M. Survival analysis of F98 glioma rat cells following minibeam or broad-beam synchrotron radiation therapy. Radiat Oncol. 2011;6:37. doi: 10.1186/1748-717X-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 75.Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9(4):299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The open reading frame (ORF) and amino acid sequences of P. deltoides PdRanBP and those for hybrid poplar (P. davidiana Dode × P. bolleana Lauche) hpPdRanBP were deposited into the National Center for Biotechnology Information (NCBI) under nucleotide numbers KU841446 and KU841447. We deposited our phylogenetic data in TreeBase, with the following URL: http://purl.org/phylo/treebase/phylows/study/TB2:S18967.


