Abstract
Background
Nasopharyngeal carcinoma (NPC) is a common malignancy in South-East Asia. NPC is characterized by distant metastasis and poor prognosis. The pathophysiological mechanism of nasopharyngeal carcinoma is unknown. This study aimed to identify the crucial miRNAs in nasopharyngeal carcinoma and their target genes, and to discover the potential mechanism of nasopharyngeal carcinoma development.
Material/Methods
Microarray expression profiling of miRNA and mRNA from the Gene Expression Omnibus database was downloaded, and we performed a significance analysis of differential expression. An interaction network of miRNAs and target genes was constructed. The underlying function of differentially expressed genes was predicted through Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses. To validate the microarray analysis data, significantly different expression levels of miRNAs and target genes were validated by quantitative real-time polymerase chain reaction.
Results
We identified 27 differentially expressed miRNAs and 982 differentially expressed mRNAs between NPC and normal control tissues. 12 miRNAs and 547 mRNAs were up-regulated and 15 miRNAs and 435 mRNAs were down-regulated in NPC samples. We found a total of 1185 negative correlation pairs between miRNA and mRNA. Differentially expressed target genes were significantly enriched in pathways in cancer, cell cycle, and cytokine-cytokine receptor interaction signaling pathways. Significantly differentially expressed miRNAs and genes, such as hsa-miR-205, hsa-miR-18b, hsa-miR-632, hsa-miR-130a, hsa-miR-34b, PIGR, SMPD3, CD22, DTX4, and CDC6, may play essential roles in the development of nasopharyngeal carcinoma.
Conclusions
hsa-miR-205, hsa-miR-18b, hsa-miR-632, hsa-miR-130a, and hsa-miR-34b may be related to the development of nasopharyngeal carcinoma by regulating the genes involved in pathways in cancer and cell cycle signaling pathways.
MeSH Keywords: Gene Regulatory Networks, MicroRNAs, Nasopharyngeal Neoplasms, Real-Time Polymerase Chain Reaction
Background
Nasopharyngeal carcinoma is a head and neck cancer characterized as highly malignancy and regional selection [1,2]. NPC is a rare cancer in Western countries, but it is common in Asian countries [3]. NPC frequently occurs in southern China, included Guangdong, Fujian, Hong Kong and Southeast Asia, including Malaysia, Indonesia, and Singapore.
The incidence rate of NPC is 2 per 100 000 worldwide [4]. The NPC incidence in southern China is 20–50 times higher than in Western countries [5]. Radiotherapy is the main curative treatment for NPC to extend patient survival time [6,7]. NPC presents highly malignant recurrence with local tissue invasion and distant metastasis, which is the dominant reason for radiotherapy failure [8].
Currently, the major etiological factors of NPC are reported to be genetic susceptibility, environmental factors, and Epstein-Barr virus (EBV) infection. Familial and large-scale case-control studies report that HLA class I genes in the MHC locus at chromosome 6p21 are notably associated with high risk of NPC. In addition, CDK5, TEL2, CELF2, and IKKB [9–12] are also reported to be associated with NPC pathogenesis. Environmental risk factors include eating salt-preserved food [13], insufficient intake of fresh vegetables and fruits [14], alcohol consumption [15], and tobacco smoking [13,16]. Epstein-Barr virus (EBV) infection is an extensively researched etiological factor for NPC. EBV belongs to the gamma herpes virus family, persistently infects B lymphocytes in more than 90% of adults, and is related to NPC tumorigenesis [17].
In addition to the above-mentioned etiological factors of NPC, mounting evidence shows that microRNAs (miRNAs) may play essential roles in NPC tumorigenesis by regulating target genes. miRNAs are small (20–25 nucleotides) non-coding RNAs that negatively regulate expression level of target gene [18]. Numerous studies have reported that miRNAs are associated with NPC cell proliferation, migration, invasion, metastasis, and irradiation sensitivity by suppressing their target genes. miR-142-3p promotes NPC cell proliferation via suppressing SOCS6 expression [19]. miR-4649-3p inhibits NPC cell proliferation by targeting protein tyrosine phosphatase SHP-1 [20]. miR-29a/b regulates SPARC and COL3A1 gene expression to promote NPC cell migration and invasion [21]. miR-23a targets IL-8/Stat3 pathway results in radio-sensitivity in NPC [22], while miR-504 down-regulates nuclear respiratory factor 1 result in radio-resistance in NPC [23]. However, the mechanism of pathogenesis in NPC remains unclear.
In this study we used bioinformatics methods to integrate miRNA and mRNA expression data, which are available in the GEO database, to identify differentially expressed miRNAs and target genes between NPC and normal control tissues, aiming to provide valuable information for use in defining the mechanism of pathogenesis in NPC.
Material and Methods
Gene expression datasets
We searched the Gene Expression Omnibus database (GEO, http://www.ncbi.nlm.nih.gov/geo) for mRNA and miRNA expression profiling of NPC, and downloaded the raw expression data. GEO is a public repository for high-throughput gene expression data [24]. We only retained the microarray studies between tumor and normal tissues. The following information was extracted from each identified study: GEO accession number, platform, number of cases and controls, time, and author.
Data processing
Different sequencing platforms and clinical samples commonly cause the heterogeneity among different microarray datasets, and make it difficult to compare microarray datasets directly. We downloaded the raw expression dataset and preprocessed it by log2 transformation and Z-score normalization.
Analysis of differentially expressed miRNA and mRNA
The miRNAs and mRNA differentially expressed between the NPC and normal control samples were identified using the limma method, which is a linear model for microarray data analysis [25]. We selected differentially expressed miRNA as false discovery rate (FDR) <0.05, and selected differentially expressed mRNA as FDR <0.001.
Identification of miRNA target genes
To obtain the target genes of miRNAs, the selected miRNAs were integrated into the miRWalk database (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/) [26], in which the correlation between target genes and miRNAs have been confirmed. miRWalk is a comprehensive database that provides predicted and experimentally validated miRNA-target interactions for humans, mice, and rats. It combines the predicted and validated information with a comparison of binding sites resulting from 12 existing miRNA-target prediction programs [26,27]. In our study, we used 6 algorithms: DIANAmT, miRanda, miRDB, miRWalk, PICTAR, and TargetScan to predict target genes of miRNA; if more than 4 of 6 algorithms predicted the same gene of miRNA, the gene was considered as a target gene of the miRNA [26]. Reverse correlations of miRNA-target gene interacting pairs were subject to construct miRNA-RNA interaction network analysis.
miRNA-target gene network
Differentially expressed miRNA and differentially expressed target genes were used to construct the interaction network by using Cytoscape software (http://cytoscape.org) [28]. In the miRNA-gene network, a circular node represented the mRNA and a diamond node represented the miRNA, and their association was represented by a line.
Functional enrichment analysis
The underlying function of differentially expressed target genes was predicted by the Gene Ontology (GO) [29] function and Kyoto Encyclopedia of Genes and Genomes (KEGG) [30] pathway enrichment analysis using the DAVID tool (Database for Annotation, Visualization, and Integrated Discovery) (http://www.david.niaid.nih.gov) [31]. We set p<0.05 and FDR <0.05 as the cut-off for selecting significantly enriched functional GO terms and KEGG pathway, respectively.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA of fresh frozen tissues was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The SuperScript III Reverse Transcription Kit (Invitrogen, Carlsbad, CA, USA) was used to synthesize the cDNA according to the manufacturer’s instructions. qRT-PCR reactions were performed using Power SYBR Green PCR Master Mix [32] (Applied Biosystems, Foster City, CA) on the Applied Biosystems 7500 (Applied Biosystems, Foster City, CA). The miRcute miRNA First-Strand cDNA Kit (Tiangen, Beijing, China) and miRcute miRNA qPCR Detection Kit (Tiangen, Beijing, China) were used for miRNA expression level detection. U6 and β-actin were used as internal control for miRNA and mRNA detected. The relative expression of target genes was calculated using the 2−ΔΔCT equation. The PCR primers were used as shown in Table 1.
Table 1.
Genes and primers for qRT-PCR.
| miRNA/mRNA | Primer sequence (5′to3′) |
|---|---|
| SMPD3 | Forward-CCAACAAGTGTAACGACGATGCC |
| Reverse-CGCTGGACGAGGAGGTAGATTTTC | |
| CD22 | Forward-ATGCCGATTCGAGAAGGAGACAC |
| Reverse-CCACGAGCACCAACTATTACAAGC | |
| DTX4 | Forward-AGAAAGGTAAAACCCCAGAGGAAGT |
| Reverse-ATGGCAACCAAGCAGTAGATGTG | |
| CDC6 | Forward-TTGAGCCAAGAAGGAGCACAAGATT |
| Reverse-CTTCCAAGAGCCCTGAAAGTGACA | |
| PIGR | Forward-AGGTGCTAGACTCTGGTTTTCGG |
| Reverse-TCTGCTCCCATCGGCTTGA | |
| β-actin | Forward-ACTTAGTTGCGTTACACCCTT |
| Reverse-GTCACCTTCACCGTTCCA | |
| hsa-Mir-205 | Forward-TCCTTCATTCCACCGGAGTCTG |
| hsa-Mir-18b | Forward-TAAGGTGCATCTAGTGCAGTTAGAA |
| hsa-Mir-632 | Forward-GTGTCTGCTTCCTGTGGG |
| hsa-Mir-130a | Forward-CAGTGCAATGTTAAAAGGGC |
| hsa-Mir-34b | Forward-CAATCACTAACTCCACTGCCAT |
| U6 | Forward-CTCGCTTCGGCAGCACA |
| Reverse-AACGCTTCACGAATTTGCGT |
Statistical analysis
At least 3 independent experiments were performed for statistical evaluation. qRT-PCR experimental data were expressed as means ±SD. The statistical significance was evaluated using the Student’s t-test and p<0.05 was considered as a significant difference.
Results
Differentially expressed miRNAs and mRNAs in the NPC
In this work, we collected a total of 3 mRNA expression profiles including 74 NPC and 31 normal control (NC) samples and 5 miRNA expression profiling including 402 NPC and 38 NC samples, as shown in Table 2. After normalization of the raw microarray data, significantly differentially expressed genes including 27 miRNA and 982 mRNA were identified in NPC compared to normal nasopharyngeal tissues; 27 miRNAs consisted of 12 up-regulated and 15 down-regulated miRNAs; 982 mRNA consisted of 547 up-regulated and 435 down-regulated mRNAs (Supplementary Table 1). hsa-miR-205, hsa-miR-196b, and hsa-miR-632 was the most significantly up-regulated miRNAs, while hsa-miR-130a, hsa-let-7a, and hsa-miR-34b were the most significantly down-regulated miRNAs in NPC compared with the normal control (Table 3).
Table 2.
Characteristics of mRNA and miRNA expression profiling of the nasopharyngeal carcinoma.
| GEO ID | Platform | Samples (N:P) | Time | Author |
|---|---|---|---|---|
| mRNA expression profiling | ||||
| GSE53819 | GPL6480Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (Probe Name version) | 18:18 | 2014 | Qian CN |
| GSE13597 | GPL96[HG-U133A] Affymetrix Human Genome U133A Array | 3:25 | 2009 | Wei W |
| GSE12452 | GPL570[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | 10:31 | 2008 | Ahlquist P |
| miRNA expression profiling | ||||
| GSE32906 | GPL11350 Illumina Custom Prostate Cancer DASL Panel miRNA | 6:16 | 2014 | Luo Z |
| GSE46172 | GPL16770Agilent-031181 Unrestricted_Human_miRNA_V16.0_Microarray (miRBase release 16.0 miRNA ID version) | 4:4 | 2013 | Bethony JM |
| GSE22587 | GPL8933Illumina Human Beta-version microRNA expression BeadChip | 4:8 | 2012 | YANG S |
| GSE32960 | GPL14722microRNA array | 18:312 | 2012 | Ma J |
| GSE36682 | Human miRNA 1K | 6:62 | 2012 | Wei R |
N – normal samples; P – patients’ samples.
Table 3.
Significantly dysregulated miRNAs.
| miRNAs | p-value | FDR |
|---|---|---|
| Up-regulated miRNAs | ||
| hsa-miR-205 | 0.00E+00 | 0.00E+00 |
| hsa-miR-196b | 1.58E-05 | 1.85E-03 |
| hsa-miR-632 | 8.13E-05 | 3.56E-03 |
| hsa-miR-18b | 7.37E-05 | 3.56E-03 |
| hsa-miR-93 | 3.81E-04 | 9.52E-03 |
| hsa-miR-326 | 3.78E-04 | 9.52E-03 |
| hsa-miR-210 | 3.18E-04 | 9.52E-03 |
| hsa-miR-376a* | 1.19E-03 | 2.46E-02 |
| hsa-miR-200c | 1.64E-03 | 3.02E-02 |
| hsa-miR-18a* | 2.26E-03 | 3.77E-02 |
| hsa-miR-542-3p | 2.60E-03 | 4.09E-02 |
| hsa-miR-9* | 3.47E-03 | 4.49E-02 |
| Down-regulated miRNAs | ||
| hsa-miR-130a | 2.02E-11 | 3.54E-09 |
| hsa-let-7a | 3.18E-05 | 2.78E-03 |
| hsa-miR-34b | 4.60E-05 | 3.22E-03 |
| hsa-let-7e | 7.38E-05 | 3.56E-03 |
| hsa-let-7d | 1.46E-04 | 5.67E-03 |
| hsa-miR-30d | 2.24E-04 | 7.83E-03 |
| hsa-miR-146a | 3.31E-04 | 9.52E-03 |
| hsa-miR-98 | 1.09E-03 | 2.46E-02 |
| hsa-miR-10b | 1.18E-03 | 2.46E-02 |
| hsa-miR-138 | 1.45E-03 | 2.83E-02 |
| hsa-miR-31 | 1.75E-03 | 3.05E-02 |
| hsa-miR-363 | 2.69E-03 | 4.09E-02 |
| hsa-miR-564 | 2.90E-03 | 4.23E-02 |
| hsa-let-7g | 3.09E-03 | 4.33E-02 |
| hsa-miR-29a | 3.31E-03 | 4.45E-02 |
FDR – false discovery rate.
The interaction network of miRNAs and target genes
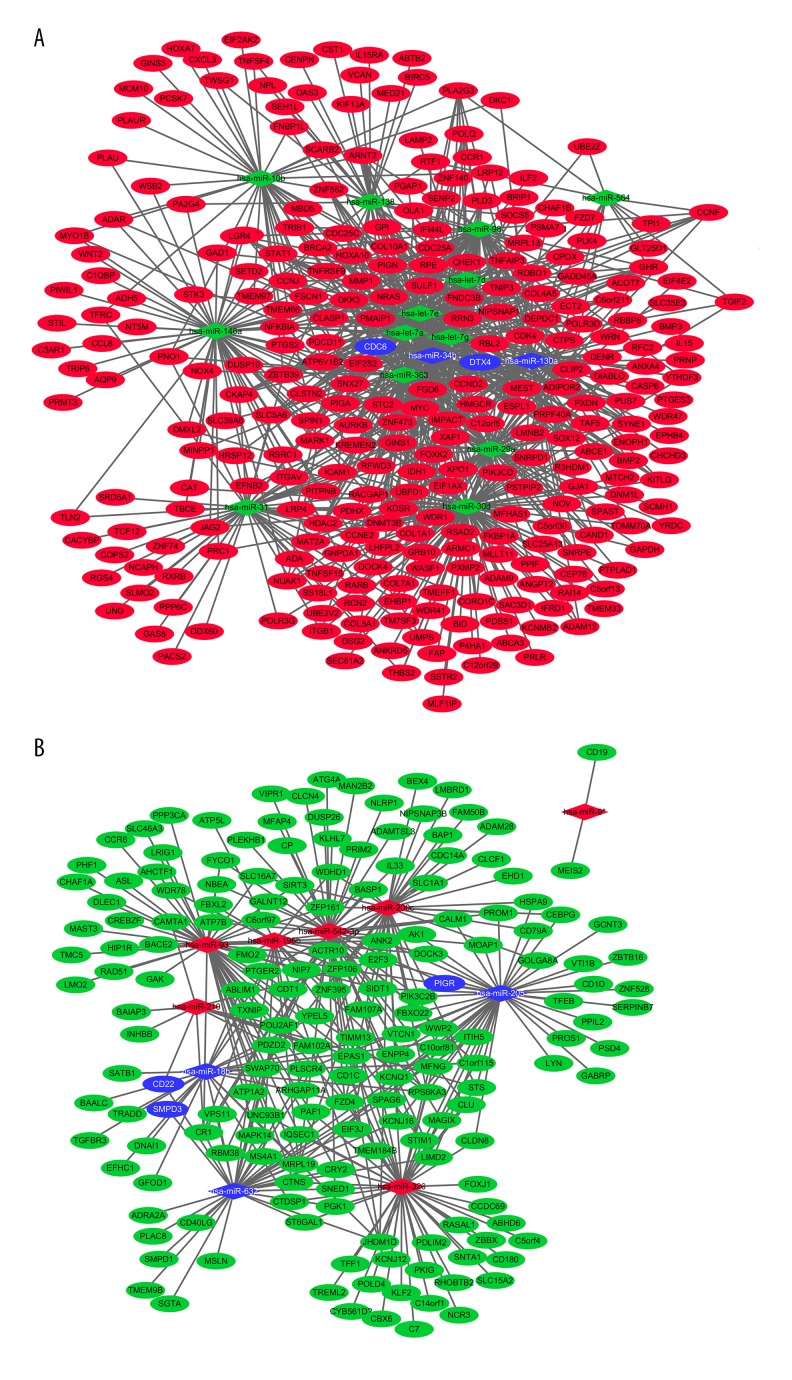
Based on the identified miRNA-target gene interaction pairs of reverse association, we compared the interaction network between miRNAs and target genes in NPC and visualized them with Cytoscape software. We used 1185 miRNA-target gene pairs of reverse correlation, including 316 pairs of up-regulated miRNA and 735 pairs of down-regulated miRNA, to construct the miRNA-target genes interaction network. The target predictions of hsa-miR-376a*and hsa-miR-18a* are not available in miRWalk databases.
In this network, the significantly differentially expressed hsa-miR-632, hsa-miR-205, hsa-miR-18b, hsa-miR-34b, and hsa-miR-130a were targeted in significantly differentially expressed PIGR, CDC6, CD22, SMPD3 and DTX4, respectively, as shown in Figure 1.
Figure 1.
miRNA-mRNA interaction network of NPC. (A) Down-regulation miRNA and up-regulation mRNA interaction network. The green and blue diamond nodes represent down-regulation, and red and blue circular nodes represent up-regulation. (B) Up-regulation miRNA and down-regulation mRNA interaction network. The green and blue circular nodes represent down-regulation, and the red and blue diamond nodes represent up-regulation. Circular nodes represent mRNAs and diamond nodes represent miRNAs. Solid lines indicate interaction associations between the miRNAs and mRNAs. The blue diamond nodes and blue circular nodes represent verified miRNA and mRNA through qRT-PCR.
GO classification of miRNA target genes
To obtain insights into the biological roles of differentially expressed miRNA target genes, we analyzed the predicted target gene of miRNAs using GO annotation. The threshold of GO terms was p-value<0.05. Nuclear division (GO: 0000280, p=3.05E-05) and cell cycle G2/M phase transition (GO: 0044839, p=4.02E-05) were the most significant enrichments of targets genes biological process. Intracellular organelle part (GO: 0044446, p=1.95E-05) and organelle part (GO: 0044422, p=3.62E-05) were the highest enrichments of cellular component. Catalytic activity (GO: 0003824, p=1.09E-04) and hydrolase activity (GO: 0016787, p=1.75E-04) were the highest enrichments of molecular function, as shown in Table 4.
Table 4.
GO function enrichment analysis of differentially expressed miRNA target genes (top 15).
| GO ID | GO Term | Count | P-value | FDR |
|---|---|---|---|---|
| Biological process | ||||
| GO: 0000280 | Nuclear division | 7 | 3.05E-05 | 1.48E-01 |
| GO: 0044839 | Cell cycle G2/M phase transition | 6 | 4.02E-05 | 9.76E-02 |
| GO: 0000086 | G2/M transition of mitotic cell cycle | 6 | 4.02E-05 | 6.50E-02 |
| GO: 0007067 | Mitotic nuclear division | 6 | 7.71E-05 | 9.37E-02 |
| GO: 0022617 | Extracellular matrix disassembly | 6 | 8.75E-05 | 8.50E-02 |
| GO: 0030574 | Collagen catabolic process | 6 | 8.75E-05 | 7.08E-02 |
| GO: 0044243 | Multicellular organismal catabolic process | 6 | 8.75E-05 | 6.07E-02 |
| GO: 0048285 | Organelle fission | 7 | 9.46E-05 | 5.74E-02 |
| GO: 0051301 | Cell division | 9 | 1.45E-04 | 7.83E-02 |
| GO: 0044763 | Single-organism cellular process | 191 | 1.87E-04 | 9.08E-02 |
| GO: 1903047 | Mitotic cell cycle process | 12 | 2.80E-04 | 1.24E-01 |
| GO: 0044772 | Mitotic cell cycle phase transition | 9 | 2.84E-04 | 1.15E-01 |
| GO: 0044770 | Cell cycle phase transition | 9 | 2.84E-04 | 1.06E-01 |
| GO: 0051726 | Regulation of cell cycle | 41 | 2.92E-04 | 1.01E-01 |
| GO: 0032963 | Collagen metabolic process | 6 | 3.49E-04 | 1.13E-01 |
| Cellular component | ||||
| GO: 0044446 | Intracellular organelle part | 159 | 1.95E-05 | 1.09E-02 |
| GO: 0044422 | Organelle part | 162 | 3.62E-05 | 1.01E-02 |
| GO: 0005581 | Collagen trimer | 5 | 6.87E-05 | 1.28E-02 |
| GO: 0005829 | Cytosol | 42 | 4.30E-04 | 6.01E-02 |
| GO: 0044424 | Intracellular part | 222 | 7.32E-04 | 8.19E-02 |
| Molecular function | ||||
| GO: 0003824 | Catalytic activity | 88 | 1.09E-04 | 1.06E-01 |
| GO: 0016787 | Hydrolase activity | 7 | 1.75E-04 | 8.55E-02 |
| GO: 0004000 | Adenosine deaminase activity | 2 | 5.17E-04 | 1.68E-01 |
| GO: 0016814 | Hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines | 2 | 5.17E-04 | 1.26E-01 |
FDR – false discovery rate.
Pathway analysis of miRNA target genes
We performed the KEGG pathway enrichment analysis for differentially expressed miRNA-target genes. FDR <0.05 was used as the criteria for pathway detection. The highest enrichment of pathways in our analysis was the pathway in cancer (FDR=1.78E-11) and cell cycle (FDR=1.48E-07) (Table 5).
Table 5.
KEGG pathway enrichment analysis of differentially expressed miRNA target genes (top 15).
| KEGG ID | KEGG term | Count | FDR | Genes |
|---|---|---|---|---|
| hsa05200 | Pathways in cancer | 27 | 1.78E-11 | CCNE2, STAT1, KITLG, RARB, BIRC5, E2F3, EPAS1, FZD7, BMP2, NRAS, COL4A5, CDK4, NFKBIA, RAD51, WNT2, BID, FZD4, MYC, PIK3CD, ITGAV, BRCA2, ITGB1, ARNT2, ZBTB16, HDAC2, PTGS2, MMP1 |
| hsa04110 | Cell cycle | 14 | 1.48E-07 | CCNE2, ESPL1, CDC14A, GADD45A, E2F3, CHEK1, CCND2, CDK4, CDC25C, CDC6, RBL2, MYC, CDC25A, HDAC2 |
| hsa05222 | Small cell lung cancer | 11 | 9.81E-07 | CCNE2, RARB, E2F3, COL4A5, CDK4, NFKBIA, MYC, PIK3CD, ITGAV, ITGB1, PTGS2 |
| hsa04060 | Cytokine-cytokine receptor interaction | 16 | 3.08E-05 | INHBB, IL15RA, KITLG, IL15, BMP2, CXCL3, CLCF1, TNFSF15, PRLR, CCR1, GHR, CD40LG, CCL8, TNFRSF9, TNFSF4, CCR6 |
| hsa04662 | B cell receptor signaling pathway | 8 | 3.15E-04 | CD22, CD79A, NRAS, NFKBIA, LYN, PIK3CD, PPP3CA, CD19 |
| hsa05162 | Measles | 10 | 3.87E-04 | CCNE2, STAT1, EIF2AK2, TNFAIP3, CCND2, CDK4, NFKBIA, PIK3CD, OAS3, ADAR |
| hsa04115 | p53 signaling pathway | 5 | 4.14E-04 | CCNE2, GADD45A, CHEK1, CCND2, CDK4 |
| hsa04640 | Hematopoietic cell lineage | 8 | 4.18E-04 | CD22, KITLG, CD1D, TFRC, CD1C, CD19, MS4A1, CR1 |
| hsa05220 | Chronic myeloid leukemia | 7 | 6.01E-04 | E2F3, NRAS, CDK4, NFKBIA, MYC, PIK3CD, HDAC2 |
| hsa04630 | Jak-STAT signaling pathway | 10 | 7.27E-04 | IL15RA, STAT1, IL15, CCND2, CLCF1, SOCS5, PRLR, MYC, PIK3CD, GHR |
| hsa05140 | Leishmaniasis | 4 | 8.58E-04 | STAT1, NFKBIA, ITGB1, PTGS2 |
| hsa05160 | Hepatitis C | 9 | 8.98E-04 | STAT1, EIF2AK2, CLDN8, NRAS, NFKBIA, TRADD, PIK3CD, MAPK14, OAS3 |
| hsa05340 | Primary immunodeficiency | 5 | 9.13E-04 | CD79A, ADA, CD40LG, CD19, UNG |
| hsa05219 | Bladder cancer | 3 | 1.22E-03 | E2F3, CDK4, MYC |
| hsa04610 | Complement and coagulation cascades | 6 | 1.77E-03 | PLAUR, PROS1, C3AR1, C7, CR1, PLAU |
FDR – false discovery rate.
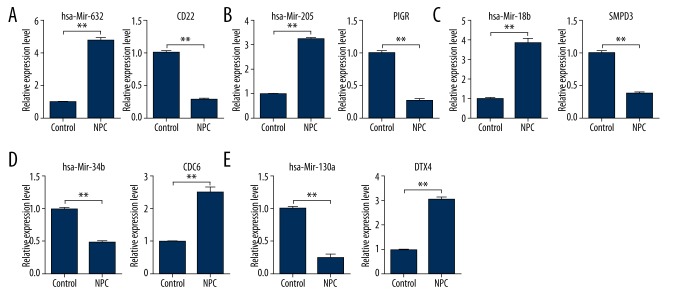
qRT-PCR validation of differentially expressed miRNAs and target genes
To validate the microarray analysis data, the levels of significantly differentially expressed miRNAs (hsa-miR-632, hsa-miR-34b, hsa-miR-130a, hsa-miR-205, and hsa -miR-18b) and target genes (CD22, CDC6, DTX4, PIGR, and SMPD3) were quantified by qRT-PCR in 2 NPC samples and 2 normal control samples. As shown in Figure 2A–2C, the expression levels of hsa-miR-632, hsa-miR-205, and hsa-miR-18b were significantly up-regulated and the respective target genes CD22, PIGR, and SMPD3 were significantly down-regulated in NPC samples (p<0.01). The expressions of hsa-miR-34b and hsa-miR-130a were significantly down-regulated and the respective target genes CDC6 and DTX4 were significantly up-regulated in NPC samples (p<0.01), as shown in Figure 2D, 2E. In conclusion, the qRT-PCR validation of differentially expressed miRNAs and target genes between NPC and normal control samples was in accordance with results of our microarray data bioinformatics analysis.
Figure 2.
miRNA and mRNA expression level in NPC and control tissues by qRT-PCR. (A) hsa-miR-632 and CD22; (B) hsa-miR-205 and PIGR; (C) hsa-miR-18b and SMPD3; (D) hsa-miR-34b and CDC6; (E) hsa-miR-130a and DTX4. Control tissues mean adjacent cancer tissues of NPC.
Discussion
miRNAs play essential roles in many fundamental biological processes, including cell proliferation, migration, invasion, and metastasis. miRNAs can function as oncogenes or oncosuppressor, depending on the targets suppressed.
In our study, we found that hsa-miR-205, hsa-miR-632, hsa-miR-196b, hsa-miR-18b, and hsa-miR-93 are the top 5 up-regulated miRNAs and we found that hsa-miR-130a, hsa-let-7a, hsa-miR-34b, hsa-let-7e, and hsa-let-7d are the top 5 down-regulated miRNAs in NPC patients. hsa-miR-205 and hsa-miR-18a are significantly up-regulated, and hsa-miR-34b is significantly down-regulated in NPC biopsy tissues [33,34], which is in accordance with our bioinformatics analysis and further was validated through qRT-PCR (Table 3, Figure 2D). Moreover, hsa-miR-205 and hsa-miR-34b expression level influence the development of NPC [33]. Functions of hsa-miR-632 up-regulation and hsa-miR-130a down-regulation in NPC are not reported.
Tang et al. reported that miR-205-5p has significant diagnostic value as a novel candidate biomarker in NPC. They performed a comprehensive analysis of microRNA expression patterns of 3 NPC biopsies and 3 normal nasopharyngeal epithelium specimens, then validated the differentially expressed miRNAs in 67 NPC and 25 normal tissues with qRT-PCR, finding that miR-205-5p is 1 of 5 significantly differentially expressed miRNAs in NPC [35]. In addition, miR-205 is related to radio-resistance of NPC, and miR-205 up-regulation results in radio-resistance of NPC through suppressing the PTEN pathway [36,37]. In our work, PIGR was predicted as target gene of miR-205, and this was validated in NPC tissues by qRT-PCR (Figure 2B). PIGR had significantly lower expression in NPC specimens, but was frequently expressed in non-tumor controls [38], which is consistent with our results (Figure 2B).
Target CDC6 was up-regulated by hsa-miR-34b in NPC tissues, as Figure 2D shows. miR-34b/c and TP-53 polymorphisms may contribute to the risk of NPC, and gene-gene interaction of miR-34b/c rs4938723 and TP-53 Arg72-Pro increases the risk of NPC [39]. It is reported that CDC6 is associated with cancer prognosis and proliferation, while CDC6 functions in NPC are not reported. CDC6 is significantly up-regulated by miR-26a/b in lung cancer specimens compared with the adjacent normal tissues, suggesting that CDC6 is associated with poorer prognosis of lung cancer [40]. Knockdown of CDC 6 effectively inhibits proliferation of tongue squamous cell carcinoma Tca8113 cells [41]. In our work, CDC6 was enriched in cell cycle, which is the top 2 KEGG enrichment pathway of differentially expressed miRNA target genes (Table 5).
hsa-miR-18b was up-regulated and consequentially caused significant down-regulation of target genes ABLIM1 and SMPD3 (NSMASE2) in NPC (Table 3, Figure 2C). SMPD3 was down-regulated by both of hsa-miR-18b and hsa-miR-632 in our study. hsa-miR-18b influences cancer cell proliferation, growth, and metastasis, while hsa-miR-18b up-regulation is mediated by loss of connective tissue growth factor through PI3K/AKT/C-Jun and C-Myc signaling to promote cell growth and cell proliferation of NPC [42]. Oxidative stress modulates NSMASE2 sub-cellular localization in plasma membranes to generate ceramide and induces apoptosis of lung carcinoma A549 cell line [43]. NSMASE2 regulates exosomal miRNA secretion and promotes angiogenesis within the tumor microenvironment, as well as in metastasis [44].
Deltex 4, E3 ubiquitin ligase is the official name of DTX4; it functions as a negative regulator of Notch signaling [45], which is crucial for the T-cell development in early stages and angiogenesis during carcinogenesis [46,47]. Dysregulation of Notch and Wnt promotes cell differentiation and tumorigenesis. In our study, DTX4 was up-regulated by hsa-miR-130a, hsa-miR-7a, hsa-miR-7g, hsa-miR-7d, and hsa-miR-7e in NPC patients.
CD22 was down-regulated by both of hsa-miR-632 and hsa-miR-210. CD22 was enriched in B cell receptor and hematopoietic cell lineage signaling pathway via KEGG pathway analysis (Table 5). It is reported that pan-B lymphocytes have scant peri-tumoral areas and are absent in 29 out of 50 NPC biopsies through using immunohistological detection [48]. hsa-miR-632 is reportedly associated with breast cancer tumorigenesis, and high expression level of hsa-miR-632 down-regulates DNAJB6, leading to significantly increased invasive and metastatic ability of breast cancer cells compared to mammary epithelial cells [49].
Pathway in cancer was identified as the most significantly enriched pathway in NPC (Table 5). Pathway in cancer is related to pathogenesis of various cancer types, such as colorectal, pancreatic, thyroid, and lung cancer [50–54], indicating that pathways in cancer may play an important role in NPC pathogenesis.
There are limitations in our work. We have constructed the regulatory network of miRNAs and mRNA inverse correlations pairs, and the pathogenesis of key miRNAs and mRNAs in NPC need to be elucidated through in vivo and in vitro experiments.
Conclusions
We identified 27 differentially expressed miRNAs and 982 differentially expressed mRNAs between NPC and normal tissues. We used 1185 miRNA-target gene pairs of inverse correlations to construct an interaction network. In this network, we found several miRNAs and genes that may play important roles in NPC, such as hsa-miR-205, hsa-miR-34b, hsa-miR-18b, hsa-miR-632, hsa-miR-130a, PIGR, CDC6, CD22, SMPD3, and DTX4. The pathway in cancer may be involved in the pathogenesis mechanism of NPC. Our findings may provide an important contribution to further elucidate the pathogenesis mechanisms of NPC.
Supplementary materials
Supplementary Table 1.
Full list of differentially expressed mRNA in nasopharyngeal carcinoma.
| Genes | FDR | Up/down regulation |
|---|---|---|
| LMNB2 | 1.00E−11 | Up |
| HDGFRP3 | 2.32E−11 | Up |
| FJX1 | 2.89E−11 | Up |
| RBBP8 | 4.91E−11 | Up |
| LHX2 | 5.18E−11 | Up |
| TNFAIP6 | 8.16E−11 | Up |
| ECT2 | 1.02E−10 | Up |
| C12orf48 | 1.74E−10 | Up |
| CHAF1B | 2.93E−10 | Up |
| VRK2 | 4.51E−10 | Up |
| NFE2L3 | 8.20E−10 | Up |
| TFRC | 1.08E−09 | Up |
| NPL | 1.08E−09 | Up |
| FAM64A | 1.42E−09 | Up |
| GALNT11 | 2.59E−09 | Up |
| TNFSF4 | 2.98E−09 | Up |
| GPSM2 | 3.06E−09 | Up |
| MAD2L1 | 4.03E−09 | Up |
| GAD1 | 4.59E−09 | Up |
| CDC6 | 5.29E−09 | Up |
| PFDN4 | 5.31E−09 | Up |
| ATF5 | 6.91E−09 | Up |
| MRPL42 | 7.85E−09 | Up |
| ZWILCH | 8.93E−09 | Up |
| FOXM1 | 9.23E−09 | Up |
| RAN | 1.00E−08 | Up |
| OLA1 | 1.31E−08 | Up |
| OIP5 | 1.99E−08 | Up |
| PSRC1 | 2.17E−08 | Up |
| FAP | 2.17E−08 | Up |
| MINPP1 | 2.86E−08 | Up |
| SEC61A2 | 3.30E−08 | Up |
| CCNF | 3.30E−08 | Up |
| C12orf5 | 4.54E−08 | Up |
| ARNT2 | 4.54E−08 | Up |
| RIF1 | 4.86E−08 | Up |
| RCN2 | 5.33E−08 | Up |
| DTX4 | 6.03E−08 | Up |
| RAD54B | 7.31E−08 | Up |
| CLASP1 | 7.31E−08 | Up |
| KIF14 | 7.46E−08 | Up |
| GNPDA1 | 7.46E−08 | Up |
| EXO1 | 7.46E−08 | Up |
| PMAIP1 | 8.23E−08 | Up |
| KCTD3 | 1.10E−07 | Up |
| GRB10 | 1.31E−07 | Up |
| GINS3 | 1.63E−07 | Up |
| PTGS2 | 1.69E−07 | Up |
| PUS7 | 1.82E−07 | Up |
| XPOT | 2.00E−07 | Up |
| PLA2G3 | 2.04E−07 | Up |
| PALB2 | 2.20E−07 | Up |
| PRMT3 | 2.36E−07 | Up |
| SAC3D1 | 2.45E−07 | Up |
| VCAN | 2.52E−07 | Up |
| FGF1 | 2.78E−07 | Up |
| ESM1 | 2.78E−07 | Up |
| C12orf11 | 2.86E−07 | Up |
| P4HA1 | 3.59E−07 | Up |
| PBK | 3.83E−07 | Up |
| RNASEH2A | 3.91E−07 | Up |
| DOCK4 | 3.91E−07 | Up |
| CNIH4 | 3.91E−07 | Up |
| NUAK1 | 3.91E−07 | Up |
| AHCY | 3.99E−07 | Up |
| TBCE | 4.26E−07 | Up |
| UBE2S | 4.33E−07 | Up |
| COL5A1 | 4.39E−07 | Up |
| NOX4 | 4.49E−07 | Up |
| GAPDH | 4.52E−07 | Up |
| EIF4E2 | 4.52E−07 | Up |
| DTL | 4.52E−07 | Up |
| STK3 | 4.94E−07 | Up |
| PSMD14 | 4.94E−07 | Up |
| DSG2 | 4.94E−07 | Up |
| CENPF | 5.08E−07 | Up |
| PTTG3P | 5.15E−07 | Up |
| FANCL | 5.16E−07 | Up |
| PPIF | 5.26E−07 | Up |
| PSMA4 | 5.52E−07 | Up |
| HDAC2 | 6.06E−07 | Up |
| MCM4 | 6.38E−07 | Up |
| CKS1B | 6.38E−07 | Up |
| UCK2 | 6.89E−07 | Up |
| EIF2S2 | 6.90E−07 | Up |
| TIPIN | 7.68E−07 | Up |
| FSCN1 | 7.68E−07 | Up |
| HSPE1 | 7.86E−07 | Up |
| MMP12 | 8.05E−07 | Up |
| TMEM194A | 8.19E−07 | Up |
| KIF18A | 8.22E−07 | Up |
| INSM1 | 8.24E−07 | Up |
| RAI14 | 8.77E−07 | Up |
| UNG | 9.44E−07 | Up |
| PIK3CB | 1.12E−06 | Up |
| TRIP6 | 1.15E−06 | Up |
| MTX2 | 1.21E−06 | Up |
| CEP152 | 1.27E−06 | Up |
| STAR | 1.30E−06 | Up |
| PLAU | 1.37E−06 | Up |
| HOMER3 | 1.38E−06 | Up |
| ME3 | 1.39E−06 | Up |
| RIC8B | 1.40E−06 | Up |
| ZFP64 | 1.56E−06 | Up |
| NUP155 | 1.58E−06 | Up |
| TOMM40 | 1.65E−06 | Up |
| PGAP1 | 1.67E−06 | Up |
| ITGAV | 1.88E−06 | Up |
| KIF4A | 1.90E−06 | Up |
| SRD5A1 | 2.03E−06 | Up |
| ZNF124 | 2.17E−06 | Up |
| RAD51AP1 | 2.17E−06 | Up |
| PTTG1 | 2.26E−06 | Up |
| BRCA2 | 2.31E−06 | Up |
| GGCT | 2.35E−06 | Up |
| ANXA4 | 2.41E−06 | Up |
| ADA | 2.74E−06 | Up |
| TMEM14A | 2.77E−06 | Up |
| PAK1IP1 | 3.08E−06 | Up |
| TMEM33 | 3.10E−06 | Up |
| HSPD1 | 3.11E−06 | Up |
| GORASP1 | 3.11E−06 | Up |
| USP18 | 3.12E−06 | Up |
| MARK1 | 3.14E−06 | Up |
| SLC39A6 | 3.17E−06 | Up |
| ZNF562 | 3.39E−06 | Up |
| CDC25C | 3.48E−06 | Up |
| RPE | 3.50E−06 | Up |
| POLQ | 3.50E−06 | Up |
| SNRPF | 3.79E−06 | Up |
| SSX2IP | 3.81E−06 | Up |
| STAP2 | 3.84E−06 | Up |
| CDC45 | 3.84E−06 | Up |
| POMGNT1 | 3.86E−06 | Up |
| BID | 3.88E−06 | Up |
| STC2 | 4.00E−06 | Up |
| BRIP1 | 4.00E−06 | Up |
| TM7SF3 | 4.21E−06 | Up |
| DUSP10 | 4.50E−06 | Up |
| PSMG1 | 4.62E−06 | Up |
| CCL4 | 4.74E−06 | Up |
| MED21 | 4.88E−06 | Up |
| NOV | 5.14E−06 | Up |
| SUMO1 | 5.21E−06 | Up |
| CPOX | 5.30E−06 | Up |
| COL7A1 | 5.30E−06 | Up |
| CAT | 5.39E−06 | Up |
| STIL | 5.84E−06 | Up |
| CCT2 | 5.93E−06 | Up |
| DNM1L | 5.95E−06 | Up |
| SLC25A13 | 6.29E−06 | Up |
| IMPACT | 6.52E−06 | Up |
| CACYBP | 6.61E−06 | Up |
| RGS4 | 6.63E−06 | Up |
| PHF14 | 7.10E−06 | Up |
| ILF2 | 7.10E−06 | Up |
| ENO1 | 7.13E−06 | Up |
| PAIP1 | 7.30E−06 | Up |
| TNFSF15 | 7.34E−06 | Up |
| EPHB4 | 7.34E−06 | Up |
| ATP6V1B2 | 7.34E−06 | Up |
| OAS3 | 7.83E−06 | Up |
| GSTO1 | 7.93E−06 | Up |
| TPI1 | 7.99E−06 | Up |
| IL15RA | 8.23E−06 | Up |
| HOXA7 | 8.40E−06 | Up |
| SETD2 | 8.66E−06 | Up |
| C1QB | 8.95E−06 | Up |
| IFRD1 | 9.15E−06 | Up |
| NT5M | 9.92E−06 | Up |
| PITPNB | 1.01E−05 | Up |
| CENPN | 1.02E−05 | Up |
| GPR137B | 1.03E−05 | Up |
| TK1 | 1.05E−05 | Up |
| TMEFF1 | 1.09E−05 | Up |
| SULF1 | 1.12E−05 | Up |
| ERCC6L | 1.13E−05 | Up |
| DENR | 1.21E−05 | Up |
| C5orf13 | 1.21E−05 | Up |
| ATP2C1 | 1.22E−05 | Up |
| STAT1 | 1.27E−05 | Up |
| TTK | 1.30E−05 | Up |
| SS18L1 | 1.30E−05 | Up |
| GABPB1 | 1.30E−05 | Up |
| BIRC5 | 1.33E−05 | Up |
| WNT2 | 1.38E−05 | Up |
| HRSP12 | 1.42E−05 | Up |
| SNRPG | 1.46E−05 | Up |
| PFDN2 | 1.48E−05 | Up |
| POLR3D | 1.54E−05 | Up |
| GJB1 | 1.57E−05 | Up |
| POGLUT1 | 1.58E−05 | Up |
| TNIP3 | 1.63E−05 | Up |
| PSMB3 | 1.64E−05 | Up |
| YWHAQ | 1.69E−05 | Up |
| NCBP2 | 1.80E−05 | Up |
| PLD3 | 1.80E−05 | Up |
| TWSG1 | 1.87E−05 | Up |
| SKP2 | 2.00E−05 | Up |
| YWHAH | 2.03E−05 | Up |
| CASP6 | 2.08E−05 | Up |
| IRF6 | 2.10E−05 | Up |
| CKAP4 | 2.12E−05 | Up |
| SLC5A6 | 2.20E−05 | Up |
| GPI | 2.27E−05 | Up |
| C1orf112 | 2.29E−05 | Up |
| MYC | 2.32E−05 | Up |
| NIT2 | 2.40E−05 | Up |
| LGR4 | 2.49E−05 | Up |
| HOXC6 | 2.53E−05 | Up |
| FPR3 | 2.53E−05 | Up |
| GLT25D1 | 2.56E−05 | Up |
| RACGAP1 | 2.62E−05 | Up |
| MIF | 2.65E−05 | Up |
| CXCL3 | 2.69E−05 | Up |
| CTPS | 2.73E−05 | Up |
| FGD6 | 2.75E−05 | Up |
| FNDC3B | 2.76E−05 | Up |
| LHFPL2 | 2.81E−05 | Up |
| CDC25A | 2.90E−05 | Up |
| CD70 | 2.97E−05 | Up |
| TOMM70A | 3.03E−05 | Up |
| COPS2 | 3.12E−05 | Up |
| CCND2 | 3.12E−05 | Up |
| MARCO | 3.17E−05 | Up |
| WDR41 | 3.40E−05 | Up |
| TAF5 | 3.40E−05 | Up |
| AP3M2 | 3.49E−05 | Up |
| TMEM39A | 3.55E−05 | Up |
| TMEM185B | 3.55E−05 | Up |
| PLK4 | 3.64E−05 | Up |
| COL10A1 | 3.64E−05 | Up |
| C11orf41 | 3.68E−05 | Up |
| KITLG | 3.70E−05 | Up |
| PLOD1 | 3.79E−05 | Up |
| UBFD1 | 3.79E−05 | Up |
| NFKBIA | 3.79E−05 | Up |
| CORO1C | 3.85E−05 | Up |
| MORC4 | 3.95E−05 | Up |
| HOXA10 | 3.98E−05 | Up |
| MRPL13 | 4.11E−05 | Up |
| GINS1 | 4.17E−05 | Up |
| ZFP112 | 4.20E−05 | Up |
| EXT1 | 4.23E−05 | Up |
| KIF13A | 4.25E−05 | Up |
| TMEM97 | 4.31E−05 | Up |
| PIK3CD | 4.44E−05 | Up |
| IFIT3 | 4.59E−05 | Up |
| ETV7 | 4.76E−05 | Up |
| NCAPD3 | 4.77E−05 | Up |
| ANGPT2 | 4.83E−05 | Up |
| SNRPD1 | 5.10E−05 | Up |
| NDUFA9 | 5.24E−05 | Up |
| TMPO | 5.33E−05 | Up |
| CD14 | 5.38E−05 | Up |
| CDK5 | 5.44E−05 | Up |
| DKC1 | 5.55E−05 | Up |
| ASB9 | 5.76E−05 | Up |
| TRMT61B | 5.82E−05 | Up |
| C1QBP | 5.91E−05 | Up |
| WRN | 5.97E−05 | Up |
| TMEM106C | 5.97E−05 | Up |
| SYNE1 | 6.22E−05 | Up |
| ICAM1 | 6.27E−05 | Up |
| FAM60A | 6.41E−05 | Up |
| SRSF9 | 6.72E−05 | Up |
| PRR5L | 6.72E−05 | Up |
| MLF1IP | 6.78E−05 | Up |
| GNL2 | 6.80E−05 | Up |
| PSMB2 | 6.94E−05 | Up |
| ASPN | 7.03E−05 | Up |
| YRDC | 7.17E−05 | Up |
| KPNA2 | 7.25E−05 | Up |
| TYROBP | 7.30E−05 | Up |
| EXOSC5 | 7.33E−05 | Up |
| TNFRSF9 | 7.34E−05 | Up |
| GAS8 | 7.37E−05 | Up |
| TBCA | 7.38E−05 | Up |
| FAM162A | 7.38E−05 | Up |
| MAPKAPK5 | 7.41E−05 | Up |
| POSTN | 7.82E−05 | Up |
| SSTR2 | 7.89E−05 | Up |
| SUPV3L1 | 8.01E−05 | Up |
| RSAD2 | 8.05E−05 | Up |
| RFC2 | 8.05E−05 | Up |
| H2AFZ | 8.05E−05 | Up |
| UBE2V2 | 8.11E−05 | Up |
| MFHAS1 | 8.29E−05 | Up |
| SYNCRIP | 8.60E−05 | Up |
| C17orf75 | 8.72E−05 | Up |
| WDR47 | 8.75E−05 | Up |
| FASTKD3 | 8.81E−05 | Up |
| NAA50 | 8.85E−05 | Up |
| ITGB1 | 8.94E−05 | Up |
| CAND1 | 9.04E−05 | Up |
| PTPN12 | 9.22E−05 | Up |
| MAT2A | 9.40E−05 | Up |
| DERA | 9.71E−05 | Up |
| KIAA0146 | 9.90E−05 | Up |
| EPCAM | 9.97E−05 | Up |
| C12orf29 | 1.05E−04 | Up |
| PYCARD | 1.06E−04 | Up |
| MDK | 1.07E−04 | Up |
| BMP3 | 1.07E−04 | Up |
| RAB17 | 1.08E−04 | Up |
| PLAUR | 1.08E−04 | Up |
| FASTKD2 | 1.09E−04 | Up |
| AIMP2 | 1.12E−04 | Up |
| WDR12 | 1.13E−04 | Up |
| ABCE1 | 1.13E−04 | Up |
| SERPINH1 | 1.15E−04 | Up |
| DEPDC1 | 1.16E−04 | Up |
| CYC1 | 1.18E−04 | Up |
| ITGB6 | 1.22E−04 | Up |
| PSMA7 | 1.23E−04 | Up |
| ZNF74 | 1.24E−04 | Up |
| SLMO2 | 1.24E−04 | Up |
| SCO2 | 1.24E−04 | Up |
| DMXL2 | 1.26E−04 | Up |
| BST2 | 1.26E−04 | Up |
| XPO1 | 1.27E−04 | Up |
| ENOPH1 | 1.27E−04 | Up |
| SCG5 | 1.28E−04 | Up |
| PAICS | 1.29E−04 | Up |
| WASF1 | 1.31E−04 | Up |
| BCL2A1 | 1.32E−04 | Up |
| LGALS1 | 1.32E−04 | Up |
| RXRB | 1.32E−04 | Up |
| RRM1 | 1.34E−04 | Up |
| PXDN | 1.37E−04 | Up |
| PTRH2 | 1.38E−04 | Up |
| JMJD4 | 1.38E−04 | Up |
| MBD5 | 1.39E−04 | Up |
| RNF19B | 1.40E−04 | Up |
| PRC1 | 1.43E−04 | Up |
| PRDM4 | 1.46E−04 | Up |
| NPM3 | 1.48E−04 | Up |
| UBE2Z | 1.48E−04 | Up |
| KIF23 | 1.53E−04 | Up |
| RRN3 | 1.55E−04 | Up |
| ZNF532 | 1.58E−04 | Up |
| PIGA | 1.58E−04 | Up |
| WBP5 | 1.59E−04 | Up |
| PIGN | 1.59E−04 | Up |
| IL13RA2 | 1.60E−04 | Up |
| GTF3C3 | 1.60E−04 | Up |
| ARMC1 | 1.63E−04 | Up |
| NOL10 | 1.64E−04 | Up |
| COL4A5 | 1.65E−04 | Up |
| GADD45A | 1.65E−04 | Up |
| FOXK2 | 1.71E−04 | Up |
| CCT3 | 1.71E−04 | Up |
| IDH1 | 1.72E−04 | Up |
| ADIPOR2 | 1.72E−04 | Up |
| PSMC4 | 1.72E−04 | Up |
| FCGR1B | 1.72E−04 | Up |
| ROBO1 | 1.72E−04 | Up |
| CST1 | 1.74E−04 | Up |
| MEST | 1.78E−04 | Up |
| CLSTN2 | 1.79E−04 | Up |
| UPF3B | 1.81E−04 | Up |
| URB2 | 1.81E−04 | Up |
| TPT1 | 1.81E−04 | Up |
| FAH | 1.81E−04 | Up |
| B4GALT2 | 1.82E−04 | Up |
| PPIA | 1.82E−04 | Up |
| NME1 | 1.89E−04 | Up |
| AQP9 | 1.95E−04 | Up |
| UQCRH | 1.96E−04 | Up |
| UBE2C | 1.96E−04 | Up |
| MRPS35 | 1.97E−04 | Up |
| ADAM9 | 1.97E−04 | Up |
| DKK3 | 2.00E−04 | Up |
| RBL2 | 2.00E−04 | Up |
| DDX60 | 2.07E−04 | Up |
| ZBTB39 | 2.09E−04 | Up |
| MCTP2 | 2.13E−04 | Up |
| LOC100499177 | 2.13E−04 | Up |
| SCMH1 | 2.15E−04 | Up |
| CUEDC2 | 2.17E−04 | Up |
| ZNF140 | 2.17E−04 | Up |
| TAP1 | 2.18E−04 | Up |
| E2F6 | 2.20E−04 | Up |
| PA2G4 | 2.26E−04 | Up |
| BMP2 | 2.29E−04 | Up |
| DIABLO | 2.35E−04 | Up |
| PSMD1 | 2.37E−04 | Up |
| CKAP5 | 2.41E−04 | Up |
| PRLR | 2.43E−04 | Up |
| ERO1LB | 2.44E−04 | Up |
| SPIN1 | 2.45E−04 | Up |
| PRNP | 2.45E−04 | Up |
| PIWIL1 | 2.45E−04 | Up |
| TLN2 | 2.47E−04 | Up |
| SRP9 | 2.49E−04 | Up |
| SOCS5 | 2.52E−04 | Up |
| CCR1 | 2.55E−04 | Up |
| CEP76 | 2.58E−04 | Up |
| SCARB2 | 2.60E−04 | Up |
| MRPL12 | 2.65E−04 | Up |
| ISYNA1 | 2.66E−04 | Up |
| RPL32 | 2.70E−04 | Up |
| PKD2 | 2.70E−04 | Up |
| KAT2B | 2.78E−04 | Up |
| TRIB1 | 2.80E−04 | Up |
| KDSR | 2.84E−04 | Up |
| IFI44L | 2.84E−04 | Up |
| UGCG | 2.86E−04 | Up |
| POLR3G | 2.89E−04 | Up |
| THBS2 | 2.97E−04 | Up |
| IGSF6 | 2.97E−04 | Up |
| STRAP | 3.12E−04 | Up |
| EIF2AK2 | 3.14E−04 | Up |
| ADAM12 | 3.14E−04 | Up |
| SNX27 | 3.15E−04 | Up |
| KREMEN2 | 3.15E−04 | Up |
| CEP192 | 3.15E−04 | Up |
| PPP6C | 3.21E−04 | Up |
| LOC730101 | 3.23E−04 | Up |
| PSMA6 | 3.25E−04 | Up |
| TGFBI | 3.25E−04 | Up |
| PSTPIP2 | 3.25E−04 | Up |
| CCL8 | 3.28E−04 | Up |
| RPS29 | 3.29E−04 | Up |
| PLA2G7 | 3.30E−04 | Up |
| PNO1 | 3.32E−04 | Up |
| RARB | 3.45E−04 | Up |
| CLIP2 | 3.45E−04 | Up |
| LAMP2 | 3.45E−04 | Up |
| CCT7 | 3.45E−04 | Up |
| ABTB2 | 3.45E−04 | Up |
| ABCA3 | 3.47E−04 | Up |
| NCK1 | 3.53E−04 | Up |
| CDK8 | 3.59E−04 | Up |
| WSB2 | 3.62E−04 | Up |
| DNMT3B | 3.63E−04 | Up |
| EVL | 3.69E−04 | Up |
| PTPLAD1 | 3.74E−04 | Up |
| GLA | 3.81E−04 | Up |
| ADH5 | 3.88E−04 | Up |
| CDK4 | 3.94E−04 | Up |
| UMPS | 3.99E−04 | Up |
| UCP2 | 3.99E−04 | Up |
| PTGES3 | 4.01E−04 | Up |
| LOC100127972 | 4.01E−04 | Up |
| FZD7 | 4.01E−04 | Up |
| TUBGCP4 | 4.04E−04 | Up |
| PPA1 | 4.07E−04 | Up |
| ARPC1A | 4.08E−04 | Up |
| PDSS1 | 4.12E−04 | Up |
| IFIT1 | 4.17E−04 | Up |
| INPP1 | 4.17E−04 | Up |
| KCNE1 | 4.22E−04 | Up |
| PXMP2 | 4.24E−04 | Up |
| IL15 | 4.28E−04 | Up |
| PCSK7 | 4.33E−04 | Up |
| PRPF40A | 4.34E−04 | Up |
| MLLT11 | 4.34E−04 | Up |
| MKKS | 4.52E−04 | Up |
| NIPSNAP1 | 4.57E−04 | Up |
| TSR2 | 4.59E−04 | Up |
| LDHA | 4.62E−04 | Up |
| CHCHD3 | 4.76E−04 | Up |
| NCAPH | 4.82E−04 | Up |
| R3HDM1 | 4.98E−04 | Up |
| SPAST | 4.98E−04 | Up |
| PDHX | 5.01E−04 | Up |
| C2orf47 | 5.01E−04 | Up |
| CBX1 | 5.02E−04 | Up |
| PACS2 | 5.10E−04 | Up |
| C3AR1 | 5.23E−04 | Up |
| ANAPC10 | 5.24E−04 | Up |
| CCNJ | 5.25E−04 | Up |
| TARS | 5.41E−04 | Up |
| ATP5G3 | 5.49E−04 | Up |
| HSPA13 | 5.52E−04 | Up |
| TCF12 | 5.54E−04 | Up |
| EIF1AX | 5.59E−04 | Up |
| CBX5 | 5.75E−04 | Up |
| YBX1 | 5.79E−04 | Up |
| CTSL1 | 5.82E−04 | Up |
| RFWD3 | 5.83E−04 | Up |
| ZNF473 | 5.84E−04 | Up |
| PDCD11 | 5.85E−04 | Up |
| TGIF2 | 5.86E−04 | Up |
| GSTK1 | 5.90E−04 | Up |
| GPR143 | 5.94E−04 | Up |
| NMD3 | 5.96E−04 | Up |
| JAG2 | 5.97E−04 | Up |
| DIAPH1 | 5.99E−04 | Up |
| GJA1 | 6.03E−04 | Up |
| KCNMB2 | 6.13E−04 | Up |
| ACOT7 | 6.15E−04 | Up |
| TMEM66 | 6.15E−04 | Up |
| REPIN1 | 6.18E−04 | Up |
| TRAP1 | 6.28E−04 | Up |
| SPDEF | 6.32E−04 | Up |
| YTHDF3 | 6.46E−04 | Up |
| TNFAIP3 | 6.63E−04 | Up |
| SNRPE | 6.63E−04 | Up |
| WDYHV1 | 6.64E−04 | Up |
| FNBP1L | 6.64E−04 | Up |
| C6orf211 | 6.65E−04 | Up |
| ATP5B | 6.65E−04 | Up |
| SEH1L | 6.81E−04 | Up |
| EFNB2 | 6.81E−04 | Up |
| C1orf109 | 6.81E−04 | Up |
| SOX12 | 6.84E−04 | Up |
| SCAI | 7.20E−04 | Up |
| ISG15 | 7.29E−04 | Up |
| RTF1 | 7.30E−04 | Up |
| ANKRD5 | 7.35E−04 | Up |
| AURKB | 7.40E−04 | Up |
| DARS | 7.52E−04 | Up |
| SUMO2 | 7.58E−04 | Up |
| WDR1 | 7.68E−04 | Up |
| LRP4 | 7.68E−04 | Up |
| MYO1B | 7.73E−04 | Up |
| MCM10 | 7.73E−04 | Up |
| BYSL | 7.73E−04 | Up |
| MMP1 | 7.81E−04 | Up |
| CCNE2 | 7.91E−04 | Up |
| ADAR | 7.96E−04 | Up |
| MTCH2 | 7.97E−04 | Up |
| CHEK1 | 8.17E−04 | Up |
| XAF1 | 8.22E−04 | Up |
| LRP12 | 8.42E−04 | Up |
| PAFAH1B3 | 8.43E−04 | Up |
| STAMBP | 8.44E−04 | Up |
| NRAS | 8.54E−04 | Up |
| GHR | 8.54E−04 | Up |
| ATP5J2 | 8.68E−04 | Up |
| HMGCR | 8.75E−04 | Up |
| SENP2 | 8.78E−04 | Up |
| SLC35E3 | 9.08E−04 | Up |
| ORC1 | 9.19E−04 | Up |
| C5orf30 | 9.42E−04 | Up |
| GSTM2 | 9.49E−04 | Up |
| RSRC1 | 9.61E−04 | Up |
| DEGS1 | 9.70E−04 | Up |
| RPS6KC1 | 9.88E−04 | Up |
| RFC3 | 9.89E−04 | Up |
| ESPL1 | 9.93E−04 | Up |
| FKBP1A | 9.96E−04 | Up |
| FBXO11 | 9.98E−04 | Up |
| EHBP1 | 9.98E−04 | Up |
| COL1A1 | 9.98E−04 | Up |
| DNALI1 | 0.00E+00 | Down |
| ALDH1L1 | 1.36E−12 | Down |
| SPAG8 | 4.00E−11 | Down |
| SCRN1 | 4.64E−11 | Down |
| FOXJ1 | 4.64E−11 | Down |
| CLU | 4.64E−11 | Down |
| KLF2 | 1.91E−10 | Down |
| SCGB1A1 | 2.34E−10 | Down |
| SCGB2A1 | 2.93E−10 | Down |
| VPS11 | 6.74E−10 | Down |
| SYBU | 8.20E−10 | Down |
| PDCD5 | 9.56E−10 | Down |
| GPR162 | 2.24E−09 | Down |
| STIM1 | 2.40E−09 | Down |
| SLC44A4 | 3.44E−09 | Down |
| PCYT2 | 3.97E−09 | Down |
| EPAS1 | 3.97E−09 | Down |
| ABLIM1 | 4.29E−09 | Down |
| MT3 | 4.59E−09 | Down |
| ATP2C2 | 5.67E−09 | Down |
| CRYL1 | 7.85E−09 | Down |
| C6orf97 | 7.93E−09 | Down |
| PROM1 | 8.64E−09 | Down |
| BLK | 8.81E−09 | Down |
| B3GALT4 | 1.31E−08 | Down |
| CD22 | 1.88E−08 | Down |
| MSLN | 1.99E−08 | Down |
| CIRBP | 2.02E−08 | Down |
| WDR78 | 2.08E−08 | Down |
| MSRA | 2.08E−08 | Down |
| PTGDS | 2.47E−08 | Down |
| SLC16A7 | 3.25E−08 | Down |
| SMPD3 | 3.41E−08 | Down |
| CD1C | 3.97E−08 | Down |
| VIPR1 | 4.54E−08 | Down |
| GNMT | 6.09E−08 | Down |
| TCTA | 7.46E−08 | Down |
| C10orf81 | 1.06E−07 | Down |
| PIGR | 1.23E−07 | Down |
| IGHD | 1.23E−07 | Down |
| FAM107A | 1.23E−07 | Down |
| ASL | 1.26E−07 | Down |
| NBEA | 1.28E−07 | Down |
| AQP5 | 1.61E−07 | Down |
| SGSM3 | 1.66E−07 | Down |
| MSMB | 1.76E−07 | Down |
| IK | 1.78E−07 | Down |
| PDLIM2 | 2.21E−07 | Down |
| DDAH2 | 2.22E−07 | Down |
| FBXO22 | 2.29E−07 | Down |
| PIK3C2B | 2.77E−07 | Down |
| VPREB3 | 3.67E−07 | Down |
| PDCD6IP | 4.01E−07 | Down |
| TREML2 | 4.33E−07 | Down |
| FCRL2 | 4.33E−07 | Down |
| FAM174B | 4.33E−07 | Down |
| DPEP2 | 4.52E−07 | Down |
| CLMN | 4.87E−07 | Down |
| RNASE4 | 5.08E−07 | Down |
| PPP3CA | 5.08E−07 | Down |
| GNG7 | 5.18E−07 | Down |
| AGR2 | 5.39E−07 | Down |
| GLTSCR2 | 5.44E−07 | Down |
| GALNT12 | 5.54E−07 | Down |
| ABHD14A | 6.19E−07 | Down |
| KLHDC2 | 6.55E−07 | Down |
| C6orf103 | 7.80E−07 | Down |
| CD72 | 7.96E−07 | Down |
| AK1 | 8.20E−07 | Down |
| LXN | 8.22E−07 | Down |
| FAM102A | 8.56E−07 | Down |
| CR1 | 8.77E−07 | Down |
| CD1D | 8.77E−07 | Down |
| MEIS3P1 | 8.84E−07 | Down |
| LMO2 | 1.01E−06 | Down |
| CYP2F1 | 1.04E−06 | Down |
| BAIAP3 | 1.05E−06 | Down |
| RPS6KA3 | 1.21E−06 | Down |
| WFDC2 | 1.24E−06 | Down |
| CD19 | 1.35E−06 | Down |
| BACE2 | 1.40E−06 | Down |
| DALRD3 | 1.41E−06 | Down |
| SIDT2 | 1.44E−06 | Down |
| PIP | 1.61E−06 | Down |
| ABCB1 | 1.88E−06 | Down |
| FAM65B | 1.90E−06 | Down |
| RRAGA | 1.92E−06 | Down |
| AGBL2 | 2.06E−06 | Down |
| C5orf45 | 2.19E−06 | Down |
| E2F3 | 2.55E−06 | Down |
| SFMBT1 | 2.56E−06 | Down |
| TUBA4A | 2.58E−06 | Down |
| BANK1 | 2.58E−06 | Down |
| TNFRSF13B | 2.64E−06 | Down |
| HMGCL | 2.67E−06 | Down |
| TTC12 | 2.84E−06 | Down |
| GCNT3 | 2.87E−06 | Down |
| VTCN1 | 2.88E−06 | Down |
| C10orf116 | 3.00E−06 | Down |
| MFNG | 3.01E−06 | Down |
| CD180 | 3.10E−06 | Down |
| AHCTF1 | 3.12E−06 | Down |
| TFF3 | 3.13E−06 | Down |
| IQSEC1 | 3.17E−06 | Down |
| POU2AF1 | 3.24E−06 | Down |
| ENPP4 | 3.24E−06 | Down |
| BEND5 | 3.51E−06 | Down |
| CLDN10 | 3.59E−06 | Down |
| CD40LG | 3.79E−06 | Down |
| ANG | 4.04E−06 | Down |
| GPR110 | 4.04E−06 | Down |
| SERHL2 | 4.17E−06 | Down |
| ACAA1 | 4.21E−06 | Down |
| KAT5 | 4.62E−06 | Down |
| TBC1D22A | 4.74E−06 | Down |
| FZD4 | 5.30E−06 | Down |
| ATP1A2 | 5.30E−06 | Down |
| KCNJ16 | 5.30E−06 | Down |
| HSPB8 | 5.99E−06 | Down |
| CCDC69 | 6.02E−06 | Down |
| CYB561D2 | 6.59E−06 | Down |
| TINF2 | 7.58E−06 | Down |
| ST6GAL1 | 7.74E−06 | Down |
| QARS | 7.89E−06 | Down |
| SELENBP1 | 9.03E−06 | Down |
| CRY2 | 9.09E−06 | Down |
| DUSP26 | 9.93E−06 | Down |
| MPG | 1.01E−05 | Down |
| COX7A1 | 1.02E−05 | Down |
| SWAP70 | 1.05E−05 | Down |
| MS4A1 | 1.05E−05 | Down |
| IFT88 | 1.09E−05 | Down |
| DND1 | 1.12E−05 | Down |
| KCNQ1 | 1.16E−05 | Down |
| CH25H | 1.21E−05 | Down |
| LYL1 | 1.22E−05 | Down |
| SRPX | 1.24E−05 | Down |
| NME5 | 1.30E−05 | Down |
| CEBPG | 1.30E−05 | Down |
| PLA2G16 | 1.32E−05 | Down |
| ZNF395 | 1.41E−05 | Down |
| PLEKHB1 | 1.47E−05 | Down |
| C11orf71 | 1.49E−05 | Down |
| RAD51 | 1.54E−05 | Down |
| BEX4 | 1.63E−05 | Down |
| C3 | 1.71E−05 | Down |
| SMPD1 | 1.75E−05 | Down |
| CLCF1 | 1.76E−05 | Down |
| GPRIN2 | 1.77E−05 | Down |
| SGSM2 | 1.79E−05 | Down |
| CAND2 | 2.00E−05 | Down |
| WDHD1 | 2.10E−05 | Down |
| ARMCX6 | 2.10E−05 | Down |
| FGD2 | 2.18E−05 | Down |
| FMO2 | 2.21E−05 | Down |
| C6 | 2.35E−05 | Down |
| LMBRD1 | 2.53E−05 | Down |
| AK2 | 2.53E−05 | Down |
| ABCA7 | 2.71E−05 | Down |
| NXF2 | 2.75E−05 | Down |
| GABRP | 2.82E−05 | Down |
| DNAH6 | 2.84E−05 | Down |
| CTDSP1 | 2.86E−05 | Down |
| PLAC8 | 2.97E−05 | Down |
| BCAS4 | 3.03E−05 | Down |
| BAALC | 3.03E−05 | Down |
| GTF2F2 | 3.05E−05 | Down |
| VILL | 3.12E−05 | Down |
| POLD4 | 3.17E−05 | Down |
| ITIH5 | 3.17E−05 | Down |
| DAZAP2 | 3.17E−05 | Down |
| SIGIRR | 3.22E−05 | Down |
| EPHB6 | 3.30E−05 | Down |
| MEIS2 | 3.35E−05 | Down |
| ICAM3 | 3.35E−05 | Down |
| CCR6 | 3.35E−05 | Down |
| PSD4 | 3.49E−05 | Down |
| EFCAB1 | 3.64E−05 | Down |
| TGFBR3 | 3.78E−05 | Down |
| TMEM184B | 3.84E−05 | Down |
| SPATA6 | 3.85E−05 | Down |
| CRLF1 | 3.95E−05 | Down |
| CLDN8 | 4.17E−05 | Down |
| ADCY2 | 4.30E−05 | Down |
| PIGH | 4.32E−05 | Down |
| TFF1 | 4.33E−05 | Down |
| TSPAN1 | 4.55E−05 | Down |
| SIDT1 | 4.63E−05 | Down |
| PHF1 | 4.65E−05 | Down |
| ARHGAP44 | 4.76E−05 | Down |
| TMEM121 | 4.97E−05 | Down |
| CRIP2 | 4.99E−05 | Down |
| APOM | 5.07E−05 | Down |
| FUCA1 | 5.18E−05 | Down |
| NEIL1 | 5.19E−05 | Down |
| LPAR1 | 5.20E−05 | Down |
| PSMA2 | 5.25E−05 | Down |
| LRIG1 | 5.32E−05 | Down |
| SRSF5 | 5.44E−05 | Down |
| ZFP106 | 5.54E−05 | Down |
| CRIP1 | 5.64E−05 | Down |
| MAST3 | 5.82E−05 | Down |
| DHX38 | 5.82E−05 | Down |
| PRKCB | 6.01E−05 | Down |
| MRPL19 | 6.11E−05 | Down |
| ABCD4 | 6.37E−05 | Down |
| HSD17B8 | 6.72E−05 | Down |
| GPR18 | 6.72E−05 | Down |
| DNAH3 | 6.72E−05 | Down |
| ADAMTSL3 | 6.72E−05 | Down |
| INHBB | 6.77E−05 | Down |
| NCR3 | 6.79E−05 | Down |
| CLDN3 | 6.79E−05 | Down |
| RNASE1 | 6.80E−05 | Down |
| CCNA1 | 6.86E−05 | Down |
| GOLGA8A | 7.00E−05 | Down |
| PDZD2 | 7.03E−05 | Down |
| CCDC19 | 7.03E−05 | Down |
| EIF1B | 7.25E−05 | Down |
| DLEC1 | 7.32E−05 | Down |
| EIF3J | 7.41E−05 | Down |
| S1PR4 | 7.82E−05 | Down |
| ACTR10 | 7.96E−05 | Down |
| CHL1 | 7.99E−05 | Down |
| PPRC1 | 8.05E−05 | Down |
| TSPAN32 | 8.06E−05 | Down |
| LRRC23 | 8.06E−05 | Down |
| CGRRF1 | 8.29E−05 | Down |
| EFCAB6 | 8.32E−05 | Down |
| LAMB2 | 8.40E−05 | Down |
| CAP2 | 8.51E−05 | Down |
| DUSP22 | 8.73E−05 | Down |
| ADAM28 | 8.76E−05 | Down |
| LMAN1 | 8.77E−05 | Down |
| TDG | 8.81E−05 | Down |
| AES | 9.04E−05 | Down |
| VNN2 | 9.22E−05 | Down |
| NUCB2 | 9.22E−05 | Down |
| TIMP4 | 9.25E−05 | Down |
| TLE2 | 9.39E−05 | Down |
| PROS1 | 9.40E−05 | Down |
| EPHX1 | 9.74E−05 | Down |
| PGK1 | 9.76E−05 | Down |
| CLCN4 | 9.78E−05 | Down |
| YPEL5 | 9.90E−05 | Down |
| ERCC1 | 1.01E−04 | Down |
| TMC6 | 1.01E−04 | Down |
| RASGRP2 | 1.05E−04 | Down |
| SERPINB7 | 1.05E−04 | Down |
| ZFYVE21 | 1.05E−04 | Down |
| GAK | 1.06E−04 | Down |
| GFOD1 | 1.07E−04 | Down |
| ARHGAP11A | 1.07E−04 | Down |
| MAGIX | 1.08E−04 | Down |
| SPAG6 | 1.16E−04 | Down |
| LRMP | 1.17E−04 | Down |
| FLII | 1.18E−04 | Down |
| SNED1 | 1.21E−04 | Down |
| PPOX | 1.24E−04 | Down |
| UBA7 | 1.24E−04 | Down |
| BASP1 | 1.24E−04 | Down |
| IGJ | 1.27E−04 | Down |
| C5orf4 | 1.32E−04 | Down |
| CTNS | 1.34E−04 | Down |
| TEX264 | 1.35E−04 | Down |
| ABHD6 | 1.36E−04 | Down |
| TIMM13 | 1.39E−04 | Down |
| CCDC28A | 1.40E−04 | Down |
| P4HTM | 1.40E−04 | Down |
| IFIH1 | 1.43E−04 | Down |
| HIP1R | 1.44E−04 | Down |
| STS | 1.46E−04 | Down |
| ATP7B | 1.46E−04 | Down |
| STAG3 | 1.54E−04 | Down |
| ZNF137P | 1.56E−04 | Down |
| ZNF528 | 1.57E−04 | Down |
| TMEM9B | 1.57E−04 | Down |
| PACRG | 1.57E−04 | Down |
| DEPDC5 | 1.63E−04 | Down |
| CCDC81 | 1.65E−04 | Down |
| PRIM2 | 1.70E−04 | Down |
| RPS6KA1 | 1.72E−04 | Down |
| C14orf1 | 1.73E−04 | Down |
| CDT1 | 1.74E−04 | Down |
| NLRP1 | 1.76E−04 | Down |
| LPPR3 | 1.77E−04 | Down |
| ANKHD1 | 1.77E−04 | Down |
| CD52 | 1.79E−04 | Down |
| TCL1A | 1.79E−04 | Down |
| ERCC5 | 1.79E−04 | Down |
| RNASET2 | 1.81E−04 | Down |
| KRT7 | 1.81E−04 | Down |
| FAM184A | 1.81E−04 | Down |
| TMEM132A | 1.81E−04 | Down |
| C11orf16 | 1.82E−04 | Down |
| CAMTA1 | 1.82E−04 | Down |
| HDC | 1.93E−04 | Down |
| NIPSNAP3B | 1.96E−04 | Down |
| LIMD2 | 1.98E−04 | Down |
| BAP1 | 2.02E−04 | Down |
| ZNF821 | 2.06E−04 | Down |
| ZBBX | 2.09E−04 | Down |
| ZCWPW1 | 2.18E−04 | Down |
| TNNC2 | 2.18E−04 | Down |
| CALM1 | 2.22E−04 | Down |
| MAPK14 | 2.23E−04 | Down |
| SEL1L3 | 2.26E−04 | Down |
| VTI1B | 2.28E−04 | Down |
| P2RX5 | 2.28E−04 | Down |
| CHAF1A | 2.29E−04 | Down |
| SGTA | 2.30E−04 | Down |
| MNS1 | 2.35E−04 | Down |
| FYCO1 | 2.35E−04 | Down |
| IL33 | 2.35E−04 | Down |
| SCUBE2 | 2.38E−04 | Down |
| TRIM24 | 2.41E−04 | Down |
| HSD17B2 | 2.42E−04 | Down |
| RASAL1 | 2.42E−04 | Down |
| ANXA11 | 2.44E−04 | Down |
| B3GNT3 | 2.44E−04 | Down |
| GLT8D1 | 2.45E−04 | Down |
| BCAR3 | 2.45E−04 | Down |
| ZBTB16 | 2.47E−04 | Down |
| ADRA2A | 2.49E−04 | Down |
| USP19 | 2.51E−04 | Down |
| RARRES2 | 2.51E−04 | Down |
| SLC46A3 | 2.60E−04 | Down |
| MAGOHB | 2.62E−04 | Down |
| SIRT3 | 2.63E−04 | Down |
| STAP1 | 2.64E−04 | Down |
| INPP4B | 2.66E−04 | Down |
| PEPD | 2.66E−04 | Down |
| JHDM1D | 2.80E−04 | Down |
| COBL | 2.80E−04 | Down |
| ITIH4 | 2.81E−04 | Down |
| DOCK3 | 2.84E−04 | Down |
| ADH1C | 2.86E−04 | Down |
| C11orf2 | 2.87E−04 | Down |
| RHOBTB2 | 3.00E−04 | Down |
| SNW1 | 3.01E−04 | Down |
| CD79A | 3.02E−04 | Down |
| TMC5 | 3.06E−04 | Down |
| ALDH3B1 | 3.14E−04 | Down |
| CDC14A | 3.22E−04 | Down |
| SNTA1 | 3.23E−04 | Down |
| LYN | 3.32E−04 | Down |
| LTF | 3.33E−04 | Down |
| MLYCD | 3.41E−04 | Down |
| DNAI1 | 3.45E−04 | Down |
| C1orf115 | 3.45E−04 | Down |
| CBX6 | 3.46E−04 | Down |
| KCNJ12 | 3.63E−04 | Down |
| ZBTB25 | 3.65E−04 | Down |
| UNC93B1 | 3.69E−04 | Down |
| C7orf44 | 3.72E−04 | Down |
| KIAA0125 | 3.80E−04 | Down |
| PARP3 | 3.94E−04 | Down |
| LOC284244 | 3.99E−04 | Down |
| FOLR1 | 3.99E−04 | Down |
| PPFIA4 | 4.13E−04 | Down |
| PKIG | 4.15E−04 | Down |
| MOAP1 | 4.17E−04 | Down |
| C17orf59 | 4.24E−04 | Down |
| EFHC1 | 4.26E−04 | Down |
| CRISP2 | 4.26E−04 | Down |
| ANK2 | 4.29E−04 | Down |
| RHOH | 4.40E−04 | Down |
| APOD | 4.45E−04 | Down |
| TFEB | 4.49E−04 | Down |
| PAIP2B | 4.49E−04 | Down |
| CREBZF | 4.52E−04 | Down |
| OCEL1 | 4.64E−04 | Down |
| SNX3 | 4.72E−04 | Down |
| TFB2M | 4.82E−04 | Down |
| ALDH6A1 | 4.90E−04 | Down |
| RPGRIP1 | 4.93E−04 | Down |
| PNMA1 | 5.02E−04 | Down |
| TCEB1 | 5.18E−04 | Down |
| ATP5L | 5.25E−04 | Down |
| LOC728855 | 5.28E−04 | Down |
| RPL36AL | 5.52E−04 | Down |
| XRCC4 | 5.69E−04 | Down |
| TMEM63A | 5.74E−04 | Down |
| C9orf9 | 5.92E−04 | Down |
| SLC15A2 | 6.00E−04 | Down |
| FAIM3 | 6.20E−04 | Down |
| VAV1 | 6.20E−04 | Down |
| CHKB | 6.24E−04 | Down |
| CP | 6.41E−04 | Down |
| PAF1 | 6.53E−04 | Down |
| MAGED2 | 6.58E−04 | Down |
| RRAD | 6.58E−04 | Down |
| TXNIP | 6.64E−04 | Down |
| ZFP161 | 6.79E−04 | Down |
| DIO1 | 6.79E−04 | Down |
| PTGER2 | 6.81E−04 | Down |
| SATB1 | 6.82E−04 | Down |
| PRR15L | 6.82E−04 | Down |
| TRADD | 6.87E−04 | Down |
| SLC1A1 | 6.88E−04 | Down |
| CHD7 | 7.19E−04 | Down |
| CRYM | 7.20E−04 | Down |
| C7 | 7.20E−04 | Down |
| RBM38 | 7.34E−04 | Down |
| WWP2 | 7.40E−04 | Down |
| DTYMK | 7.48E−04 | Down |
| OR7E47P | 7.52E−04 | Down |
| NIP7 | 7.54E−04 | Down |
| FAM50B | 7.57E−04 | Down |
| XPA | 7.58E−04 | Down |
| KLHL7 | 7.73E−04 | Down |
| ZNF839 | 7.81E−04 | Down |
| SLC34A1 | 7.81E−04 | Down |
| FBXL2 | 7.85E−04 | Down |
| PLSCR4 | 7.92E−04 | Down |
| PBXIP1 | 7.93E−04 | Down |
| NAT6 | 7.94E−04 | Down |
| PPIL2 | 7.97E−04 | Down |
| KLHL36 | 8.19E−04 | Down |
| MAN2B2 | 8.24E−04 | Down |
| EHD1 | 8.42E−04 | Down |
| GPR183 | 8.75E−04 | Down |
| MFAP4 | 8.84E−04 | Down |
| HCLS1 | 8.99E−04 | Down |
| APEH | 9.01E−04 | Down |
| LOC100129973 | 9.17E−04 | Down |
| DPAGT1 | 9.23E−04 | Down |
| TREX1 | 9.39E−04 | Down |
| PPP3CC | 9.39E−04 | Down |
| ATG4A | 9.44E−04 | Down |
| LY9 | 9.49E−04 | Down |
| TERF2IP | 9.53E−04 | Down |
| HSPA9 | 9.53E−04 | Down |
| ITGA7 | 9.61E−04 | Down |
| CAPNS1 | 9.63E−04 | Down |
FDR: false discovery rate
Footnotes
Source of support: Departmental sources
References
- 1.Chin D, Boyle GM, Porceddu S, et al. Head and neck cancer: Past, present and future. Expert Rev Anticancer Ther. 2006;6:1111–18. doi: 10.1586/14737140.6.7.1111. [DOI] [PubMed] [Google Scholar]
- 2.Kulkarni P, Saxena U. Head and neck cancers, the neglected malignancies: Present and future treatment strategies. Expert Opin Ther Targets. 2014;18:351–54. doi: 10.1517/14728222.2014.888059. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15(10):1765–77. doi: 10.1158/1055-9965.EPI-06-0353. [DOI] [PubMed] [Google Scholar]
- 5.Tsao SW, Yip YL, Tsang CM, et al. Etiological factors of nasopharyngeal carcinoma. Oral Oncol. 2014;50:330–38. doi: 10.1016/j.oraloncology.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Yang W, Lan X, Li D, et al. MiR-223 targeting MAFB suppresses proliferation and migration of nasopharyngeal carcinoma cells. BMC Cancer. 2015;15:461. doi: 10.1186/s12885-015-1464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei WI, Sham JST. Nasopharyngeal carcinoma. Lancet. 2005;365:2041–54. doi: 10.1016/S0140-6736(05)66698-6. [DOI] [PubMed] [Google Scholar]
- 8.Ren XY, Zhou G-Q, Jiang W, et al. Low SFRP1 expression correlates with poor prognosis and promotes cell invasion by activating the Wnt/β-catenin signaling pathway in NPC. Cancer Prev Res. 2015;8(10):968–77. doi: 10.1158/1940-6207.CAPR-14-0369. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Zhong T, Dang Y, et al. Aberrant expression of CDK5 infers poor outcomes for nasopharyngeal carcinoma patients. Int J Clin Exp Pathol. 2015;8(7):8066–74. [PMC free article] [PubMed] [Google Scholar]
- 10.Sang Y, Chen MY, Luo D, et al. TEL2 suppresses metastasis by down-regulating SERPINE1 in nasopharyngeal carcinoma. Oncotarget. 2015;6(30):29240–53. doi: 10.18632/oncotarget.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo YM, Sun MX, Li J, et al. Association of CELF2 polymorphism and the prognosis of nasopharyngeal carcinoma in southern Chinese population. Oncotarget. 2015;6(29):27176–86. doi: 10.18632/oncotarget.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phoon YP, Cheung AK, Cheung FM, et al. IKBB tumor suppressive role in nasopharyngeal carcinoma via NF-kappaB-mediated signalling. Int J Cancer. 2016;138(1):160–70. doi: 10.1002/ijc.29702. [DOI] [PubMed] [Google Scholar]
- 13.Jia WH, Qin HD. Non-viral environmental risk factors for nasopharyngeal carcinoma: A systematic review. Semin Cancer Biol. 2012;22:117–26. doi: 10.1016/j.semcancer.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Jia WH, Luo XY, Feng BJ, et al. Traditional Cantonese diet and nasopharyngeal carcinoma risk: A large-scale case-control study in Guangdong, China. BMC Cancer. 2010;10:446. doi: 10.1186/1471-2407-10-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji X, Zhang W, Xie C, et al. Nasopharyngeal carcinoma risk by histologic type in central China: impact of smoking, alcohol and family history. Int J Cancer. 2011;129:724–32. doi: 10.1002/ijc.25696. [DOI] [PubMed] [Google Scholar]
- 16.Hsu W-L, Chen J-Y, Chien Y-C, et al. Independent effect of EBV and cigarette smoking on nasopharyngeal carcinoma: A 20-year follow-up study on 9,622 males without family history in Taiwan. Cancer Epidemiol Biomarkers Prev. 2009;18:1218–26. doi: 10.1158/1055-9965.EPI-08-1175. [DOI] [PubMed] [Google Scholar]
- 17.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–68. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 18.Ohtsuka M, Ling H, Doki Y, et al. MicroRNA processing and human cancer. J Clin Med. 2015;4:1651–67. doi: 10.3390/jcm4081651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi X, Li J, Zhou C, et al. MiR-142-3p suppresses SOCS6 expression and promotes cell proliferation in nasopharyngeal carcinoma. Cell Physiol Biochem. 2015;36:1743–52. doi: 10.1159/000430147. [DOI] [PubMed] [Google Scholar]
- 20.Pan X, Peng G, Liu S, et al. MicroRNA-4649-3p inhibits cell proliferation by targeting protein tyrosine phosphatase SHP-1 in nasopharyngeal carcinoma cells. Int J Mol Med. 2015;36:559–64. doi: 10.3892/ijmm.2015.2245. [DOI] [PubMed] [Google Scholar]
- 21.Qiu F, Sun R, Deng N, et al. miR-29a/b enhances cell migration and invasion in nasopharyngeal carcinoma progression by regulating SPARC and COL3A1 gene expression. PloS One. 2015;10:e0120969. doi: 10.1371/journal.pone.0120969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu JQ, Yi HM, Ye H, et al. MiR-23a sensitizes nasopharyngeal carcinoma to irradiation by targeting IL-8/Stat3 pathway. Oncotarget. 2015;6(29):28341–56. doi: 10.18632/oncotarget.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao L, Tang M, Hu Z, et al. miR-504 mediated down-regulation of nuclear respiratory factor 1 leads to radio-resistance in nasopharyngeal carcinoma. Oncotarget. 2015;6:15995–6018. doi: 10.18632/oncotarget.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diboun I, Wernisch L, Orengo CA, Koltzenburg M. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics. 2006;7:252. doi: 10.1186/1471-2164-7-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839–47. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 28.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hulsegge I, Kommadath A, Smits MA. Globaltest and GOEAST: Two different approaches for Gene Ontology analysis. BMC Proc. 2009;3(Suppl 4):S10. doi: 10.1186/1753-6561-3-S4-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M. The KEGG database. Novartis Foundation symposium 2002; 2002. pp. 91–101. discussion 103, 119–28, 244–52. [PubMed] [Google Scholar]
- 31.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Min W, Wang B, Li J, et al. The expression and significance of five types of miRNAs in breast cancer. Med Sci Monit Basic Res. 2014;20:97–104. doi: 10.12659/MSMBR.891246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo Z, Zhang L, Li Z, et al. An in silico analysis of dynamic changes in microRNA expression profiles in stepwise development of nasopharyngeal carcinoma. BMC Med Genomics. 2012;5:3. doi: 10.1186/1755-8794-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li T, Chen JX, Fu XP, et al. microRNA expression profiling of nasopharyngeal carcinoma. Oncol Rep. 2011;25:1353–63. doi: 10.3892/or.2011.1204. [DOI] [PubMed] [Google Scholar]
- 35.Tang J-F, Yu Z-H, Liu T, et al. Five miRNAs as novel diagnostic biomarker candidates for primary nasopharyngeal carcinoma. Asian Pac J Cancer Prev. 2013;15:7575–81. doi: 10.7314/apjcp.2014.15.18.7575. [DOI] [PubMed] [Google Scholar]
- 36.Qu C, Liang Z, Huang J, et al. MiR-205 determines the radioresistance of human nasopharyngeal carcinoma by directly targeting PTEN. Cell Cycle. 2012;11:785–96. doi: 10.4161/cc.11.4.19228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D, Wang S, Liu Q, Yang, et al. SZ-685C exhibits potent anticancer activity in both radiosensitive and radioresistant NPC cells through the miR-205-PTEN-Akt pathway. Oncol Rep. 2013;29:2341–47. doi: 10.3892/or.2013.2376. [DOI] [PubMed] [Google Scholar]
- 38.Chang Y, Lee TC, Li JC, et al. Differential expression of osteoblast-specific factor 2 and polymeric immunoglobulin receptor genes in nasopharyngeal carcinoma. Head Neck. 2005;27:873–82. doi: 10.1002/hed.20253. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Wu J, Sima X, et al. Interactions of miR-34b/c and TP-53 polymorphisms on the risk of nasopharyngeal carcinoma. Tumour Biol. 2013;34:1919–23. doi: 10.1007/s13277-013-0736-9. [DOI] [PubMed] [Google Scholar]
- 40.Zhang X, Xiao D, Wang Z, et al. MicroRNA-26a/b regulate DNA replication licensing, tumorigenesis, and prognosis by targeting CDC6 in lung cancer. Mol Cancer Res. 2014;12:1535–46. doi: 10.1158/1541-7786.MCR-13-0641. [DOI] [PubMed] [Google Scholar]
- 41.Feng CJ, Lu XW, Luo DY, et al. Knockdown of Cdc6 inhibits proliferation of tongue squamous cell carcinoma Tca8113 cells. Technol Cancer Res Treat. 2013;12:173–81. doi: 10.7785/tcrt.2012.500302. [DOI] [PubMed] [Google Scholar]
- 42.Yu X, Zhen Y, Yang H, et al. Loss of connective tissue growth factor as an unfavorable prognosis factor activates miR-18b by PI3K/AKT/C-Jun and C-Myc and promotes cell growth in nasopharyngeal carcinoma. Cell Death Dis. 2013;4:e634. doi: 10.1038/cddis.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levy M, Castillo SS, Goldkorn T. nSMase2 activation and trafficking are modulated by oxidative stress to induce apoptosis. Biochem Biophys Res Commun. 2006;344:900–5. doi: 10.1016/j.bbrc.2006.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kosaka N, Iguchi H, Hagiwara K, et al. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. 2013;288:10849–59. doi: 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui J, Li Y, Zhu L, et al. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol. 2012;13:387–95. doi: 10.1038/ni.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radtke F, Wilson A, Mancini SJ, MacDonald HR. Notch regulation of lymphocyte development and function. Nat Immunol. 2004;5:247–53. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- 47.Katoh M. Networking of WNT, FGF, Notch, BMP, and Hedgehog signaling pathways during carcinogenesis. Stem Cell Rev. 2007;3:30–38. doi: 10.1007/s12015-007-0006-6. [DOI] [PubMed] [Google Scholar]
- 48.Poe JC, Fujimoto Y, Hasegawa M, et al. CD22 regulates B lymphocyte function in vivo through both ligand-dependent and ligand-independent mechanisms. Nat Immunol. 2004;5:1078–87. doi: 10.1038/ni1121. [DOI] [PubMed] [Google Scholar]
- 49.Mitra A, Rostas JW, Dyess DL, et al. Micro-RNA-632 downregulates DNAJB6 in breast cancer. Lab Invest. 2012;92:1310–17. doi: 10.1038/labinvest.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Behrens J. The role of the Wnt signalling pathway in colorectal tumorigenesis. Biochem Soc Trans. 2005;33:672–75. doi: 10.1042/BST0330672. [DOI] [PubMed] [Google Scholar]
- 51.DeArmond D, Brattain MG, Jessup JM, et al. Autocrine-mediated ErbB-2 kinase activation of STAT3 is required for growth factor independence of pancreatic cancer cell lines. Oncogene. 2003;22:7781–95. doi: 10.1038/sj.onc.1206966. [DOI] [PubMed] [Google Scholar]
- 52.Reddi HV, McIver B, Grebe SK, Eberhardt NL. The paired box-8/peroxisome proliferator-activated receptor-γ oncogene in thyroid tumorigenesis. Endocrinology. 2007;148:932–95. doi: 10.1210/en.2006-0926. [DOI] [PubMed] [Google Scholar]
- 53.Aviel-Ronen S, Blackhall FH, Shepherd FA, Tsao MS. K-ras mutations in non-small-cell lung carcinoma: a review. Clin Lung Cancer. 2006;8:30–38. doi: 10.3816/CLC.2006.n.030. [DOI] [PubMed] [Google Scholar]
- 54.Tascedda F, Malagoli D, Accorsi A, et al. Molluscs as models for translational medicine. Med Sci Monit Basic Res. 2015;21:96–99. doi: 10.12659/MSMBR.894221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Full list of differentially expressed mRNA in nasopharyngeal carcinoma.
| Genes | FDR | Up/down regulation |
|---|---|---|
| LMNB2 | 1.00E−11 | Up |
| HDGFRP3 | 2.32E−11 | Up |
| FJX1 | 2.89E−11 | Up |
| RBBP8 | 4.91E−11 | Up |
| LHX2 | 5.18E−11 | Up |
| TNFAIP6 | 8.16E−11 | Up |
| ECT2 | 1.02E−10 | Up |
| C12orf48 | 1.74E−10 | Up |
| CHAF1B | 2.93E−10 | Up |
| VRK2 | 4.51E−10 | Up |
| NFE2L3 | 8.20E−10 | Up |
| TFRC | 1.08E−09 | Up |
| NPL | 1.08E−09 | Up |
| FAM64A | 1.42E−09 | Up |
| GALNT11 | 2.59E−09 | Up |
| TNFSF4 | 2.98E−09 | Up |
| GPSM2 | 3.06E−09 | Up |
| MAD2L1 | 4.03E−09 | Up |
| GAD1 | 4.59E−09 | Up |
| CDC6 | 5.29E−09 | Up |
| PFDN4 | 5.31E−09 | Up |
| ATF5 | 6.91E−09 | Up |
| MRPL42 | 7.85E−09 | Up |
| ZWILCH | 8.93E−09 | Up |
| FOXM1 | 9.23E−09 | Up |
| RAN | 1.00E−08 | Up |
| OLA1 | 1.31E−08 | Up |
| OIP5 | 1.99E−08 | Up |
| PSRC1 | 2.17E−08 | Up |
| FAP | 2.17E−08 | Up |
| MINPP1 | 2.86E−08 | Up |
| SEC61A2 | 3.30E−08 | Up |
| CCNF | 3.30E−08 | Up |
| C12orf5 | 4.54E−08 | Up |
| ARNT2 | 4.54E−08 | Up |
| RIF1 | 4.86E−08 | Up |
| RCN2 | 5.33E−08 | Up |
| DTX4 | 6.03E−08 | Up |
| RAD54B | 7.31E−08 | Up |
| CLASP1 | 7.31E−08 | Up |
| KIF14 | 7.46E−08 | Up |
| GNPDA1 | 7.46E−08 | Up |
| EXO1 | 7.46E−08 | Up |
| PMAIP1 | 8.23E−08 | Up |
| KCTD3 | 1.10E−07 | Up |
| GRB10 | 1.31E−07 | Up |
| GINS3 | 1.63E−07 | Up |
| PTGS2 | 1.69E−07 | Up |
| PUS7 | 1.82E−07 | Up |
| XPOT | 2.00E−07 | Up |
| PLA2G3 | 2.04E−07 | Up |
| PALB2 | 2.20E−07 | Up |
| PRMT3 | 2.36E−07 | Up |
| SAC3D1 | 2.45E−07 | Up |
| VCAN | 2.52E−07 | Up |
| FGF1 | 2.78E−07 | Up |
| ESM1 | 2.78E−07 | Up |
| C12orf11 | 2.86E−07 | Up |
| P4HA1 | 3.59E−07 | Up |
| PBK | 3.83E−07 | Up |
| RNASEH2A | 3.91E−07 | Up |
| DOCK4 | 3.91E−07 | Up |
| CNIH4 | 3.91E−07 | Up |
| NUAK1 | 3.91E−07 | Up |
| AHCY | 3.99E−07 | Up |
| TBCE | 4.26E−07 | Up |
| UBE2S | 4.33E−07 | Up |
| COL5A1 | 4.39E−07 | Up |
| NOX4 | 4.49E−07 | Up |
| GAPDH | 4.52E−07 | Up |
| EIF4E2 | 4.52E−07 | Up |
| DTL | 4.52E−07 | Up |
| STK3 | 4.94E−07 | Up |
| PSMD14 | 4.94E−07 | Up |
| DSG2 | 4.94E−07 | Up |
| CENPF | 5.08E−07 | Up |
| PTTG3P | 5.15E−07 | Up |
| FANCL | 5.16E−07 | Up |
| PPIF | 5.26E−07 | Up |
| PSMA4 | 5.52E−07 | Up |
| HDAC2 | 6.06E−07 | Up |
| MCM4 | 6.38E−07 | Up |
| CKS1B | 6.38E−07 | Up |
| UCK2 | 6.89E−07 | Up |
| EIF2S2 | 6.90E−07 | Up |
| TIPIN | 7.68E−07 | Up |
| FSCN1 | 7.68E−07 | Up |
| HSPE1 | 7.86E−07 | Up |
| MMP12 | 8.05E−07 | Up |
| TMEM194A | 8.19E−07 | Up |
| KIF18A | 8.22E−07 | Up |
| INSM1 | 8.24E−07 | Up |
| RAI14 | 8.77E−07 | Up |
| UNG | 9.44E−07 | Up |
| PIK3CB | 1.12E−06 | Up |
| TRIP6 | 1.15E−06 | Up |
| MTX2 | 1.21E−06 | Up |
| CEP152 | 1.27E−06 | Up |
| STAR | 1.30E−06 | Up |
| PLAU | 1.37E−06 | Up |
| HOMER3 | 1.38E−06 | Up |
| ME3 | 1.39E−06 | Up |
| RIC8B | 1.40E−06 | Up |
| ZFP64 | 1.56E−06 | Up |
| NUP155 | 1.58E−06 | Up |
| TOMM40 | 1.65E−06 | Up |
| PGAP1 | 1.67E−06 | Up |
| ITGAV | 1.88E−06 | Up |
| KIF4A | 1.90E−06 | Up |
| SRD5A1 | 2.03E−06 | Up |
| ZNF124 | 2.17E−06 | Up |
| RAD51AP1 | 2.17E−06 | Up |
| PTTG1 | 2.26E−06 | Up |
| BRCA2 | 2.31E−06 | Up |
| GGCT | 2.35E−06 | Up |
| ANXA4 | 2.41E−06 | Up |
| ADA | 2.74E−06 | Up |
| TMEM14A | 2.77E−06 | Up |
| PAK1IP1 | 3.08E−06 | Up |
| TMEM33 | 3.10E−06 | Up |
| HSPD1 | 3.11E−06 | Up |
| GORASP1 | 3.11E−06 | Up |
| USP18 | 3.12E−06 | Up |
| MARK1 | 3.14E−06 | Up |
| SLC39A6 | 3.17E−06 | Up |
| ZNF562 | 3.39E−06 | Up |
| CDC25C | 3.48E−06 | Up |
| RPE | 3.50E−06 | Up |
| POLQ | 3.50E−06 | Up |
| SNRPF | 3.79E−06 | Up |
| SSX2IP | 3.81E−06 | Up |
| STAP2 | 3.84E−06 | Up |
| CDC45 | 3.84E−06 | Up |
| POMGNT1 | 3.86E−06 | Up |
| BID | 3.88E−06 | Up |
| STC2 | 4.00E−06 | Up |
| BRIP1 | 4.00E−06 | Up |
| TM7SF3 | 4.21E−06 | Up |
| DUSP10 | 4.50E−06 | Up |
| PSMG1 | 4.62E−06 | Up |
| CCL4 | 4.74E−06 | Up |
| MED21 | 4.88E−06 | Up |
| NOV | 5.14E−06 | Up |
| SUMO1 | 5.21E−06 | Up |
| CPOX | 5.30E−06 | Up |
| COL7A1 | 5.30E−06 | Up |
| CAT | 5.39E−06 | Up |
| STIL | 5.84E−06 | Up |
| CCT2 | 5.93E−06 | Up |
| DNM1L | 5.95E−06 | Up |
| SLC25A13 | 6.29E−06 | Up |
| IMPACT | 6.52E−06 | Up |
| CACYBP | 6.61E−06 | Up |
| RGS4 | 6.63E−06 | Up |
| PHF14 | 7.10E−06 | Up |
| ILF2 | 7.10E−06 | Up |
| ENO1 | 7.13E−06 | Up |
| PAIP1 | 7.30E−06 | Up |
| TNFSF15 | 7.34E−06 | Up |
| EPHB4 | 7.34E−06 | Up |
| ATP6V1B2 | 7.34E−06 | Up |
| OAS3 | 7.83E−06 | Up |
| GSTO1 | 7.93E−06 | Up |
| TPI1 | 7.99E−06 | Up |
| IL15RA | 8.23E−06 | Up |
| HOXA7 | 8.40E−06 | Up |
| SETD2 | 8.66E−06 | Up |
| C1QB | 8.95E−06 | Up |
| IFRD1 | 9.15E−06 | Up |
| NT5M | 9.92E−06 | Up |
| PITPNB | 1.01E−05 | Up |
| CENPN | 1.02E−05 | Up |
| GPR137B | 1.03E−05 | Up |
| TK1 | 1.05E−05 | Up |
| TMEFF1 | 1.09E−05 | Up |
| SULF1 | 1.12E−05 | Up |
| ERCC6L | 1.13E−05 | Up |
| DENR | 1.21E−05 | Up |
| C5orf13 | 1.21E−05 | Up |
| ATP2C1 | 1.22E−05 | Up |
| STAT1 | 1.27E−05 | Up |
| TTK | 1.30E−05 | Up |
| SS18L1 | 1.30E−05 | Up |
| GABPB1 | 1.30E−05 | Up |
| BIRC5 | 1.33E−05 | Up |
| WNT2 | 1.38E−05 | Up |
| HRSP12 | 1.42E−05 | Up |
| SNRPG | 1.46E−05 | Up |
| PFDN2 | 1.48E−05 | Up |
| POLR3D | 1.54E−05 | Up |
| GJB1 | 1.57E−05 | Up |
| POGLUT1 | 1.58E−05 | Up |
| TNIP3 | 1.63E−05 | Up |
| PSMB3 | 1.64E−05 | Up |
| YWHAQ | 1.69E−05 | Up |
| NCBP2 | 1.80E−05 | Up |
| PLD3 | 1.80E−05 | Up |
| TWSG1 | 1.87E−05 | Up |
| SKP2 | 2.00E−05 | Up |
| YWHAH | 2.03E−05 | Up |
| CASP6 | 2.08E−05 | Up |
| IRF6 | 2.10E−05 | Up |
| CKAP4 | 2.12E−05 | Up |
| SLC5A6 | 2.20E−05 | Up |
| GPI | 2.27E−05 | Up |
| C1orf112 | 2.29E−05 | Up |
| MYC | 2.32E−05 | Up |
| NIT2 | 2.40E−05 | Up |
| LGR4 | 2.49E−05 | Up |
| HOXC6 | 2.53E−05 | Up |
| FPR3 | 2.53E−05 | Up |
| GLT25D1 | 2.56E−05 | Up |
| RACGAP1 | 2.62E−05 | Up |
| MIF | 2.65E−05 | Up |
| CXCL3 | 2.69E−05 | Up |
| CTPS | 2.73E−05 | Up |
| FGD6 | 2.75E−05 | Up |
| FNDC3B | 2.76E−05 | Up |
| LHFPL2 | 2.81E−05 | Up |
| CDC25A | 2.90E−05 | Up |
| CD70 | 2.97E−05 | Up |
| TOMM70A | 3.03E−05 | Up |
| COPS2 | 3.12E−05 | Up |
| CCND2 | 3.12E−05 | Up |
| MARCO | 3.17E−05 | Up |
| WDR41 | 3.40E−05 | Up |
| TAF5 | 3.40E−05 | Up |
| AP3M2 | 3.49E−05 | Up |
| TMEM39A | 3.55E−05 | Up |
| TMEM185B | 3.55E−05 | Up |
| PLK4 | 3.64E−05 | Up |
| COL10A1 | 3.64E−05 | Up |
| C11orf41 | 3.68E−05 | Up |
| KITLG | 3.70E−05 | Up |
| PLOD1 | 3.79E−05 | Up |
| UBFD1 | 3.79E−05 | Up |
| NFKBIA | 3.79E−05 | Up |
| CORO1C | 3.85E−05 | Up |
| MORC4 | 3.95E−05 | Up |
| HOXA10 | 3.98E−05 | Up |
| MRPL13 | 4.11E−05 | Up |
| GINS1 | 4.17E−05 | Up |
| ZFP112 | 4.20E−05 | Up |
| EXT1 | 4.23E−05 | Up |
| KIF13A | 4.25E−05 | Up |
| TMEM97 | 4.31E−05 | Up |
| PIK3CD | 4.44E−05 | Up |
| IFIT3 | 4.59E−05 | Up |
| ETV7 | 4.76E−05 | Up |
| NCAPD3 | 4.77E−05 | Up |
| ANGPT2 | 4.83E−05 | Up |
| SNRPD1 | 5.10E−05 | Up |
| NDUFA9 | 5.24E−05 | Up |
| TMPO | 5.33E−05 | Up |
| CD14 | 5.38E−05 | Up |
| CDK5 | 5.44E−05 | Up |
| DKC1 | 5.55E−05 | Up |
| ASB9 | 5.76E−05 | Up |
| TRMT61B | 5.82E−05 | Up |
| C1QBP | 5.91E−05 | Up |
| WRN | 5.97E−05 | Up |
| TMEM106C | 5.97E−05 | Up |
| SYNE1 | 6.22E−05 | Up |
| ICAM1 | 6.27E−05 | Up |
| FAM60A | 6.41E−05 | Up |
| SRSF9 | 6.72E−05 | Up |
| PRR5L | 6.72E−05 | Up |
| MLF1IP | 6.78E−05 | Up |
| GNL2 | 6.80E−05 | Up |
| PSMB2 | 6.94E−05 | Up |
| ASPN | 7.03E−05 | Up |
| YRDC | 7.17E−05 | Up |
| KPNA2 | 7.25E−05 | Up |
| TYROBP | 7.30E−05 | Up |
| EXOSC5 | 7.33E−05 | Up |
| TNFRSF9 | 7.34E−05 | Up |
| GAS8 | 7.37E−05 | Up |
| TBCA | 7.38E−05 | Up |
| FAM162A | 7.38E−05 | Up |
| MAPKAPK5 | 7.41E−05 | Up |
| POSTN | 7.82E−05 | Up |
| SSTR2 | 7.89E−05 | Up |
| SUPV3L1 | 8.01E−05 | Up |
| RSAD2 | 8.05E−05 | Up |
| RFC2 | 8.05E−05 | Up |
| H2AFZ | 8.05E−05 | Up |
| UBE2V2 | 8.11E−05 | Up |
| MFHAS1 | 8.29E−05 | Up |
| SYNCRIP | 8.60E−05 | Up |
| C17orf75 | 8.72E−05 | Up |
| WDR47 | 8.75E−05 | Up |
| FASTKD3 | 8.81E−05 | Up |
| NAA50 | 8.85E−05 | Up |
| ITGB1 | 8.94E−05 | Up |
| CAND1 | 9.04E−05 | Up |
| PTPN12 | 9.22E−05 | Up |
| MAT2A | 9.40E−05 | Up |
| DERA | 9.71E−05 | Up |
| KIAA0146 | 9.90E−05 | Up |
| EPCAM | 9.97E−05 | Up |
| C12orf29 | 1.05E−04 | Up |
| PYCARD | 1.06E−04 | Up |
| MDK | 1.07E−04 | Up |
| BMP3 | 1.07E−04 | Up |
| RAB17 | 1.08E−04 | Up |
| PLAUR | 1.08E−04 | Up |
| FASTKD2 | 1.09E−04 | Up |
| AIMP2 | 1.12E−04 | Up |
| WDR12 | 1.13E−04 | Up |
| ABCE1 | 1.13E−04 | Up |
| SERPINH1 | 1.15E−04 | Up |
| DEPDC1 | 1.16E−04 | Up |
| CYC1 | 1.18E−04 | Up |
| ITGB6 | 1.22E−04 | Up |
| PSMA7 | 1.23E−04 | Up |
| ZNF74 | 1.24E−04 | Up |
| SLMO2 | 1.24E−04 | Up |
| SCO2 | 1.24E−04 | Up |
| DMXL2 | 1.26E−04 | Up |
| BST2 | 1.26E−04 | Up |
| XPO1 | 1.27E−04 | Up |
| ENOPH1 | 1.27E−04 | Up |
| SCG5 | 1.28E−04 | Up |
| PAICS | 1.29E−04 | Up |
| WASF1 | 1.31E−04 | Up |
| BCL2A1 | 1.32E−04 | Up |
| LGALS1 | 1.32E−04 | Up |
| RXRB | 1.32E−04 | Up |
| RRM1 | 1.34E−04 | Up |
| PXDN | 1.37E−04 | Up |
| PTRH2 | 1.38E−04 | Up |
| JMJD4 | 1.38E−04 | Up |
| MBD5 | 1.39E−04 | Up |
| RNF19B | 1.40E−04 | Up |
| PRC1 | 1.43E−04 | Up |
| PRDM4 | 1.46E−04 | Up |
| NPM3 | 1.48E−04 | Up |
| UBE2Z | 1.48E−04 | Up |
| KIF23 | 1.53E−04 | Up |
| RRN3 | 1.55E−04 | Up |
| ZNF532 | 1.58E−04 | Up |
| PIGA | 1.58E−04 | Up |
| WBP5 | 1.59E−04 | Up |
| PIGN | 1.59E−04 | Up |
| IL13RA2 | 1.60E−04 | Up |
| GTF3C3 | 1.60E−04 | Up |
| ARMC1 | 1.63E−04 | Up |
| NOL10 | 1.64E−04 | Up |
| COL4A5 | 1.65E−04 | Up |
| GADD45A | 1.65E−04 | Up |
| FOXK2 | 1.71E−04 | Up |
| CCT3 | 1.71E−04 | Up |
| IDH1 | 1.72E−04 | Up |
| ADIPOR2 | 1.72E−04 | Up |
| PSMC4 | 1.72E−04 | Up |
| FCGR1B | 1.72E−04 | Up |
| ROBO1 | 1.72E−04 | Up |
| CST1 | 1.74E−04 | Up |
| MEST | 1.78E−04 | Up |
| CLSTN2 | 1.79E−04 | Up |
| UPF3B | 1.81E−04 | Up |
| URB2 | 1.81E−04 | Up |
| TPT1 | 1.81E−04 | Up |
| FAH | 1.81E−04 | Up |
| B4GALT2 | 1.82E−04 | Up |
| PPIA | 1.82E−04 | Up |
| NME1 | 1.89E−04 | Up |
| AQP9 | 1.95E−04 | Up |
| UQCRH | 1.96E−04 | Up |
| UBE2C | 1.96E−04 | Up |
| MRPS35 | 1.97E−04 | Up |
| ADAM9 | 1.97E−04 | Up |
| DKK3 | 2.00E−04 | Up |
| RBL2 | 2.00E−04 | Up |
| DDX60 | 2.07E−04 | Up |
| ZBTB39 | 2.09E−04 | Up |
| MCTP2 | 2.13E−04 | Up |
| LOC100499177 | 2.13E−04 | Up |
| SCMH1 | 2.15E−04 | Up |
| CUEDC2 | 2.17E−04 | Up |
| ZNF140 | 2.17E−04 | Up |
| TAP1 | 2.18E−04 | Up |
| E2F6 | 2.20E−04 | Up |
| PA2G4 | 2.26E−04 | Up |
| BMP2 | 2.29E−04 | Up |
| DIABLO | 2.35E−04 | Up |
| PSMD1 | 2.37E−04 | Up |
| CKAP5 | 2.41E−04 | Up |
| PRLR | 2.43E−04 | Up |
| ERO1LB | 2.44E−04 | Up |
| SPIN1 | 2.45E−04 | Up |
| PRNP | 2.45E−04 | Up |
| PIWIL1 | 2.45E−04 | Up |
| TLN2 | 2.47E−04 | Up |
| SRP9 | 2.49E−04 | Up |
| SOCS5 | 2.52E−04 | Up |
| CCR1 | 2.55E−04 | Up |
| CEP76 | 2.58E−04 | Up |
| SCARB2 | 2.60E−04 | Up |
| MRPL12 | 2.65E−04 | Up |
| ISYNA1 | 2.66E−04 | Up |
| RPL32 | 2.70E−04 | Up |
| PKD2 | 2.70E−04 | Up |
| KAT2B | 2.78E−04 | Up |
| TRIB1 | 2.80E−04 | Up |
| KDSR | 2.84E−04 | Up |
| IFI44L | 2.84E−04 | Up |
| UGCG | 2.86E−04 | Up |
| POLR3G | 2.89E−04 | Up |
| THBS2 | 2.97E−04 | Up |
| IGSF6 | 2.97E−04 | Up |
| STRAP | 3.12E−04 | Up |
| EIF2AK2 | 3.14E−04 | Up |
| ADAM12 | 3.14E−04 | Up |
| SNX27 | 3.15E−04 | Up |
| KREMEN2 | 3.15E−04 | Up |
| CEP192 | 3.15E−04 | Up |
| PPP6C | 3.21E−04 | Up |
| LOC730101 | 3.23E−04 | Up |
| PSMA6 | 3.25E−04 | Up |
| TGFBI | 3.25E−04 | Up |
| PSTPIP2 | 3.25E−04 | Up |
| CCL8 | 3.28E−04 | Up |
| RPS29 | 3.29E−04 | Up |
| PLA2G7 | 3.30E−04 | Up |
| PNO1 | 3.32E−04 | Up |
| RARB | 3.45E−04 | Up |
| CLIP2 | 3.45E−04 | Up |
| LAMP2 | 3.45E−04 | Up |
| CCT7 | 3.45E−04 | Up |
| ABTB2 | 3.45E−04 | Up |
| ABCA3 | 3.47E−04 | Up |
| NCK1 | 3.53E−04 | Up |
| CDK8 | 3.59E−04 | Up |
| WSB2 | 3.62E−04 | Up |
| DNMT3B | 3.63E−04 | Up |
| EVL | 3.69E−04 | Up |
| PTPLAD1 | 3.74E−04 | Up |
| GLA | 3.81E−04 | Up |
| ADH5 | 3.88E−04 | Up |
| CDK4 | 3.94E−04 | Up |
| UMPS | 3.99E−04 | Up |
| UCP2 | 3.99E−04 | Up |
| PTGES3 | 4.01E−04 | Up |
| LOC100127972 | 4.01E−04 | Up |
| FZD7 | 4.01E−04 | Up |
| TUBGCP4 | 4.04E−04 | Up |
| PPA1 | 4.07E−04 | Up |
| ARPC1A | 4.08E−04 | Up |
| PDSS1 | 4.12E−04 | Up |
| IFIT1 | 4.17E−04 | Up |
| INPP1 | 4.17E−04 | Up |
| KCNE1 | 4.22E−04 | Up |
| PXMP2 | 4.24E−04 | Up |
| IL15 | 4.28E−04 | Up |
| PCSK7 | 4.33E−04 | Up |
| PRPF40A | 4.34E−04 | Up |
| MLLT11 | 4.34E−04 | Up |
| MKKS | 4.52E−04 | Up |
| NIPSNAP1 | 4.57E−04 | Up |
| TSR2 | 4.59E−04 | Up |
| LDHA | 4.62E−04 | Up |
| CHCHD3 | 4.76E−04 | Up |
| NCAPH | 4.82E−04 | Up |
| R3HDM1 | 4.98E−04 | Up |
| SPAST | 4.98E−04 | Up |
| PDHX | 5.01E−04 | Up |
| C2orf47 | 5.01E−04 | Up |
| CBX1 | 5.02E−04 | Up |
| PACS2 | 5.10E−04 | Up |
| C3AR1 | 5.23E−04 | Up |
| ANAPC10 | 5.24E−04 | Up |
| CCNJ | 5.25E−04 | Up |
| TARS | 5.41E−04 | Up |
| ATP5G3 | 5.49E−04 | Up |
| HSPA13 | 5.52E−04 | Up |
| TCF12 | 5.54E−04 | Up |
| EIF1AX | 5.59E−04 | Up |
| CBX5 | 5.75E−04 | Up |
| YBX1 | 5.79E−04 | Up |
| CTSL1 | 5.82E−04 | Up |
| RFWD3 | 5.83E−04 | Up |
| ZNF473 | 5.84E−04 | Up |
| PDCD11 | 5.85E−04 | Up |
| TGIF2 | 5.86E−04 | Up |
| GSTK1 | 5.90E−04 | Up |
| GPR143 | 5.94E−04 | Up |
| NMD3 | 5.96E−04 | Up |
| JAG2 | 5.97E−04 | Up |
| DIAPH1 | 5.99E−04 | Up |
| GJA1 | 6.03E−04 | Up |
| KCNMB2 | 6.13E−04 | Up |
| ACOT7 | 6.15E−04 | Up |
| TMEM66 | 6.15E−04 | Up |
| REPIN1 | 6.18E−04 | Up |
| TRAP1 | 6.28E−04 | Up |
| SPDEF | 6.32E−04 | Up |
| YTHDF3 | 6.46E−04 | Up |
| TNFAIP3 | 6.63E−04 | Up |
| SNRPE | 6.63E−04 | Up |
| WDYHV1 | 6.64E−04 | Up |
| FNBP1L | 6.64E−04 | Up |
| C6orf211 | 6.65E−04 | Up |
| ATP5B | 6.65E−04 | Up |
| SEH1L | 6.81E−04 | Up |
| EFNB2 | 6.81E−04 | Up |
| C1orf109 | 6.81E−04 | Up |
| SOX12 | 6.84E−04 | Up |
| SCAI | 7.20E−04 | Up |
| ISG15 | 7.29E−04 | Up |
| RTF1 | 7.30E−04 | Up |
| ANKRD5 | 7.35E−04 | Up |
| AURKB | 7.40E−04 | Up |
| DARS | 7.52E−04 | Up |
| SUMO2 | 7.58E−04 | Up |
| WDR1 | 7.68E−04 | Up |
| LRP4 | 7.68E−04 | Up |
| MYO1B | 7.73E−04 | Up |
| MCM10 | 7.73E−04 | Up |
| BYSL | 7.73E−04 | Up |
| MMP1 | 7.81E−04 | Up |
| CCNE2 | 7.91E−04 | Up |
| ADAR | 7.96E−04 | Up |
| MTCH2 | 7.97E−04 | Up |
| CHEK1 | 8.17E−04 | Up |
| XAF1 | 8.22E−04 | Up |
| LRP12 | 8.42E−04 | Up |
| PAFAH1B3 | 8.43E−04 | Up |
| STAMBP | 8.44E−04 | Up |
| NRAS | 8.54E−04 | Up |
| GHR | 8.54E−04 | Up |
| ATP5J2 | 8.68E−04 | Up |
| HMGCR | 8.75E−04 | Up |
| SENP2 | 8.78E−04 | Up |
| SLC35E3 | 9.08E−04 | Up |
| ORC1 | 9.19E−04 | Up |
| C5orf30 | 9.42E−04 | Up |
| GSTM2 | 9.49E−04 | Up |
| RSRC1 | 9.61E−04 | Up |
| DEGS1 | 9.70E−04 | Up |
| RPS6KC1 | 9.88E−04 | Up |
| RFC3 | 9.89E−04 | Up |
| ESPL1 | 9.93E−04 | Up |
| FKBP1A | 9.96E−04 | Up |
| FBXO11 | 9.98E−04 | Up |
| EHBP1 | 9.98E−04 | Up |
| COL1A1 | 9.98E−04 | Up |
| DNALI1 | 0.00E+00 | Down |
| ALDH1L1 | 1.36E−12 | Down |
| SPAG8 | 4.00E−11 | Down |
| SCRN1 | 4.64E−11 | Down |
| FOXJ1 | 4.64E−11 | Down |
| CLU | 4.64E−11 | Down |
| KLF2 | 1.91E−10 | Down |
| SCGB1A1 | 2.34E−10 | Down |
| SCGB2A1 | 2.93E−10 | Down |
| VPS11 | 6.74E−10 | Down |
| SYBU | 8.20E−10 | Down |
| PDCD5 | 9.56E−10 | Down |
| GPR162 | 2.24E−09 | Down |
| STIM1 | 2.40E−09 | Down |
| SLC44A4 | 3.44E−09 | Down |
| PCYT2 | 3.97E−09 | Down |
| EPAS1 | 3.97E−09 | Down |
| ABLIM1 | 4.29E−09 | Down |
| MT3 | 4.59E−09 | Down |
| ATP2C2 | 5.67E−09 | Down |
| CRYL1 | 7.85E−09 | Down |
| C6orf97 | 7.93E−09 | Down |
| PROM1 | 8.64E−09 | Down |
| BLK | 8.81E−09 | Down |
| B3GALT4 | 1.31E−08 | Down |
| CD22 | 1.88E−08 | Down |
| MSLN | 1.99E−08 | Down |
| CIRBP | 2.02E−08 | Down |
| WDR78 | 2.08E−08 | Down |
| MSRA | 2.08E−08 | Down |
| PTGDS | 2.47E−08 | Down |
| SLC16A7 | 3.25E−08 | Down |
| SMPD3 | 3.41E−08 | Down |
| CD1C | 3.97E−08 | Down |
| VIPR1 | 4.54E−08 | Down |
| GNMT | 6.09E−08 | Down |
| TCTA | 7.46E−08 | Down |
| C10orf81 | 1.06E−07 | Down |
| PIGR | 1.23E−07 | Down |
| IGHD | 1.23E−07 | Down |
| FAM107A | 1.23E−07 | Down |
| ASL | 1.26E−07 | Down |
| NBEA | 1.28E−07 | Down |
| AQP5 | 1.61E−07 | Down |
| SGSM3 | 1.66E−07 | Down |
| MSMB | 1.76E−07 | Down |
| IK | 1.78E−07 | Down |
| PDLIM2 | 2.21E−07 | Down |
| DDAH2 | 2.22E−07 | Down |
| FBXO22 | 2.29E−07 | Down |
| PIK3C2B | 2.77E−07 | Down |
| VPREB3 | 3.67E−07 | Down |
| PDCD6IP | 4.01E−07 | Down |
| TREML2 | 4.33E−07 | Down |
| FCRL2 | 4.33E−07 | Down |
| FAM174B | 4.33E−07 | Down |
| DPEP2 | 4.52E−07 | Down |
| CLMN | 4.87E−07 | Down |
| RNASE4 | 5.08E−07 | Down |
| PPP3CA | 5.08E−07 | Down |
| GNG7 | 5.18E−07 | Down |
| AGR2 | 5.39E−07 | Down |
| GLTSCR2 | 5.44E−07 | Down |
| GALNT12 | 5.54E−07 | Down |
| ABHD14A | 6.19E−07 | Down |
| KLHDC2 | 6.55E−07 | Down |
| C6orf103 | 7.80E−07 | Down |
| CD72 | 7.96E−07 | Down |
| AK1 | 8.20E−07 | Down |
| LXN | 8.22E−07 | Down |
| FAM102A | 8.56E−07 | Down |
| CR1 | 8.77E−07 | Down |
| CD1D | 8.77E−07 | Down |
| MEIS3P1 | 8.84E−07 | Down |
| LMO2 | 1.01E−06 | Down |
| CYP2F1 | 1.04E−06 | Down |
| BAIAP3 | 1.05E−06 | Down |
| RPS6KA3 | 1.21E−06 | Down |
| WFDC2 | 1.24E−06 | Down |
| CD19 | 1.35E−06 | Down |
| BACE2 | 1.40E−06 | Down |
| DALRD3 | 1.41E−06 | Down |
| SIDT2 | 1.44E−06 | Down |
| PIP | 1.61E−06 | Down |
| ABCB1 | 1.88E−06 | Down |
| FAM65B | 1.90E−06 | Down |
| RRAGA | 1.92E−06 | Down |
| AGBL2 | 2.06E−06 | Down |
| C5orf45 | 2.19E−06 | Down |
| E2F3 | 2.55E−06 | Down |
| SFMBT1 | 2.56E−06 | Down |
| TUBA4A | 2.58E−06 | Down |
| BANK1 | 2.58E−06 | Down |
| TNFRSF13B | 2.64E−06 | Down |
| HMGCL | 2.67E−06 | Down |
| TTC12 | 2.84E−06 | Down |
| GCNT3 | 2.87E−06 | Down |
| VTCN1 | 2.88E−06 | Down |
| C10orf116 | 3.00E−06 | Down |
| MFNG | 3.01E−06 | Down |
| CD180 | 3.10E−06 | Down |
| AHCTF1 | 3.12E−06 | Down |
| TFF3 | 3.13E−06 | Down |
| IQSEC1 | 3.17E−06 | Down |
| POU2AF1 | 3.24E−06 | Down |
| ENPP4 | 3.24E−06 | Down |
| BEND5 | 3.51E−06 | Down |
| CLDN10 | 3.59E−06 | Down |
| CD40LG | 3.79E−06 | Down |
| ANG | 4.04E−06 | Down |
| GPR110 | 4.04E−06 | Down |
| SERHL2 | 4.17E−06 | Down |
| ACAA1 | 4.21E−06 | Down |
| KAT5 | 4.62E−06 | Down |
| TBC1D22A | 4.74E−06 | Down |
| FZD4 | 5.30E−06 | Down |
| ATP1A2 | 5.30E−06 | Down |
| KCNJ16 | 5.30E−06 | Down |
| HSPB8 | 5.99E−06 | Down |
| CCDC69 | 6.02E−06 | Down |
| CYB561D2 | 6.59E−06 | Down |
| TINF2 | 7.58E−06 | Down |
| ST6GAL1 | 7.74E−06 | Down |
| QARS | 7.89E−06 | Down |
| SELENBP1 | 9.03E−06 | Down |
| CRY2 | 9.09E−06 | Down |
| DUSP26 | 9.93E−06 | Down |
| MPG | 1.01E−05 | Down |
| COX7A1 | 1.02E−05 | Down |
| SWAP70 | 1.05E−05 | Down |
| MS4A1 | 1.05E−05 | Down |
| IFT88 | 1.09E−05 | Down |
| DND1 | 1.12E−05 | Down |
| KCNQ1 | 1.16E−05 | Down |
| CH25H | 1.21E−05 | Down |
| LYL1 | 1.22E−05 | Down |
| SRPX | 1.24E−05 | Down |
| NME5 | 1.30E−05 | Down |
| CEBPG | 1.30E−05 | Down |
| PLA2G16 | 1.32E−05 | Down |
| ZNF395 | 1.41E−05 | Down |
| PLEKHB1 | 1.47E−05 | Down |
| C11orf71 | 1.49E−05 | Down |
| RAD51 | 1.54E−05 | Down |
| BEX4 | 1.63E−05 | Down |
| C3 | 1.71E−05 | Down |
| SMPD1 | 1.75E−05 | Down |
| CLCF1 | 1.76E−05 | Down |
| GPRIN2 | 1.77E−05 | Down |
| SGSM2 | 1.79E−05 | Down |
| CAND2 | 2.00E−05 | Down |
| WDHD1 | 2.10E−05 | Down |
| ARMCX6 | 2.10E−05 | Down |
| FGD2 | 2.18E−05 | Down |
| FMO2 | 2.21E−05 | Down |
| C6 | 2.35E−05 | Down |
| LMBRD1 | 2.53E−05 | Down |
| AK2 | 2.53E−05 | Down |
| ABCA7 | 2.71E−05 | Down |
| NXF2 | 2.75E−05 | Down |
| GABRP | 2.82E−05 | Down |
| DNAH6 | 2.84E−05 | Down |
| CTDSP1 | 2.86E−05 | Down |
| PLAC8 | 2.97E−05 | Down |
| BCAS4 | 3.03E−05 | Down |
| BAALC | 3.03E−05 | Down |
| GTF2F2 | 3.05E−05 | Down |
| VILL | 3.12E−05 | Down |
| POLD4 | 3.17E−05 | Down |
| ITIH5 | 3.17E−05 | Down |
| DAZAP2 | 3.17E−05 | Down |
| SIGIRR | 3.22E−05 | Down |
| EPHB6 | 3.30E−05 | Down |
| MEIS2 | 3.35E−05 | Down |
| ICAM3 | 3.35E−05 | Down |
| CCR6 | 3.35E−05 | Down |
| PSD4 | 3.49E−05 | Down |
| EFCAB1 | 3.64E−05 | Down |
| TGFBR3 | 3.78E−05 | Down |
| TMEM184B | 3.84E−05 | Down |
| SPATA6 | 3.85E−05 | Down |
| CRLF1 | 3.95E−05 | Down |
| CLDN8 | 4.17E−05 | Down |
| ADCY2 | 4.30E−05 | Down |
| PIGH | 4.32E−05 | Down |
| TFF1 | 4.33E−05 | Down |
| TSPAN1 | 4.55E−05 | Down |
| SIDT1 | 4.63E−05 | Down |
| PHF1 | 4.65E−05 | Down |
| ARHGAP44 | 4.76E−05 | Down |
| TMEM121 | 4.97E−05 | Down |
| CRIP2 | 4.99E−05 | Down |
| APOM | 5.07E−05 | Down |
| FUCA1 | 5.18E−05 | Down |
| NEIL1 | 5.19E−05 | Down |
| LPAR1 | 5.20E−05 | Down |
| PSMA2 | 5.25E−05 | Down |
| LRIG1 | 5.32E−05 | Down |
| SRSF5 | 5.44E−05 | Down |
| ZFP106 | 5.54E−05 | Down |
| CRIP1 | 5.64E−05 | Down |
| MAST3 | 5.82E−05 | Down |
| DHX38 | 5.82E−05 | Down |
| PRKCB | 6.01E−05 | Down |
| MRPL19 | 6.11E−05 | Down |
| ABCD4 | 6.37E−05 | Down |
| HSD17B8 | 6.72E−05 | Down |
| GPR18 | 6.72E−05 | Down |
| DNAH3 | 6.72E−05 | Down |
| ADAMTSL3 | 6.72E−05 | Down |
| INHBB | 6.77E−05 | Down |
| NCR3 | 6.79E−05 | Down |
| CLDN3 | 6.79E−05 | Down |
| RNASE1 | 6.80E−05 | Down |
| CCNA1 | 6.86E−05 | Down |
| GOLGA8A | 7.00E−05 | Down |
| PDZD2 | 7.03E−05 | Down |
| CCDC19 | 7.03E−05 | Down |
| EIF1B | 7.25E−05 | Down |
| DLEC1 | 7.32E−05 | Down |
| EIF3J | 7.41E−05 | Down |
| S1PR4 | 7.82E−05 | Down |
| ACTR10 | 7.96E−05 | Down |
| CHL1 | 7.99E−05 | Down |
| PPRC1 | 8.05E−05 | Down |
| TSPAN32 | 8.06E−05 | Down |
| LRRC23 | 8.06E−05 | Down |
| CGRRF1 | 8.29E−05 | Down |
| EFCAB6 | 8.32E−05 | Down |
| LAMB2 | 8.40E−05 | Down |
| CAP2 | 8.51E−05 | Down |
| DUSP22 | 8.73E−05 | Down |
| ADAM28 | 8.76E−05 | Down |
| LMAN1 | 8.77E−05 | Down |
| TDG | 8.81E−05 | Down |
| AES | 9.04E−05 | Down |
| VNN2 | 9.22E−05 | Down |
| NUCB2 | 9.22E−05 | Down |
| TIMP4 | 9.25E−05 | Down |
| TLE2 | 9.39E−05 | Down |
| PROS1 | 9.40E−05 | Down |
| EPHX1 | 9.74E−05 | Down |
| PGK1 | 9.76E−05 | Down |
| CLCN4 | 9.78E−05 | Down |
| YPEL5 | 9.90E−05 | Down |
| ERCC1 | 1.01E−04 | Down |
| TMC6 | 1.01E−04 | Down |
| RASGRP2 | 1.05E−04 | Down |
| SERPINB7 | 1.05E−04 | Down |
| ZFYVE21 | 1.05E−04 | Down |
| GAK | 1.06E−04 | Down |
| GFOD1 | 1.07E−04 | Down |
| ARHGAP11A | 1.07E−04 | Down |
| MAGIX | 1.08E−04 | Down |
| SPAG6 | 1.16E−04 | Down |
| LRMP | 1.17E−04 | Down |
| FLII | 1.18E−04 | Down |
| SNED1 | 1.21E−04 | Down |
| PPOX | 1.24E−04 | Down |
| UBA7 | 1.24E−04 | Down |
| BASP1 | 1.24E−04 | Down |
| IGJ | 1.27E−04 | Down |
| C5orf4 | 1.32E−04 | Down |
| CTNS | 1.34E−04 | Down |
| TEX264 | 1.35E−04 | Down |
| ABHD6 | 1.36E−04 | Down |
| TIMM13 | 1.39E−04 | Down |
| CCDC28A | 1.40E−04 | Down |
| P4HTM | 1.40E−04 | Down |
| IFIH1 | 1.43E−04 | Down |
| HIP1R | 1.44E−04 | Down |
| STS | 1.46E−04 | Down |
| ATP7B | 1.46E−04 | Down |
| STAG3 | 1.54E−04 | Down |
| ZNF137P | 1.56E−04 | Down |
| ZNF528 | 1.57E−04 | Down |
| TMEM9B | 1.57E−04 | Down |
| PACRG | 1.57E−04 | Down |
| DEPDC5 | 1.63E−04 | Down |
| CCDC81 | 1.65E−04 | Down |
| PRIM2 | 1.70E−04 | Down |
| RPS6KA1 | 1.72E−04 | Down |
| C14orf1 | 1.73E−04 | Down |
| CDT1 | 1.74E−04 | Down |
| NLRP1 | 1.76E−04 | Down |
| LPPR3 | 1.77E−04 | Down |
| ANKHD1 | 1.77E−04 | Down |
| CD52 | 1.79E−04 | Down |
| TCL1A | 1.79E−04 | Down |
| ERCC5 | 1.79E−04 | Down |
| RNASET2 | 1.81E−04 | Down |
| KRT7 | 1.81E−04 | Down |
| FAM184A | 1.81E−04 | Down |
| TMEM132A | 1.81E−04 | Down |
| C11orf16 | 1.82E−04 | Down |
| CAMTA1 | 1.82E−04 | Down |
| HDC | 1.93E−04 | Down |
| NIPSNAP3B | 1.96E−04 | Down |
| LIMD2 | 1.98E−04 | Down |
| BAP1 | 2.02E−04 | Down |
| ZNF821 | 2.06E−04 | Down |
| ZBBX | 2.09E−04 | Down |
| ZCWPW1 | 2.18E−04 | Down |
| TNNC2 | 2.18E−04 | Down |
| CALM1 | 2.22E−04 | Down |
| MAPK14 | 2.23E−04 | Down |
| SEL1L3 | 2.26E−04 | Down |
| VTI1B | 2.28E−04 | Down |
| P2RX5 | 2.28E−04 | Down |
| CHAF1A | 2.29E−04 | Down |
| SGTA | 2.30E−04 | Down |
| MNS1 | 2.35E−04 | Down |
| FYCO1 | 2.35E−04 | Down |
| IL33 | 2.35E−04 | Down |
| SCUBE2 | 2.38E−04 | Down |
| TRIM24 | 2.41E−04 | Down |
| HSD17B2 | 2.42E−04 | Down |
| RASAL1 | 2.42E−04 | Down |
| ANXA11 | 2.44E−04 | Down |
| B3GNT3 | 2.44E−04 | Down |
| GLT8D1 | 2.45E−04 | Down |
| BCAR3 | 2.45E−04 | Down |
| ZBTB16 | 2.47E−04 | Down |
| ADRA2A | 2.49E−04 | Down |
| USP19 | 2.51E−04 | Down |
| RARRES2 | 2.51E−04 | Down |
| SLC46A3 | 2.60E−04 | Down |
| MAGOHB | 2.62E−04 | Down |
| SIRT3 | 2.63E−04 | Down |
| STAP1 | 2.64E−04 | Down |
| INPP4B | 2.66E−04 | Down |
| PEPD | 2.66E−04 | Down |
| JHDM1D | 2.80E−04 | Down |
| COBL | 2.80E−04 | Down |
| ITIH4 | 2.81E−04 | Down |
| DOCK3 | 2.84E−04 | Down |
| ADH1C | 2.86E−04 | Down |
| C11orf2 | 2.87E−04 | Down |
| RHOBTB2 | 3.00E−04 | Down |
| SNW1 | 3.01E−04 | Down |
| CD79A | 3.02E−04 | Down |
| TMC5 | 3.06E−04 | Down |
| ALDH3B1 | 3.14E−04 | Down |
| CDC14A | 3.22E−04 | Down |
| SNTA1 | 3.23E−04 | Down |
| LYN | 3.32E−04 | Down |
| LTF | 3.33E−04 | Down |
| MLYCD | 3.41E−04 | Down |
| DNAI1 | 3.45E−04 | Down |
| C1orf115 | 3.45E−04 | Down |
| CBX6 | 3.46E−04 | Down |
| KCNJ12 | 3.63E−04 | Down |
| ZBTB25 | 3.65E−04 | Down |
| UNC93B1 | 3.69E−04 | Down |
| C7orf44 | 3.72E−04 | Down |
| KIAA0125 | 3.80E−04 | Down |
| PARP3 | 3.94E−04 | Down |
| LOC284244 | 3.99E−04 | Down |
| FOLR1 | 3.99E−04 | Down |
| PPFIA4 | 4.13E−04 | Down |
| PKIG | 4.15E−04 | Down |
| MOAP1 | 4.17E−04 | Down |
| C17orf59 | 4.24E−04 | Down |
| EFHC1 | 4.26E−04 | Down |
| CRISP2 | 4.26E−04 | Down |
| ANK2 | 4.29E−04 | Down |
| RHOH | 4.40E−04 | Down |
| APOD | 4.45E−04 | Down |
| TFEB | 4.49E−04 | Down |
| PAIP2B | 4.49E−04 | Down |
| CREBZF | 4.52E−04 | Down |
| OCEL1 | 4.64E−04 | Down |
| SNX3 | 4.72E−04 | Down |
| TFB2M | 4.82E−04 | Down |
| ALDH6A1 | 4.90E−04 | Down |
| RPGRIP1 | 4.93E−04 | Down |
| PNMA1 | 5.02E−04 | Down |
| TCEB1 | 5.18E−04 | Down |
| ATP5L | 5.25E−04 | Down |
| LOC728855 | 5.28E−04 | Down |
| RPL36AL | 5.52E−04 | Down |
| XRCC4 | 5.69E−04 | Down |
| TMEM63A | 5.74E−04 | Down |
| C9orf9 | 5.92E−04 | Down |
| SLC15A2 | 6.00E−04 | Down |
| FAIM3 | 6.20E−04 | Down |
| VAV1 | 6.20E−04 | Down |
| CHKB | 6.24E−04 | Down |
| CP | 6.41E−04 | Down |
| PAF1 | 6.53E−04 | Down |
| MAGED2 | 6.58E−04 | Down |
| RRAD | 6.58E−04 | Down |
| TXNIP | 6.64E−04 | Down |
| ZFP161 | 6.79E−04 | Down |
| DIO1 | 6.79E−04 | Down |
| PTGER2 | 6.81E−04 | Down |
| SATB1 | 6.82E−04 | Down |
| PRR15L | 6.82E−04 | Down |
| TRADD | 6.87E−04 | Down |
| SLC1A1 | 6.88E−04 | Down |
| CHD7 | 7.19E−04 | Down |
| CRYM | 7.20E−04 | Down |
| C7 | 7.20E−04 | Down |
| RBM38 | 7.34E−04 | Down |
| WWP2 | 7.40E−04 | Down |
| DTYMK | 7.48E−04 | Down |
| OR7E47P | 7.52E−04 | Down |
| NIP7 | 7.54E−04 | Down |
| FAM50B | 7.57E−04 | Down |
| XPA | 7.58E−04 | Down |
| KLHL7 | 7.73E−04 | Down |
| ZNF839 | 7.81E−04 | Down |
| SLC34A1 | 7.81E−04 | Down |
| FBXL2 | 7.85E−04 | Down |
| PLSCR4 | 7.92E−04 | Down |
| PBXIP1 | 7.93E−04 | Down |
| NAT6 | 7.94E−04 | Down |
| PPIL2 | 7.97E−04 | Down |
| KLHL36 | 8.19E−04 | Down |
| MAN2B2 | 8.24E−04 | Down |
| EHD1 | 8.42E−04 | Down |
| GPR183 | 8.75E−04 | Down |
| MFAP4 | 8.84E−04 | Down |
| HCLS1 | 8.99E−04 | Down |
| APEH | 9.01E−04 | Down |
| LOC100129973 | 9.17E−04 | Down |
| DPAGT1 | 9.23E−04 | Down |
| TREX1 | 9.39E−04 | Down |
| PPP3CC | 9.39E−04 | Down |
| ATG4A | 9.44E−04 | Down |
| LY9 | 9.49E−04 | Down |
| TERF2IP | 9.53E−04 | Down |
| HSPA9 | 9.53E−04 | Down |
| ITGA7 | 9.61E−04 | Down |
| CAPNS1 | 9.63E−04 | Down |
FDR: false discovery rate