Abstract
There is a need for a rapid, accurate serodiagnostic test useful for both women and men infected by Trichomonas vaginalis, which causes the number one sexually transmitted infection (STI). Women and men exposed to T. vaginalis make serum antibody to fructose-1,6-bisphosphate aldolase (ALD), α-enolase (ENO), and glyceraldehyde-3-phosphate dehydrogenase (GAP). We identified, by epitope mapping, the common and distinct epitopes of each protein detected by the sera of women patients with trichomonosis and by the sera of men highly seropositive to the immunogenic protein α-actinin (positive control sera). We analyzed the amino acid sequences to determine the extent of identity of the epitopes of each protein with other proteins in the databanks. This approach identified epitopes unique to T. vaginalis, indicating these peptide-epitopes as possible targets for a serodiagnostic test. Individual or combinations of 15-mer peptide epitopes with low to no identity with other proteins were reactive with positive control sera from both women and men but were unreactive with negative control sera. These analyses permitted the synthesis of a recombinant His6 fusion protein of 111 amino acids with an Mr of ~13.4 kDa, which consisted of 15-mer peptides of two distinct epitopes each for ALD, ENO, and GAP. This recombinant protein was purified by affinity chromatography. This composite protein was detected by enzyme-linked immunosorbent assay (ELISA), dot blots, and immunoblots, using positive control sera from women and men. These data indicate that it is possible to identify epitopes and that either singly, in combination, or as a composite protein represent targets for a point-of-care serodiagnostic test for T. vaginalis.
Keywords: diagnostics, point-of-care, targets, trichomonosis
Introduction
The literature is replete with the negative health outcomes to women that result from trichomonosis, the sexually transmitted infection (STI) caused by Trichomonas vaginalis.1 Although published reports frequently state that men clear this STI, there have been no rigorous clinical studies to support this contention. A remarkable feature of this STI among women and men is persistence,1 where in men, the parasite has been detected in the prostate and where recently T. vaginalis DNA was detected in hyperplastic prostate tissue.2,3 Indeed, this hallmark for T. vaginalis may relate to the fact that in recent population studies, most women are asymptomatic4,5 despite high prevalence rates of infection in women.4,6 That the host–parasite interrelationship is exceedingly complex is now appreciated, as evidenced by the role of environmental factors regulating the expression of virulence attributes7–10 and the identification of preparatory factors for colonization and pathogenesis.11–19
Epidemiological studies indicating the relation between seropositivity and prostate cancer20–22 and the recent report showing T. vaginalis DNA at high rates in prostatic tissue3 have made evident the need for a better point-of-care (POC) serodiagnostic test that is applicable to men with trichomonosis and that would be also useful for women patients with trichomonosis. There is currently a commercially available POC diagnostic test for trichomonosis23–25 (OSOM® Trichomonas Rapid Test, Sekisui Diagnostics, Lexington, MA, USA), but this test is useful only for women patients with trichomonosis. Nucleic acid amplification tests (NAATs) have proven useful and effective in the detection of T. vaginalis infections in both women and men.26–29 However, these tests require trained personnel, expensive equipment, and shipment of possibly labile samples to special facilities for processing. Thus, an inexpensive, rapid, sensitive, noninvasive POC serodiagnostic test for women and men may provide a platform for screening large at-risk cohorts. Such a test would be especially suitable for examining men if the connection between trichomonosis and prostate cancer is validated. Recently, the highly immunogenic trichomonad protein α-actinin30 (ACT) was analyzed and found to have potential as a serodiagnostic target.31 However, we hypothesize that surface protein immunogens of T. vaginalis have equal if not more utility to form the basis of POC antibody-based serological tests for women and men.
We present data on the characterization of three metabolic enzyme immunogens, fructose-1,6-bisphosphate aldolase (ALD), α-enolase (ENO),32 and glyceraldehyde-3-phosphate dehydrogenase (GAP).19 The sera from women patients with trichomonosis and the sera from men, which are both highly seropositive for ACT20–22,31 (the target protein for screening men for seropositivity), are immunoreactive with these proteins (positive control sera). Therefore, we identified the epitopes detected by positive control sera from women and men, and sequence identity analysis permitted identification of epitopes unique to T. vaginalis, based on minimal or absent identity to amino acid sequences of other proteins in databanks. Synthesis of 15-mer peptides containing the epitope sequences were screened individually or in combination and found to be immunoreactive with seropositive sera. Lastly, a plasmid was constructed that contained a synthesized DNA that encoded for a recombinant protein consisting of six sequential 15-mer peptides referred to as series of epitopes (SOE), within which were epitopes for ALD, ENO, and GAP. This recombinant SOE (rSOE) was detected by the positive control sera from women and men, indicating its potential as a composite target for the serodiagnosis of trichomonosis.
Materials and methods
SPOTs membrane synthesis for ALD, ENO, and GAP proteins
Oligopeptides (Custom Peptide Arrays) that have been immobilized on membranes (SPOTs system; Sigma-Aldrich Corp, St Louis, MO, USA) are routinely used for epitope identification, and this technology was used recently on a trichomonad immunogenic protein.31 Thereby, oligopeptides derived from the sequences of T. vaginalis ALD (GenBank® accession number AAW78351), ENO31 (GenBank accession number AAK73099), and GAP19 (GenBank accession number AAA30325) were synthesized on activated membranes. ALD is comprised of 328 amino acids (36.3 kDa), ENO is comprised of 472 amino acids (51.3 kDa), and GAP is comprised of 350 amino acids (38.1 kDa). Five to 10 nmol of each peptide was covalently bound to a Whatman 50 cellulose support (Whatman International Ltd, Maidstone, UK) by the C-terminus, using Fmoc-L amino acid chemistry, and had an acetylated N-terminus. The oligopeptides, which spanned the entire sequence of the protein, were 11-mer amino acids in length and had a sequential overlap of eight amino acids.
Probing the ALD, ENO, and GAP SPOTs membrane with sera from women and men and monoclonal antibodies (MAbs)
During the course of our research on T. vaginalis, we have examined, for reactivity to the trichomonad immunogenic protein ACT, hundreds of sera samples of women patients with trichomonosis as well as thousands of sera samples of men, in studies determining the relation between serostatus and prostate cancer.19–22,31–36 Unreactive sera (negative control sera) and highly reactive sera (positive control sera) with ACT were recently used to identify the ACT epitopes.31 Importantly, individual ACT-seropositive sera of women and men gave similar reactivity to trichomonad proteins and ACT, and this permitted us to pool sera, to have sufficient amounts for conducting epitope mapping experiments, as outlined below.
Each ALD, ENO, and GAP membrane was washed with a small volume of 100% MeOH for 5 minutes to avoid precipitation of hydrophobic peptides during the following procedure. After a 10-minute wash performed three times (3×) in 25 mL of tris(hydroxymethyl)aminomethane (Tris)-buffered saline (TBS; 50 mM Tris-HCl, pH 8.0, 137 mM NaCl, and 2.7 mM KCl), the SPOTs membrane was incubated in blocking buffer (TBS-5% bovine serum albumin [BSA]) for 2 hours. All reactions with the SPOTs membrane were conducted at room temperature (RT), with gentle agitation on a shaker platform. The membrane was first incubated with a 1:10 dilution of negative control sera from women or men. No spots were detected for each protein membrane. Next, the membranes were incubated similarly with a hybridoma supernatant containing monoclonal antibody (MAb) B4432 and MAb B4319 reactive with epitopes to each of the ENO and GAP proteins, respectively, in addition to MAbs newly generated to the proteins (Tables 1–3). This was done to determine whether the reactions of these MAbs were identical to those seen using positive control sera from women and men. Finally, the membranes were incubated with positive control sera from women or men. All incubations with sera and MAbs were for 3 hours prior to washing and processing (described below). Importantly, the sera from women and men were categorized as either unreactive (negative control sera) or highly reactive (positive control sera) with ACT, a highly immunogenic trichomonad protein,30,31 as before.20–22
Table 1.
Epitopesa,b of Trichomonas vaginalis fructose-1,6-bisphosphate aldolase are reactive with MAbs and human sera
| SPOTsi no | Amino acid sequence | Epitope sequence | Positive control women sera | Positive control men sera | MAb reactions |
|---|---|---|---|---|---|
| 13–14 | 40–47 | AIITASVK | A-W1c | ||
| 19–21 | 58–65 | AGARKYAN | A-M1c,d | ||
| 21 | 61–71 | RKYANQTMLRY | A-W2c,d | ||
| 31 | 91–101 | PIVLHLDHGDS | A-W3c,d,e | ||
| 47–49 | 142–149 | RPDYVTVE | ALD12c | ||
| 55–57 | 166–173 | KHTYTRPE | ALD64c,f | ||
| 57 | 169–179 | YTRPEEVQDFV | A-W4c | ||
| 64–65 | 193–203 | TSHGAYKF | A-W5c,f,g,h | ||
| 78 | 232–242 | SIPQEYVEMVN | A-W6c | ||
| 93 | 277–287 | RMVMTGTIRRL | A-M2c | ||
| 99–100 | 298–305 | RQYLGEAR | A-W7c,d,h | A-M3c,d,h | |
| 101–103 | 304–311 | ARTKLTEM | A-W8 |
Notes:
This refers to epitopes that have ≥50% amino acid sequence identity with the indicated human and microbial pathogen homologues;
there is minimal amino acid sequence identity of these epitopes with the human homologue;
Treponema pallidum;
Neisseria gonorrhoeae;
Candida albicans, Saccharomyces cerevisiae, Streptococcus pyogenes, S. pneumoniae, Staphylococcus aureus, and Escherichia coli;
C. albicans and S. cerevisiae;
C. albicans, S. aureus, and E. coli;
S. aureus; inumbers of amino acids for the immobilized peptides (Materials and methods).
Abbreviations: A, aldolase; M, positive control men sera; MAb, monoclonal antibody; W, positive control women sera; ALD12 and ALD64, designate MAbs to aldolase.
Table 2.
Epitopesa of Trichomonas vaginalis α-enolase are reactive with MAbs and human sera
| SPOTsm no | Amino acid sequence | Epitope sequence | Positive control women sera | Positive control men sera | MAb reactions |
|---|---|---|---|---|---|
| 2–3 | 7–14 | AIVKECIA | ALD13 | ||
| 21–23 | 64–71 | YLGRVTLA | E-W1 | ALD55/97 | |
| 23–25 | 70–77 | LAARSSAP | E-M1d | ||
| 31–33 | 94–101 | DKARYGGK | E-W2b,f–l | E-M2b,f–l | |
| 46–47 | 139–146 | TVLKKNIG | E-W3h | E-M3h | |
| 62 | 184–194 | VPKKFKLPSPF | E-W4d | E-M4d | |
| 67 | 199–209 | NGGKHAGGNLK | E-M5b,d–f,h,l | ||
| 76 | 226–236 | QLRMVAEVYQK | E-M6 | ||
| 79–81 | 238–245 | GGLLVKKY | E-W5d | ||
| 81 | 241–251 | LVKKYGLSAKN | E-M7d,e | ||
| 99–100 | 298–305 | FYDEEKKL | E-M8d | ||
| 110 | 328–338 | KKHPAIVSIED | E-W6c,f | ||
| 114–116 | 343–350 | LDYENWTK | E-W7 | ||
| 117–118 | 352–359 | NARLGQRV | E-M9f,i–k | ||
| 121–123 | 364–371 | DDLYTTNPb | ALD11c,f | ||
| 123–124 | 370–377 | NPITIKKG | E-W8d | E-M10d | |
| 149–151 | 448–455 | ERIQKYTR | E-W9b,i–k | E-M11b,h–j | B44b,i–k |
| 154–155 | 463–470 | LKEHDMLA | ALD25 |
Notes:
This refers to epitopes that have ≥50% amino acid sequence identity with the indicated human and microbial pathogen homologues;
human homologue;
microbial pathogens listed in Table 1;
Candida Albicans;
Saccharomyces cerevisiae;
Chlamydia Trachomatis;
Treponema Pallidum;
Neisseria gonorrhoeae;
Streptococcus Pneumoniae;
as per “a” above except for Streptococcus pyogenes;
as per “a” above except for Staphylococcus aureus;
as per “a” above except for Escherichia coli;
numbers of amino acids for the immobilized peptides (Materials and methods).
Abbreviations: E, α-enolase; M, positive control men sera; MAb, monoclonal antibody; W, positive control women sera; ALD13, ALD55/97, ALD11, and ALD25, designate MAbs to enolase.
Table 3.
Epitopesa of Trichomonas vaginalis glyceraldehyde-3-phosphate dehydrogenase are reactive with MAbs and human sera
| SPOTsl no | Amino acid sequence | Epitope sequence | Positive control women seral | Positive control men seral | MAb reactions |
|---|---|---|---|---|---|
| 3–5 | 13–17 | LYPKD | G-M1 | ||
| 12 | 34–44 | YLLKYDTAHRA | G-W1e,h,i,k | ALD32Ce,h,i,k | |
| 20 | 58–68 | FTVGEGADKWV | G-M2 | ||
| 23–24 | 70–77 | KSIGGRLG | G-W2 | ALD30A | |
| 25–27 | 79–83 | SQLPW | G-M3b,d–g | ||
| 31–33 | 94–101 | STGIFRTK | G-W3c–e,j | ||
| 35–37 | 106–113 | AEGKIKKD | G-W4 | ||
| 39–41 | 118–125 | HLVSGAKK | G-W5c–k | G-M4c–k | |
| 51–53 | 157–161 | SNASC | G-M5c–k | ||
| 57–59 | 175–182 | NAFGIRNG | G-W6b–e,i–k | ||
| 67–69 | 205–212 | RRARAAGM | G-W7h,i | B43i,j | |
| 72–74 | 217–224 | TSTGAAIA | G-W8b–k | ||
| 75–77 | 229–233 | CHGLP | G-M6 | ||
| 84 | 250–260 | SLVDLTVNVNA | G-W9c–e,j,k | ||
| 97–98 | 292–299 | VSSDIIGC | G-M7b–e,h,i,k | ||
| 99–100 | 298–305 | GCQYSSIV | G-M8b–f | ||
| 101 | 301–311 | YSSIVDALSTK | G-W10e,f,h,i | ||
| 109 | 325–335 | VLSWYDNEWMY | G-W11b–k | G-M9b–k | |
| 111–113 | 337–341 | CRCAD | G-M10 |
Notes:
This refers to epitopes that have ≥50% amino acid sequence identity with the indicated human and microbial pathogen homologues;
human homologue;
Candida Albicans;
Saccharomyces Cerevisiae;
Chlamydia Trachomatis;
Treponema Pallidum;
Neisseria Gonorrhoeae;
Streptococcus Pneumoniae;
Streptococcus Pyogenes;
Staphylococcus Aureus;
Escherichia Coli;
numbers of amino acids for the immobilized peptides (Materials and methods).
Abbreviations: G, glyceraldehyde-3-phosphate dehydrogenase; M, positive control men sera; MAb, monoclonal antibody; W, positive control women sera; ALD32C and ALD30A, designate MAbs to glyceraldehyde-3-phosphate dehydrogenase.
After washing the SPOTs membranes 3× for 5 minutes in TBS, the membrane was processed for color development and SPOTs analysis, as detailed recently.31 Images of the membranes were obtained with an Apple iPhone 4 (Apple Inc, Cupertino, CA, USA) and for assessment of the extent of reactivity and determination of the epitope amino acid sequences (Tables 1–3). The epitope sequences were compared with other proteins by using the protein-protein basic local alignment search tool (BLAST [http://www.ncbi.nlm.nih.gov/BLAST]). Amino acid sequence alignments of the proteins were performed with CLC Protein Workbench, Version 6.5, (Aarhus, Denmark). Hydrophobicity plots37 and antigenicity plots38 were constructed using Lasergene® MegAlign® (DNASTAR Inc, Madison, WI, USA). Finally, the membranes were regenerated through a series of washings, also as previously described.31
Synthesis of individual 15-mer epitopes
Peptide epitopes from each protein were selected, based on low percent identity of the amino acid protein sequence and solubility in aqueous solutions, for handling of the oligopeptides. The 15-mer amino acid sequences (Table 4) were sent to Sigma-Aldrich Corp and synthesized using their PEPscreen® Custom Peptide service. Each individual 15-mer peptide was acetylated at the amino terminus and was amidylated at the carboxy terminus. The 15-mer peptide was screened by mass spectrometry to determine the yield and purity of each product. Peptide epitopes were received with a pass/fail designation and the amount of peptide was provided. Three ACT 15-mer peptides, as described recently,31 were used as positive controls in dot blots (below). These peptides were designated ACT1, ACT2, and ACT3 and corresponded to the amino acid sequences AQPLYDEAIAFKEEV, FKDTFKYFDKDKSNS, and SVNRHHSQLITYIKH, respectively. The underlined amino acids represent the epitopes, as characterized before.31
Table 4.
List of synthetic 15-mer peptides of representative epitopes of ALD, ENO, GAP, and ACT that are reactive with positive control women and men sera
| No | Women/men epitope designationa | Peptide name | Amino acid numbers | Amino acid sequenceb |
|---|---|---|---|---|
| 1 | A-W1 | ALD1 | 36–50 | EQLQAIITASVKTES |
| 2 | A-ALD12 | ALD2 | 138–152 | EAHSRPDYVTVEGEL |
| 3 | A-ALD64 | ALD3 | 159–173 | EDDVKAEKHTYTRPE |
| 4 | A-W4 | ALD4 | 167–181 | HTYTRPEEVODFVSK |
| 5 | A-W6 | ALD5 | 230–244 | SSSIPOEYVEMVNKY |
| 6 | A-M2 | ALD6 | 275–289 | DGRMVMTGTIRRLFV |
| 7 | A-W8 | ALD7 | 301–315 | LGEARTKLTEMYMRK |
| 8 | E-W1 | ENO1 | 57–71 | VKYLGRVTLAARSSA |
| 9 | E-M1 | ENO2 | 70–84 | VTLAARSSAPSGAST |
| 10 | E-W4, E-M4 | ENO3 | 182–196 | TDGTVLKKNIGGNAC |
| 11 | E-M6 | ENO4 | 224–238 | DKVPKKFKLPSPFFN |
| 12 | E-W5 | ENO5 | 236–250 | KLGGLLVKKYGLSAK |
| 13 | E-M8 | ENO6 | 295–309 | SSEFYDEEKKLYEVE |
| 14 | E-W7 | ENO7 | 344–358 | DYENWTKLNARLGQR |
| 15 | E-W8, E-M10 | ENO8 | 366–380 | LYTTNPITIKKGLEG |
| 16 | G-M1 | GAP1 | 8–22 | RACRKLYPKDIQVVA |
| 17 | G-M2 | GAP2 | 56–70 | QEFTVGEGADKWVVK |
| 18 | G-W2 | GAP3 | 67–81 | WVVKSIGGRLGPSQL |
| 19 | G-W4 | GAP4 | 104–118 | KDAEGKIKKDDGYDGH |
| 20 | G-M6 | GAP5 | 224–238 | ALPKVCHGLPPKSLD |
| 21 | G-M10 | GAP6 | 332–346 | EWMYSCRCADIFHRL |
| 22 | ACT-W9, M1 | ACT1 | 463–467 | SVNRHHSQLITYIKH |
| 23 | ACT-W10, M2 | ACT2 | 492–506 | AQPLYDEAIAFKEEV |
| 24 | ACT-W13, M5 | ACT3 | 803–817 | FKDTFKYFDKDKSNS |
Notes:
underlined amino acids represent epitope sequences, as determined by the SPOTs assay (Materials and methods).
Abbreviations: A and ALD1 through ALD7, aldolase peptides; ACT and ACT1 through ACT3, alpha-actinin peptides; E and ENO1 through ENO8, alpha-enolase peptides; G and GAP1 through GAP6, glyceraldehyde-3-phosphate dehydrogenase peptides; M, positive control men sera; W, positive control women sera.
Dot-blot detection of individual and combined 15-mer epitopes with women and men sera
Approximately 1 µg of individual and/or a combination of synthetic peptides were dot-blotted onto a nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA) diluted and allowed to air dry for 30 minutes at 37°C. These dot blots were fit into individual wells of a 96-well microtiter enzyme-linked immunosorbent assay (ELISA) plate. Then, 100 µL of 2% ELISA-grade BSA (Sigma-Aldrich Corp) in phosphate-buffered saline (PBS) (eBSA-PBS), pH 7.4, was added and incubated for 2 hours at RT, after which 5 µL of a 1:1 dilution (v/v) of sera (from T. vaginalisc –negative-control and –positive-control women or men) in PBS, pH 7.4, was added and incubated for 30 minutes at RT. The remainder of the procedure was as detailed recently.31 Densitometric scans were produced using the ImageJ software (rsbweb.nih.gov/ij).
Recombinant SOEprotein expression and purification
The SOE encoding for six 15-mer amino acid sequences, two each of which contained epitopes for ALD, ENO, and GAP, consisted of 111 amino acids for an Mr of 13.35 kDa. These epitopes were chosen because of minimal or no amino acid sequence identity to other proteins in databanks. The DNA encoding for the SOE with a His6 tag at the carboxy terminus was cloned into a pET23b expression plasmid construct containing an ampicillin (amp) resistance locus obtained from Gen-way Biotech, Inc (San Diego, CA, USA) that was transformed into Escherichia coli Bl21DE3. Recombinant E. coli (rE. coli) was stored as glycerol stocks at −70°C until used, which were thawed and streaked onto Luria Broth (LB) agar plates (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl, and 15 g/L agar) containing 25 µg amp. Isolated colonies were inoculated into 200 mL fresh LB containing amp and incubated in a shaker incubator at 37°C and 220 rpm. After overnight (o/n) growth, rE. coli were inoculated into fresh LB medium with amp and incubated for 3 hours at 37°C at 220 rpm, prior to the addition of 1 mM isopropylthiogalactoside (IPTG) (Sigma-Aldrich Corp, St. Louis, MO, USA) and incubation for an additional 3 hours. The rE. coli were centrifuged using a Sovall SLA-1500 rotor (Thermo Fisher Scientific Inc, Waltham, MA, USA) at 8,000 rpm and 4°C for 15 minutes. The supernatant was decanted, and the pellet was stored at −80°C until used.
At different time intervals prior to and after the IPTG addition, 1 mL of rE. coli was microfuged at 10,000 rpm for 15 minutes, and pellets were prepared for sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (34, 35), for analysis of recombinant SOE::His6 fusion protein (12.2 kDa) expression after IPTG addition. Immunoblot analysis after SDS-PAGE confirmed the synthesis of SOE::His6, using as probe-positive control, the sera of women and men who were seropositive to ACT, as defined above.
For purification of recombinant SOE::His6 (rSOE), pelleted rE. coli were thawed for 15 minutes on ice and suspended in 10 to 20 mL lysis buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10 mM β-mercaptoethanol (β-ME), and 0.1% Triton-X100). Lysates were sonicated 3× at RT for 30 seconds, with samples placed on ice at 30-second intervals. Sonicates were centrifuged using a Sorvall SS-34 rotor (Thermo Fisher Scientific Inc) at 8,000 rpm and 4°C for 20 minutes to remove insoluble debris. Clarified supernatant was applied to a nickel-nitrilotriacetic acid (Ni2+-NTA) Superflow affinity column that binds the His6 tag on the protein, according to the manufacturer’s instructions (Qiagen Inc, Venlo, The Netherlands). Pooled, purified SOE protein preparations were concentrated and dialyzed against PBS using a stirred Amicon™ Ultrafiltration Cell according to the manufacturer’s instructions (EMD Millipore, Billerica, MA, USA). The protein concentration was determined by the Lowry protein assay39, with BSA as a standard. Purification of the rSOE::His6 protein was confirmed by SDS-PAGE and immunoblot, as above. After SDS-PAGE, acrylamide gels were stained with Coomassie brilliant blue for visualization of the purified fusion protein used for immunoblotting. Positive efficient blotting of the recombinant protein was determined by staining the nitrocellulose membrane with Ponceau S solution. Both Coomassie brilliant blue and Ponceau S stains were from Sigma-Aldrich.
ELISA using rSOE
ELISA was performed by immobilizing purified rSOE protein onto 96-well, flat-bottom Nunc polystyrene plates (Thermo Fisher Scientific, Rochester, NY), as detailed recently.31 Briefly, each well was coated with 100 µL containing 1 µg of rSOE diluted in sodium carbonate (28.6 mM):sodium bicarbonate (71.4 mM) buffer, pH 9.6, prepared in PBS, and the plates were incubated o/n at RT with gentle agitation. Each plate was then washed 3× with PBS-1% Tween 20 (PBS-T). On the third wash, the plates were incubated in PBS-T for 5 minutes at RT with gentle agitation, prior to removing the PBS-T. The plates were then incubated upside down o/n at RT on paper towels before being covered with plastic wrap and stored at 4°C until used. For testing, plates were washed twice with PBS-T. On the second wash, the plates were incubated in PBS-T at RT for 5 minutes with gentle agitation and slap dried. Each well was then blocked with 200 µL of eBSA for 2 hours at 37°C. Plates were then washed twice with PBS-T. On the second wash, the plates were incubated in PBS-T at RT for 5 minutes with gentle agitation, followed by removing the PBS-T. Next, 100 µL of a 1:25 dilution of the women and men sera in eBSA-PBS was added to each well in duplicate and incubated at RT for 5 minutes with gentle agitation before incubation for 4 hours at 37°C. The plates were washed 3× with PBS-T. On the third wash, the plates were incubated in PBS-T for 5 minutes at RT with gentle agitation. After removal of the PBS-T, 100 µL of secondary horseradish peroxidase-conjugated goat-anti-human immunoglobulin (Ig)G (Fc-specific) immunoglobulin IgG (Fc-specific) (Bio-Rad Laboratories, Hercules, CA), diluted 1:1,500 in eBSA-PBS, was added to each well and incubated at RT with gentle agitation for 5 minutes, before incubation for 1 hour at 37°C. The plates were washed 3× with PBS-T, as above, prior to addition of 100 µL of color development solution (phosphate-citrate buffer with sodium perborate and 2,2’-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt) prepared according to the manufacturer’s (Sigma-Aldrich Corp) instructions and incubated at RT with gentle agitation for 15 minutes. Absorbance values at 405 nm (A405 mm) were obtained using a Bio-Tek Synergy HT plate reader (Bio-Tek Instruments, Inc, Winooski, VT, USA).
Results
Epitope characterization of ALD, ENO, and GAP by reactive sera of women and men
Tables 1–3 list the epitopes detected for each of the three proteins. There were 13, 18, and 19 epitopes detected for ALD, ENO, and GAP, respectively. Available MAbs also detected unique epitopes for each protein. Sequence identity comparisons with other microbial and fungal pathogens as well as human homologues revealed that some epitopes had >50% identity at the amino acid–protein sequence level, as indicated. It is noteworthy that some epitopes had identity to other pathogens causing STIs and to human homologues. Importantly, positive control sera from both women and men recognized the same epitopes. Interestingly but perhaps not surprisingly, MAbs to ALD and ENO, but not GAP, detected epitopes not recognized by the positive control sera from the women and men. These data suggest that it is possible to identify epitopes unique to ALD, ENO, and GAP and with little to no identity to other proteins. Of interest is the fact that inclusion of other neighboring amino acids for each of the epitopes significantly decreased the percent identity to each of the homologues examined. Finally, only ALD had almost threefold more epitopes recognized by the women sera (eight epitopes) versus the men sera (three epitopes). Both ENO and GAP had approximately the same number of epitopes each. Lastly and interestingly, women and men detected both identical and distinct linear epitopes throughout the protein.
Sequence alignment of the T. vaginalis ALD proteins with proteins of other representative organisms and Homo sapiens
We have previously presented amino acid sequence alignments for ENO and GAP.19,32 In Figure 1 we now show the amino acid sequence comparison of ALD with the homo-logues indicated in Table 1. The boxed amino acids contained in the T. vaginalis sequence are the epitopes presented in Table 1. The order from top to bottom of the sequences represents the highest to lowest percent identities.
Figure 1.
Sequence analyses of Trichomonas vaginalis fructose-1,6-bisphosphate aldolase.
Notes: The amino acid sequence identity of T. vaginalis enzyme was compared with the ALD proteins of Treponema pallidum, Neisseria gonorrhoeae, Streptococcus pyogenes, Streptococcus pneumoniae, Staphylococcus aureus, Escherichia coli, Candida albicans, Saccharomyces cerevisiae, and Homo sapiens. The percent identity was found to be 68%, 39%, 30%, 30%, 38%, 32%, 23%, 21%, and 10%, respectively. Boxed amino acids indicate epitopes of T. vaginalis detected by women and men sera and MAbs ALD12 and ALD64 (Table 1). Blue-colored amino acid residues represent high sequence identity ranging from 68% to 100%. Red residues are low sequence identity ranging from 0 to 33%, and dashed lines are gaps within the sequences of the corresponding ALD proteins of other organisms. Black lettering signifies approximately 34% to 67% identity. A-W1 through A-W8 are epitopes detected by women sera; A-M1 through A-M3 are epitopes detected by men sera.
Abbreviations: ALD, aldolase; Ca, Candida albicans; Ec, Escherichia coli; Hs, Homo sapiens; M, men; MAbs, monoclonal antibodies; Ng, Neisseria gonorrhoeae; Sa, Staphylococcus aureus; Sc, Saccharomyces cerevisiae; Spn, S. pneumoniae; Spy, Streptococcus pyogenes; Tp, Treponema pallidum; W, women; ALD12 and ALD64, designate MAbs to aldolase as per Table 1.
Hydrophobicity and antigenicity profiles of the ALD, ENO, and GAP sequences
Figure 2 presents analyses of hydrophobicity37 and antigenicity38 alignments relative to the epitopes along the protein (top of each profile). Of interest is that with few exceptions, the epitopes represent hydrophilic regions contained within the protein (values <0), perhaps consistent with the presentation of amino acids for antibody synthesis and recognition. The hydrophilic domains correspond to increased antigenicity (values >0), as shown in the bottom profiles. Based on these features of the epitopes, we then selected seven epitopes for ALD, eight for ENO, and six for GAP for synthesis of 15-mer peptides encoding the epitopes, as shown in Table 4, and the individual amino acid sequence encoding the epitope within the 15-mer peptide is shown with underlining.
Figure 2.
Hydrophobicity and antigenicity profiles of Trichomonas vaginalis α-enolase, aldolase, and glyceraldehyde-3-phosphate dehydrogenase proteins.
Notes: The individual epitopes detected by women and men sera and the MAbs are labeled above the numbered linear protein. Hydrophilicity is presented as values <0, and the corresponding antigenicity is >0.
Abbreviations: A and ALD, fructose-1,6-bisphosphate aldolase; E and ENO, α-enolase; G and GAP, glyceraldehyde-3-phosphate dehydrogenase; M, men; MAb, monoclonal antibody; W, women; ALD13, ALD55, ALD11, ALD25, and B44, designate MAbs to alpha-enolase, ALD12/64 designates a MAb to aldolase, and ALD32, ALD30, and B43 designate MAbs to glyceraldehyde-3-phosphate dehydrogenase.
Reaction of 15-mer peptides with epitopes with women and men sera
Figure 3 presents results from representative dot-blot reactions, using the positive control sera from women and men, of 15-mer peptides (Table 4) for ALD (Figure 3A), ENO (Figure 3B), and GAP (Figure 3C). Peptides derived from ACT (Figure 3D) were used as a positive control.31 Reactivity was detected for each 15-mer peptide, albeit at different levels of spot intensities. No peptides were detectable using the negative control sera for both women and men that was determined to be unreactive with the full-length proteins. These data suggest that some peptide epitopes have potential as serodiagnostic targets.
Figure 3.

Representative experiment showing dot blots of individual 15-mer peptide epitopes.
Notes: The peptides were derived from the proteins fructose-1,6-bisphosphate aldolase (A), α-enolase (B), glyceraldehyde-3-phosphate dehydrogenase (C), and α-actinin (D). Individual 15-mer peptides were spotted onto nitrocellulose prior to reacting with the sera of women and men (indicated below the spots), as detailed in Materials and methods. The individual epitopes are those indicated in Table 4.
Abbreviations: ACT, α-actinin; ALD, fructose-1,6-bisphosphate aldolase; ENO, α-enolase; GAP, glyceraldehyde-3-phosphate dehydrogenase.
An important finding is that while epitope mapping revealed epitopes unique to women and men (Tables 1–3), in fact, individual 15-mer peptides with epitopes were seen by the positive control sera from both women and men. For example, ALD1 (epitope A-W1, Table 1) and ALD5 (epitope A-W6) were initially identified with the positive control women sera, but these epitopes were also positive as 15-mer peptides when tested with men sera. Likewise, ENO7 (epitope E-W7) and GAP3 (G-W2) were detected by men sera, and GAP (epitope G-M6) was seen by women sera. These results indicate that serum reactivity of epitopes requires examination under different experimental conditions.
We next wanted to perform dot blots using random combinations of the 15-mer peptides (Table 4), to determine whether an increased extent of reactivity was seen for women and men sera. We further wanted combinations of peptides that might give equal reactivity for both sera. Figure 4A shows, in duplicate, the intensity of signal for each combination with the positive control women and men sera (labeled 4+/5+), while no detection of the peptide cocktails was evident with negative control sera (labeled -0-). Densitometric scans (bars 1–7) revealed overall better extent of reactions with the combination of peptides compared with the individual peptides for both women and men sera (data not shown). The combined peptides ACT2 and ACT3 (bars 8) served as positive controls with positive control sera from men and women reactive with ACT,31 and the pooled peptides GAP1, GAP6, and ENO3 (bars 9) as well as GAP5, ENO5, and ALD6 (bars 10) showed no reactivity with the negative control sera of both women and men.
Figure 4.

Representative experiments in duplicate dot blots of combinations of 15-mer peptide epitopes (A) and densitometric scans of reactive dot blots (B).
Notes: (A) shows duplicate dot blots of combinations of peptides (A1 though A10) incubated with men and women sera that were highly seroreactive (labeled 4+/5+) with α-actinin by ELISA (34) as well as with the negative control nonreactive sera (labeled -0-). Only one dot blot each is shown for the negative control reaction. (B) presents the densitometric scans performed on the dot blots from experiment 1 for the combinations of peptides (bars numbered 1 through 7) as well as for the positive control ACT2 plus ACT3 (bar numbered 8). Bars 9 and 10 show the absence of any reactivity, using the negative control women and/or men sera. The individual epitopes are those indicated in Table 4.
Abbreviations: ACT, α-actinin; ALD, fructose-1,6-bisphosphate aldolase; con, control; ELISA, enzyme-linked immunosorbent assay; ENO, α-enolase; GAP, glyceraldehyde-3-phosphate dehydrogenase glyceraldehyde; neg, negative.
Expression, purification, and immunodetection of recombinant SOE
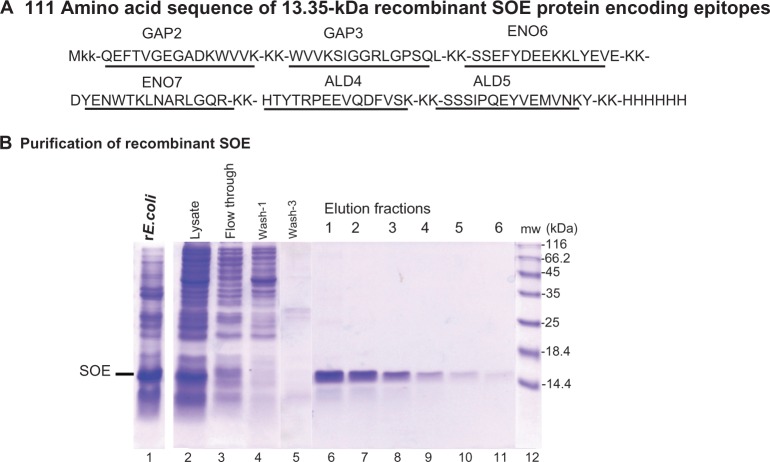
We hypothesized that a novel protein, comprised of a series of 15-mer peptides immunoreactive with positive control sera from women and men, could be synthesized for analysis as a serodiagnostic target. We, therefore, constructed a plasmid encoding for a His6 fusion recombinant protein of 111 amino acids (Mr 13.35 kDa), within which were the epitopes for GAP2, GAP3, ENO6, ENO7, ALD4, and ALD5 (Figure 5A). Underlined amino acids indicate the 15-mer peptides (Table 4). These 15-mer amino acids were chosen because of little or no identity to other proteins in databanks (Tables 1–3). Recombinant E. coli expressing the SOE (lane 1) was synthesized and found to be soluble in lysates (lane 2) (Materials and methods), which permitted purification in Ni2+-NTA affinity chromatography, as shown in Figure 5B (lanes 6–11). The relative mobility of the SOE compared with molecular weight standards showed an Mr slightly greater than the calculated 13.35 kDa.
Figure 5.
The 111 amino acid sequence and expression and purification of the rSOE encoding epitopes of GAP, ENO, and ALD.
Notes: (A) The sequence of two 15-mer epitopes (underlined) for each protein, as shown in Table 4. The six H residues at the carboxy terminus represent the hexahistidine added for both purification using Ni2+-NTA affinity chromatography and detection using MAb. (B) The SDS-PAGE and Coomassie-brilliant blue stained gels of recombinant Escherichia coli expressing the rSOE (lane 1) and the lysate prepared for chromatography (lane 2), as described in Materials and methods. Flow through is the lysate obtained after chromatography (lane 3), followed by two representative washes of the column (lanes 4 and 5). Finally, lanes 6–11 show the rSOE eluted from the column, and the single band indicates specific purification of the rSOE::His6.
Abbreviations: ALD, fructose-1,6-bisphosphate aldolase; ENO, α-enolase; GAP, glyceraldehyde-3-phosphate dehydrogenase; His6, hexahistidine; MAb, monoclonal antibody; mw, molecular weight; Ni2+, nickel; NTA, nitrilotriacetic acid; rSOE, recombinant protein of sequences of epitopes; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; SOE, sequences of epitopes.
Lastly, we performed assays to assess whether this novel recombinant protein was detectable with positive control sera from women and men, as above. Figure 6A shows representative reactions by ELISA, of rSOE immobilized onto individual wells of 96-well microtiter plates. Negative control sera of women and men had little to no reactivity, as evidenced by low A405 nm with the rSOE in comparison with the strong signals (high A405 nm) obtained with the positive control sera from women and men. A very strong reaction was seen with rSOE using the positive control MAb to hexahistidine in the fusion recombinant protein. As expected, the MAb HA423231 that was reactive with the ACT used as a negative control gave no reactivity with the rSOE. Further, Figure 6 demonstrates that the positive control sera from women and men detected the rSOE by dot blots (Figure 6B) and by immunoblot after SDS-PAGE and immunoblotting onto nitrocellulose (Figure 6C). The Coomassie-brilliant blue stained gel is a duplicate from the SDS-PAGE used for immunoblotting of the rSOE, and the Ponceau S-stained nitrocellulose is a duplicate included to show the transfer of the rSOE protein onto nitrocellulose.
Figure 6.

Immunodetection of the rSOE by ELISA (A), dot blot (B), and immunoblotting after SDS-PAGE (C).
Notes: (A) shows a representative ELISA for detection of rSOE immobilized onto individual wells of 96-well microtiter plates (Materials and methods) by sera of women and men highly reactive to the trichomonad protein α-actinin (34) (labeled 4+) compared with the negative control, unreactive sera (labeled 0). HA423 is a MAb to α-actinin, which was unreactive to the rSOE. In contrast, the MAb to hexahistidine of the fusion protein readily bound to the rSOE, as evidenced by the very high absorbance values (A405 nm). (C) Representative immunoblot after SDS-PAGE and blotting onto nitrocellulose, of rSOE detected by pooled sera of women and men reactive with α-actinin, as above in (A) (lane 1). Lane 2 is a duplicate of the nitrocellulose blot stained with Ponceau S to show efficient transfer onto nitrocellulose, and lane 3 is the Coomassie-brilliant blue-stained gel of a duplicate sample of the rSOE used for blotting.
Abbreviations: ELISA, enzyme-linked immunosorbent assay; His6, hexahistidine; MAb, monoclonal antibody; mw, molecular weight; rSOE, recombinant protein of sequences of epitopes; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Discussion
In this study, we have successfully identified epitopes of T. vaginalis ALD, ENO,31 and GAP,19 reactive with positive control sera from women and men, as serodiagnostic targets. Indeed, ACT is a target for a serodiagnostic test that would be worthwhile for both women and men exposed to this STI compared with the currently used POC OSOM® test sold by Sekisui Diagnostics, Inc, which works only for women patients with trichomonosis.24,25 The utility of this serum antibody target prompted us to examine for other immunogenic proteins, to which women patients and ACT-seropositive men make serum antibody and which might serve in future serodiagnostics. We, therefore, felt it important to perform a stepwise approach that would identify epitopes of immuno-genic proteins studied previously, such as ENO32 and GAP.19 More recently, we determined that another immunogenic protein is ALD (Alderete, unpublished data, 2013). In this study, we showed that 15-mer peptides containing the epitopes were readily detected by the positive control sera of women and men but not the negative control sera. These findings reinforce the notion that epitopes with little sequence identity to other known proteins, especially those of microbial pathogens and humans, can be identified to serve as serodiagnostic targets.
Interestingly, other microbial pathogens, including sexually transmitted disease agents, have high amino acid sequence identity to some of the epitopes detected by the positive control sera from women and men reactive to T. vaginalis (Tables 1–3). Although we have no evidence that the sera of patients infected by these other microbial pathogens used for comparison of sequence identity are cross-reactive with the epitopes identified in our study, we feel that this method and analysis would select for protein sequences of T. vaginalis that would be specific targets for the serodiagnosis of this STI. The fact that increasing the number of amino acids surrounding the epitopes significantly decreased the percent sequence identity further affirms the diminished possibility of any immuno-cross-reactivity. Nonetheless, it is conceivable that in fact, during infections by these other pathogens, patients may have antibody to similar sequences of their respective proteins.
An interesting finding is that the epitopes detected by the MAbs were distinct from those seen by the positive control sera from women and men, suggesting different presentations during immunization compared with natural infections. Further, women and men did not react with the identical epitopes, and this may be the result of the uniqueness of host responses, due to the distinct anatomy of the urogenital regions and immune surveillance. Although speculative, this may indicate that future serodiagnostics will need to take into account those epitopes detected by both women and men.
It is noteworthy that the trichomonad ENO has three epitopes (Table 2) and GAP has six epitopes (Table 3) with >50% sequence identity to the human homologue. Recently, it has been shown that patients with Streptococcus pyogenes infections had serum antibody to streptococcal ENO, and this antibody was cross-reactive with human tissues, such as the liver.40,41 While the adverse consequences of this remain undefined, such host responses may lead to autoimmune manifestations. Indeed, if infections result in tissue damage, this may be the result of such immuno-cross-reactivity and not just microbial toxins. Given this possibility, this requires further consideration.
As a direct result of epitope identification and characterization, we reasoned that it might be possible to synthesize a recombinant hybrid protein (rSOE) comprised of epitopes from the three immunogenic proteins of ALD, ENO, and GAP (Figure 5). Indeed, this rSOE was readily purified and served as an immunotarget detected by various assays (Figure 6). We feel this may be a novel way to produce such an amalgam of epitopes unique to a particular pathogen, such as T. vaginalis, and recognized by both women and men, regardless of the distinct natures of infection and protein presentations during immune surveillance. Such an rSOE may lead to novel platforms for rapid and accurate POC serodiagnostics for infectious diseases.
An important finding is that epitope mapping initially detected epitopes unique to positive control sera from women versus men (Tables 1−3). However, when tested individually, 15-mer peptides with epitopes were, in fact, seen by sera of both women and men (Figure 3). For example, ALD1 (epitope A-W1) and ALD5 (epitope A-W6) were initially identified with positive control women sera (Table 4), but these epitopes were also positive as 15-mer peptides when tested with men sera (Figure 3). Likewise, ENO7 (epitope E-W7) and GAP3 (G-W2) were detected by men sera, and GAP5 (epitope G-M6) was seen by women sera. These results indicate that serum reactivity of epitopes requires examination under different experimental conditions. Further, these findings inform that 11-mer cellulose-immobilized linear peptides for SPOTs analysis (Tables 1–3) present epitopes in a way that is different from 15-mer peptides immobilized on alternative surfaces, like nitrocellulose or microtiter wells. Likewise, serum antibody appears to distinguish the epitopes depending on the immobilizing surface, which may permit certain conformations accessible to antibody.
In summary, both women and men exposed to T. vaginalis make serum antibody to numerous trichomonad proteins.19,30–36 As in an earlier recent study for the immunogenic trichomonad protein ACT,31 we have identified epitopes of ALD, ENO, and GAP that are unique to this organism and showed their usefulness as serodiagnostic targets. We have also established the ability to synthesize a novel hybrid recombinant protein composed of numerous epitopes of ALD, ENO, and GAP proteins (called SOE) that forms a platform for a future serodiagnostic test. We have shown that such a hybrid rSOE can be the basis of a test for both women and men.
Acknowledgments
We thank Laurence N Kolonel, MD, PhD of the University of Hawaii Cancer Center and Siobhan Sutcliffe, PhD of Washington University at St Louis for their generous contribution of additional male sera that was highly seropositive for ACT-P2 and that permitted us to screen for epitopes of ACT. We also want to thank Patrick Joyce, Adan Medina, and Clare Tang, undergraduate student members of the laboratory, for their assistance during the course of these experiments and for their thoughtful discussions. This work was supported in part by the Washington Research Foundation.
Footnotes
Disclosure
The authors report no conflict of interest in this work.
References
- 1.Hobbs MM, Seña AC, Swygard H, Schwebke JR. Trichomonas vagi-nalis and trichomoniasis. In: Holmes KK, Sparling PF, Stamm WE, Piot P, Wasserheit JN, Corey L, Cohen MS, Watts DH, editors. Sexually Transmitted Diseases. 4th ed. New York, NY: McGraw Hill Medical; 2008. pp. 771–794. [Google Scholar]
- 2.Gardner WA, Jr, Culberson DE, Bennett BD. Trichomonas vaginalis in the prostate gland. Arch Pathol Lab Med. 1986;110(5):430–432. [PubMed] [Google Scholar]
- 3.Mitteregger D, Aberle SW, Makristathis A, et al. High detection rate of Trichomonas vaginalis in benign hyperplastic prostatic tissue. Med Microbiol Immunol. 2012;201(1):113–116. doi: 10.1007/s00430-011-0205-2. [DOI] [PubMed] [Google Scholar]
- 4.Bachmann LH, Hobbs MM, Seña AC, et al. Trichomonas vaginalis genital infections: progress and challenges. Clin Infect Dis. 2011;53(Suppl 3):S160–S172. doi: 10.1093/cid/cir705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huppert JS. Trichomoniasis in teens: an update. Curr Opin Obstet Gynecol. 2009;21(5):371–378. doi: 10.1097/GCO.0b013e32832e0827. [DOI] [PubMed] [Google Scholar]
- 6.Krashin JW, Koumans EH, Bradshaw-Sydnor AC, et al. Trichomonas vaginalis prevalence, incidence, risk factors and antibiotic-resistance in an adolescent population. Sex Transm Dis. 2010;37(7):440–444. doi: 10.1097/OLQ.0b013e3181cfcd8c. [DOI] [PubMed] [Google Scholar]
- 7.Lehker MW, Alderete JF. Biology of trichomonosis. Curr Opin Infect Dis. 2000;13(1):37–45. doi: 10.1097/00001432-200002000-00007. [DOI] [PubMed] [Google Scholar]
- 8.Figueroa-Angulo EE, Rendón-Gandarilla FJ, Puente-Rivera J, et al. The effects of environmental factors on the virulence of Trichomonas vaginalis. Microbes Infect. 2012;14(15):1411–1427. doi: 10.1016/j.micinf.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Huang KY, Huang PJ, Ku FM, Lin R, Alderete JF, Tang P. Comparative transcriptomic and proteomic analyses of Trichomonas vaginalis following adherence to fibronectin. Infect Immun. 2012;80(11):3900–3911. doi: 10.1128/IAI.00611-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horváthová L, Šafaríková L, Basler M, et al. Transcriptomic identifi-cation of iron-regulated and iron-independent gene copies within the heavily duplicated Trichomonas vaginalis genome. Genome Biol Evol. 2012;4(10):1017–1029. doi: 10.1093/gbe/evs078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muñoz C, Pérez M, Orrego PR, et al. A protein phosphatase 1 gamma (PP1γ) of the human protozoan parasite Trichomonas vaginalis is involved in proliferation and cell attachment to the host cell. Int J Parasitol. 2012;42(8):715–727. doi: 10.1016/j.ijpara.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Rendón-Gandarilla FJ, Ramón-Luing Lde L, Ortega-López J, Rosa de Andrade I, Benchimol M, Arroyo R. The TvLEGU-1, a legumain-like cysteine proteinase, plays a key role in Trichomonas vaginalis cytoadherence. Biomed Res Int. 2013;2013:561979. doi: 10.1155/2013/561979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casta e Silva Filho F, de Souza W, Lopes JD. Presence of laminin-binding proteins in trichomonads and their role in adhesion. Proc Natl Acad Sci U S A. 1988;85(21):8042–8046. doi: 10.1073/pnas.85.21.8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arroyo R, González-Robles A, Martínez-Palomo A, Alderete JF. Signalling of Trichomonas vaginalis for amoeboid transformation and adhesion synthesis follows cytoadherence. Mol Microbiol. 1993;7(2):299–309. doi: 10.1111/j.1365-2958.1993.tb01121.x. [DOI] [PubMed] [Google Scholar]
- 15.Garcia AF, Chang TH, Benchimol M, Klumpp DJ, Lehker MW, Alderete JF. Iron and contact with host cells induce expression of adhesins on surface of Trichomonas vaginalis. Mol Microbiol. 2003;47(5):1207–1224. doi: 10.1046/j.1365-2958.2003.03366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia AF, Benchimol M, Alderete JF. 2005. Trichomonas vaginalis polyamine metabolism is linked to host cell adherence and cytotoxicity. Infect Immun. 2005;73(5):2602–2610. doi: 10.1128/IAI.73.5.2602-2610.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kucknoor AS, Mundodi V, Alderete JF. Adherence to human vaginal epithelial cells signals for increased expression of Trichomonas vaginalis genes. Infect Immun. 2005;73(10):6472–6478. doi: 10.1128/IAI.73.10.6472-6478.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia AF, Alderete J. Characterization of the Trichomonas vaginalis surface-associated AP65 and binding domain interacting with trichomonads and host cells. BMC Microbiol. 2007;7:116. doi: 10.1186/1471-2180-7-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lama A, Kucknoor A, Mundodi V, Alderete JF. Glyceraldehyde-3-phosphate dehydrogenase is a surface-associated, fibronectin-binding protein of Trichomonas vaginalis. Infect Immun. 2009;77(7):2703–2711. doi: 10.1128/IAI.00157-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutcliffe S, Giovannucci E, Alderete JF, et al. Plasma antibodies against Trichomonas vaginalis and subsequent risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2006;15(5):939–945. doi: 10.1158/1055-9965.EPI-05-0781. [DOI] [PubMed] [Google Scholar]
- 21.Stark JR, Judson G, Alderete JF, et al. Prospective study of Trichomonas vaginalis infection and prostate cancer incidence and mortality: Physicians’ Health Study. J Natl Cancer Inst. 2009;101(20):1406–1411. doi: 10.1093/jnci/djp306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sutcliffe S, Alderete JF, Till C, et al. Trichomonosis and subsequent risk of prostate cancer in the Prostate Cancer Prevention Trial. Int J Cancer. 2009;124(9):2082–2087. doi: 10.1002/ijc.24144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones HE, Lippman SA, Caiaffa-Filho HH, Young T, van de Wijgert JH. Performance of a rapid self-test for detection of Trichomonas vaginalis in South Africa and Brazil. J Clin Microbiol. 2013;51(3):1037–1039. doi: 10.1128/JCM.01547-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pillay A, Lewis J, Ballard RC. Evaluation of Xenostrip-Tv, a rapid diagnostic test for Trichomonas vaginalis infection. J Clin Microbiol. 2004;42(8):3853–3856. doi: 10.1128/JCM.42.8.3853-3856.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hegazy MM, El-Tantawy NL, Soliman MM, El-Sadeek ES, El-Nagar HS. Performance of rapid immunochromatographic assay in the diagnosis of Trichomoniasis vaginalis. Diagn Microbiol Infect Dis. 2012;74(1):49–53. doi: 10.1016/j.diagmicrobio.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Lee JJ, Moon HS, Lee TY, Hwang HS, Ahn MH, Ryu JS. PCR for diagnosis of male Trichomonas vaginalis infection with chronic prostatitis and urethritis. Korean J Parasitol. 2012;50(2):157–159. doi: 10.3347/kjp.2012.50.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ginocchio CC, Chapin K, Smith JS, et al. Prevalence of Trichomonas vaginalis and coinfection with Chlamydia trachomatis and Neisseria gonorrhoeae in the United States as determined by the Aptima Trichomonas vaginalis nucleic acid amplification assay. J Clin Microbiol. 2012;50(8):2601–2608. doi: 10.1128/JCM.00748-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munson E, Miller C, Napierala M, et al. Assessment of screening practices in a subacute clinical setting following introduction of Trichomonas vaginalis nucleic acid amplification testing. WMJ. 2012;111(5):233–236. [PubMed] [Google Scholar]
- 29.Munson KL, Napierala M, Munson E, et al. Screening of male patients for Trichomonas vaginalis with transcription-mediated amplification in a community with a high prevalence of sexually transmitted infections. J Clin Microbiol. 2013;51(1):101–104. doi: 10.1128/JCM.02526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Addis MF, Rappelli P, Pinto De Andrade AM, et al. Identification of Trichomonas vaginalis alpha-actinin as the most common immunogen recognized by sera of women exposed to the parasite. J Infect Dis. 1999;180(5):1727–1730. doi: 10.1086/315095. [DOI] [PubMed] [Google Scholar]
- 31.Neace CJ, Alderete JF. Epitopes of the highly immunogenic Trichomonas vaginalis α-actinin are serodiagnostic targets for both women and men. J Clin Microbiol. 2013;51(8):2483–2490. doi: 10.1128/JCM.00582-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mundodi V, Kucknoor AS, Alderete JF. Immunogenic and plasminogen-binding surface-associated alpha-enolase of Trichomonas vaginalis. Infect Immun. 2008;76(2):523–531. doi: 10.1128/IAI.01352-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehker MW, Alderete JF. Iron regulates growth of Trichomonas vaginalis and the expression of immunogenic trichomonad proteins. Mol Microbiol. 1992;6(1):123–132. doi: 10.1111/j.1365-2958.1992.tb00844.x. [DOI] [PubMed] [Google Scholar]
- 34.Alderete JF, Newton E, Dennis C, Neale KA. The vagina of women infected with Trichomonas vaginalis has numerous proteinases and antibody to trichomonad proteinases. Genitourin Med. 1991;67(6):469–474. doi: 10.1136/sti.67.6.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alderete JF, Newton E, Dennis C, Engbring J, Neale KA. Vaginal antibody of patients with trichomoniasis is to a prominent surface immunogen of Trichomonas vaginalis. Genitourin Med. 1991;67(3):220–225. doi: 10.1136/sti.67.3.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alderete JF, Newton E, Dennis C, Neale KA. Antibody in sera of patients infected with Trichomonas vaginalis is to trichomonad proteinases. Genitourin Med. 1991;67(4):331–334. doi: 10.1136/sti.67.4.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 38.Hopp TP, Woods KR. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–275. [PubMed] [Google Scholar]
- 40.Sunblad V, Bussmann L, Chiauzzi VA, Pancholi V, Charreau EH. Alpha-enolase: a novel autoantigen in patients with premature ovarian failure. Clin Endocrinol (Oxf) 2006;65(6):745–751. doi: 10.1111/j.1365-2265.2006.02661.x. [DOI] [PubMed] [Google Scholar]
- 41.Fontán PA, Pancholi V, Nociari MM, Fischetti VA. Antibodies to streptococcal surface enolase react with human alpha-enolase: implications in poststreptococcal sequelae. J Infect Dis. 2000;182(6):1712–1721. doi: 10.1086/317604. [DOI] [PubMed] [Google Scholar]