Abstract
BRCA1 and BRCA2 are prominently associated with inherited breast and ovarian cancer. The encoded proteins function in DNA damage responses, but no functional link between BRCA1 and BRCA2 has been established. We show here that PALB2 physically and functionally connects BRCA1 and BRCA2 into a DNA damage response network that also includes the RAD51 recombinase. PALB2 directly binds BRCA1, as determined with bacterially expressed fragments of each protein. Furthermore, PALB2 independently interacts with BRCA1 and BRCA2 through its NH2 and COOH termini, respectively. Critically, two point mutants (L21P and L24P) of the PALB2 coiled-coil domain or an NH2-terminal deletion (Δ1–70) disrupt its interaction with BRCA1. We have reconstituted PALB2-deficient cells with PALB2Δ1–70, PALB2-L21P, or PALB2-L24P, or with COOH-terminally truncated PALB2 that is deficient for interaction with BRCA2. Using extracts from these cells, we find that PALB2 mediates the physical interaction of BRCA2 with a COOH-terminal fragment of BRCA1. Analysis of the assembly of foci in these cells by BRCA1, PALB2, BRCA2, and RAD51 suggests that BRCA1 recruits PALB2, which in turn organizes BRCA2 and RAD51. Resistance to mitomycin C and the repair of DNA double-strand breaks by homologous recombination require the interaction of PALB2 with both BRCA1 and BRCA2. These results suggest that BRCA1 and BRCA2 cooperate in DNA damage responses in a PALB2-dependent manner, and have important implications for the genesis of breast/ovarian cancer and for chemotherapy with DNA interstrand cross-linking agents.
Introduction
BRCA1 and BRCA2 are the major genes associated with inherited susceptibility to breast and ovarian cancer (1–4). Cells that are deficient for either protein share similar phenotypes, including hypersensitivity to DNA interstrand cross-linkers, such as mitomycin C (MMC), and defective repair of DNA double-strand breaks (DSB) by homologous recombination (HR; reviewed in refs. 5, 6). These observations suggest that BRCA1 and BRCA2 function in cellular responses to DNA damage. Importantly, BRCA1 and BRCA2 have not been functionally linked (reviewed in refs. 5–7). Although it has been reported that BRCA1 and BRCA2 coimmunopurify (8–10), the interaction may be indirect and may involve only a small proportion of either protein (6, 7).
PALB2 (partner and localizer of BRCA2) is also a breast cancer susceptibility gene (11–13) and was first identified by its interaction with BRCA2 protein (14). PALB2 is required for the localization of BRCA2 to sites of DNA damage (14). BRCA2, in turn, regulates the recruitment of RAD51 to DNA damage foci and its assembly into nucleoprotein filaments that initiate HR through strand invasion (15–17). How PALB2 is localized has not been determined, however.
PALB2 has also been identified as the Fanconi anemia gene FANCN (18, 19). Fanconi anemia is associated with chromosome instability and a predisposition to cancer (reviewed in ref. 20). EUFA1341 cells, derived from a Fanconi anemia patient, lack PALB2 and are hypersensitive to MMC (18).
Here, we show that an NH2-terminal coiled-coil domain of PALB2 is required for coimmunoprecipitation of PALB2 with BRCA1 and for the localization of PALB2. Furthermore, PALB2 directly binds BRCA1. Importantly, numerous BRCA2-dependent functions require the capacity of PALB2 to interact with both BRCA1 and BRCA2, including the assembly of BRCA2 foci, the assembly of RAD51 foci, HR, and resistance to MMC. These results show that BRCA1, PALB2, BRCA2, and RAD51 function in a DNA damage response pathway that culminates in HR. Together, our results suggest that PALB2 serves as a physical and functional linker between BRCA1 and BRCA2. Defects at any step in this pathway may increase genetic instability, thereby leading to cancer susceptibility.
Results
Interaction with BRCA1 Regulates PALB2 Behavior
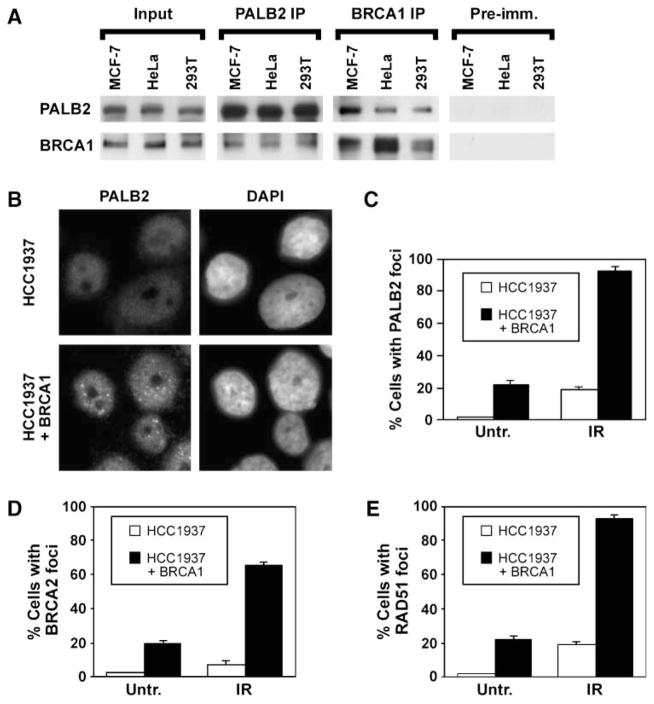
Because PALB2 localizes BRCA2 to DNA damage foci (14), we sought to determine how PALB2 itself is recruited to sites of DNA damage. Given that BRCA1 scaffolds DNA damage responses (21), we considered whether BRCA1 may have a role in this process. First, we examined whether PALB2 and BRCA1 associate using a coimmunoprecipitation assay (Fig. 1A). PALB2 and BRCA1 coimmunoprecipitated from extracts of MCF7 mammary adenocarcinoma cells, HeLa, and 293T cells using antibodies against either protein.
FIGURE 1.
PALB2 and BRCA1 coimmunoprecipitate and BRCA1 regulates PALB2 behavior. A. Levels of PALB2 and BRCA1 in extracts from undamaged MCF7, HeLa, or 293T cells are indicated by immunoblotting (input). These samples represent 2% of the amount of protein used for immunoprecipitation with anti-PALB2 or anti-BRCA1 antibodies. Preimmune serum for the PALB2 antibody was used as a control. B. Representative i mages of the assembly of PALB2 foci in HCC1937 cells, which are deficient for BRCA1, or their counterparts corrected by expression of BRCA1. Cells were fixed at 16 h following exposure to 15 Gy IR, and the position of nuclei was determined by counterstaining with 4′,6-diamidino-2-phenylindole (DAPI). C to E. Quantification of the assembly of PALB2 (C), BRCA2 (D), and RAD51 (E) foci in HCC1937 or their corrected counterparts. Cells were exposed to 15 Gy IR and were fixed after 16 h, or were left untreated (Untr.), and average values ± SD were determined from three independent counts of 150 or more cells each.
Knowing that the proteins associate in a complex, we then determined whether BRCA1 localizes PALB2 in HCC1937 cells (Fig. 1B–C). HCC1937 cells express a BRCA1 mutant truncated in its COOH terminus (8). PALB2 foci did not form in HCC1937 cells but did assemble in their counterparts corrected by expression of BRCA1, as shown in images in Fig. 1B. As shown by quantification, assembly of PALB2 foci was defective both in untreated populations of HCC1937 cells and following exposure to ionizing radiation (IR; Fig. 1C).
There are conflicting reports about the assembly of RAD51 foci in HCC1937 cells (16, 22), and localization of BRCA2 to laser-induced DSBs, but not foci, has been examined previously in these cells (23). Thus, we tested the same HCC1937 cells for the assembly of BRCA2 (Fig. 1D) and RAD51 (Fig. 1E) nuclear foci. In our hands, the assembly of BRCA2 and RAD51 foci was deficient in HCC1937 cells, either with or without exposure to IR. These defects were corrected by restoration of BRCA1. Thus, BRCA1 appears to regulate the assembly of nuclear foci by PALB2, BRCA2, and RAD51, all of which interact (14).
Leu 21 and Leu 24 in the PALB2 Coiled-Coil Domain Mediate PALB2 Interaction with BRCA1
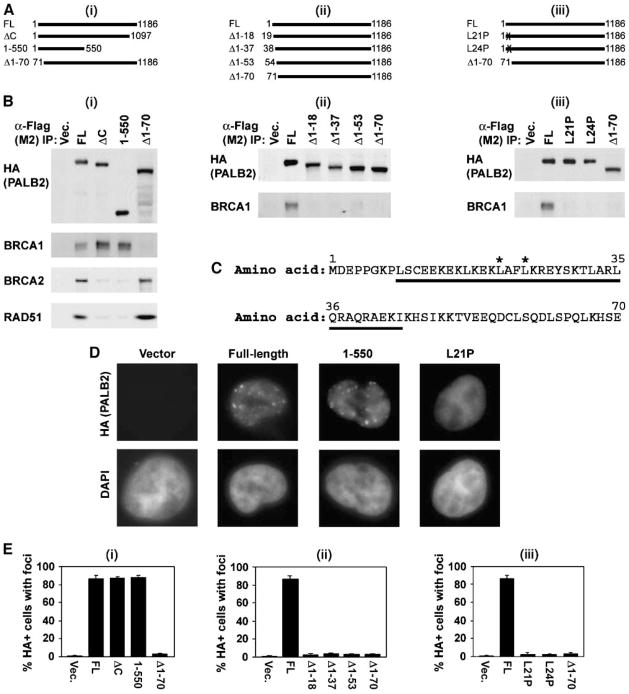
Next, we transiently expressed HA-Flag epitope-tagged mutants of PALB2 in 293T cells to identify the region of PALB2 that interacts with BRCA1 (Fig. 2). The COOH terminus of PALB2 contains four WD40 motifs. A truncation that eliminates all four WD40 repeats of PALB2 disrupts the PALB2-BRCA2 interaction and has been linked to breast cancer (12). To more precisely define the interaction of PALB2 with BRCA2, and its potential interaction with BRCA1, we truncated PALB2 after P1097 (ΔC), just before the third WD40 repeat (Fig. 2Ai). We also expressed a mutant truncated in the middle of the protein (1–550), as described previously (18). Exon 4, which encodes amino acids 71 to 561, is not required for resistance to MMC (18). Thus, we instead tested the role of the first 70 amino acids of PALB2 in binding to BRCA1 by expressing a mutant lacking this domain (Δ1–70; Fig. 2Ai). We found that binding of PALB2 to BRCA2 or RAD51 required only WD40 repeats 3 to 4 (Fig. 2Bi). These repeats were not required for PALB2 to bind BRCA1, however. BRCA1 also coimmunoprecipitated with PALB2/1–550, indicating that BRCA1 binds to the NH2-terminal half of PALB2. In contrast, PALB2Δ1–70 coimmunoprecipitated with BRCA2 and RAD51, but not BRCA1. BRCA1 and BRCA2 therefore interact independently with different regions of PALB2.
FIGURE 2.
An NH2-terminal domain of PALB2, distinct from the BRCA2-interacting domain of PALB2, mediates interaction with BRCA1 and assembly of PALB2 nuclear foci. A. Diagram of sets of PALB2 mutants used to examine the interaction of PALB2 with BRCA1. These included large NH2- or COOH-terminal truncations (i), a series of smaller NH2-terminal truncations (ii), and two different point mutants (*) within the NH2-terminal coiled-coil domain of PALB2 (iii). Each PALB2 construct contained a Flag-HA epitope tag. In the case of NH2-terminal deletion mutants, the epitope tags were at the COOH terminus of the protein. All other constructs contained NH2-terminal epitope tags. FL, PALB2-full-length. B. Immunoprecipitation with anti-Flag antibodies was done for each experimental set. Antibodies against BRCA1, and BRCA2 or RAD51 in i, were used to determine whether the protein was present in immunoprecipitates. C. Diagram of amino acids 1 to 70 of PALB2. The predicted coiled-coil domain is underlined (amino acids 9–44); *, point mutants that were tested (L21P and L24P). D. Examples of PALB2 foci assembled by wild-type protein, PALB2/1–550, or PALB2-L21P following expression in 293T cells. Assembly of expressed PALB2 and its mutants, all of which contained an HA-Flag epitope tag, was detected with anti-HA antibodies. E. Quantification of the assembly of PALB2 foci, detected with anti-HA antibodies against the epitope tag for each set of mutants. Values for wild-type PALB2 and PALB2Δ1–70 are repeated in each data set for the purpose of comparison. Cells were fixed at 16 h following treatment with 15 Gy IR. Average values ± SD were determined from three independent counts of 150 or more cells each.
To directly test whether the interaction of PALB2 with BRCA1 is required for the assembly of PALB2 foci, we analyzed 293T cells that transiently expressed HA-Flag-tagged PALB2, and its mutants, by immunofluorescence microscopy (Fig. 2D and E). PALB2ΔC and PALB2/1–550 assembled into nuclear foci in the absence of its interaction with BRCA2 or RAD51, following exposure of cells to IR, as assayed with anti-HA antibodies (Fig. 2D and Ei). But PALB2Δ1–70 did not assemble into foci, suggesting that the interaction of PALB2 with BRCA1, but not BRCA2 or RAD51, is required for the recruitment of PALB2 to DNA damage foci.
We then compared mutants with truncations of the NH2-terminal 18, 37, 53, or 70 amino acids of PALB2 to better define the role of this region in PALB2 localization and its interaction with BRCA1 (Fig. 2Aii). Each of these mutants was defective for coimmunoprecipitation with BRCA1 (Fig. 2Bii) and for the assembly of PALB2 (HA) foci (Fig. 2Eii).
PALB2 contains a predicted coiled-coil domain from amino acids 9 to 44 (Fig. 2C). Such domains mediate protein-protein interactions (24). A portion, or all, of this domain was eliminated in the PALB2Δ1–70 mutants. We therefore sought to more specifically test the role of the PALB2 coiled-coil domain and its interaction with BRCA1 in localizing PALB2. For this purpose, we mutated conserved sites within the PALB2 coiled-coil domain. The PALB2-L21P and PALB2-L24P mutants (Fig. 2Aiii) coimmunoprecipitated with BRCA2 and RAD51, but not BRCA1 (Fig. 2Biii). Thus, the coiled-coil domain of PALB2 is required for its interaction with BRCA1. Furthermore, PALB2-L21P and PALB2-L24P localized to nuclei but were deficient for assembly into nuclear foci (Fig. 2D and Eiii). These results suggest that the interaction of PALB2 with BRCA1, through the coiled-coil domain at the NH2 terminus of PALB2, mediates recruitment of PALB2 into DNA damage foci.
Recruitment of BRCA2 and RAD51 into Foci Requires the Interaction of PALB2 and BRCA1
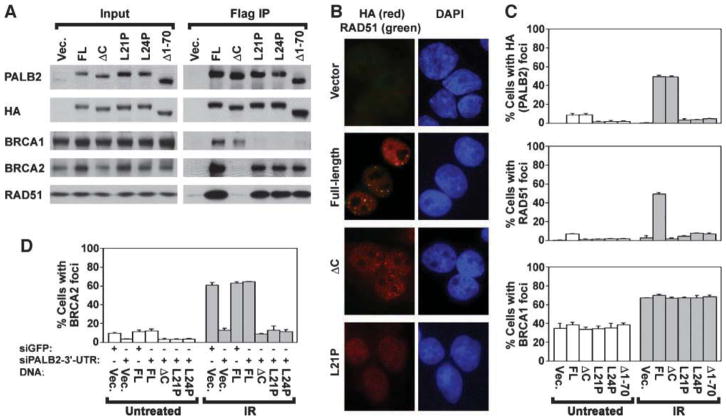
To test the functional importance of the capacity of PALB2 to bind to BRCA1, we stably expressed selected PALB2 mutants in EUFA1341 cells along with an HA-Flag epitope tag (Fig. 3A). Consistent with results obtained in Fig. 2, BRCA2 and RAD51 did not coimmunoprecipitate with PALB2ΔC, which lacks WD40 repeats 3 and 4 (Fig. 3A). However, BRCA1 did coimmunoprecipitate with PALB2ΔC. In contrast, the NH2 terminus and the coiled-coil domain of PALB2 mediated its interaction with BRCA1. BRCA2 and RAD51, but not BRCA1, coimmunoprecipitated with PALB2Δ1–70, PALB2-L21P, and PALB2-L24P (Fig. 3A).
FIGURE 3.
The interaction of PALB2 with BRCA1 is required for the assembly of PALB2, BRCA2, and RAD51 nuclear foci. A. Levels of stable expression of wild-type PALB2 (FL), PALB2ΔC, PALB2Δ1–70, PALB2-L21P, and PALB2-L24P with an HA-Flag epitope tag in EUFA1341 fibroblasts are indicated for each cell line (input). Coimmunoprecipitation of BRCA1, BRCA2, and RAD51 was determined by immunoblotting. Blots for input extracts represent 2.5% of the amount of protein used for immunoprecipitation. B. Examples of PALB2 foci assembled by full-length (wild-type) PALB2, PALB2ΔC, PALB2-L21P, or pMMP vector alone. In each case, the expressed protein contained HA-Flag epitope tags. PALB2 foci detected with anti-HA antibodies (red) and RAD51 foci detected with anti-RAD51 antibodies (green) are shown in a merged image. Colocalization is indicated by yellow foci, whereas red foci indicate the presence of PALB2 but not RAD51 foci. C. Quantification of the assembly of each form of PALB2 into nuclear foci (top), and of the assembly of RAD51 (center) or BRCA1 (bottom) into foci in EUFA1341 fibroblasts reconstituted with each form of PALB2. D. Quantification of the assembly of BRCA2 foci in U2OS cells exogenously expressing wild-type PALB2, PALB2ΔC, PALB2Δ1–70, PALB2-L21P, or PALB2-L24P that were depleted of endogenous PALB2 using a siRNA against its 3′-UTR. Controls trans-fected with a siRNA directed against GFP are indicated. Cells were analyzed at 4 d after transfection with the siRNA. Cells were fixed at 16 h following treatment with 15 Gy IR or were left untreated, and average values ± SD were determined from three independent counts of 150 or more cells each (C–D).
Representative images show that mutation of the coiled-coil domain of PALB2 disrupted the assembly of PALB2 and RAD51 foci in EUFA1341 cells (Fig. 3B). Wild-type PALB2 assembled into foci, which colocalized with RAD51 foci, however. In contrast, assembly of PALB2, but not RAD51 foci, was normal in EUFA1341 cells expressing PALB2ΔC (Fig. 3B). Effects on the assembly of PALB2 and RAD51 foci are quantified in Fig. 3C. In particular, cells expressing each of the NH2-terminal mutants of PALB2, PALB2Δ1–70, PALB2-L21P, and PALB2-L24P, were deficient for the assembly of PALB2 and RAD51 foci, either with or without exposure to IR. In summary, the recruitment of PALB2 to nuclear foci depends on its interaction with BRCA1, whereas the recruitment of RAD51 is disrupted by mutation of either the NH2 or COOH termini of PALB2.
We next analyzed the assembly of BRCA1 foci in EUFA1341 cells reconstituted with PALB2 mutants that were defective for interaction with either BRCA1 or BRCA2 (examples are shown in Supplementary Fig. S1). The vast majority of foci assembled by wild-type PALB2 colocalized with BRCA1 foci. Because it has been reported that endogenous PALB2 strongly colocalizes with BRCA1 (14), the above result shows normal localization of epitope-tagged PALB2. Furthermore, PALB2-L21P did not assemble into nuclear foci but supported normal assembly of BRCA1 foci. The assembly of BRCA1 foci in cells reconstituted with each form of BRCA1 is quantified in Fig. 3C. None of the mutants, including PALB2Δ1–70, PALB2-L21P, PALB2-L24P, and PALB2ΔC, were associated with abnormal assembly of BRCA1 foci. Thus, our results suggest that BRCA1 acts upstream of PALB2.
We could not clearly distinguish BRCA2 foci in EUFA1341 cells using available antibodies. Instead, to assay requirements for the assembly of BRCA2 foci, we examined U2OS cells that expressed various forms of PALB2. Cells were depleted of endogenous PALB2 with a siRNA against the 3′-untranslated region (3′-UTR) of PALB2, as determined on immunoblots (Supplementary Fig. S2A). Examples of images obtained by immunofluorescence microscopy confirm that this siRNA depleted endogenous PALB2 but did not affect exogenously expressed PALB2 (Supplementary Fig. S2B).
As quantified in Fig. 3D, we found, using this system, that both the NH2 and COOH termini of PALB2 were required for the assembly of BRCA2 foci. The assembly of BRCA2 foci was defective, either with or without exposure to IR, in cells containing vector alone, PALB2Δ1–70, PALB2-L21P, PALB2-L24P, and PALB2ΔC (Fig. 3D). Examples of defects in the assembly of BRCA2 foci are shown in Supplementary Fig. S2B. Together, our results suggest that BRCA1, PALB2, BRCA2, and RAD51 cooperate in a pathway in which BRCA1 organizes PALB2, which in turn localizes BRCA2.
PALB2 Directly Binds BRCA1 and Functionally Links BRCA1 to BRCA2
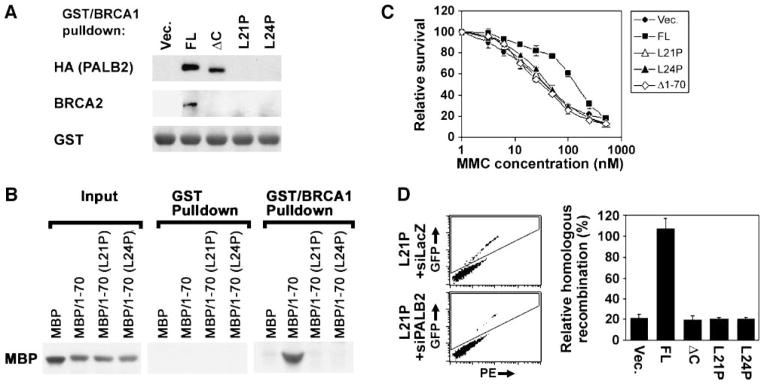
Given that PALB2 independently interacts with BRCA1 and BRCA2, we sought to determine whether it physically links these proteins. We detected coimmunoprecipitation of BRCA1 and BRCA2 in HeLa extracts, but were unable to observe an interaction in extracts from EUFA1341 cells corrected with PALB2. Thus, we instead incubated a fusion protein containing glutathione S-transferase (GST) and amino acids 1293 to 1863 of human BRCA1 with extracts from EUFA1341 cells containing various forms of PALB2 (Fig. 4A). This COOH-terminal fragment of BRCA1 has been reported to bind BRCA2 in a similar in vitro assay (8). GST/BRCA1-1293–1863 associated with BRCA2 in extracts from EUFA1341 cells corrected with wild-type PALB2 but not cells that contained NH2- or COOH-terminal mutants of PALB2, including PALB2ΔC, PALB2-L21P, and PALB2-L24P (Fig. 4A). Critically, each of these extracts had detectable levels of BRCA2 protein (Fig. 3A). The association of PALB2 with BRCA1 in vitro required a functional coiled-coil in the NH2 terminus of PALB2 (Fig. 4A). Thus, PALB2 does indeed seem to mediate the physical interaction of BRCA1 and BRCA2.
FIGURE 4.
Direct binding of the coiled-coil domain of PALB2 to BRCA1 is required for the association of BRCA2 with BRCA1, for resistance to MMC, and for DSB-initiated HR. A. GST protein fused with the 1293–1863 fragment of BRCA1 was incubated with extracts from EUFA1341 fibroblasts exogenously expressing wild-type PALB2, PALB2ΔC, PALB2-L21P, or PALB2-L24P. Associated protein complexes were isolated on glutathione beads. The presence of BRCA2 or PALB2 in isolated protein complexes was determined by immunoblotting. GST-BRCA1-1293–1863 was detected with anti-GST antibodies. B. MBP, MBP/PALB2-1–70, MBP/PALB2-1–70(L21P), or MBP/PALB2-1–70(L24P), isolated using maltose beads (input), were incubated with GST or GST/ BRCA1-1293–1863 beads. Binding of each MBP fusion protein to GST or GST/BRCA1-1293–1863 was determined by immunoblotting with anti-MBP antibodies. The blot for input protein represents 10% of the amount of protein used for the pulldown assay. C. Survival of EUFA1341 fibroblasts stably expressing wild-type PALB2, PALB2-L21P, PALB2-L24P, or PALB2Δ1–70 with HA-Flag epitope tags was determined using a colorimetric assay following treatment with a range of concentrations of MMC (0–500 nmol/L). Relative survival for each cell line ± SD was normalized to the value for that cell line without treatment. D. Assay of HR in U2OS-DR cells stably expressing wild-type PALB2, PALB2ΔC, PALB2-L21P, or PALB2-L24P (with Flag-HA epitope tags), which were depleted of endogenous PALB2 using a siRNA against its 3′-UTR. Examples of dot plots demonstrating the level of GFP (inside box) versus phycoerytherin (PE) for cells expressing PALB2-L21P and I-SCEI. Cells were transfected with a control siRNA against LacZ or with a siRNA directed against the 3′-UTR of endogenous PALB2. At right is quantification of HR for each cell type, relative to controls for the corresponding cell line transfected with siLacZ instead of siPALB2–3′-UTR. The amount of GFP-positive cells ranged from 0% to 2% in each experiment. Columns, average from three independent experiments; bars, SD.
Next, we sought to determine whether PALB2 directly binds BRCA1. For this purpose, we determined whether maltose-binding protein (MBP) fused with amino acids 1 to 70 of PALB2 pulls down with GST/BRCA1-1293–1863 (Fig. 4B). Both proteins were isolated from bacteria using their respective tags. MBP/PALB2-1–70, but not MBP/PALB2-1–70(L21P) or MBP/PALB2-1–70(L24P), strongly bound to GST/BRCA1-1293–1863. MBP/PALB2-1–70 did not bind to GST alone. These results suggest that PALB2 directly binds to BRCA1 and may physically link BRCA2 to BRCA1 by independently binding each protein.
We then sought to evaluate the functional importance of PALB2 as a linker of BRCA1 and BRCA2, in addition to its role in regulating the assembly of BRCA2 and RAD51 foci. Because PALB2 and BRCA1 are both required for cellular resistance to DNA interstrand cross-linking agents, we determined whether the interaction of these proteins is required for cellular resistance to MMC. Critically, mutants of PALB2 that did not interact with BRCA1, including PALB2Δ1–70, PALB2-L21P, and PALB2-L24P, conferred no resistance to MMC, compared with cells containing the vector alone (Fig. 4C).
Next, we sought to determine whether PALB2 links BRCA1 and BRCA2 into a pathway of HR. For this purpose, we used U2OS cells with an integrated copy of the pDR reporter (25). Depletion of endogenous PALB2 with a siRNA against its 3′-UTR similarly inhibited HR in cells containing exogenous PALB2-L21P, PALB2-L24P, or the empty vector alone (Fig. 4D). Interestingly, cells that contained PALB2ΔC, which were defective for interaction with BRCA2, yielded similar levels of DSB-initiated HR as cells that contained PALB2-L21P or PALB2-L24P. Our results suggest that binding of PALB2 to both BRCA1 and BRCA2 is required for HR.
Discussion
We find that PALB2 physically links BRCA1 to BRCA2. This network seems to mediate several BRCA2-dependent functions, including the assembly of RAD51 foci, HR, and resistance to MMC. Strikingly, different domains of PALB2, the NH2 and COOH termini, are required for its interaction with BRCA1 and BRCA2, respectively. BRCA1 seems to act upstream because BRCA1 nuclear foci assemble in cells that lack PALB2 or in which mutant PALB2 does not bind to BRCA1. Given that the assembly of PALB2 nuclear foci is dependent on its interaction with BRCA1, we suggest that BRCA1 recruits PALB2 to sites of DNA damage. It has been shown previously that PALB2 colocalizes with γ-H2AX foci at DSBs (14).
Consistent with previous results, we find that the COOH terminus of PALB2, which contains four WD40 domains, is required for the interaction of PALB2 with BRCA2 (12, 18). However, we have further dissected the COOH terminus of PALB2 and find that deletion of WD40 repeats 3 and 4 is sufficient to disrupt the interaction with BRCA2. We also find, using PALB2-deficient cells reconstituted with PALB2 mutants defective for interaction with either BRCA1 or BRCA2, that the assembly of BRCA2 and RAD51 nuclear foci is deficient. Thus, in our model (Fig. 5), we propose that once PALB2 is positioned, it recruits BRCA2 to sites of DNA damage, which in turn regulates the assembly of RAD51 foci. Consistent with this possibility, we show that BRCA1-deficient HCC1937 cells, which were originally derived from a breast carcinoma (26), display defective assembly of foci by each of the downstream proteins, PALB2, BRCA2, and RAD51. Furthermore, it was already known BRCA2 is required for the assembly of RAD51 foci at the end of the apparent BRCA1-PALB2-BRCA2-RAD51 pathway (16, 17).
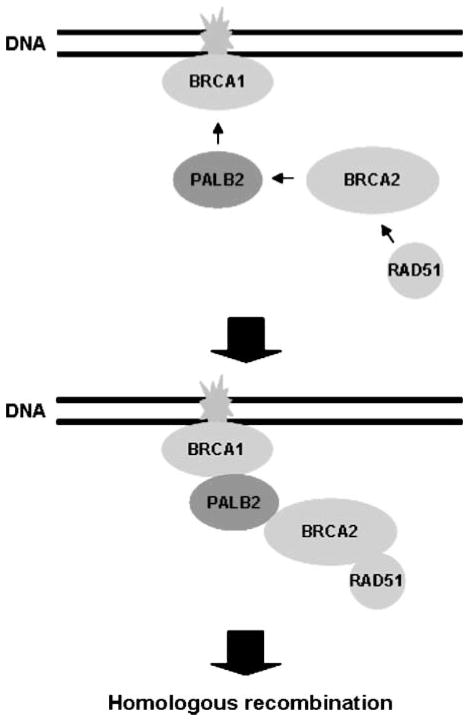
FIGURE 5.
Proposed model for the role of PALB2 in linking BRCA1 with BRCA2 and RAD51 in a pathway of HR. We propose that BRCA1 binds to the NH2 terminus of PALB2 following its own recruitment to sites of DNA damage. We suggest that the COOH terminus of positioned PALB2 then binds to BRCA2, which in turn regulates the assembly of the RAD51 nucleoprotein filament that initiates HR. In this manner, PALB2 links BRCA1, BRCA2, and RAD51 into a network that is required for HR. Our results suggest that BRCA1 and BRCA2 do not directly interact, but associate using PALB2 as an intermediary.
During the submission of this work, a study with consistent findings was published online (27). Both studies find that the coiled-coil domain present at the NH2 terminus of PALB2 directly binds to BRCA1 and that this interaction is required for the assembly of BRCA2 foci. In contrast to the other paper, however, we define and characterize two point mutants of the coiled-coil domain that disrupt binding to BRCA1. In this manner, we more specifically implicate PALB2 as a linker of BRCA1 and BRCA2. Furthermore, we directly compare PALB2-deficient cells reconstituted with mutants defective for interaction with either BRCA1 or BRCA2. This has led to a more definitive demonstration that PALB2 links BRCA1, BRCA2, and RAD51 into a DNA damage response pathway. Additionally, using the set of reconstituted cells, we have determined that PALB2 is positioned between BRCA1 and BRCA2 in this pathway. Unlike the other article, we assay the assembly of RAD51 foci and resistance to the DNA interstrand cross-linking agent, MMC, as functional end points of this pathway. We conclude, on the basis of these assays, that PALB2 serves as a functional linker of BRCA1 and BRCA2. PALB2 mutants that disrupt interaction with either protein result in defective assembly of RAD51 foci and defects in HR.
It was reported previously that PALB2 both stabilizes and localizes BRCA2 (14). NH2-terminal PALB2 mutants, which did not localize correctly (Fig. 3B–C), still bound to BRCA2 and largely stabilized it (Fig. 3A). Thus, the deficiency for BRCA2 foci in cells containing PALB2 mutants that did not bind to BRCA1 (Fig. 3D) is likely due to a defect in localization rather than a failure to stabilize BRCA2.
BRCA1 seems to act as a large scaffolding protein that is involved in signaling the cellular response to DSBs (5, 28, 29). The interaction of PALB2 with BRCA1 may thereby coordinate the regulation of BRCA2 and RAD51, which are centrally required for HR, with DNA damage signaling. It has recently been reported that the Chk1 and Chk2 protein kinases phosphorylate the COOH terminus of BRCA2, and that this regulates the interaction of BRCA2 with RAD51 (30). It will be interesting to determine whether Chk1/Chk2-dependent phosphorylation of BRCA2 is dependent on the interaction of PALB2 with BRCA1. The function of PALB2 as a linker of BRCA1 and BRCA2 may permit more sensitive regulation of HR and may allow BRCA1 to mediate other functions, such as checkpoint signaling, independently from HR.
Deficiencies in BRCA1, BRCA2, or PALB2 result in genomic instability (20). Deficient assembly of PALB2, BRCA2, and RAD51 nuclear foci in untreated cells (Figs. 1 and 3) might reflect defective recruitment of the machinery for HR to sites of endogenous DNA damage. It will be interesting to determine, in the future, whether this leads to increased spontaneous genomic instability that could drive the growth of cancerous cells.
BRCA1, PALB2, BRCA2, and RAD51 are all required for HR (14, 31–33). We find that disruption of the interaction of PALB2 with either BRCA1 or BRCA2 results in equivalent defects in HR. This suggests that PALB2 links BRCA1 and BRCA2 into a pathway of HR. Previous results have shown that BRCA2 regulates the assembly of RAD51 into nucleoprotein filaments with 3′ DNA overhangs. These nucleoprotein filaments initiate HR through strand invasion (15). Thus, RAD51 seems to function at the end of the BRCA1-PALB2-BRCA2-RAD51 pathway.
DNA interstrand cross-linking agents are extensively used for chemotherapy (34, 35). We find that the capacity of PALB2 to bind BRCA1 through its NH2-terminal coiled-coil domain is required for resistance to the DNA interstrand cross-linking agent MMC (Fig. 4C). The COOH terminus of PALB2, and its interaction with BRCA2, is also required for resistance to MMC (11, 12, 18). Together, these results suggest that resistance to MMC requires the capacity of PALB2 to independently bind both BRCA1 and BRCA2. Thus, PALB2 seems to link BRCA1 and BRCA2 into a pathway required for resistance to MMC. The BRCA1-PALB2-BRCA2-RAD51 network could be a critical determinant of the responsiveness of specific tumors and individuals to DNA interstrand cross-linking agents. A defect anywhere in this pathway would be expected to result in defective assembly of RAD51 foci, which might be predictive of the responsiveness of a particular tumor to DNA interstrand cross-linking agents.
In summary, BRCA1 and BRCA2 are the most frequently mutated genes that have been identified in inherited breast and ovarian cancer (1–4). This may stem from disruption of the DNA damage response pathway that both the BRCA1 and BRCA2 proteins function in.
Materials and Methods
Cell Culture
MCF-7, HeLa, U2OS, and 293T cells were obtained from American Type Culture Collection. EUFA1341 fibroblasts were a gift from Dr. Hans Joenje (Vrije Universiteit Medical Center, Amsterdam, The Netherlands). EUFA1341 cells were cultured in F10 (HAM) and DMEM (1:1) containing 10% fetal bovine serum and penicillin-streptomycin. Other cell lines were grown in DMEM containing 10% FBS, L-glutamine, and penicillin-streptomycin. Cells were cultured at 37°C in a 5% CO2 environment.
MMC was added from a 3 mmol/L stock in ethanol kept at −20°C. Irradiation was done with a Mark I-68 Cesium 137 apparatus (J. L. Shepherd and Associates).
Antibodies
Rabbit anti-RAD51 (H-92; Santa Cruz Biotechnology) and mouse anti-BRCA2 (Ab-1; Calbiochem) antibodies were used for immunofluorescence microscopy, immunoprecipitation, and immunoblotting. IgG-purified rabbit antisera 2599 and 2600 generated against amino acids 601–880 of PALB2 were used for immunoprecipitation, and for immunoblotting and immunofluorescence microscopy, respectively. Mouse anti-HA antibody (HA.11; Covance) was used for immunoblotting and immunofluorescence microscopy, whereas mouse anti-Flag antibody (M2; Sigma) was used for immunoprecipitation of tagged proteins. Rabbit anti-BRCA1 (Cell Signaling Technology) was used for immunoprecipitation and immunofluorescence microscopy. Mouse anti-BRCA1 (D9; Santa Cruz Biotechnology) was used for immunofluorescence microscopy. Rabbit anti-BRCA1 (Millipore) and mouse anti-actin antibodies (gift from Dr. James Lessard, Cincinnati Children’s Research Foundation, Cincinnati, OH) were used for immunoblotting.
Cloning and Mutagenesis
The cDNA for human PALB2 was purchased from Origene Technologies, Inc. PALB2 containing Flag and HA epitope tags in tandem at its NH2 or COOH termini were generated by cloning PALB2 into pOZ obtained from Yoshihiro Nakatani (Dana-Farber Cancer Institute). PALB2 containing either NH2- or COOH-terminal Flag-HA tags was subcloned into pCDNA3.1 as a vector for transient expression. NH2-terminal truncation mutants were generated from pOZ-PALB2-Flag-HA using appropriate primers to generate a Flag-HA fusion in pCDNA3.1 with PALB2 starting at amino acids 19, 38, 54, or 71. pCDNA3-Flag-HA-PALB2 was used for the generation of L21P and L24P mutants, and COOH-terminal truncation mutants, using a Quik-Change II Site-directed mutagenesis kit (Stratagene). COOH-terminal truncation mutants were generated by the introduction of a stop codon. Wild-type PALB2, and various PALB2 mutants, were subcloned into the pMMP retroviral vector (36) for stable expression of proteins.
The 1293–1863 fragment of BRCA1 was cloned into pGEX-2TK for the production of a fusion protein with GST. The 1–70 fragment of PALB2, and its mutants, was cloned into pMAL-C2X to generate fusion proteins with MBP.
Transfection and Transduction
Transfection of target cells with pCDNA3.1 plasmids or with siRNAs, and virus production in Phoenix cells, transduction of target cells, and selection with puromycin were as described previously (37).
Immunoprecipitation
Extracts were prepared from 293T cells at 48 h after transfection with various forms of PALB2 or from EUFA1341 cells stably expressing PALB2 and its mutants. Extracts that contained both detergent-soluble and chromatin-associated PALB2 in NETN [20 mmol/L Tris (pH 7.5) containing 420 mmol/L NaCl, 1 mmol/L EDTA, and 0.5% NP40] were prepared using a modification of a previously described protocol (14). Extracts were precleared, incubated with anti-Flag or anti-BRCA1 antibody, and washed as described previously (38).
In vitro Analysis of Binding to a COOH-Terminal Fragment of BRCA1
GST/BRCA1-1293–1863 and MBP/PALB2-1–70 fusion proteins in 50 mmol/L Tris (pH 8.0) were isolated from Escherichia coli using reagents from GE Healthcare and New England BioLabs, incubated at 4°C for 2 h, and washed, as described previously (39).
Extracts from EUFA1341 cells stably expressing various forms of PALB2 were prepared in NETN buffer. Extracts were precleared with GST beads, incubated with GST/BRCA1-1293–1863, and washed with NETN buffer as described previously (39).
Immunoblotting
Lysates were electrophoresed by SDS-PAGE and then transferred to nitrocellulose, blocked, incubated with primary antibodies and horseradish peroxidase–linked secondary antibodies, and revealed by chemiluminescence (Amersham) as previously described (37). Gradient gels (4–12%) were used for analysis of immunoprecipitates. In all other experiments, 6% gels were used.
Immunofluorescence Microscopy
Unless otherwise noted, cells were fixed before permeabilization, washed, and incubated with primary and secondary antibodies as previously described (37). For detection of BRCA2, cells were prepermeabilized and fixed as described previously (36). Coverslips were mounted over Vectashield containing 4′,6-diamidino-2-phenylindole, and were sealed with fingernail polish.
Microscopy, collection of images, and the generation of figures was as described previously (37).
MMC Sensitivity Assay
Measurement of relative growth was done in triplicate for each cell line, as described previously (40). Survival was calculated relative to the average corrected absorbance for each untreated cell line following subtraction of background values determined from wells not containing cells.
Assay of DSB-Initated HR
U2OS-DR cells with an integrated reporter construct for HR-mediated repair of GFP (25) were kindly provided by Drs. Maria Jasin and Koji Nakanishi (Memorial Sloan-Kettering Cancer Center, New York, NY). U2OS-DR cells that stably expressed pMMP vector, wild-type PALB2, PALB2ΔC, PALB2-L21P, or PALB2-L24P with NH2-terminal HA-Flag epitope tags were selected with puromycin. To deplete endogenous PALB2, cells were transfected with 1.8 μg of a siRNA directed against its 3′-UTR (5′-GGAGAATATCTGAATGACA-3′). Alternatively, cells were transfected with siLacZ as a control. At 48 h after transfection with siRNAs, cells were transfected with pCBASce encoding I-SCEI to induce DSBs. Cells were collected 72 h later by trypsinization and placed in fresh medium for analysis.
The percentage of GFP-positive cells was determined using a FacsCalibur flow cytometer. Background measurements obtained from U2OS-DR cells containing the empty vector, and transfected with siLacZ but not siPALB2-3′-UTR, were subtracted from each value. The efficiency of HR, relative to cells transfected with the siLacZ control, was then calculated from three independent experiments for cells expressing each form of PALB2.
Supplementary Material
Acknowledgments
Grant support: NIH R01 HL085587 (P.R. Andreassen).
We thank Dr. Hans Joenje (Vrije Universiteit Medical Center), Dr. James Lessard (Cincinnati Children’s Research Foundation), Dr. Yoshihiro Nakatani (Dana-Farber Cancer Institute), and Drs. Maria Jasin and Koji Nakanishi (Memorial Sloan-Kettering Cancer Center), for EUFA1341 fibroblasts, anti-actin antibodies, pOZ vectors, and U2OS-DR cells and pCBASce, respectively.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note: Supplementary materials for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
References
- 1.Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science (New York NY) 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Wooster R, Neuhausen SL, Mangion J, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12–13. Science (New York, NY) 1994;265:2088–90. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- 3.Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–92. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 4.Hall JM, Lee MK, Newman B, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science (New York, NY) 1990;250:1684–9. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 5.Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–74. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- 6.Venkitaraman AR. Functions of BRCA1 and BRCA2 in the biological response to DNA damage. J Cell Sci. 2001;114:3591–8. doi: 10.1242/jcs.114.20.3591. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, West SC. Distinct functions of BRCA1 and BRCA2 in double-strand break repair. Breast Cancer Res. 2002;4:9–13. doi: 10.1186/bcr417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Silver DP, Walpita D, et al. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell. 1998;2:317–28. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 9.Kumaraswamy E, Shiekhattar R. Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol Cell Biol. 2007;27:6733–41. doi: 10.1128/MCB.00961-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong Y, Hakimi MA, Chen X, et al. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell. 2003;12:1087–99. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 11.Erkko H, Xia B, Nikkila J, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–9. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 12.Tischkowitz M, Xia B, Sabbaghian N, et al. Analysis of PALB2/ FANCN-associated breast cancer families. Proc Natl Acad Sci U S A. 2007;104:6788–93. doi: 10.1073/pnas.0701724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahman N, Seal S, Thompson D, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–7. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia B, Sheng Q, Nakanishi K, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–29. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 15.Davies AA, Masson JY, McIlwraith MJ, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–82. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 16.Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–51. [PubMed] [Google Scholar]
- 17.Tutt A, Bertwistle D, Valentine J, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–16. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia B, Dorsman JC, Ameziane N, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–61. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 19.Reid S, Schindler D, Hanenberg H, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–4. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 20.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev. 2007;8:735–48. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–39. [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Willers H, Feng Z, et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol. 2004;24:708–18. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM. Multifactorial contributions to an acute DNA damage response by BRCA1/ BARD1-containing complexes. Genes Dev. 2006;20:34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–82. [PubMed] [Google Scholar]
- 25.Nakanishi K, Yang YG, Pierce AJ, et al. Human Fanconi anemia monoubi-quitination pathway promotes homologous DNA repair. Proc Natl Acad Sci U S A. 2005;102:1110–5. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomlinson GE, Chen TT, Stastny VA, et al. Characterization of a breast cancer cell line derived from a germ-line BRCA1 mutation carrier. Cancer Res. 1998;58:3237–42. [PubMed] [Google Scholar]
- 27.Zhang F, Ma J, Wu J, et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19:524–9. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1–1 pathway. Genes Dev. 2004;18:1423–38. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foray N, Marot D, Gabriel A, et al. A subset of ATM- and ATR-dependent phosphorylation events requires the BRCA1 protein. EMBO J. 2003;22:2860–71. doi: 10.1093/emboj/cdg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bahassi EM, Ovesen JL, Riesenberg AL, Bernstein WZ, Hasty PE, Stambrook PJ. The checkpoint kinases Chk1 and Chk2 regulate the functional associations between hBRCA2 and Rad51 in response to DNA damage. Oncogene. 2008;27:3977–85. doi: 10.1038/onc.2008.17. [DOI] [PubMed] [Google Scholar]
- 31.Stark JM, Hu P, Pierce AJ, Moynahan ME, Ellis N, Jasin M. ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J Biol Chem. 2002;277:20185–94. doi: 10.1074/jbc.M112132200. [DOI] [PubMed] [Google Scholar]
- 32.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–8. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 33.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–72. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 34.Wang W, Figg WD. Secondary BRCA1 and BRCA2 alterations and acquired chemoresistance. Cancer Biol Ther. 2008;7:1004–5. doi: 10.4161/cbt.7.7.6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andreassen PR, Ren K. Fanconi anemia proteins, DNA interstrand crosslink repair pathways, and cancer therapy. Curr Cancer Drug Targets. 2009;9:101–17. doi: 10.2174/156800909787314011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Andreassen PR, D’Andrea AD. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol Cell Biol. 2004;24:5850–62. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan Q, Zhang F, Barrett B, Ren K, Andreassen PR. A role for monoubiquitinated FANCD2 at telomeres in ALT cells. Nucleic Acids Res. 2009;37:1740–54. doi: 10.1093/nar/gkn995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andreassen PR, Margolis RL. Microtubule dependency of p34cdc2 inactivation and mitotic exit in mammalian cells. J Cell Biol. 1994;127:789–802. doi: 10.1083/jcb.127.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Einarson MB, Pugacheva EN, Orlinick JR. GST pull-down. CSH Protocols. 2007 doi: 10.1101/pdb.prot4757. [DOI] [PubMed] [Google Scholar]
- 40.Litman R, Peng M, Jin Z, et al. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–65. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.