Abstract
Several types of pediatric cancers reportedly contain high frequency missense mutations in histone H3, yet the underlying oncogenic mechanism remains poorly characterized. Here, we report that the H3 lysine 36 to methionine (H3K36M) mutation impairs the differentiation of mesenchymal progenitor cells and generates undifferentiated sarcoma in vivo. H3K36M mutant nucleosomes inhibit the enzymatic activities of several H3K36 methyltransferases. Depleting H3K36 methyltransferases, or expressing an H3K36I mutant that similarly inhibits H3K36 methylation, is sufficient to phenocopy the H3K36M mutation. Following the loss of H3K36 methylation, a genome-wide gain in H3K27 methylation leads to a redistribution of Polycomb Repressive Complex 1 and de-repression of its target genes known to block mesenchymal differentiation. Our findings are mirrored in human undifferentiated sarcomas where novel K36M/I mutations in H3.1 are identified.
Somatic missense mutations in histone H3 genes were recently identified in pediatric brain and bone malignancies (1-3). These include lysine 27 to methionine (K27M) and glycine 34 to arginine/valine (G34R/V) mutations found in pediatric gliomas, glycine 34 to tryptophan/leucine (G34W/L) mutations found in giant cell tumors of the bone, and lysine 36 to methionine (K36M) mutations reported in ~95% of chondroblastomas. These cancer-associated H3 mutations (so-called "oncohistones") map to, or close to, well-known sites of histone post-translational modifications (PTMs) such as methylation of H3K27 and H3K36. The highly clustered mutational patterns and only 1 of the 32 alleles encoding human histone H3 being mutated in tumors imply that the mutations are dominant or neomorphic. Moreover, the exquisite tumor type specificity suggests that oncohistones derived from different tissues-of-origin are likely to be functionally and mechanistically distinct. While progress has been made in understanding the oncogenic effect of glioma-associated histone mutations (4-8), their role in bone and cartilage neoplasms remains unclear.
We focused on the chondroblastoma-derived H3K36M mutation. Chondroblastoma is characterized by the accumulation of immature chondroblasts (9), suggesting that the neoplasm may arise from uncontrolled proliferation and/or incomplete differentiation of mesenchymal progenitor cells (MPCs). We created isogenic C3H10T1/2 murine MPCs that stably express either wild-type or K36M mutant H3.3 at pathologically relevant levels (hereafter referred to as H3.3WT or H3.3K36M cells, respectively) (fig. S1, A and B). Differentially expressed genes between H3.3WT and H3.3K36M cells were highly enriched for pathways involved in cellular and developmental processes (fig. S1, C and D and table S1), suggesting that H3.3K36M mutation induces aberrant expression of genes involved in cell differentiation. While H3.3WT and H3.3K36M cells displayed comparable proliferation rates (fig. S1E), H3.3K36M cells exhibited significantly diminished chondrocytic differentiation capacity (Fig. 1A). H3.3K36M cells also showed a decrease in mRNA expression of mature chondrocyte markers (Col2a1, Col9a1, Col11a1 and Acan) before and after differentiation induction (Fig. 1B). Expressing the H3.3K36M mutation in murine pre-chondrocytic ATDC5 cells (10) also significantly inhibited chondrocyte differentiation (fig. S1F).
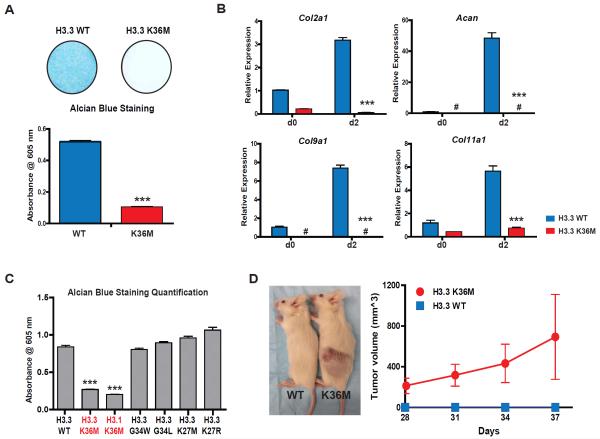
Fig. 1. H3K36M mutation impairs mesenchymal differentiation and generates undifferentiated sarcoma.
(A) Representative Alcian blue staining images of cell culture wells after 9 days of chondrocyte differentiation. The bar graph shows the quantification of staining in cell extract. (B) The mRNA expression of Acan, Col2a1, Col9a1 and Col11a1 in cells before (d0) and two days after (d2) chondrocyte differentiation induction. (C) Quantification of Alcian blue staining in cell extract after 9 days of chondrocyte differentiation. (D) Measurement of subcutaneous tumor growth of H3.3WT or H3.3K36M cells. A representative image of mice implanted with cells at the time of sacrifice is shown. In (D), error bars indicate standard deviation (n=10 mice in each group). In other experiments, error bars indicate standard deviation from three biological replicates. #, undetectable; ***, p<0.001.
Oncohistones found in other cancer types (H3.3K27M or H3.3G34W/L) had little impact on chondrocyte differentiation of MPCs (Fig. 1C and fig. S2A). These results are consistent with the tissue specificity of histone mutations identified in human cancers. Expression of H3.1K36M in MPCs altered the transcriptome and inhibited chondrocyte differentiation to a similar extent as H3.3K36M (Fig. 1C and fig. S2, A and B). The presence of the K36M mutation did not affect the genome-wide deposition profiles of H3.1 and H3.3 (fig. S2, C to E) (11).
Whole-genome sequencing of chondroblastomas revealed H3.3K36M as the only recurrent mutation (3). We sought to determine whether the differentiation arrest induced by the H3K36M mutation is sufficient to facilitate tumor development in vivo. Subcutaneous injection of MPCs stably expressing H3.3 or H3.1 K36M, but not wild-type H3, generated tumors in severe combined immunodeficiency (SCID) mice (Fig. 1D and fig. S3A). These tumors resembled human undifferentiated sarcomas (fig. S3B), retained the expression of mutant histones and shared a similar gene expression profile with cells cultured in vitro (fig. S2B and fig. S3C).
Although H3K36M mutations are uncommon in bone and cartilage tumors other than chondroblastomas (3), our in vivo modeling results suggest that K36M mutations in H3.3 or H3.1 may exist in poorly differentiated soft tissue tumors. Pediatric undifferentiated soft tissue sarcomas are rare but aggressive tumors with poor prognosis and undefined mutational landscape (12). Through screening a panel of ten pediatric undifferentiated soft tissue sarcomas, we identified one tumor carrying an H3.1K36M mutation and confirmed its protein expression (fig. S4).
The development of undifferentiated sarcomas by H3K36M mutant MPCs prompted us to determine whether the mutation interferes with the differentiation of these cells towards other lineages. H3.3K36M cells exhibited a significant block of differentiation to adipocytes and osteocytes (fig. S5, A and B). Moreover, the expression of master regulators of adipogenesis and osteogenesis was decreased in H3.3K36M cells, in addition to regulators of chondrocyte differentiation. H3.3K36M cells also displayed enhanced expression of transcription factors involved in the maintenance of mesenchymal multipotency (fig. S5C).
We found that the H3K36M transgene caused a marked reduction in H3K36me2/3 and a concomitant increase in H3K27me2/3 in various cell types (Fig. 2A and fig. S6, A to C). Immunoblots of purified heterotypic mononucleosomes containing epitope-tagged H3.3 and endogenous H3 revealed that K36M-containing nucleosomes displayed decreased H3K36me2/3 and increased H3K27me3 on the endogenous wild-type H3 (fig. S6D). Co-immunostaining of murine H3.3K36M tumors demonstrated an inverse correlation between H3.3K36M expression and levels of H3K36me3 (fig. S6E). Chondroblastoma samples carrying the H3.3K36M mutation exhibited decreased H3K36me2/3 and increased H3K27me3 compared to H3 wild-type chondroblastoma or chondrosarcomas (fig. S7, A and B). IHC staining revealed that H3.3K36M-expressing chondroblastoma cells, but not concomitant non-neoplastic multinuclear osteoclasts (3), H3 wild-type chondroblastoma or chondrosarcomas, were negative for H3K36me3 (fig. S7, C and D).
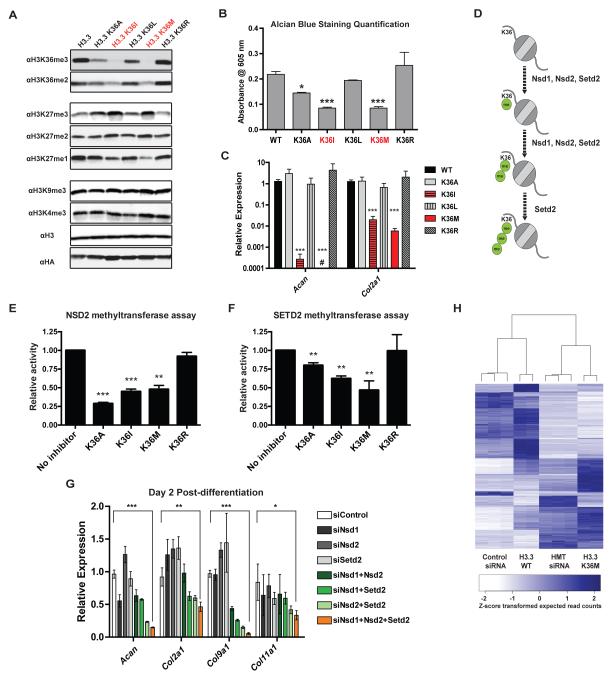
Fig. 2. H3K36 mutations dominantly inhibit H3K36 methyltransferases.
(A) Immunoblots of lysates generated from 293T cells expressing FLAG and HA-tagged wild-type (WT) or K36A/I/L/M/R mutant H3.3. (B) Quantification of Alcian blue staining in cell extract after 9 days of chondrocyte differentiation. (C) The mRNA expression of Acan and Col2a1 in cells after two days of chondrocyte differentiation. (D) Schematic introduction of mammalian H3K36 methyltransferases. (E) and (F) In vitro methyltransferase assays with recombinant human full-length NSD2 (E) or SET domain of SETD2 (F), recombinant substrate nucleosomes and tritiated SAM. Various K36 mutant H3(27-47) peptides were added to the reaction mixture. For NSD2 assays, the final concentration of inhibitor peptides was 30 μM, for SETD2 assays 50 μM. Methyltransferase activity was quantified via scintillation and normalized. (G) MPCs were transfected with siRNA for 3 days, followed by chondrocyte differentiation induction for 2 days. The mRNA expression of Acan, Col2a1, Col9a1 and Col11a1 was measured. (H) Heatmap showing the expression pattern of greater than 4-fold differentially expressed genes between cells treated with siRNAs against Nsd1/2 and Setd2 (HMT siRNA) or control siRNA. For all experiments, error bars indicate standard deviation from three biological replicates. #, undetectable; *, p<0.05; **, p<0.01; ***, p<0.001.
The capacity of various H3K36 mutants to impair chondrocyte differentiation of MPCs was correlated to the magnitude of changes in H3K36 and H3K27 methylation (Fig. 2, A to C and fig. S8A). In particular, the H3K36I mutation recapitulated H3K36M's impact on histone methylation and gene expression (Fig. 2A and fig. S8, A to C). Furthermore, an H3.1K36I mutation was identified in a pediatric undifferentiated soft tissue sarcoma from the same patient cohort where we found the H3.1K36M mutation (fig. S4 and table S2). Therefore, H3K36 hypomethylation and H3K27 hypermethylation appear to be characteristic features shared by the oncogenic H3K36M/I mutations.
Gliomas containing the H3K27M mutation exhibit low levels of H3K27 methylation, and we demonstrated that this loss of methylation occurred primarily through inhibition of the Polycomb Repressive Complex 2 (PRC2) histone methyltransferase (6). As the catalytic mechanism is highly conserved among SET domain-containing histone methyltransferases, we hypothesized that H3K36M/I mutations inhibit their cognate methyltransferases. Unlike H3K27, methylation of H3K36 is catalyzed by several methyltransferases including Nsd1 and Nsd2, which catalyze H3K36me1/2, and Setd2, which catalyzes H3K36me1/2/3 (13) (Fig. 2D). Methyltransferase assays with peptides or purified nucleosomes demonstrated that H3K36M/I, but not H3K36R, were potent inhibitors of SETD2 and NSD2 activity (Fig. 2, E and F and fig. S9, A to C). H3K36A significantly inhibited NSD2 activity while exerting modest inhibition of SETD2 (Fig. 2, E and F and fig. S9D), consistent with its observed effects on cellular H3K36me2/3 (Fig. 2A and fig. S8A). While expression of the H3K36 methyltransferases was not reduced in H3.3K36M cells, we found these proteins markedly enriched in immunoprecipitates from H3K36M-containing mononucleosomes (fig. S10, A and B), suggesting that the global H3K36 hypomethylation in H3.3K36M cells results from the dominant sequestration and inhibition of methyltransferases by mutant nucleosomes.
To assess the contribution of methyltransferase inhibition to the H3K36M-induced differentiation arrest, Nsd1, Nsd2 and Setd2 were depleted separately or in combination in MPCs using small interfering RNA (siRNA). Depletion of Nsd1, Nsd2 and Setd2 in combination, but not individually, impaired the chondrocyte differentiation of MPCs, with the most dramatic effect observed in cells treated with siRNAs against all three methyltransferases (Fig. 2G and fig. S10C). In addition, clustering analysis revealed that transcriptome changes induced by H3.3K36M could be largely recapitulated via the combined knockdown of Nsd1/2 and Setd2 (Fig. 2H). As anticipated, the individual knockdown of Nsd1/2 and Setd2 led to specific decreases in H3K36me2 and H3K36me3, respectively (fig. S10C). Cells depleted for all H3K36-directed methyltransferases not only displayed reduced H3K36me2/3, but also showed elevated levels of H3K27me3 comparable to that in H3.3K36M cells, indicating that the gain of H3K27me3 in H3.3K36M cells only occurs upon global loss of both H3K36me2 and H3K36me3. Nucleosomes containing H3K36me2/3 are poor substrates for PRC2 in vitro (14, 15). Our data are consistent with a model whereby K36M-mediated loss of H3K36 methylation provides new nucleosomal substrates for PRC2, which results in increased H3K27me3 levels.
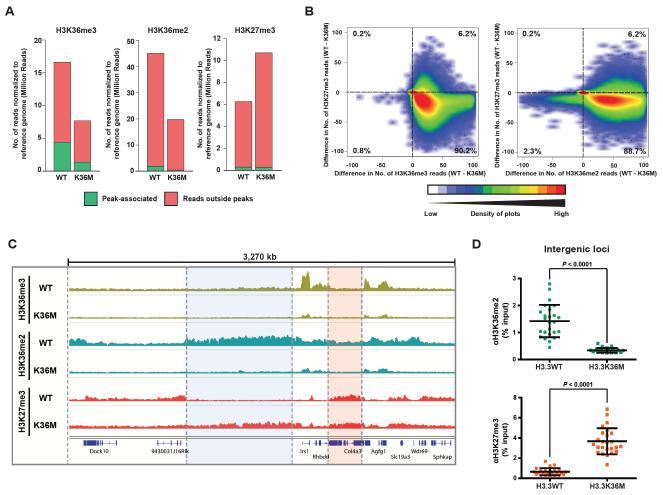
We next used chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq) to profile the chromatin landscape in H3.3K36M cells. Because traditional ChIP-seq methods preclude direct comparison of amplitudinal signals between samples, we adopted a strategy that utilizes exogenous reference chromatin (ChIP-Rx) as internal normalization control to achieve quantitative analysis of H3K27me3 and H3K36me2/3 (fig. S11A) (16). We observed a genome-wide loss of H3K36me2/3 and a concomitant increase in H3K27me3 in H3.3K36M cells (Fig. 3, A and B). While the levels of gene-associated H3K27me3 remained unchanged, we observed a specific gain of H3K27me3 at intergenic regions previously devoid of H3K27me3 and marked by H3K36me2/3 (Fig. 3, C and D and fig. S11B). Highly similar genome-wide changes in H3K36me2 and H3K27me3 were observed in H3.1K36M cells (fig. S11, C and D), which were independent of the mutant nucleosome localization (fig. S12A). Depletion of Nsd1/2 and Setd2 was sufficient to mimic K36M’s effects on the intergenic loss of H3K36me2 and gain of H3K27me3 (fig. S12B). As a consequence of increased nucleosomal substrate availability, levels of chromatin-bound Ezh2 and Suz12 were elevated in H3.3K36M cells (fig. S12, C and D).
Fig. 3. Genome-wide profiling of H3K36me2/3 and H3K27me3 in H3.3WT and H3.3K36M cells.
(A) After normalization, the number of H3K36me2/3 and H3K27me3 reads that are peak-associated or outside peaks are shown. (B) Scatter plot showing the correlation between genome-wide changes in H3K27me3 and H3K36me2/3. The genome was divided into 10,000 bp bins, and the difference in number of normalized ChIP-seq reads of H3K36me2/3 or H3K27me3 between H3.3WT and H3.3K36M cells was plotted. The percentage of bins found in each quadrant is shown. (C) Genome viewer representations of normalized ChIP-seq reads for H3K36me2/3 and H3K27me3 at a 3,270 kb region. Refseq genes are annotated at the bottom. (D) The enrichment (% input) of H3K36me2 and H3K27me3 at various intergenic regions was measured with ChIP-qPCR. Each data point represents a genomic locus.
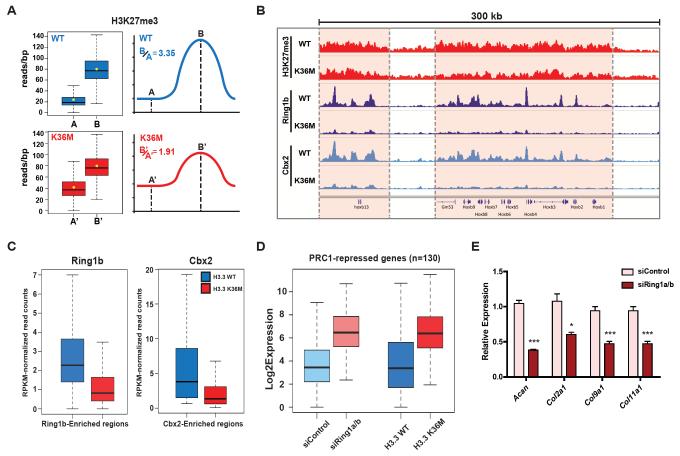
The increase in H3K27me3 at intergenic regions led to a decreased ratio of gene-associated to intergenic H3K27me3 (Fig. 4A). Genes marked by high H3K27me3 enrichment showed loss of H3K27me3 relative to the genome-wide average and increased expression (fig. S13). We hypothesized that recruitment of the H3K27me3 "readers" may be altered in H3.3K36M cells due to the gain of intergenic H3K27me3 and the relative loss of H3K27me3 enrichment at genes. The localization of Ring1b and Cbx2, integral components of the canonical PRC1 complex that binds to H3K27me3 and suppresses gene expression, was markedly decreased at their bound genes in H3.3K36M cells or following knockdown of H3K36 methyltransferases (Fig. 4, B and C and fig. S14, A and B), even though protein levels and chromatin association of PRC1 complex remained unchanged (fig. S12C). Increased enrichment of H2AK119ub1 was found at intergenic loci that gained H3K27me3 in H3.3K36M cells, suggesting a spreading of PRC1 activity to these regions (fig. S14C).
Fig. 4. Intergenic gain of H3K27me3 leads to redistribution of PRC1 and aberrant gene activation.
(A) Box plots (left) and schematics (right) displaying the ratio of average H3K27me3 reads per base-pair between gene-associated peaks (B or B') and intergenic regions (A or A'). (B) Genome viewer representations of normalized ChIP-seq reads for H3K27me3, Ring1b and Cbx2 at Hoxb gene cluster. Refseq genes are annotated at the bottom. (C) Box plots of normalized Ring1b and Cbx2 ChIP-seq peak reads. P-value <2.2 × 10−16 determined by Wilcoxon’s rank sum test. (D) Box plots of expression levels of PRC1-repressed genes are shown for control or Ring1a/b siRNA knockdown cells and for H3.3WT and H3.3K36M cells. P-value < 2.2 × 10−16 determined by Wilcoxon’s rank sum test. (E) MPCs were transfected with siRNA for 3 days, followed by chondrocyte differentiation induction for 2 days. The mRNA expression of Acan, Col2a1, Col9a1 and Col11a1 was measured. Error bars indicate standard deviation from three biological replicates. *, p<0.05; ***, p<0.001.
We postulate a model in which "dilution" of the PRC1 complex away from repressed genic loci leads to ectopic gene expression contributing to the differentiation blockade by H3.3K36M (fig. S14D). We found that the majority of PRC1-dependent gene expression changes (93 of 130 up-regulated and 60 of 72 down-regulated genes upon Ring1a/b knockdown) were recapitulated by H3.3K36M expression (Fig. 4D, fig. S15, A to C and table S3). A specific upregulation of PRC1-repressed genes was observed in H3.3K36M cells (fig. S15D). Many of these genes (Wnt6, Sox6) are implicated in the self-renewal and lineage-specification of mesenchymal stem cells (fig. S15E) (17, 18). Consistently, Ring1a/b knockdown was sufficient to impair the chondrocyte differentiation of MPCs (Fig. 4E and fig. S15F). Transcriptome analyses of human chondroblastoma samples revealed a similar de-repression of polycomb-bound genes associated with H3.3K36M mutation (fig. S16).
We present evidence that the H3K36M mutation plays a driver role in the development of mesenchymal neoplasms through impaired differentiation of MPCs. Although H3.1 and H3.3 K36M are found at different genomic locations, they result in the same genome-wide changes in chromatin landscape, gene expression profile, and tumorigenic capacity. These data imply that the specific genomic locations of H3K36M-containing nucleosomes are relatively unimportant for K36M's function. While other non-mutually exclusive mechanisms may be involved (19), the dominant inhibition of H3K36 methyltransferases is a critical downstream event mediating H3K36M's differentiation-arresting potential. The concurrent inactivation of multiple methyltransferases via a single missense mutation in histone H3 provides an efficient means to "lock" cells into an aberrant chromatin state that, in the context of mesenchymal progenitors, promotes neoplastic transformation. Our study highlights an underappreciated role of intergenic H3K36 methylation in polycomb complex recruitment through the antagonization of H3K27me3 propagation (20, 21) and demonstrate that modest alterations to the intergenic to gene-associated H3K27me3 ratio can have profound outcomes on polycomb-mediated gene silencing. We speculate that similar mechanisms may underlie other human cancers where histone H3K27 and K36-directed enzymes are frequently dysregulated.
Supplementary Material
Acknowledgments
We thank D. Weinberg, B. Carey, B. Sabari and R. Phillips for help in manuscript preparation. This research was supported by funding from funds of Rockefeller University (to C.D.A.), startup funds provided by the Wisconsin Institute for Discovery (to P.W.L.), Starr Cancer Consortium (grant SCC I6-A614 to C.D.A.), Sidney Kimmel Foundation (Kimmel Scholar Award to P.C. and P.W.L.) and NIH grants (P01CA196539 to P.W.L., C.D.A., T.W.M., N.J., J.M. and B.A.G.; Innovator grant DP2OD007447 and R01GM110174 to B.A.G.; DP2CA174499 and K08CA151660 to P.C.; K08 CA181475 to S.V.). This work was performed within the context of the I-CHANGE consortium and supported by funding from Genome Canada, Genome Quebec, The Institute for Cancer Research of the Canadian Institutes for Health Research (CIHR) McGill University and the Montreal Children’s Hospital Foundation. C.L. is the Kandarian Family Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2195-14). N.J. is a member of the Penny Cole lab and the recipient of a Chercheur Clinician Senior Award. J.M. holds a Canada Research Chair (tier 2). D.H. is supported by a Boehringer Ingelheim Fonds Predoctoral fellowship. D.B. is supported by a studentship from the T.D trust/Montreal Children’s Hospital Foundation. We thank the molecular cytology core facility of MSKCC for technical help and EMD Millipore for H3.3K36M antibody generation. The sequencing data reported in this paper are deposited at GEO database (accession number: GSE69291).
References and Notes
- 1.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 2.Wu G, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behjati S, et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet. 2013;45:1479–1482. doi: 10.1038/ng.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjerke L, et al. Histone H3.3 Mutations Drive Pediatric Glioblastoma through Upregulation of MYCN. Cancer Discov. 2013;3:512–519. doi: 10.1158/2159-8290.CD-12-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan K-M, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27:985–990. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bender S, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Funato K, Major T, Lewis PW, Allis CD, Tabar V. Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science. 2014;346:1529–1533. doi: 10.1126/science.1253799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher CDM, World Health Organization; International Agency for Research on Cancer, editor. WHO classification of tumours of soft tissue and bone. 4th ed. IARC Press; Lyon: 2013. World Health Organization classification of tumours. [Google Scholar]
- 10.Yao Y, Wang Y. ATDC5: an excellent in vitro model cell line for skeletal development. J. Cell. Biochem. 2013;114:1223–1229. doi: 10.1002/jcb.24467. [DOI] [PubMed] [Google Scholar]
- 11.Banaszynski LA, Allis CD, Lewis PW. Histone variants in metazoan development. Dev. Cell. 2010;19:662–674. doi: 10.1016/j.devcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Somers GR, et al. Pediatric undifferentiated sarcoma of the soft tissues: a clinicopathologic study. Pediatr. Dev. Pathol. 2006;9:132–142. doi: 10.2350/08-05-0098.1. [DOI] [PubMed] [Google Scholar]
- 13.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan W, et al. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitges FW, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 16.Orlando DA, et al. Quantitative ChIP-Seq Normalization Reveals Global Modulation of the Epigenome. Cell Reports. 2014;9:1163–1170. doi: 10.1016/j.celrep.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev. Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 18.Smits P, Dy P, Mitra S, Lefebvre V. Sox5 and Sox6 are needed to develop and maintain source, columnar, and hypertrophic chondrocytes in the cartilage growth plate. J. Cell Biol. 2004;164:747–758. doi: 10.1083/jcb.200312045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herz H-M, et al. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science. 2014;345:1065–1070. doi: 10.1126/science.1255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaydos LJ, Rechtsteiner A, Egelhofer TA, Carroll CR, Strome S. Antagonism between MES-4 and Polycomb repressive complex 2 promotes appropriate gene expression in C. elegans germ cells. Cell Rep. 2012;2:1169–1177. doi: 10.1016/j.celrep.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popovic R, et al. Histone methyltransferase MMSET/NSD2 alters EZH2 binding and reprograms the myeloma epigenome through global and focal changes in H3K36 and H3K27 methylation. PLoS Genet. 2014;10:e1004566. doi: 10.1371/journal.pgen.1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun G, Chung D, Liang K, Keleş S. Statistical analysis of ChIP-seq data with MOSAiCS. Methods Mol. Biol. 2013;1038:193–212. doi: 10.1007/978-1-62703-514-9_12. [DOI] [PubMed] [Google Scholar]
- 23.Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leng N, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin S, Garcia BA. Examining histone posttranslational modification patterns by highresolution mass spectrometry. Meth. Enzymol. 2012;512:3–28. doi: 10.1016/B978-0-12-391940-3.00001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.