Abstract
Silver–Russell syndrome is a clinically and genetically heterogeneous disorder, characterized by prenatal and postnatal growth restriction, relative macrocephaly, body asymmetry and characteristic facial features. It is one of the imprinting disorders, which results as a consequence of aberrant imprinted gene expressions. Currently, maternal uniparental disomy of chromosome 7 accounts for approximately 10% of Silver–Russell syndrome cases, while ˜50% of patients have hypomethylation at imprinting control region 1 at chromosome 11p15.5 locus, leaving ˜40% of cases with unknown etiologies. This review aims to provide a comprehensive list of molecular defects in Silver–Russell syndrome reported to date and to highlight the importance of multiple-loci/tissue testing and trio (both parents and proband) screening. The epigenetic and phenotypic overlaps with other imprinting disorders will also be discussed.
Keywords: : discordant twins, epigenetics, epigenomic editing, fetal growth, genomic imprinting, multilocus imprinting disturbance, Silver–Russell syndrome
Silver–Russell syndrome
Silver–Russell syndrome (SRS; OMIM ID: 180860) is an imprinting disorder affecting prenatal and postnatal growth. To date, more than 400 cases have been reported since its first description by Silver et al. in 1953 and Russell in 1954 who independently reported a subset of children with low birth weight, short stature, body asymmetry and characteristic facial features [1,2].
Genomic imprinting is an epigenetic modification process, which allows a gene to be expressed in a monoallelic, parent-of-origin specific manner. This phenomenon which is almost exclusive to the placental mammals was identified in the 1980s through a set of mouse experiments [3,4]. Since then more than 100 imprinted genes have been identified in mice and approximately half of these are conserved in humans [5]. The imprinted expression is regulated by differential methylation of imprinting control regions (ICRs) which are established in germline during gametogenesis [6]. Disturbance in the expression dosage of imprinted genes due to uniparental disomy (UPD), epi/mutation and copy number variation (CNV) could lead to imprinting disorders, mainly characterized by growth, metabolic and neurological abnormalities [7]. There are currently 10 reported imprinting disorders in humans: Angelman syndrome (OMIM 105830), Prader–Willi syndrome (OMIM 176270), transient neonatal diabetes mellitus 1 (OMIM 601410), pseudohypoparathyroidism Type Ib (OMIM 603233), Silver–Russell syndrome, Beckwith–Wiedemann syndrome (OMIM 130650), Temple syndrome (OMIM 616222), Kagami–Ogata syndrome [OMIM 608149], maternal UPD of chromosome 20 syndrome and precocious puberty syndrome [8].
Diagnosis of SRS is relatively difficult as it has many clinical features (summarized in Table 1). To date, six clinical scoring systems of SRS have been suggested [9–14]. Majority of them diagnose SRS by the presence of three or four out of the following characteristics: birth weight ≤-2SD, postnatal growth restriction, relative macrocephaly, facial characteristics and body asymmetry (Table 1). Accurate diagnosis is a challenge since postnatal growth measurements require follow-up, and facial features become less obvious with age. The most recently described ‘Netchine–Harbison clinical scoring system’ includes feeding difficulties in addition to the aforementioned features, and having four out of these six features diagnose ‘Likely SRS’ [9]. Screening 69 patients with this system identified 98% of the SRS patients with SRS-associated molecular anomalies. It is interesting to note that, in their ‘Likely SRS double negative’ group which have no ICR1 hypomethylation or upd(7)mat, one patient was found to have upd(20)mat. It is a molecular defect found in patients with upd(20)mat syndrome, a newly defined imprinting disorder characterized by pre/postnatal growth restriction and feeding difficulties [15]. Another patient had hypomethylation at DLK1/MEG3 intergenic DMR (IG-DMR), which is a characteristic of Temple syndrome (TS). TS is another imprinting disorder sharing clinical features with SRS. In addition, a recent review by Fokstuen and Kotzot pointed out that none of the systems include delayed bone age, even though it is observed in most of the upd(7)mat patients [16]. Therefore, a consensus in diagnostic criteria of SRS would be useful, and a European-wide consensus is due to be published in 2016 [Moore G, Pers. Inform.].
Table 1. . Silver–Russell syndrome clinical characteristics with their frequencies and diagnostics scoring system comparison.
| Clinical features |
Molecular subgroups |
Reported diagnostics criteria for SRS |
||||||
|---|---|---|---|---|---|---|---|---|
| ICR1 hypo (n = 44) (%) | upd(7)mat (n = 20) (%) | Lai et al. | Price et al. | Netchine et al. | Bartholdi et al. | Dias et al. | Azzi et al.† | |
| Growth: | ||||||||
| – Birth weight ≤-2 SD | 82 | 70 | ✓ | ✓ | ✓ (SGA) | ✓ (BW) ✓ (BL) ≤10th centile |
✓ | ✓ (SGA) |
| – Height at examination ≤-2 SD |
57 | 65 | ✓ | ✓ | ✓ | ✓ ≤3rd centile |
✓ (after 2 years) | ✓ |
| – Relative macrocephaly | 70 | 90 | ✓ | ✓ | ✓ ✓ (normal HC) |
✓ | ✓ | |
| – Body asymmetry |
68 |
30 |
✓ |
✓ |
✓ |
✓ (face) ✓ (body) ✓ (limbs) |
✓ |
✓ |
| Craniofacial features: | ✓ | ✓ | ✓ (others) | |||||
| – Triangular face | 59 | 90 | ✓ | |||||
| – Frontal bossing | 60 | 60 | ✓ | ✓ | ✓ | |||
| – Micro/retrognathia | 64 | 35 | ||||||
| – Dental crowding | 36 | 45 | ||||||
| – Downturned corners of the mouth | 30 | 20 | ||||||
| – Thin upper lip | 27 | 30 | ||||||
| – Low-set/posteriorly rotated ears |
36 |
75 |
|
|
|
|
|
|
| Developmental: | ✓ (normal) | |||||||
| – Global delay | 20 | 65 | ||||||
| – Speech delay |
39 |
50 |
|
|
|
|
|
|
| Other clinical signs: | ||||||||
| – 5th finger clinodactyly | 75 | 45 | ✓ | ✓ | ||||
| – 2/3 toe syndactyly | 23 | 20 | ||||||
| – Camptodactyly | 16 | 25 | ||||||
| – Café-au-lait macules | 14 | 15 | ||||||
| – Excessive sweating | 64 | 75 | ||||||
| – Hypoglycemia | 24 (n = 37) | 29 (n = 17) | ||||||
| – Feeding difficulties | 84 | 90 | ✓ | ✓ | ||||
| – Gastro-oesophageal reflux | 14 | 10 | ||||||
| – Otitis media | 14 | 20 | ||||||
| – Delayed closure of fontanelles | 43 (n = 28) | 36 (n = 11) | ||||||
| – Movement disorder |
0 |
15 |
|
|
|
|
|
|
| Congenital abnormalities: | ✓ (others) | |||||||
| – Cleft palate/bifid uvula | 7 | 0 | ||||||
| – Congenital heart defect | 9 | 0 | ||||||
| – Male genital anomaly | 23 (n = 22) | 0 (n = 5) | ✓ | |||||
| – Renal anomaly | 0 | 10 | ||||||
| – Scoliosis |
9 |
5 |
|
|
|
|
|
|
| Scores required for diagnosis of SRS | ≥3/5 | ≥4/5 | SGA + ≥3/5 | ≥8/15 | ≥3/4 | ≥4/6 | ||
Frequencies (%) are based on n = 64 SRS patients (n = 44 with ICR1 hypomethylation and n = 20 upd(7)mat) studied by Wakeling et al. [17]. To date, six diagnostic scoring systems have been reported.
†The Netchine–Harbison clinical scoring system reported by Azzi et al. [9] is based on the Netchine et al. [13] scoring system, where SGA is no longer mandatory (having four out of six of the features diagnose them as ‘Likely SRS’).
BW: Birth weight; BL: Birth length; ICR1: Imprinting control region 1; SD: Standard deviation; SGA: Small for gestational age (defined as birth weight and/or length ≤-2 SD score); SRS: Silver–Russell syndrome; Upd(7)mat: Maternal uniparental disomy of chromosome 7.
Modified with permission from Wakeling et al. [17].
Molecular anomalies of SRS
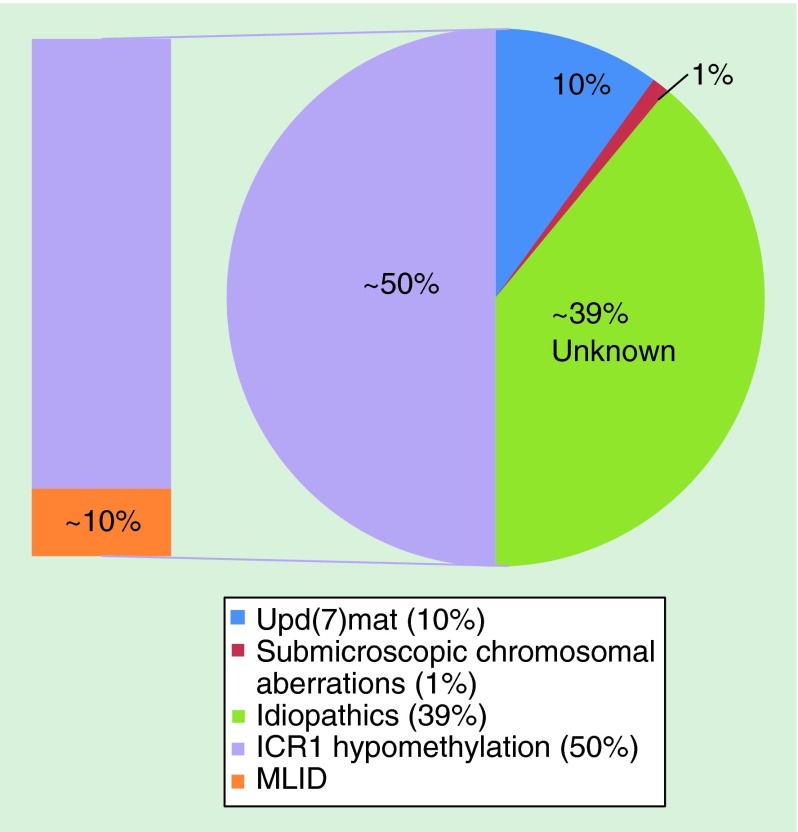
Majority of SRS cases are sporadic, but various modes of inheritance including recessive, dominant and X-linked have been suggested [18]. To date, numerous chromosomal aberrations have been associated with SRS, including chromosome 1, 2, 7, 8, 11, 12, 13, 14, 15, 16, 17, 18, 20, 21, 22 and X (summarized in Supplementary Table 1), with majority of anomalies attributed to chromosome 7 and 11 (Figure 1). One has to be cautious that some of these reported cases include SRS-like patients who might not fit the definitive criteria.
Figure 1. . Summary of reported molecular anomalies of Silver–Russell syndrome.
ICR1: Imprinting control region 1; MLID: Multilocus imprinting disturbance; Upd(7)mat: Maternal uniparental disomy of chromosome 7.
Chromosome 7
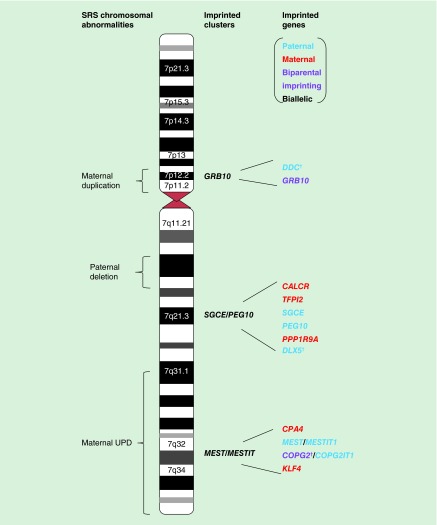
Cytogenetic abnormalities of chromosome 7 have been reported in several SRS cases (Supplementary Table 1 & Figure 2). UPD refers to the inheritance of two copies of a chromosome pair from a single parent, where heterodisomy is the inheritance of both parental homologues (grandparental pair) and isodisomy is the transmission of two identical homologue of one parent. Maternal UPD for chromosome 7 (upd(7)mat) occurs in approximately 10% of SRS cases [19]. Chromosome 7 was first linked to SRS etiology though identification of a cystic fibrosis patient with SRS-like features [20], followed by reports of growth restricted upd(7)mat patients born to CFTR heterozygous mothers but carried homozygous mutations, resulted from isodisomy for chromosome 7 [21,22]. Therefore, exposure of recessive mutations has been previously suggested as the cause of SRS in upd(7)mat patients. However, no common isodisomic region on chromosome 7 was identified in five SRS cases investigated [23], making it unlikely to be the causative mechanism for SRS. The same study also indicated that most SRS upd(7)mat are likely to arise from meiosis I nondisjunction error, followed by trisomic rescue as evident by heterodisomy at the centromeres. This is in contrast to the Beckwith–Wiedemann syndrome (BWS) cases with upd(11)pat where all reported cases are isodisomic resulting in upd(11)pat and normal cell mosaic, implying postzygotic mitotic nondisjunction error [24]. Trisomy 7 mosaicism as a consequence of trisomy rescue, has also been ruled out as the possible mechanism for SRS, since no trisomic cells have been detected in upd(7)mat patients’ lymphocytes or fibroblasts [25], and subjects with confined placental mosaicism for trisomy 7 have normal birth weight [26].
Figure 2. . Human chromosome 7 ideogram with imprinted genes and chromosomal aberrations reported in Silver–Russell syndrome patients.
Letters (red = maternal expression, blue = paternal expression, purple = biparental imprinting expression and black = biallelic expression). *The imprinting statuses of DDC, DLX5 and COPG2 in humans have conflicting results.
SRS: Silver–Russell syndrome; UPD: Uniparental disomy.
It is therefore most likely that the pathogenesis of SRS with upd(7)mat is attributed to the disrupted expression of imprinted genes on chromosome 7. Reduced expression of growth promoting paternal gene(s) or overexpression of maternally expressed gene(s) which has growth limiting function could lead to the disorder. Importantly, the four reported cases with paternal isodisomy for chromosome 7 (upd(7)pat) do not exhibit growth and developmental phenotypes, except for one patient who became overweight at 3 months of age, and another who showed growth restriction at 6 months of age but this was likely to be owing to his recessive disease conditions [27–30]. Therefore, rather than the loss of paternally expressed growth-promoting genes, overexpression of growth-limiting maternal genes on chromosome 7 may be more relevant for SRS growth restriction phenotype. Although there is a SRS-like case with paternal deletion of Chr7q32, including the paternally expressed gene MEST [31].
The candidate SRS regions within chromosome 7 were narrowed down following identification of SRS patients with maternal segmental duplications affecting 7p11.2-p13 locus and maternal segmental UPD involving 7q31-qter region [32–38]. The 7p11.2-p13 region encompasses an imprinted gene (GRB10) and biallelically expressed growth-related genes (IGFBP1, IGFBP3 and EGFR). The 7q31-qter region harbors four imprinted genes (CPA4, MEST, COPG2 [conflicting reports] and KLF4) and two imprinted noncoding RNAs (MESTIT1 and COPG2IT1). However, screening the coding regions of these candidate genes have so far failed to identify pathogenic mutations although it should be noted that some of these patients might have carried ICR1 hypomethylation [39]. Though, one study reported a patient with 7p11.2-p12 maternal duplication without SRS phenotypes and the duplication did not include GRB10, lending the possibility of involvement of this gene in SRS pathogenesis [35]. In addition, a recent report presented a case with GRB10 hypermethylation with an additional Chr20p13 microdeletion [40].
It is possible that there are more imprinted genes that have not been discovered yet within this region. Recently, Hannula et al. [41] searched for novel DMRs and imprinted genes on chromosome 7 by comparing nine upd(7)mat, one upd(7)pat and ten control whole blood DNA samples using the Infinium 450K HumanMethylation array. Two of the identified DMRs mapped to the previously predicted imprinted genes HOXA4 (7p15.2) and GLI3 (7p13), and one at SVOPL gene (7q34) and monoallelic expression of HOXA4 and maternal expression of SVOPL were confirmed in blood. A recent large-scale study assessing allele-specific expressions in 1678 human RNA-seq samples from 45 tissues in 178 individuals, also revealed interesting results [42]. DDC gene adjacent to GRB10, which was previously shown to be imprinted in mice and biallelically expressed in multiple human fetal tissues [43], was shown to be imprinted in multiple adult human tissues including brains. Another disputed imprinted gene, DLX5 which maps to the PEG10/SGCE imprinted cluster at 7q21.3, was shown to be biallelically expressed in mice but imprinted in humans. The observed discrepancies could possibly be explained by the difference in the types and developmental stages of tissues used. In addition, some genes were missed in their study due to their stringent inclusion criteria such as requiring four expressed heterozygous SNPs per gene for parent-of-origin analysis. Interestingly, the same study also identified the mouse Copg2 to be biparentally imprinted, a phenomenon previously reported for Igf2 and Grb10 [44]. Although it was not assessed in human samples, Copg2 showed paternal expression in E15 brain and maternal expression in E17.5 brain, adding further complexity. In conclusion, the roles of imprinted genes on chromosome 7 in SRS etiology require further investigation.
Chromosome 11p15.5
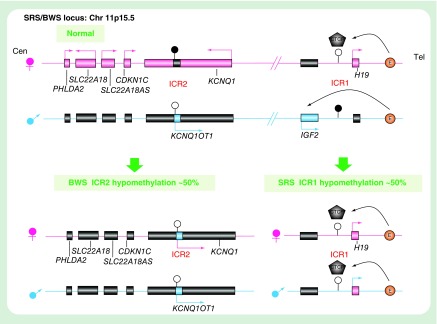
The imprinted cluster at chromosome 11p15.5 contains two independent regulatory domains: the telomeric IGF2/H19 domain controlled by the paternally methylated ICR1 and the centromeric KCNQ1 domain controlled by the maternally methylated ICR2 (also known as KvDMR1) (Figure 3). Both ICR1 and ICR2 acquire differential methylation in the germline, which resets in each generation to lay sex-specific imprints during gametogenesis [6].
Figure 3. . Schematic representation of the human imprinted cluster at chromosome 11p15.5 implicated in Silver–Russell syndrome and Beckwith–Wiedemann syndrome.
Maternal and paternal chromosomes are represented in pink and blue, respectively. Paternal (blue), maternal (pink) and biallelic (black) and the direction of the transcription are indicated by arrows. Figure orientations: left = centromeric and right = telomeric. The CTCF protein can bind to the unmethylated ICR1, blocking the IGF2 from accessing the downstream enhancers. The methylated ICR1 do not allow CTCF binding, resulting in IGF2 promoter to interact with the enhancers.
BWS: Beckwith–Wiedemann syndrome; ICR: Imprinting control region; SRS: Silver–Russell syndrome.
Hypomethylation of the ICR1 is observed in 31–63% of SRS cases (Supplementary Table 1). The ICR1 is located 2–4kb upstream of H19, and it controls the reciprocal imprinted expression of the paternally expressed growth promoter IGF2 and maternally expressed growth repressing noncoding RNA gene H19 [45]. There are seven binding sites for the CTCF zinc finger protein at ICR1. The binding of the CTCF protein to the unmethylated maternal ICR1 blocks the IGF2 promoter from interacting with the shared enhancers downstream of H19, resulting in maternal H19 expression. The protein cannot bind to the paternally methylated ICR1 which allows the paternal IGF2 expression (Figure 3). Therefore, hypomethylation of ICR1 is thought to cause suppression and derepression of paternal IGF2 and H19, respectively, by permitting the CTCF protein to bind to the paternal ICR1, leading to a growth restriction phenotype [46]. Strikingly, hypermethylation at ICR1 and hypomethylation of ICR2 is observed in approximately 5 and 50% of BWS patients respectively, which is an overgrowth syndrome suggested to be phenotypically and genotypically opposite to SRS.
SRS patients have been shown to carry variable ICR1 methylation levels in different tissues. This epigenetically mosaic pattern strongly indicates postzygotic epigenetic modification error, which is manifested as body asymmetry phenotype observed in most patients. Further evidence comes from twin studies. To date, nine discordant monozygotic SRS twins and only one concordant twin have been reported [46–55]. Notably, discordant BWS MZ twin cases are predominantly females with the reported male to female ratio of 1:12 [56]. Similarly, discordant SRS MZ twins show a modest skew with male to female ratio of 2:7. Investigations of the ICR1 methylation levels have been carried out on three of the nine discordant MZ twins so far. Two of the twin pairs showed ICR1 hypomethylation in leucocyte DNA of both the affected and unaffected pair but in other tissues such as buccal cells and skin fibroblast, only the affected individual showed hypomethylation for the ICR1 [46,48]. In the third discordant MZ twin, however, hypomethylation was detected only in the affected twin's blood [55]. This discrepancy could be attributed to the presence or absence of shared circulation in the placenta. All cases were monochorionic-diamniotic twins, 70% of which is estimated to share blood circulation [57]. This would results in transferring of hypomethylated hematopoietic stem cells from the affected to the unaffected twin, resulting in similar methylation levels in the leucocyte. The third case showed no obvious shared circulation in the placenta, consistent with the distinguished methylation pattern between the twins in their blood cells [55].
DNA methylation & mRNA expression
ICR1 hypomethylation is assumed to lead to a decrease in the overall IGF2 expression and increase in H19. Several studies have tested the correlation between the ICR1 methylation level and IGF2 or H19 expression levels in different tissues. Gicquel et al. showed that the IGF2 expression level is lower in the affected discordant twin's fibroblast, and that H19 was biallelically expressed, indicating loss of imprinting [46]. More recently, Azzi et al. reported that ICR1 hypomethylation levels in fibroblasts correlated with the IGF2 expression, but when compared with controls, the difference was not significant [58]. Allelic expression of the H19 was not assessed in these patients and the H19 level did not correlate with the degree of ICR1 methylation. The same study also assessed the tissue-specific epigenotypes, including blood, fibroblasts, right and left buccal cells of the patients with semihypertrophy, showing variable levels of the methylation. Eleven percent of the patients showed discordance between leukocytes and buccal cells, strongly suggestive of postzygotic epigenetic variation leading to mosaic epigenetic profiles. On a protein level, ICR1 hypomethylation does not cause reduced IGF-II serum level, although this might not represent its function in prenatal period and in other tissues [59].
MLID & SRS
A subset of patients with imprinting disorders has been found to carry more than one DNA methylation abnormality, termed multilocus imprinting disturbance (MLID) [60]. Of the 10 imprinting disorders described, 5 of them have been reported with MLID: 50–75% of transient neonatal diabetes mellitus type 1 (TNDM1) patients with PLAGL1/HYMAI DMR hypomethylation, 4.5–50% of pseudohypoparathyroidism type Ib (PHP-Ib) with epimutation at one of the Chr20q13 DMRs, 20–45% of BWS with ICR2 hypomethylation, 7–36% of SRS with ICR1 hypomethylation and a single case with Prader–Willi syndrome (PWS)/BWS features (summarized in [61,62]). The wide frequency range of MLID in these patients may reflect the differences in the sample sizes, ethnicities, techniques, clinical scoring systems and number of loci investigated. Unlike the TNDM1 MLID cases, SRS patients with MLID are not significantly clinically distinguished from patients with isolated epimutation at ICR1, although an increased developmental delay in the SRS MLID groups was reported [63].
Strikingly, SRS and BWS cases with nearly identical epimutations have been reported [48,54,63–64]. Azzi et al. identified three SRS and one BWS carrying both ICR1 and ICR2 hypomethylation [64]. Moreover, one of these SRS patients and the BWS case also had PLAGL1 DMR hypomethylation despite the absence of TNDM1 phenotypes, adding further complexity to the molecular aetiology. From these studies it is estimated that approximately 4% of ICR1 hypomethylated SRS cases also have additional ICR2 hypomethylation. Of note, ICR2 hypomethylation in some of these cases were restricted to leucocytes but absent in their buccal cells, showing evidence of epigenetic mosaicism [63]. Interestingly, ICR2 hypomethylation has previously been identified in growth restricted cases [54,65], and in clinically normal ART-conceived individuals [66]. Since ICR2 hypomethylation has been considered a hallmark for the overgrowth disorder BWS, the reason for such shared epimutations in phenotypically normal or opposite disorders is currently unknown. It has been suggested that there is an ‘epidominance’ effects, where the phenotype is dominated by the most severely affected loci [58]. Also, since the majority of the study is carried out in blood, the mosaic nature of epimutation makes it difficult to determine the degree to which the most functionally relevant tissue is affected.
Despite the shared epimutations identified in SRS and BWS, additional epimutations at the Chr14q32 imprinting locus is so far predominant in SRS MLID patients. Hypomethylation of paternally methylated IG-DMR and MEG3-DMR at DLK1/MEG3 loci on Chr14q32 is associated with TS (previously referred to as upd(14)mat syndrome) [67]. TS shares characteristic features with other imprinting disorders. Pre/postnatal growth restriction, hypotonia and small hands are observed in both TS and PWS [68]. Pre/postnatal growth restriction, intellectual disability, speech delay are seen in approximately equal frequency amongst SRS and TS patients. Other symptoms such as body asymmetry, relative macrocephaly, prominent forehead, fifth finger clinodactyly, feeding difficulties, hypoglycemia, ear abnormalities and micrognathia are described in both syndromes but at higher frequency in SRS. Body asymmetry especially was suggested to be a useful SRS indicator, whereas precocious puberty and obesity are specific to TS. TS patients also have different characteristic facies from SRS, such as broad forehead, short nose with wide nasal tip and relatively short philtrum [67]. There are four SRS cases reported either having isolated 14q32 epi/mutations or in combination with upd(7)mat (Supplementary Table 1). These patients exhibit either SRS phenotypes only or characteristics for both SRS and TS. On the other hand, SRS MLID patients with both ICR1 hypomethylation and Chr14q32 epimutation generally have typical SRS phenotypes [64]. Although these cases constitute a small proportion of SRS it does suggest that an additional test for Chr14q32, especially in atypical or idiopathic cases is worth considering.
Fetal genetic causes of the epigenetic errors
Genetic causes for MLID is being explored and mutations in several trans-acting factors have been reported in MLID patients and their mothers [69–72]. Homozygous or compound heterozygous mutations within a trans-acting ZFP57 gene have been shown to be associated with hypomethylation at multiple imprinted loci in approximately 5% of TNDM1 patients [71]. ZFP57 encodes a Krüppel-associated box domain zinc finger protein which has been shown to bind to most germline imprinted DMRs in a parental allele-specific manner, and is required for DNA methylation maintenance at these loci in early embryonic stages [73]. Despite this, screening of 30 SRS patients with ICR1 hypomethylation and three SRS with MLID did not reveal any pathogenic mutation within ZFP57, therefore the involvement of this gene in the etiology of SRS is currently unknown [48,74].
Maternal effect genes
Another mechanism for MLID has been suggested through maternal-effect mutations. Three members of the NLRP family have been associated with imprinting disorders and aberrant DNA methylation. A single case of homozygous frameshift mutation in NLRP2 was reported in a mother with two BWS children and ICR2 hypomethylation in a consanguineous family [72]. Homozygous or compound heterozygous mutations in NLRP7 and KHDC3L have been identified in women who had recurrent biparental hydatidiform moles, with evidence of multiple imprinted DMR-specific methylation defects [69,75]. More recently, maternal rare variants in NLRP5 have been associated with MLID and imprinting disorders [70]. They have screened 39 MLID cases (SRS, BWS, TNDM and undiagnosed cases) and 33 mothers identifying five mothers carrying mutations, 2 of whom had SRS children. One mother was a compound heterozygote (c.1664G>T; c.2320T>C) and had atypical SRS child with cleft lip and palate, had a BWS child with a different partner and six miscarriages in total. The second mother was heterozygous for c.1156_1158dupCCT and had discordant monozygotic SRS twins. The authors suggest that the biallelic mutation (compound heterozygous) may have more severe effects than monoallelic mutations (heterozygous), although one mother was homozygous for a mutation (c.1699A>G) and had three healthy children. Since the mutations occur in heterozygous, compound heterozygous and homozygous forms, it is difficult to assess the functional model, but possibly suggesting a dominant negative or near loss-of-function mutations manifested by variable phenotypic outcome.
Familial SRS involving Chr11 gene mutations
Mutations in two imprinted genes at Chr11p15.5, CDKN1C and IGF2, have recently been reported in familial SRS cases [76,77]. CDKN1C is a preferentially maternally expressed tumor suppressor gene under the control of ICR2 (Figure 3), and is mutated in approximately 10% of BWS [78]. Most BWS cases are sporadic but approximately 15% are familial, 40% of which carry CDKN1C mutations. CDKN1C protein has three domains: a cyclin-dependent kinase inhibitory domain (CKI), a proline and alanine (PAPA) repeat domain and a QT domain which contains a proliferating cell nuclear antigen (PCNA) binding domain [79]. The BWS-CDKN1C pathogenic variants are maternally transmitted dominant loss-of-function missense mutations clustered to the CKI domain or truncation mutations spread across the gene [80].
Remarkably, gain-of-function mutations in CDKN1C have been identified in IMAGe syndrome (intrauterine growth restriction, metaphyseal dysplasia, congenital adrenal hypoplasia and genital anomalies) patients, who carried missense mutations localized to the PCNA-binding domain [79]. Since CDKN1C is thought to act as the growth suppressor, it is likely that loss-of-function leads to overgrowth (BWS) while gain-of-function results in growth restriction (IMAGe). Moreover, Brioude, et al. reported that screening 97 idiopathic SRS cases identified a missense mutation (c.836G>T) in a family with nine affected SRS members, six of whom was tested and shown to carry maternally inherited mutation [77]. The c.836G>T mutation is also within the PCNA domain, and at the same amino acid position as one of the reported IMAGe patients with different amino acid substitutions (IMAGe: p.Arg279Pro and SRS: Arg279Leu). The authors suggested that the increased stability of the mutant protein implies gain-of-function mutation. No further mutations were identified in the rest of 91 cases. More recently, paternally inherited IGF2 nonsense mutation (c.191C>A) was reported in a family with four members affected with pre/postnatal growth failure with SRS-like phenotype, possibly suggesting the role of IGF2 in postnatal growth [76]. This is an interesting observation since IGF2 is mainly produced in the liver after birth where it is expressed biallelically [13], with its main role is generally considered to be at prenatal growth period.
CNV & structural variation at Chr11
A number of CNVs at 11p15.5 imprinting cluster have been reported in SRS patients, affecting approximately 1% of all SRS cases (Supplementary Table 1). There are two paternally expressed and six maternally expressed imprinted genes under the control of ICR1 and ICR2, and the majority of CNVs involve maternal duplication of both ICR1 and ICR2, which is anticipated to result in excess of maternally expressed growth suppressor genes (Figure 3). One of the interesting observations is that maternal duplication of the whole H19/IGF2 domain including IGF2 enhancer, H19, ICR1 and IGF2 does not cause SRS but maternal duplication of the H19/IGF2 domain excluding the IGF2 gene does result in SRS [81], possibly indicating that doubling the H19 expression may be sufficient to cause SRS.
Molecular diagnostics & investigations for SRS
Current diagnostics process
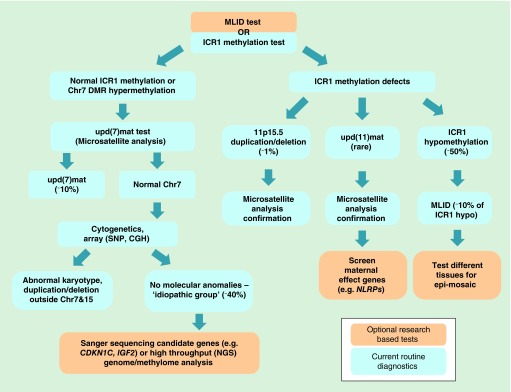
For the molecular diagnosis of SRS patients, the most thorough test would be to carry out the trio whole genome bisulfite/sequencing (WGS and WGBS) to simultaneously identify UPDs including upd(7)mat, ICR1 hypomethylation and MLID, de novo and inherited CNV, genomic mutations in maternal (e.g., NLRPs) and fetal (e.g., CDKN1C, IGF2) candidate genes and any novel epi/mutations. However, it is currently not a cost effective method and the data analysis is still not hands-on for the routine clinical diagnostics settings. A suggested diagnostic flow path is shown in Figure 4. In many diagnostics laboratories, methylation-specific multiplex ligation-dependent probe amplification is used routinely. This allows simultaneous detection of methylation levels at ICR1 and ICR2, as well as CNV at 11p15.5 in a single experiment. The resulting CNV could be further confirmed by microsatellite typing or qPCR. For those who are ICR1 hypomethylation negative, microsatellite analysis (MSA) for upd(7)mat test is also typically performed. Together these would identify approximately 60% of established molecular anomalies of SRS. The advantage and disadvantages of the currently available techniques for the diagnostics and for research studies are summarized in the Table 2.
Figure 4. . Suggested Silver–Russell syndrome diagnostics flow chart.
To test for all known SRS molecular anomalies, combination of different diagnostics techniques should be performed. Blue shade indicates the techniques used in the routine diagnostics (only widely used techniques shown here). Orange shade indicates the techniques that can be used optionally or in the research studies.
CGH: Comparative genomic hybridization; ICR1: Imprinting control region 1; MLID: Multilocus imprinting disturbance; NGS: Next-generation sequencing; SRS: Silver–Russell syndrome; Upd(7)mat: Maternal uniparental disomy of chromosome 7.
Table 2. . A summary of techniques used for Silver–Russell syndrome diagnostics and research investigations.
| Genomic/DNA methylation | Methods | upd(7)mat (˜10%) | Chromosomal aberrations | Candidate gene mutation screen | New candidate gene/region detection | ICR1 hypo (˜50%) | MLID | Pros and cons |
|---|---|---|---|---|---|---|---|---|
| Genomic | Microsatellite analysis† | ◯ | ◯ | X | X | X | X | Pros: fast and only routine molecular biology techniques required. Cons: targets one locus at a time. Does not distinguish UPD and CNV. Could be difficult for mosaic sample |
| Cytogenetic analysis† | X | ◯ | X | Δ | X | X | Pros: investigates genome-wide. Cons: low resolution (>5 Mb) | |
| FISH† | X | ◯ | X | X | X | X | Pros: investigates genome-wide. Cons: low resolution. Requires prior knowledge of the affected locus for the probe design | |
| Array (e.g., CGH/SNP)† | ◯ | ◯ | X | Δ | X | X | Pros: investigates genome-wide in high resolution. Cons: balanced rearrangements not detected | |
| Sanger sequencing‡ | X | X | ◯ | X | X | X | Pros: useful for familial SRS cases (e.g., IGF2, CDKN1C). Cons: not suitable for a large gene | |
| Exome sequencing‡ | Δ | Δ | ◯ | ◯ | X | X | Pros: cost and time effective. Cons: misses non-exonic regions | |
| |
Whole genome sequencing‡ |
◯ |
◯ |
◯ |
◯ |
X |
X |
Pros: investigates genome-wide in single-base resolution. Cons: currently not cost effective for routine diagnostic use |
| DNA methylation | Bisulfite sequencing† | Δ | X | X | X | ◯ | Δ | Pros: quantitative and easy. Cons: takes a long time, cloning required. Not suitable for testing many loci |
| MS PCR† | Δ | X | X | X | ◯ | Δ | Pros: quick and easy. Cons: semi-quantitative. Does not distinguish UPD and CNV | |
| MS pyrosequencing† | Δ | X | X | X | ◯ | Δ | Pros: quantitative. Cons: requires pyrosequencing platform. Not suitable for testing many loci | |
| MS qPCR† | Δ | X | X | X | ◯ | Δ | Pros: quantitative. Cons: not single-base resolution | |
| MS SNuPE† | Δ | X | X | X | ◯ | ◯ | Pros: cost and time effective. Cons: only tests selected regions | |
| High-density methylation array‡ | Δ | X | X | ◯ | ◯ | ◯ | Pros: cost- and time-effective method to test multiple loci. Could be used to check UPD if iDMRs are involved. Cons: bioinformatics analysis still not hands-on for routine diagnostics | |
| Enrichment NGS (e.g., MeDIP-seq)‡ | Δ | X | X | ◯ | ◯ | ◯ | Pros: investigates genome-wide. Distinguish 5mC and 5hmC. Cons: not single-base resolution. Bioinformatics analysis still not hands-on for routine diagnostics | |
| |
Whole genome bisulfite sequencing‡ |
Δ |
◯ |
Δ |
◯ |
◯ |
◯ |
Pros: investigates genome-wide. Cons: currently not cost effective for diagnostic use |
| Genomic and DNA methylation | MS-MLPA† | X | Δ | X | X | ◯ | Δ | Pros: simultaneously detect ICR1 and ICR2 methylation and CNV at Chr11p15.5. Cons: misses other DMRs |
Currently no single technique could test for all known SRS molecular anomalies. Therefore, combinations of different techniques should be performed.
†Indicates the techniques used in the routine diagnostics (only widely used techniques shown here).
‡Indicates the techniques that can be used optionally or in the research studies.
◯ = suitable, Δ = partially suitable and X = not suitable.
CGH: Comparative genome hybridization; CNV: Copy number variation; FISH: Fluorescent in situ hybridization; ICR1: Imprinting control region 1; MeDIP-seq: Methylated DNA immunoprecipitation sequencing; MLID: Multilocus methylation disturbance; MS: Methylation-specific; MS-MLPA: Methylation-specific multiplex ligation probe-dependent amplification; MS SNuPE: Methylation-specific single nucleotide primer extension; NGS: Next-generation sequencing; SRS: Silver–Russell syndrome; UPD: Uniparental disomy; qPCR: Quantitative PCR.
Molecular investigations for research studies
The importance of multilocus testing in imprinting disorders is becoming more apparent. Therefore, it is preferred that the initial methylation testing include several imprinting DMRs. We and others have recently used high-density methylation array to search genome-wide for methylation defects in SRS patients [82–85]. Interestingly, by using Illumina's 450K Infinium HumanMethylation array to look for epimutations in the idiopathic SRS patients, we have identified two SRS patients previously shown to carry normal ICR1 levels by restriction digestion method testing a single CpG, to have moderate ICR1 hypomethylation [84]. This suggests an advantage of using different and more sensitive techniques for the DNA methylation quantification and also having multiple probes at the target loci. To overcome the heterogeneous nature of SRS, we have used a round-robin statistical method so that the control methylation levels are compared with individual SRS patients rather than comparing against the SRS as a group. One problem with the 450K array, as well as with the newer 850K MethylationEPIC BeadChip, is that they do not have probes targeting at Chr14 DLK1/MEG3 IG-DMR loci, which is implicated in the TS as discussed earlier. Therefore, if 450K/850K EPIC array were used, this region has to be investigated by an alternative method.
The majority of methylation analyses use conventional bisulfite conversion method, which means that it will not be able to distinguish 5-methylcytosine (5mC) and 5-hydroxymethycytosine (5hmC). This would result in overestimation of the methylation levels. Tet enzyme can convert 5mC to 5hmC, which is thought to be an intermediate of the active demethylation step [86]. Its distribution profile in the imprinted regions, and possible correlation with imprinting disorders remains to be addressed. Recently, Matsubara et al. reported low levels of 5hmC at IG-DMR and MEG3-DMR in peripheral blood of Kagami–Ogata syndrome (KOS14) patients who have hypermethylation at these loci, concluding that the 5hmC has only minor contribution to the increased methylation [87]. Also, higher 5hmC abundance was observed in their neural samples, consistent with the previous observation that 5hmC is enriched in the brain tissues and in embryonic stem cells [88]. Further characterization of 5hmC profile in other imprinting disorders will increase the understanding of the molecular pathology.
Current treatment practice
Due to the heterogeneous nature of the SRS phenotypes, treatments from different clinical specialists may be required according to their specific symptoms. These include clinical geneticists for clinical and molecular diagnosis alongside pediatric endocrinologists for observation of growth trajectory and decision on growth hormone administration. The multispecialist team also need to include dieticians and gastroenterologists for feeding problems, pediatric surgeons if intervention required for the body asymmetry, dental specialists, speech therapists, psychologists and clinical genetic counselors for advice on family planning.
Feeding therapy
The majority of SRS patients exhibit feeding difficulties in early childhood, seen as lack of appetite, food fussiness and slow eating [89]. Gastrointestinal complications are common in SRS children, including gastroesophageal reflux, esophagitis and food aversion [90,91]. These unpleasant experiences are likely to be contributing factors for their reduced appetite. It is important to monitor and maintain the adequate caloric intake for the patients in order to prevent associated problems such as hypoglycemia, and to promote their growth. A recent study reported that malnutrition affects 70% of SRS children, and suggested considerations for providing enriched diet, enteral feeding and nutritional supplements for improvement of nutritional status [91]. Nevertheless, the feeding difficulties have been reported to gradually improve with age, and around 6 years of age, food refusal and fussiness are not significantly different from normal children [89].
Growth hormone treatment
Children born small for gestational age typically experience catch-up growth during the first year of life which completes around 2 years of age [92]. This is not generally the case for the SRS patients; a study of 386 SRS patients showed that mean adult height was 151.2 cm in males and 139.9 cm in females [93]. Recombinant GH therapy is available for small for gestational age patients including SRS, who have not demonstrated adequate catch-up growth by age two, which was approved by US FDA in 2001 [94]. In Europe, the use of GH was officially indicated by European Medicines Agency in 2003 (formally known as European Agency for the Evaluation of Medicinal Products in 1995–2004), for children with growth failure who are ≥4 years old and have height less than -2.5 SD and 1 SD below mid-parental height SDS. GH therapy in SRS children has shown an evidence of beneficial outcome. A long-term follow-up study of 26 SRS children treated with GH for the median of 9.8 years showed a significant increment of median height from -2.7 SD at the start to -1.3 SD final height [95]. Whether the effect of GH is influenced by the different molecular subtypes of SRS is currently unknown. One study reported a trend that the GH response was better in the upd(7)mat group compared with the ICR1 hypomethylation group (upd(7)mat n = 5 and ICR1 hypo n = 19) [96]. However, these studies are still in the preliminary stage and it should be investigated further in a larger sample size, as it would influence the decision on whether to give or when to give GH. Nevertheless, the first focus on management of growth should focus on adequate caloric intake as GH cannot work without adequate glycogen stores.
Long-term follow-ups
There is a paucity of information available on the long-term health issues with SRS patients, although a small number of reports indicate unremarkable medical problems. As a consequence, they are rarely followed up to the adulthood. The rare familial cases of SRS (e.g., IGF2, CDKN1C and Chr15p15.5 CNV) implies that SRS patients are fertile, although precise reproductive fitness of other molecular groups (ICR1 hypomethylation/upd(7)mat/idiopathics) are not well documented. The DNA methylation patterns are reset in every generation therefore the recurrence risk is low, unless it is caused by underlying genetic mutations (e.g., maternal effect mutation). However, Bartholdi et al. have reported a father to daughter transmission of ICR1 hypomethylation in a single family [10]. Upd(7) mat are most likely caused by nondisjunction followed by trisomic rescue so the recurrent risk is also low [23].
In a recent follow-up study, three Japanese male SRS adults in their early 20s were reported who have developed Type 2 diabetes, hypertension and obesity [97]. They were all male patients and had ICR1 hypomethylation. This finding is consistent with the frequently reported observations that low birth weight is associated with increased risk of metabolic abnormalities and cardiac diseases as adults [98]. Such observations have led to the hypothesis commonly known as the Developmental Origins of Health and Disease, which suggests that suboptimal intrauterine or early life environment, such as low maternal calorie intake during pregnancy, would cause long-term programming, possibly via epigenetic modifications, to have increased susceptibility to adulthood diseases. One of the ongoing studies to follow-up SRS patients include the ‘Study of Adults and Adolescents with Russell–Silver Syndrome in the UK’ to observe the long-term health issues and the effects of growth hormone. This would benefit the patients in terms of lifestyle choice and parental decision regarding growth hormone treatment.
Looking forward development of technology
Recent advances in epi/genome engineering technology hold a potential for targeted epigenetic therapy in the future. Present epigenetic drugs (epidrugs) such as DNMT inhibitors (g.a. 5-aza-2′-deoxycytidine), lead to genome-wide modification which is not desirable especially for the imprinting disorders with specific epimutations. Zinc finger nucleases and transcription-activator-like effector nucleases systems are early methods developed for targeted in vivo genetic modifications, both of which are based on protein–DNA binding interaction [99]. More recently, the clustered regularly interspaced short palindromic repeats/Cas systems, based on guide RNA–DNA paring, has demonstrated several advantages over the ZNF and transcription-activator-like effector nucleases methods in terms of simplicity of the design, efficiency and the ability to target multiple sites at once [99]. For site-specific DNA methylation modification, a catalytically inactive Cas9 (dCas9) variant, which can bind but cannot cleave DNA, could be fused to de novo methyltransferase DNMT3a for methylation and to TET1 enzyme for demethylation. So far, locus-specific DNA methylation modification have been successfully demonstrated by the ZFP coupled with the DNMT3a catalytic domain for methylating Maspin and SOX2 gene promoters in breast cancer cells [100], and TALE-TET1 fusion protein for demethylation of endogenous gene promoters (KLF4, HBB and RHOXF2) in three human cell lines [101]. Further improvement on off-target effects and delivery methods are required for therapeutic use.
Conclusion & future perspective
This review aimed to summarize the molecular defects associated with SRS to provide an up-to-date overview, and to highlight the importance of multilocus testing. Due to the heterogeneous presentation of SRS, a consensus in diagnostic criteria would be required. This problem was discussed in a recent meeting in Barcelona October 2015 among SRS experts, and the consensus is currently being prepared to publish in 2016 (european molecular genetics quality network, Pers. Comm.). Although ICR1 hypomethylation is observed in more than half of SRS patients, the underlying mechanisms of how the loss of methylation leads to the phenotype have not been fully explained. In addition, how the upd(7)mat and ICR1 hypomethylation could both lead to same disorder remains unsolved. Also, majority of the studies correlating ICR epimutation and gene expression levels only investigate the immediate neighboring genes – IGF2 and H19 in case of SRS patients. Therefore, it would be interesting to investigate the transcriptome and proteome of ICR1 and upd(7)mat patients to elucidate the whole picture of molecular network in the growth pathway. Due to the phenotypic overlaps with other imprinting disorders and growth restriction disorders, final diagnosis of SRS should be based on both, thorough genetic and epigenetic testing, which would also assist the genetic counseling and family planning. Finally, follow-up studies on SRS patients could possibly improve prognosis of the patients through tailored therapy based on each individual's molecular defects.
Executive summary.
We need a consensus in Silver–Russell syndrome (SRS) diagnosis criteria.
More than half of SRS patients have ICR1 hypomethylation and about 10% have upd(7)mat.
SRS patients require multilocus testing and trio screening.
SRS epimutation shows mosaic pattern; therefore, multitissue testing is recommended.
Phenotypic/epigenetic overlap with other imprinting disorders including Temple syndrome and upd(20)mat are becoming apparent.
There is a modest skew in male to female ratio in SRS monozygotic twin with the ratio of 2:7.
ICR1 hypomethylation group have more ‘classic’ SRS phenotypes compared with upd(7)mat group.
There are not significant phenotypic differences between multilocus imprinting disturbance SRS and ICR1 hypomethylation, except that developmental delay was more frequent in multilocus imprinting disturbance group.
Supplementary Material
Acknowledgements
The authors thank G Moore and S Abu-Amero for reading and editing the manuscript.
Footnotes
Financial & competing interests disclosure
M Ishida is funded by Medical Research Council (MRC). The fetal growth and development research group is funded by the MRC, Wellbeing of Women and supported by the National Institute for Health Research Biomedical Research Centre at Great Ormond Street Hospital for Children NHS Foundation Trust and University College London. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Russell A. A syndrome of intra-uterine dwarfism recognizable at birth with cranio-facial dysostosis, disproportionately short arms, and other anomalies (5 examples) Proc. R. Soc. Med. 1954;47(12):1040–1044. [PubMed] [Google Scholar]
- 2.Silver HK, Kiyasu W, George J, Deamer WC. Syndrome of congenital hemihypertrophy, shortness of stature, and elevated urinary gonadotropins. Pediatrics. 1953;12(4):368–376. [PubMed] [Google Scholar]
- 3.Mcgrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37(1):179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- 4.Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308(5959):548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- 5.www.har.mrc.ac.uk/ MRC Harwell: an International Centre for Mouse Genetics.
- 6.Lewis A, Reik W. How imprinting centres work. Cytogenet. Genome Res. 2006;113(1–4):81–89. doi: 10.1159/000090818. [DOI] [PubMed] [Google Scholar]
- 7.Ishida M, Moore GE. The role of imprinted genes in humans. Mol. Aspects Med. 2013;34(4):826–840. doi: 10.1016/j.mam.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 8.www.imprinting-disorders.eu/ Imprinting disorders.
- 9.Azzi S, Salem J, Thibaud N, et al. A prospective study validating a clinical scoring system and demonstrating phenotypical-genotypical correlations in Silver–Russell syndrome. J. Med. Genet. 2015;52(7):446–453. doi: 10.1136/jmedgenet-2014-102979. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes the latest clinical scoring system for Silver–Russell syndrome diagnosis with some advantage over previously described systems.
- 10.Bartholdi D, Krajewska-Walasek M, Ounap K, et al. Epigenetic mutations of the imprinted IGF2-H19 domain in Silver–Russell syndrome (SRS): results from a large cohort of patients with SRS and SRS-like phenotypes. J. Med. Genet. 2009;46(3):192–197. doi: 10.1136/jmg.2008.061820. [DOI] [PubMed] [Google Scholar]
- 11.Dias RP, Nightingale P, Hardy C, et al. Comparison of the clinical scoring systems in Silver–Russell syndrome and development of modified diagnostic criteria to guide molecular genetic testing. J. Med. Genet. 2013;50(9):635–639. doi: 10.1136/jmedgenet-2013-101693. [DOI] [PubMed] [Google Scholar]
- 12.Lai KY, Skuse D, Stanhope R, Hindmarsh P. Cognitive abilities associated with the Silver–Russell syndrome. Arch. Dis. Child. 1994;71(6):490–496. doi: 10.1136/adc.71.6.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Netchine I, Rossignol S, Dufourg MN, et al. 11p15 imprinting center region 1 loss of methylation is a common and specific cause of typical Russell–Silver syndrome: clinical scoring system and epigenetic-phenotypic correlations. J. Clin. Endocrinol. Metab. 2007;92(8):3148–3154. doi: 10.1210/jc.2007-0354. [DOI] [PubMed] [Google Scholar]
- 14.Price SM, Stanhope R, Garrett C, Preece MA, Trembath RC. The spectrum of Silver–Russell syndrome: a clinical and molecular genetic study and new diagnostic criteria. J. Med. Genet. 1999;36(11):837–842. [PMC free article] [PubMed] [Google Scholar]
- 15.Mulchandani S, Bhoj EJ, Luo M, et al. Maternal uniparental disomy of chromosome 20: a novel imprinting disorder of growth failure. Genet. Med. 2016;18(4):309–315. doi: 10.1038/gim.2015.103. [DOI] [PubMed] [Google Scholar]
- 16.Fokstuen S, Kotzot D. Chromosomal rearrangements in patients with clinical features of Silver–Russell syndrome. Am. J. Med. Genet. A. 2014;164A(6):1595–1605. doi: 10.1002/ajmg.a.36464. [DOI] [PubMed] [Google Scholar]
- 17.Wakeling EL, Amero SA, Alders M, et al. Epigenotype-phenotype correlations in Silver–Russell syndrome. J. Med. Genet. 2010;47(11):760–768. doi: 10.1136/jmg.2010.079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abu-Amero S, Monk D, Frost J, Preece M, Stanier P, Moore GE. The genetic aetiology of Silver–Russell syndrome. J. Med. Genet. 2008;45(4):193–199. doi: 10.1136/jmg.2007.053017. [DOI] [PubMed] [Google Scholar]
- 19.Kotzot D, Schmitt S, Bernasconi F, et al. Uniparental disomy 7 in Silver–Russell syndrome and primordial growth retardation. Hum. Mol. Genet. 1995;4(4):583–587. doi: 10.1093/hmg/4.4.583. [DOI] [PubMed] [Google Scholar]
- 20.Spence JE, Perciaccante RG, Greig GM, et al. Uniparental disomy as a mechanism for human genetic disease. Am. J. Hum. Genet. 1988;42(2):217–226. [PMC free article] [PubMed] [Google Scholar]
- 21.Eggerding FA, Schonberg SA, Chehab FF, Norton ME, Cox VA, Epstein CJ. Uniparental isodisomy for paternal 7p and maternal 7q in a child with growth retardation. Am. J. Hum. Genet. 1994;55(2):253–265. [PMC free article] [PubMed] [Google Scholar]
- 22.Voss R, Ben-Simon E, Avital A, et al. Isodisomy of chromosome 7 in a patient with cystic fibrosis: could uniparental disomy be common in humans? Am. J. Hum. Genet. 1989;45(3):373–380. [PMC free article] [PubMed] [Google Scholar]
- 23.Preece MA, Abu-Amero SN, Ali Z, et al. An analysis of the distribution of hetero- and isodisomic regions of chromosome 7 in five mUPD7 Silver–Russell syndrome probands. J. Med. Genet. 1999;36(6):457–460. [PMC free article] [PubMed] [Google Scholar]
- 24.Romanelli V, Meneses HN, Fernandez L, et al. Beckwith-Wiedemann syndrome and uniparental disomy 11p: fine mapping of the recombination breakpoints and evaluation of several techniques. Eur. J. Hum. Genet. 2011;19(4):416–421. doi: 10.1038/ejhg.2010.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monk D, Hitchins M, Russo S, Preece M, Stanier P, Moore GE. No evidence for mosaicism in Silver–Russell syndrome. J. Med. Genet. 2001;38(4):E11. doi: 10.1136/jmg.38.4.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalousek DK, Langlois S, Robinson WP, et al. Trisomy 7 CVS mosaicism: pregnancy outcome, placental and DNA analysis in 14 cases. Am. J. Med. Genet. 1996;65(4):348–352. doi: 10.1002/(SICI)1096-8628(19961111)65:4<348::AID-AJMG19>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 27.Fares F, David M, Lerner A, et al. Paternal isodisomy of chromosome 7 with cystic fibrosis and overgrowth. Am. J. Med. Genet. A. 2006;140(16):1785–1788. doi: 10.1002/ajmg.a.31380. [DOI] [PubMed] [Google Scholar]
- 28.Hoglund P, Holmberg C, De La Chapelle A, Kere J. Paternal isodisomy for chromosome 7 is compatible with normal growth and development in a patient with congenital chloride diarrhea. Am. J. Hum. Genet. 1994;55(4):747–752. [PMC free article] [PubMed] [Google Scholar]
- 29.Le Caignec C, Isidor B, De Pontbriand U, et al. Third case of paternal isodisomy for chromosome 7 with cystic fibrosis: a new patient presenting with normal growth. Am. J. Med. Genet. A. 2007;143A(22):2696–2699. doi: 10.1002/ajmg.a.31999. [DOI] [PubMed] [Google Scholar]
- 30.Pan Y, McCaskill CD, Thompson KH, et al. Paternal isodisomy of chromosome 7 associated with complete situs inversus and immotile cilia. Am. J. Hum. Genet. 1998;62(6):1551–1555. doi: 10.1086/301857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eggermann T, Spengler S, Begemann M, et al. Deletion of the paternal allele of the imprinted MEST/PEG1 region in a patient with Silver–Russell syndrome features. Clin. Genet. 2012;81(3):298–300. doi: 10.1111/j.1399-0004.2011.01719.x. [DOI] [PubMed] [Google Scholar]
- 32.Eggermann T, Schonherr N, Jager S, et al. Segmental maternal UPD(7q) in Silver–Russell syndrome. Clin. Genet. 2008;74(5):486–489. doi: 10.1111/j.1399-0004.2008.01057.x. [DOI] [PubMed] [Google Scholar]
- 33.Hannula K, Lipsanen-Nyman M, Kontiokari T, Kere J. A narrow segment of maternal uniparental disomy of chromosome 7q31-qter in Silver–Russell syndrome delimits a candidate gene region. Am. J. Hum. Genet. 2001;68(1):247–253. doi: 10.1086/316937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joyce CA, Sharp A, Walker JM, Bullman H, Temple IK. Duplication of 7p12.1-p13, including GRB10 and IGFBP1, in a mother and daughter with features of Silver–Russell syndrome. Hum. Genet. 1999;105(3):273–280. doi: 10.1007/s004390051101. [DOI] [PubMed] [Google Scholar]
- 35.Leach NT, Chudoba I, Stewart TV, Holmes LB, Weremowicz S. Maternally inherited duplication of chromosome 7, dup(7)(p11.2p12), associated with mild cognitive deficit without features of Silver–Russell syndrome. Am. J. Med. Genet. A. 2007;143A(13):1489–1493. doi: 10.1002/ajmg.a.31794. [DOI] [PubMed] [Google Scholar]
- 36.Monk D, Bentley L, Hitchins M, et al. Chromosome 7p disruptions in Silver Russell syndrome: delineating an imprinted candidate gene region. Hum. Genet. 2002;111(4–5):376–387. doi: 10.1007/s00439-002-0777-4. [DOI] [PubMed] [Google Scholar]
- 37.Monk D, Wakeling EL, Proud V, et al. Duplication of 7p11.2-p13, including GRB10, in Silver–Russell syndrome. Am. J. Hum. Genet. 2000;66(1):36–46. doi: 10.1086/302717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reboul MP, Tandonnet O, Biteau N, et al. Mosaic maternal uniparental isodisomy for chromosome 7q21-qter. Clin. Genet. 2006;70(3):207–213. doi: 10.1111/j.1399-0004.2006.00664.x. [DOI] [PubMed] [Google Scholar]
- 39.Eggermann T, Begemann M, Binder G, Spengler S. Silver–Russell syndrome: genetic basis and molecular genetic testing. Orphanet J. Rare Dis. 2010;5:19. doi: 10.1186/1750-1172-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eggermann T, Schneider-Ratzke B, Begemann M, Spengler S. Isolated hypermethylation of GRB10 (7p12.2) in a Silver–Russell syndrome patient carrying a 20p13 microdeletion. Clin. Genet. 2014;85(4):399–400. doi: 10.1111/cge.12186. [DOI] [PubMed] [Google Scholar]
- 41.Hannula-Jouppi K, Muurinen M, Lipsanen-Nyman M, et al. Differentially methylated regions in maternal and paternal uniparental disomy for chromosome 7. Epigenetics. 2014;9(3):351–365. doi: 10.4161/epi.27160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Babak T, Deveale B, Tsang EK, et al. Genetic conflict reflected in tissue-specific maps of genomic imprinting in human and mouse. Nat. Genet. 2015;47(5):544–549. doi: 10.1038/ng.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hitchins MP, Bentley L, Monk D, et al. DDC and COBL, flanking the imprinted GRB10 gene on 7p12, are biallelically expressed. Mamm. Genome. 2002;13(12):686–691. doi: 10.1007/s00335-002-3028-z. [DOI] [PubMed] [Google Scholar]
- 44.Gregg C, Zhang J, Butler JE, Haig D, Dulac C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science. 2010;329(5992):682–685. doi: 10.1126/science.1190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tremblay KD, Saam JR, Ingram RS, Tilghman SM, Bartolomei MS. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat. Genet. 1995;9(4):407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- 46.Gicquel C, Rossignol S, Cabrol S, et al. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver–Russell syndrome. Nat. Genet. 2005;37(9):1003–1007. doi: 10.1038/ng1629. [DOI] [PubMed] [Google Scholar]
- 47.Bailey W, Popovich B, Jones KL. Monozygotic twins discordant for the Russell–Silver syndrome. Am. J. Med. Genet. 1995;58(2):101–105. doi: 10.1002/ajmg.1320580202. [DOI] [PubMed] [Google Scholar]
- 48.Begemann M, Spengler S, Kanber D, et al. Silver–Russell patients showing a broad range of ICR1 and ICR2 hypomethylation in different tissues. Clin. Genet. 2011;80(1):83–88. doi: 10.1111/j.1399-0004.2010.01514.x. [DOI] [PubMed] [Google Scholar]
- 49.Nyhan WL. Genetic and Malformation Syndromes in Clinical Medicine. Year Book Medical Publishers; Chicago, USA: 1976. Silver syndrome: Silver–Russell Syndrome, Russell–Silver Syndrome; pp. 298–300. [Google Scholar]
- 50.Rimoin DL. The Silver syndrome in twins. Birth Defects. 1969;5(2):183–187. [Google Scholar]
- 51.Sagot P, David A, Talmant C, Pascal O, Winer N, Boog G. Russell–Silver syndrome: an explanation for discordant growth in monozygotic twins. Fetal Diagn. Ther. 1996;11(1):72–78. doi: 10.1159/000264283. [DOI] [PubMed] [Google Scholar]
- 52.Samn M, Lewis K, Blumberg B. Monozygotic twins discordant for the Russell–Silver syndrome. Am. J. Med. Genet. 1990;37(4):543–545. doi: 10.1002/ajmg.1320370424. [DOI] [PubMed] [Google Scholar]
- 53.Teran CG, Villarroel P, Teran-Escalera CN. Russell–Silver syndrome: twin presentation. BMJ Case Rep. 2009;2009 doi: 10.1136/bcr.06.2008.0202. pii: bcr06.2008.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turner CL, Mackay DM, Callaway JL, et al. Methylation analysis of 79 patients with growth restriction reveals novel patterns of methylation change at imprinted loci. Eur. J. Hum. Genet. 2010;18(6):648–655. doi: 10.1038/ejhg.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamazawa K, Kagami M, Fukami M, Matsubara K, Ogata T. Monozygotic female twins discordant for Silver–Russell syndrome and hypomethylation of the H19-DMR. J. Hum. Genet. 2008;53(10):950–955. doi: 10.1007/s10038-008-0329-4. [DOI] [PubMed] [Google Scholar]
- 56.Bliek J, Alders M, Maas SM, et al. Lessons from BWS twins: complex maternal and paternal hypomethylation and a common source of haematopoietic stem cells. Eur. J. Hum. Genet. 2009;17(12):1625–1634. doi: 10.1038/ejhg.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hall JG. Twinning. Lancet. 2003;362(9385):735–743. doi: 10.1016/S0140-6736(03)14237-7. [DOI] [PubMed] [Google Scholar]
- 58.Azzi S, Blaise A, Steunou V, et al. Complex tissue-specific epigenotypes in Russell–Silver Syndrome associated with 11p15 ICR1 hypomethylation. Hum. Mutat. 2014;35(10):1211–1220. doi: 10.1002/humu.22623. [DOI] [PubMed] [Google Scholar]
- 59.Binder G, Seidel AK, Weber K, et al. IGF-II serum levels are normal in children with Silver–Russell syndrome who frequently carry epimutations at the IGF2 locus. J. Clin. Endocrinol. Metab. 2006;91(11):4709–4712. doi: 10.1210/jc.2006-1127. [DOI] [PubMed] [Google Scholar]
- 60.Eggermann T, Netchine I, Temple IK, et al. Congenital imprinting disorders: EUCID.net – a network to decipher their aetiology and to improve the diagnostic and clinical care. Clin. Epigenetics. 2015;7(1):23. doi: 10.1186/s13148-015-0050-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes the EUCID.net (European network of congenital imprinting disorders) consortium, which is aimed to promote better clinical care and scientific investigation of imprinted disorders. This also have a good summary of imprinting disorders.
- 61.Eggermann T, Heilsberg AK, Bens S, et al. Additional molecular findings in 11p15-associated imprinting disorders: an urgent need for multi-locus testing. J. Mol. Med. (Berl.) 2014;92(7):769–777. doi: 10.1007/s00109-014-1141-6. [DOI] [PubMed] [Google Scholar]
- 62.Fuke T, Mizuno S, Nagai T, et al. Molecular and clinical studies in 138 Japanese patients with Silver–Russell syndrome. PLoS ONE. 2013;8(3):e60105. doi: 10.1371/journal.pone.0060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poole RL, Docherty LE, Al Sayegh A, et al. Targeted methylation testing of a patient cohort broadens the epigenetic and clinical description of imprinting disorders. Am. J. Med. Genet. A. 2013;161A(9):2174–2182. doi: 10.1002/ajmg.a.36049. [DOI] [PubMed] [Google Scholar]
- 64.Azzi S, Rossignol S, Steunou V, et al. Multilocus methylation analysis in a large cohort of 11p15-related foetal growth disorders (Russell Silver and Beckwith Wiedemann syndromes) reveals simultaneous loss of methylation at paternal and maternal imprinted loci. Hum. Mol. Genet. 2009;18(24):4724–4733. doi: 10.1093/hmg/ddp435. [DOI] [PubMed] [Google Scholar]
- 65.Murphy R, Mackay D, Mitchell EA. Beckwith Wiedemann imprinting defect found in leucocyte but not buccal DNA in a child born small for gestational age. BMC Med. Genet. 2012;13:99. doi: 10.1186/1471-2350-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gomes MV, Huber J, Ferriani RA, Amaral Neto AM, Ramos ES. Abnormal methylation at the KvDMR1 imprinting control region in clinically normal children conceived by assisted reproductive technologies. Mol. Hum. Reprod. 2009;15(8):471–477. doi: 10.1093/molehr/gap038. [DOI] [PubMed] [Google Scholar]
- 67.Ioannides Y, Lokulo-Sodipe K, Mackay DJ, Davies JH, Temple IK. Temple syndrome: improving the recognition of an underdiagnosed chromosome 14 imprinting disorder: an analysis of 51 published cases. J. Med. Genet. 2014;51(8):495–501. doi: 10.1136/jmedgenet-2014-102396. [DOI] [PubMed] [Google Scholar]
- 68.Buiting K. Prader–Willi syndrome and Angelman syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2010;154C(3):365–376. doi: 10.1002/ajmg.c.30273. [DOI] [PubMed] [Google Scholar]
- 69.Djuric U, El-Maarri O, Lamb B, et al. Familial molar tissues due to mutations in the inflammatory gene, NALP7, have normal postzygotic DNA methylation. Hum. Genet. 2006;120(3):390–395. doi: 10.1007/s00439-006-0192-3. [DOI] [PubMed] [Google Scholar]
- 70.Docherty LE, Rezwan FI, Poole RL, et al. Mutations in NLRP5 are associated with reproductive wastage and multilocus imprinting disorders in humans. Nat. Commun. 2015;6:8086. doi: 10.1038/ncomms9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mackay DJ, Callaway JL, Marks SM, et al. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat. Genet. 2008;40(8):949–951. doi: 10.1038/ng.187. [DOI] [PubMed] [Google Scholar]
- 72.Meyer E, Lim D, Pasha S, et al. Germline mutation in NLRP2 (NALP2) in a familial imprinting disorder (Beckwith-Wiedemann syndrome) PLoS Genet. 2009;5(3):e1000423. doi: 10.1371/journal.pgen.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Strogantsev R, Krueger F, Yamazawa K, et al. Allele-specific binding of ZFP57 in the epigenetic regulation of imprinted and non-imprinted monoallelic expression. Genome Biol. 2015;16:112. doi: 10.1186/s13059-015-0672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spengler S, Gogiel M, Schonherr N, Binder G, Eggermann T. Screening for genomic variants in ZFP57 in Silver–Russell syndrome patients with 11p15 epimutations. Eur. J. Med. Genet. 2009;52(6):415–416. doi: 10.1016/j.ejmg.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 75.Parry DA, Logan CV, Hayward BE, et al. Mutations causing familial biparental hydatidiform mole implicate c6orf221 as a possible regulator of genomic imprinting in the human oocyte. Am. J. Hum. Genet. 2011;89(3):451–458. doi: 10.1016/j.ajhg.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Begemann M, Zirn B, Santen G, et al. Paternally inherited IGF2 mutation and growth restriction. N. Engl. J. Med. 2015;373(4):349–356. doi: 10.1056/NEJMoa1415227. [DOI] [PubMed] [Google Scholar]
- 77.Brioude F, Oliver-Petit I, Blaise A, et al. CDKN1C mutation affecting the PCNA-binding domain as a cause of familial Russell Silver syndrome. J. Med. Genet. 2013;50(12):823–830. doi: 10.1136/jmedgenet-2013-101691. [DOI] [PubMed] [Google Scholar]
- 78.Choufani S, Shuman C, Weksberg R. Beckwith-Wiedemann syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2010;154C(3):343–354. doi: 10.1002/ajmg.c.30267. [DOI] [PubMed] [Google Scholar]
- 79.Arboleda VA, Lee H, Parnaik R, et al. Mutations in the PCNA-binding domain of CDKN1C cause IMAGe syndrome. Nat. Genet. 2012;44(7):788–792. doi: 10.1038/ng.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Soejima H, Higashimoto K. Epigenetic and genetic alterations of the imprinting disorder Beckwith-Wiedemann syndrome and related disorders. J. Hum. Genet. 2013;58(7):402–409. doi: 10.1038/jhg.2013.51. [DOI] [PubMed] [Google Scholar]
- 81.Begemann M, Spengler S, Gogiel M, et al. Clinical significance of copy number variations in the 11p15.5 imprinting control regions: new cases and review of the literature. J. Med. Genet. 2012;49(9):547–553. doi: 10.1136/jmedgenet-2012-100967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Court F, Tayama C, Romanelli V, et al. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014;24(4):554–569. doi: 10.1101/gr.164913.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kannenberg K, Urban C, Binder G. Increased incidence of aberrant DNA methylation within diverse imprinted gene loci outside of IGF2/H19 in Silver–Russell syndrome. Clin. Genet. 2012;81(4):366–377. doi: 10.1111/j.1399-0004.2012.01844.x. [DOI] [PubMed] [Google Scholar]
- 84.Prickett AR, Ishida M, Bohm S, et al. Genome-wide methylation analysis in Silver–Russell syndrome patients. Hum. Genet. 2015;134(3):317–332. doi: 10.1007/s00439-014-1526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rezwan FI, Docherty LE, Poole RL, et al. A statistical method for single sample analysis of HumanMethylation450 array data: genome-wide methylation analysis of patients with imprinting disorders. Clin. Epigenetics. 2015;7(1):48. doi: 10.1186/s13148-015-0081-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 2012;13(1):7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 87.Matsubara K, Kagami M, Nakabayashi K, et al. Exploration of hydroxymethylation in Kagami-Ogata syndrome caused by hypermethylation of imprinting control regions. Clin. Epigenetics. 2015;7(1):90. doi: 10.1186/s13148-015-0124-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Blissett J, Harris G, Kirk J. Feeding problems in Silver–Russell syndrome. Dev. Med. Child Neurol. 2001;43(1):39–44. doi: 10.1017/s0012162201000068. [DOI] [PubMed] [Google Scholar]
- 90.Anderson J, Viskochil D, O'Gorman M, Gonzales C. Gastrointestinal complications of Russell–Silver syndrome: a pilot study. Am. J. Med. Genet. 2002;113(1):15–19. doi: 10.1002/ajmg.10667. [DOI] [PubMed] [Google Scholar]
- 91.Marsaud C, Rossignol S, Tounian P, Netchine I, Dubern B. Prevalence and management of gastrointestinal manifestations in Silver–Russell syndrome. Arch. Dis. Child. 2015;100(4):353–358. doi: 10.1136/archdischild-2013-305864. [DOI] [PubMed] [Google Scholar]
- 92.Clayton PE, Cianfarani S, Czernichow P, Johannsson G, Rapaport R, Rogol A. Management of the child born small for gestational age through to adulthood: a consensus statement of the International Societies of Pediatric Endocrinology and the Growth Hormone Research Society. J. Clin. Endocrinol. Metab. 2007;92(3):804–810. doi: 10.1210/jc.2006-2017. [DOI] [PubMed] [Google Scholar]
- 93.Wollmann HA, Kirchner T, Enders H, Preece MA, Ranke MB. Growth and symptoms in Silver–Russell syndrome: review on the basis of 386 patients. Eur. J. Pediatr. 1995;154(12):958–968. doi: 10.1007/BF01958638. [DOI] [PubMed] [Google Scholar]
- 94.Lee PA, Chernausek SD, Hokken-Koelega AC, Czernichow P International Small for Gestational Age Advisory B. International Small for Gestational Age Advisory Board consensus development conference statement: management of short children born small for gestational age, April 24-October 1, 2001. Pediatrics. 2003;111(6 Pt 1):1253–1261. doi: 10.1542/peds.111.6.1253. [DOI] [PubMed] [Google Scholar]
- 95.Toumba M, Albanese A, Azcona C, Stanhope R. Effect of long-term growth hormone treatment on final height of children with Russell–Silver syndrome. Horm. Res. Paediatr. 2010;74(3):212–217. doi: 10.1159/000295924. [DOI] [PubMed] [Google Scholar]
- 96.Binder G, Seidel AK, Martin DD, et al. The endocrine phenotype in Silver–Russell syndrome is defined by the underlying epigenetic alteration. J. Clin. Endocrinol. Metab. 2008;93(4):1402–1407. doi: 10.1210/jc.2007-1897. [DOI] [PubMed] [Google Scholar]
- 97.Takenouchi T, Awazu M, EggermannT, Kosaki K. Adult phenotype of Russell–Silver syndrome: a molecular support for Barker-Brenner's theory. Congenit. Anom. (Kyoto) 2015;55(3):167–169. doi: 10.1111/cga.12105. [DOI] [PubMed] [Google Scholar]
- 98.Barker DJ. The origins of the developmental origins theory. J. Intern. Med. 2007;261(5):412–417. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- 99.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rivenbark AG, Stolzenburg S, Beltran AS, et al. Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics. 2012;7(4):350–360. doi: 10.4161/epi.19507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Maeder ML, Angstman JF, Richardson ME, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE–TET1 fusion proteins. Nat. Biotechnol. 2013;31(12):1137–1142. doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.