Abstract
Background & objectives:
Mycobacterium tuberculosis (M. tuberculosis) has four homologous mammalian cell entry (mce) operons (mce1-4) that encode exported proteins and have a possible role in the virulence mechanism of this pathogen. The expression of mce operon is considered to be complex and not completely understood. Although expression of mce operon at different in vitro growth phases has been studied earlier, its expression in different M. tuberculosis isolates under different growth phases is not yet studied. The present preliminary study was conducted on a limited number of isolates to know the trend of expression pattern of mce operon genes in different M. tuberculosis isolates under different growth stages.
Methods:
In this study, we monitored the transcriptional profile of selected mce operon genes (mce1A, mce1D, mce2A, mce2D, mce3A, mce3C) in different M. tuberculosis isolates (MDR1, MDR2, and sensitive isolate) at early exponential and stationary phases using real-time quantitative PCR.
Results:
The expression ratio of all selected mce operon genes in all M. tuberculosis isolates was reduced at the initial phase and increased substantially at a later phase of growth. Higher expression of mce1 operon genes was found in all M. tuberculosis isolates as compared to other mce operon genes (mce2 and mce3 operons) at stationary growth phase.
Interpretation & conclusions:
The higher expression of mce operon genes at stationary phase (as compared to early exponential phase) suggested growth phase dependent expression of mce operon genes. This indicated that the mce operon genes might have a role in M. tuberculosis survival and adaptation on the onset of adverse condition like stationary phase. Identification of differentially expressed genes will add to our understanding of the bacilli involved in adaptation to different growth conditions.
Keywords: Drug resistance, gene expression, mammalian cell entry operon, Mycobacterium tuberculosis, real-time PCR
The emergence of HIV-TB co-infection and multi-drug resistant cases of TB has increased the severity and magnitude of the TB epidemic. There is limited information about the behaviour and virulence factors of drug resistant isolates of Mycobacterium tuberculosis (M. tuberculosis). In the pathogenesis of M. tuberculosis the most critical step is the entry into the host cells (macrophages) and its intracellular growth and survival1,2. The survival of bacterium in host cells and also its adaptation to different physiological conditions (in vivo and in vitro) depend on expression of different sets of genes3. Therefore, study of M. tuberculosis genes responsible for bacilli adaptation to different physiological conditions (virulence property) would be of great importance to understand the pathogenesis of tuberculosis.
The mammalian cell entry (mce) genes encode for invasive/adhesive cell surface proteins and possibly have a role in invasion of host cells during the early events of infection and survival of M. tuberculosis in macrophages4,5,6,7,8. The whole genome sequencing of M. tuberculosis revealed the presence of four dispersed but homologous mce operons (mce1-4) arranged in a nearly identical manner and having two integral proteins (yrb EA-B) and six mce genes (mceA-F)5,8,9. The presence or absence of mce operon does not largely determine the pathogenicity9,10,11 however, its characteristic expression may be of importance in the virulence. Earlier, inactivation of mce2 operon in M. bovis BCG has been shown to reduce its ability to invade non-phagocytic HeLa cells indicating a role of mce2 operon-encoded proteins in virulence12. mce operons are known to be expressed as polycistronic transcripts and their expression profiles vary in a growth phase specific manner both in liquid and solid culture13,14.
The genetic basis of drug resistance has largely been studied and candidate genes along with mutations have been identified15. However, it has also been found that a proportion of drug resistant isolates of M. tuberculosis do not have the expected mutations suggesting the involvement of some other mechanism(s) responsible for resistance16,17. Moreover, M. tuberculosis grows under various stress conditions within cells, and these conditions may provide a hypermutable background, making resistance more likely. The genetic variations in mce operon genes (mce1 and 4) have been studied in clinical isolates of M. tuberculosis and high level of polymorphism of operon genes has been reported in drug sensitive isolates as compared to resistant18. As the levels of expression of the mce operon genes in different M. tuberculosis isolates during different growth (in vitro) stages might be necessary for evaluating their putative functions, virulence mechanism, etc., therefore, we explored the mce operon genes (mce1A, mce1D, mce2A, mce2D, mce3A, mce3C) expression analysis in different M. tuberculosis isolates at early exponential and stationary growth phases (in vitro) by quantitative real-time PCR.
Material & Methods
Bacterial strains: This study was conducted in the Microbiology and Molecular Biology department of National JALMA Institute for Leprosy and Other Mycobacterial Diseases (NJILOMD), Agra, India. A total of four M. tuberculosis isolates referred to as ‘MDR1’, ‘MDR2’, ‘S’ and a standard strain ‘H37Rv’ (TMC102, taken from our Institute repository) were included in the study. All isolates were confirmed as M. tuberculosis by standard biochemical tests15, and were reconfirmed by PCR-RFLP (restriction fragment length polymorphism) detection based on primers targeting hsp 65 KD19 and 1.8kb20. All M. tuberculosis isolates were subjected to drug susceptibility testing by standard L-J (Lowenstein-Jensen) proportion method21 against four first-line antitubercular drugs [0.2 µg/ml of isoniazid (INH),4 µg/ml of streptomycin (STR), 40 µg/ml of rifampicin (RIF) and 2 µg/ml of ethambutol (EMB)]. Based on the drug susceptibility testing ‘MDR1’ was resistant to rifampicin, isoniazid, ethambutol and streptomycin whereas ‘MDR2’ was resistant to only rifampicin and isoniazid. Similar to standard H37Rv strain, ‘S’ was also susceptible to all four drugs, and was used as absolute control and expression of targeted operon genes in H37Rv was used to normalize the expression of the same genes in other three selected isolates of different drug sensitivity profile.
Culture and harvesting: All M. tuberculosis isolates were grown in Middlebrook 7H9 broth supplemented with albumin dextrose catalase (ADC supplement) (Difco, USA) and 1 per cent glycerol at 37°C as a primary culture. Using the 10 per cent of primary culture, secondary culture was cultivated into 500 ml screw cap conical flasks, each having 100 ml of medium in shaking condition. Cells were grown below an OD600 of 0.3. The bacilli were recovered at early exponential phase (OD600 of 0.8) and stationary phase (OD600 of 2.0) and centrifuged at 6000 g for 10 min at 4°C. The pellet was washed twice with chilled 1x phosphate buffer saline (PBS) before processing for RNA isolation.
RNA isolation and cDNA synthesis: Total RNA was obtained from growing cultures of M. tuberculosis isolates in early exponential and stationary phases of growth using commercially available Trizol reagent (Invitrogen, Darmstadt, Germany) as per manufacturer's instructions. From different growth phases, appropriate amount of culture was harvested in duplicate from three independent experiments. Complementary DNA (cDNA) was synthesized from isolated RNA in a reaction volume of 20 µl by using first strand cDNA synthesis kit (Fermentas Life sciences, Leon-Rot, Germany) using 500 ng of RNA, random hexamer (0.5 µg/µl) and avian myeloblastosis virus- reverse transcriptase (AMV-RT) (10 U/µl).
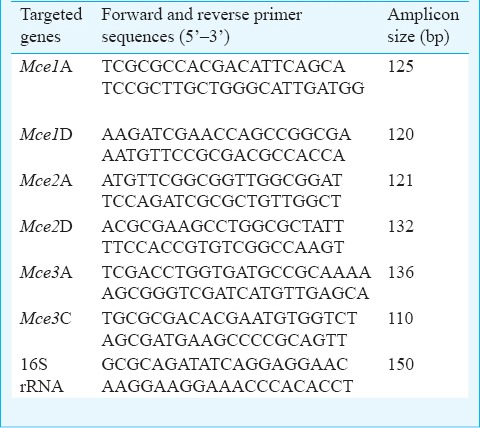
Quantitative real-time PCR and data analysis: A total of six genes (mce1A, mce1D, mce2A, mce2D, mce3A, mce3C) of mce 1, 2 and 3 operons were studied using self designed primers (using Primer 3 software, www.frodo.wi.mit.edu/primer3/) commercially procured from Sigma Aldrich, Bengaluru, India. The nucleotide sequences of each primer are given in the Table. Each 10 μl real-time PCR reaction in SYBR Green I format consisted of 5 μl of SYBR green mix, 0.5 μl forward primer (0.5 μM), 0.5 μl reverse primer (0.5 μM), 2 μl nuclease-free PCR grade water and 2 μl cDNA (0.5 μg) was added in a DEPC (diethylpyrocarbonate) treated microcentrifuge tube. The reaction mixture was further subjected to LightCycler 480II instrument (Roche Diagnostics, Mannheim, Germany) and fluorescent data were acquired during each extension phase. PCR cycling conditions were similar as 95°C, 10 min (95°C 10 sec, 62°C 15 sec, 72°C 12 sec) X 45 cycles 72°C, 2 min annealing temperature for each primer. In each set of reactions, fold changes were described in the form of threshold cycle (Ct) and the transcript levels of the target genes of the three M. tuberculosis isolates were normalized with the reference gene 16S rRNA and then normalized to the values obtained from control strain H37Rv. To analyse the quantitative differences in gene expression in all the selected genes at both phases and also among different isolates of M. tuberculosis in comparison to control strain, logarithmic graphics of the fold change expression of each gene were created.
Table.
Primers used for quantitative real-time PCR
The relative expression ratio was calculated from triplicate normalized ratios for each gene, and quantitative real-time PCR data were analyzed by using the 2-(ΔΔCt) method as described by Livak and Schmittgen22. Relative expression (in fold change) of targeted gene of ‘MDR 1’, ‘MDR 2’ and ‘S’ was estimated using 16S rRNA gene as a control (for calculation of Ct value) and same gene of H37Rv strain as calibrator (for calculation of ΔΔCt). The logarithmic graphs depicting fold change in expression of each targeted gene of different isolates were prepared. The significant difference of gene expression during exponential and stationary phase was compared using student t test and significant differences in the expression of the gene in the isolate were analysed using one way ANOVA by Graph pad prism (Version 5) software (San Diego, California, USA).
Results
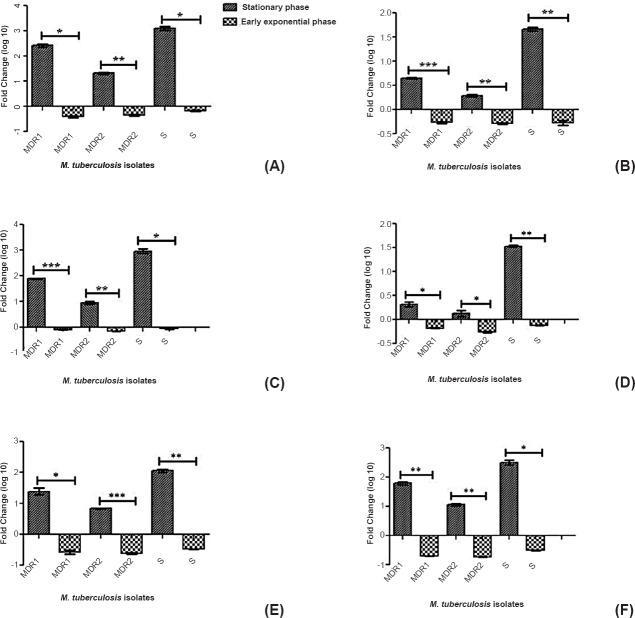
The relative expression of all selected mce operon genes (mce1A, mce1D, mce2A, mce2D, mce3A, mce3C) was significantly higher at stationary phase as compared to early exponential phase in all M. tuberculosis isolates (MDR1, MDR2 and sensitive isolates) (Fig. 1). We selected first (A) and middle genes (C, D) of each mce operon (mce1-3) as a representative of operon to evaluate their level of expression by quantitative methods.
Fig. 1.
Relative expression (Fold change in logarithmic graphics) analysis of mce1A gene (A), mce1D gene (B), mce2A gene (C), mce2D gene (D), mce3A gene (E), mce3C gene (F) of M. tuberculosis isolates (multi drug resistant and sensitive) during two different growth phases. The relative expression (analyzed by student t test) of all mce operon genes was significantly higher in stationary phase as compared to early exponential phase (P<0.001) for all the three isolates. MDR1-Multi-drug resistant isolate (resistant to rifampicin, isoniazid, ethambutol streptomycin), MDR 2- Multi drug resistant isolate (resistant to both isoniazid and rifampicin), S, sensitive isolate. P*<0.05, **<0.01, ***<0.001. Values are mean ±SD of three experiments.
The relative expression of mce1 operon genes was significantly lower during early exponential phase (mce1A gene-0.394 ± 0.0403, 0.454± 0.031, 0.676 ± 0.029 and mce1D gene-0.543 ± 0.021, 0.521 ± 0.015, and 0.535 ± 0.060 in MDR1, MDR2, sensitive isolates, respectively) as compared to stationary phase (mce1A gene-262.9 ± 41.08, 21.8 ± 1.174, 1257 ± 227 fold and mce1D gene-4.48 ± 0.155, 1.93 ± 0.113, 46.3 ± 3.627 fold in MDR1, MDR2, sensitive isolates, respectively) and both genes differed significantly for both phase in all the three isolates. (P<0.01, <0.01, <0.05, respectively). The relative expression of mce2 operon genes was lower in early exponential phase (mce2A gene-0.824 ± 0.040, 0.729 ± 0.004, 0.882 ± 0.008 fold and mce2D gene- 0.659 ± 0.006, 0.547 ± 0.021, 0.760 ± 0.011 fold in MDR1, MDR2, sensitive isolates, respectively) compared to the stationary phase (mce2A gene -79.4 ± 3.111, 9.03 ± 1.146, 932.6 ± 190.9 fold and mce2D gene- 2.07± 0.247, 1.33 ± 0.190, 33.9 ± 1.499 fold in MDR1, MDR2, sensitive isolates, respectively) and both genes differed significantly for both phase in all the three isolates. (P<0.05 for all). The relative expression of mce3 operon genes was lower in early exponential phase (mce3A gene-0.267 ± 0.0417, 0.240 ± 0.008, 0.333 ± 0.001 and mce3C gene-0.203 ± 0.0021, 0.185 ± 0.0035, 0.316 ± 0.0056 in MDR1, MDR2, sensitive isolates, respectively) as compared to stationary phase (mce3A-25.06 ± 6.20, 6.84 ± 0.0989, 115.3 ± 12.97 and mce3C gene-62.4 ± 6.718, 11.5 ± 1.188, 318.5 ± 54.39 in MDR1, MDR2, sensitive isolates, respectively) and both genes differed significantly for both phase in all the three isolates (P<0.05, <0.001 and <0.01, respectively).
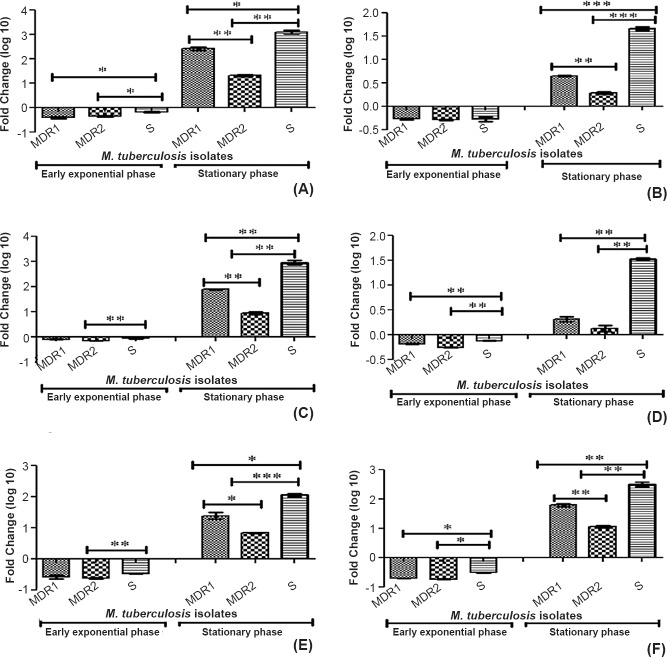
The ANOVA analysis showed a significantly higher expression of all the studied genes (mce1A, mce1D, mce2A, mce2D, mce3A, mce3C) in stationary phase in all the three selected isolates (MDR1, MDR2, sensitive isolates) (P<0.001 for all the three isolates). Further, to analyse the significance of the genes of a particular isolate (MDR1, MDR2, sensitive isolate separately) one way ANOVA analysis was performed, which showed a significant difference in expression between the genes of the same operon and genes of different operon in a single isolate. The mce1A gene (mce1 operon) showed a significantly higher expression in extracellular growth at stationary phase as compared to mce2A (P<0.001) and mce3A (P<0.001) in all the three M. tuberculosis isolates (MDR1, MDR2, sensitive isolates). The mce3C gene (mce3 operon) showed a higher expression (non significant) in extracellular growth as compared to mce1D and mce2D genes at stationary growth phase in MDR1 and sensitive M. tuberculosis isolates. The ANOVA analysis showed a significantly lower expression of all the studied genes (mce1A, mce1D, mce2A, mce2D, mce3A, mce3C) in early exponential phase in all the three selected isolates (MDR1, MDR2, sensitive isolate) (P<0.001 for all the three isolates). Higher expression was observed for the first gene of each operon as compared to middle gene of the same operon (except mce3 operon genes). There was also found higher expression of all the selected mce operon genes (mce1A, mce1D, mce2A, mce2D, mce3A, mce3C) in sensitive isolate among all M. tuberculosis isolates (Fig. 2).
Fig. 2.
Relative expression (fold change in logarithmic graphics) analysis of mce1A gene (A), mce1D gene (B), mce2A gene (C), mce2D gene (D), mce3A gene (E), mce3C gene (F) of M. tuberculosis isolates (multi-drug resistant and sensitive) during two different growth stages (early-exponential and stationary phases) using gene specific primers by real time PCR. MDR1-Multi-drug resistant isolate (resistant to rifampicin, isoniazid, ethambutol), streptomycin, MDR 2- multi-drug resistant isolate (resistant to both isoniazid and rifampicin), S, sensitive isolate. P*<0.05, **<0.01, ***<0.001.
Discussion
In this study, we analysed the relative expression of mce operon genes (mce1A, mce1D, mce2A, mce2D, mce3A, mce3C) in different M. tuberculosis isolates (MDR1, MDR2, sensitive isolates) at early exponential and stationary in vitro growth conditions. We used quantitative real-time PCR to detect the transcript level more accurately as compared to previous studies3,14,23,24 where all the observations were relied mostly on the results of semi-quantitative RT-PCR and only expression of mce operon was described in terms of presence or absence of mce gene transcripts in M. tuberculosis H37Rv. Although the expression of all genes of mce1 operon during in vitro-grown M. tuberculosis in Dubos broth media (during exponential phase) through RT-PCR was demonstrated by Harboe et al23, but Kumar et al13 could not detect the transcript of mce1 in L-J slant at stationary growth phase of M. tuberculosis. This discrepancy may be due to higher inoculum size, use of different culture media as well shaking of culture broth. The impact of cultivation medium, its condition (shaking and standing), inoculum size and stress/stimulus has been reported earlier14.
In general, at an early growth stage, the expression of all selected mce operon genes showed suppression but after that increased at stationary phase of growth. Transcriptionally and metabolically activeness of stationary phase bacilli was suggested by Voskuil et al25. The higher expression of mce operon genes may also be due to possible over-expression under nutritionally limiting conditions (such as the stationary phase of culture) to take up the available nutrients and also to effectively eliminate the secondary metabolites14. Kumar et al13 have demonstrated that during simple ageing of logarithmically growing cultures (stress conditions), the metabolic activity of M. tuberculosis can be slowed down in vitro, and during these stress conditions bacilli have to be adapted to this situation by differential expression of different genes. Thus, our results indicate that mce1, mce2, mce3 operon genes might have a role in M. tuberculosis survival and adaptation on the onset of adverse condition like nutritionally depleted condition at stationary phase.
In the present study, significant variation was observed in expression of six mce genes at stationary growth stage, suggesting the levels of expression of the different mce genes might be important for evaluating their putative function. There was also higher expression of mce1A in comparison to other genes of the same operon (mce1D) or genes of other operon (mce2 and mce3 operons), suggesting that mce1 operon (specially mce1A gene) has major functional role extracellularly. A previous study also revealed that mce1 operon has higher expression in the extracellular environment as compared to within macrophages6. Studies on mce operon showed a recombinant surface protein (mce1A) of M. tuberculosis conferred upon a non-pathogenic Escherichia coli, which had the ability to enter inside the mammalian cells (HeLa cells) and survival4,7 suggesting roles of mce1 operon both in cell entry and survival. Harboe et al23 also demonstrated the expression of all six genes of mce1 operon in in vitro-grown M. tuberculosis by RT-PCR.
The differential expression profile of mce operon genes at different growth phases in this study was expected as an earlier study also showed growth phase dependent expression pattern of mce operon13. At stationary growth phase, the relative expression of all mce operon (mce1, mce2, mce3) genes remained higher for sensitive isolate among all M. tuberculosis isolates. The significant difference was observed in the expression profile of different mce operon in all the three M. tuberculosis isolates (MDR1, MDR2, sensitive isolates) at both growth phases, indicating complex regulation of mce operon genes expression. Pasrich et al18 investigated the extent of polymorphism in eight genes in the mce1 and 4 operon in 112 clinical isolates of M. tuberculosis varying in their drug susceptible profile (more polymorphic mutations in mce genes in the drug sensitive isolate as compared to resistant isolates). Since drug resistance provides an extra edge to resistant isolates, the more expression (as in present investigation) or polymorphic mutation18 of mce operon genes in susceptible isolates may also relate to virulence mechanisms after adaptability to the environment. It was further supported by Shimono et al26 who demonstrated the transformation of wild type M. tuberculosis strain into hypervirulent by disruption of mce1 operon.
In the present study, relative expression of the mce3A gene was lower at both early exponential and stationary phases as compared to mce1A and mce2A genes. This may be due to dominant role of mce1A and mce2A genes (over mce3A) in in vitro growth and stationary phase adaptation. The mce3C gene (mce3 operon) showed a higher expression (non significant) in extracellular growth as compared to mce1D and mce2D genes at stationary growth phase of bacilli in MDR1 and sensitive M. tuberculosis isolates. Overall, mce 1 operon showed significantly higher expression in extracellular growth as compared to other mce operons (mce2 and mce3). There was also higher expression for the first gene of each operon as compared to middle gene of the same operon (except mce3 operon genes). With respect to expression of two genes of the same operon, no marked variation was observed for the selected genes of mce1, 2 and 3 operons at early exponential phase. At the stationary growth stage different mRNA transcript levels were measured for the same two genes of each mce1, 2 and 3 operons. Though our experiment resulted into an unexpected expression pattern of genes of the same mce operon but all genes of any operon will express similarly, is not necessary due to varying stability of different segments of mRNA and also because some operons have internal promoters or differential regulation of mRNA stability27. In M. tuberculosis system the effect of two promoters on regulation of an operon has been demonstrated earlier that resulted into different transcripts28.
Joon et al29 reported the presence of two functional promoters for mce1 operon in M. tuberculosis that could potentially segregate different functions of a single operon. Casali et al6 also showed that the expression of mce operon might be tuned under multiple negative regulators (mce1R, negative transcriptional regulator of mce1 operon). It has been reported that Mce3R (a TetR family transcriptional regulator), downregulates the expression of mce3 operon of M. tuberculosis in vitro 30. Santangelo et al31 showed that the expression of mce3 operon of M. tuberculosis was regulated by Mce3R together with two transcriptional units, indicating a functional relation between the products encoded in the three operons. A previous study suggested differential expression of mce operon under different environmental and experimental conditions13. These findings suggest that mce operon regulation for M. tuberculosis may be more complex than one would expect for a prokaryotic system. Therefore, genes of the same mce operon, probably under the control of different regulators can express differentially.
In conclusion, the differential expression of mce operon genes at two different growth stages suggested growth phase dependent expression of mce genes, serving in adaptation of bacilli in different environmental conditions. The differences in expression of mce operon genes among M. tuberculosis isolates also indicated the possible association of mce genes expression with genetic context of the isolates (including drug resistance profile). Variation in expression of different mce operons at different growth phases is thought to be essential for bacterial survival and also variation in mce operon gene expression among M. tuberculosis clinical isolates has implications for pathogencity. Identification of such differentially expressed mce genes may be important for drug targets, vaccine antigens. The expression analysis of mce operon genes in different M. tuberculosis isolates at different growth stages may enrich our understanding about its role in its virulence.
Acknowledgment
The first author (PS) thanks Lady TATA Memorial Trust, Mumbai, for providing senior research fellowship (SRF). Authors acknowledge the support and help of Drs Ekata Sinha, Pravin Kumar Singh, PVJ Reddy and Shri Ravi.
Conflicts of Interest: None.
References
- 1.Aderem A, Underhill DM. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 2.Glickman MS, Jacobs WR., Jr Microbial pathogenesis of Mycobacterium tuberculosis: dawn of a discipline. Cell. 2000;104:477–85. doi: 10.1016/s0092-8674(01)00236-7. [DOI] [PubMed] [Google Scholar]
- 3.Timm J, Post FA, Bekker LG, Walther GB, Wainwright HC, Manganelli R, et al. Differential expression of iron -, carbon -, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc Natl Acad Sci USA. 2003;100:14321–6. doi: 10.1073/pnas.2436197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arruda S, Bomfim G, Knights R, Huima-Byron T, Riley LW. Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science. 1993;261:1454–7. doi: 10.1126/science.8367727. [DOI] [PubMed] [Google Scholar]
- 5.Casali N, Konieczny M, Schmidt MA, Riley LW. Invasion activity of a Mycobacterium tuberculosis peptide presented by the Escherichia coli AIDA autotransporter. Infect Immun. 2002;70:6846–52. doi: 10.1128/IAI.70.12.6846-6852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casali N, White AM, Riley LW. Regulation of the Mycobacterium tuberculosis mce1 Operon. J Bacteriol. 2006;188:441–9. doi: 10.1128/JB.188.2.441-449.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chitale S, Ehrt S, Kawamura I, Fujimura T, Shimono N, Anand N, et al. Recombinant Mycobacterium tuberculosis protein associated with mammalian cell entry. Cell Microbiol. 2001;3:247–54. doi: 10.1046/j.1462-5822.2001.00110.x. [DOI] [PubMed] [Google Scholar]
- 8.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 9.Tekaia F, Gordon SV, Garnier T, Brosch R, Barrell BG, Cole ST. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber Lung Dis. 1999;79:329–42. doi: 10.1054/tuld.1999.0220. [DOI] [PubMed] [Google Scholar]
- 10.Haile Y, Caugant DA, Bjune G, Wiker HG. Mycobacterium tuberculosis mammalian cell entry operon (mce) homologs in Mycobacterium other than tuberculosis (MOTT) FEMS Immunol Med Microbiol. 2002;33:125–32. doi: 10.1111/j.1574-695X.2002.tb00581.x. [DOI] [PubMed] [Google Scholar]
- 11.Wiker HG, Spierings E, Kolkman MA, Ottenhoff TH, Harboe M. The mammalian cell entry operon 1 (mce1) of Mycobacterium leprae and Mycobacterium tuberculosis. Microb Pathog. 1999;27:173–7. doi: 10.1006/mpat.1999.0298. [DOI] [PubMed] [Google Scholar]
- 12.Flesselles B, Anand NN, Remani J, Loosmore SM, Klein MH. Disruption of the mycobacterial cell entry gene of Mycobacterium bovis BCG results in a mutant that exhibits a reduced invasiveness for epithelial cells. FEMS Microbiol Lett. 1999;177:237–42. doi: 10.1111/j.1574-6968.1999.tb13738.x. [DOI] [PubMed] [Google Scholar]
- 13.Kumar A, Bose M, Brahmachari V. Analysis of expression profile of mammalian cell entry (mce) operon of Mycobacterium tuberculosis. Infect Immun. 2003;71:6083–7. doi: 10.1128/IAI.71.10.6083-6087.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar A, Chandolia A, Chaudhry U, Brahmachari V, Bose M. Comparison of mammalian cell entry operon of mycobacteria: in silico analysis and expression profiling. FEMS Immunol Med Microbiol. 2005;43:185–95. doi: 10.1016/j.femsim.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 15.Vestal AL. Procedures for isolation and identification of mycobacteria. CDC Atlanta, Georgia: US Department of Health, Education and Welfare; 1977. Isolation procedures and incubation and microscopy and staining; pp. 23–39. Publication No. 77-8230. [Google Scholar]
- 16.Cole ST, Telenti A. Drug resistance in Mycobacterium tuberculosis. Eur Respir J. 1995;20 (Suppl):701s–13s. [PubMed] [Google Scholar]
- 17.Morris S, Bai GH, Suffys P, Portillo-Gomez L, Fairchok M, Rouse D. Molecular mechanisms of multiple drug resistance in clinical isolates of Mycobacterium tuberculosis. J Infect Dis. 1995;171:954–60. doi: 10.1093/infdis/171.4.954. [DOI] [PubMed] [Google Scholar]
- 18.Pasricha R, Chandolia A, Ponnan P, Saini NK, Sharma S, Chopra M, et al. Single nucleotide polymorphism in the genes of mce1 and mce4 operon of Mycobacterium tuberculosis: analysis of clinical isolates and standard reference strains. BMC Microbiol. 2011;11:41. doi: 10.1186/1471-2180-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Telenti A, Marchesi F, Balz M, Bally F, Böttger EC, Bodmer T. Rapid identification of Mycobacteria to the species level by PCR and restriction enzyme analysis. J Clin Microbiol. 1993;31:175–8. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katoch VM, Parashar D, Chauhan DS, Singh D, Sharma VD, Ghosh S. Rapid identification of mycobacteria by gene amplification restriction analysis technique targeting 16S - 23S ribosomal RNA internal transcribed spacer & flanking region. Indian J Med Res. 2007;125:155–62. [PubMed] [Google Scholar]
- 21.Canetti G, Fox W, Khomenko A, Mahler HT, Menon NK, Mitchison DA, et al. Advances in techniques of testing mycobacterial drug sensitivity, and the use of sensitivity tests in tuberculosis control programmes. Bull World Health Organ. 1969;41:21–43. [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-delta delta C (T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Harboe M, Christensen A, Haile Y, Ulvund G, Ahmad S, Mustafa AS, et al. Demonstration of expression of six proteins of the mammalian cell entry (mce1) operon of Mycobacterium tuberculosis by anti-peptide antibodies, enzyme-linked immunosorbent assay and reverse transcription-polymerase chain reaction. Scand J Immunol. 1999;50:519–27. doi: 10.1046/j.1365-3083.1999.00632.x. [DOI] [PubMed] [Google Scholar]
- 24.Ahmad S, El-Shazly S, Mustafa AS, Al-Attiyah R. The six mammalian cell entry proteins (Mce3A-F) encoded by the mce3 operon are expressed during in-vitro growth of Mycobacterium tuberculosis. Scand J Immunol. 2005;62:16–24. doi: 10.1111/j.1365-3083.2005.01639.x. [DOI] [PubMed] [Google Scholar]
- 25.Voskuil MI, Visconti KC, Schoolnik GK. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis. 2004;84:218–27. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Shimono N, Morici L, Casali N, Cantrell S, Sidders B, Ehrt S, et al. Hypervirulent mutant of Mycobacterium tuberculosis resulting from disruption of the mce1 operon. Proc Natl Acad Sci USA. 2003;100:15918–23. doi: 10.1073/pnas.2433882100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adhya S. Suboperonic Regulatory Signals. Sci STKE 2003. 2003:pe 22. doi: 10.1126/stke.2003.185.pe22. [DOI] [PubMed] [Google Scholar]
- 28.Master S, Zahrt TC, Song J, Deretic V. Mapping of Mycobacterium tuberculosis katG promoters and their differential expression in infected macrophages. J Bacteriol. 2001;183:4033–9. doi: 10.1128/JB.183.13.4033-4039.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joon M M, Bhatia S, Pasricha R, Bose M, Brahmachari V. Functional analysis of an intergenic non-coding sequence within mce1 operon of M. tuberculosis. BMC Microbiol. 2010;10:128. doi: 10.1186/1471-2180-10-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santangelo MP, Goldstein J, Alito A, Gioffre A, Caimi K, Zabal O, et al. Negative transcriptional regulation of the mce3 operon in Mycobacterium tuberculosis. Microbiology. 2002;148:2997–3006. doi: 10.1099/00221287-148-10-2997. [DOI] [PubMed] [Google Scholar]
- 31.Santangelo MP, Klepp L, Nunez-Garcia J, Blanco FC, Soria M, Garcıa-Pelayo MC, et al. Mce3R, a TetR-type transcriptional repressor, controls the expression of a regulon involved in lipid metabolism in Mycobacterium tuberculosis. Microbiology. 2009;155:2245–55. doi: 10.1099/mic.0.027086-0. [DOI] [PubMed] [Google Scholar]