Abstract
Optogenetic approaches promise to revolutionize neuroscience by using light to manipulate neural activity in genetically or functionally defined neurons with millisecond precision. Harnessing the full potential of optogenetic tools, however, requires light to be targeted to the right neurons at the right time. Here we discuss some barriers and potential solutions to this problem. We review methods for targeting the expression of light-activatable molecules to specific cell types, under genetic, viral or activity-dependent control. Next we explore new ways to target light to individual neurons to allow for their precise activation and inactivation. These techniques provide a level of precision in the temporal and spatial activation of neurons that was not achievable in previous experiments. In combination with simultaneous recording and imaging techniques, these strategies will allow us to mimic the natural activity patterns of neurons in vivo, enabling previously impossible ‘dream experiments’.
Introduction
The introduction of optogenetic tools - light-activated proteins that can activate or inactivate neural activity – is transforming the field of neuroscience. For the first time it is now possible to use light to both trigger and silence activity in genetically defined populations of neurons with millisecond precision. In principle, this enables fundamental experiments that probe the causal role of specific neurons in controlling circuit activity and behaviour with unprecedented power and precision. Over the past decade, optogenetic tools have become a mature technology. A wide variety of different opsins are readily available, and the ‘optogenetic toolkit’ is already part of the standard repertoire for investigating the functional properties of neurons at the molecular, cellular, circuit, and behavioural levels1–3. While the adoption of optogenetics by thousands of laboratories worldwide has led to many new scientific insights, it has also exposed some of the weaknesses of current optogenetic approaches. These include a lack of specificity for the cell types being targeted, imprecise control of the number and spatial location of cells being manipulated, variability in the level of optogenetic modulation across a neuronal population, and the synchronous activation (or inactivation) of cells expressing optogenetic probes. In short, these are targeting problems: they reflect the inability to precisely deliver optogenetic probes, and the light that controls them, to the right neurons at the right time. In this review, we discuss the nature of these problems, explore various strategies for solving them (Fig. 1), and give examples of “dream experiments” that will become possible with the application of these new approaches.
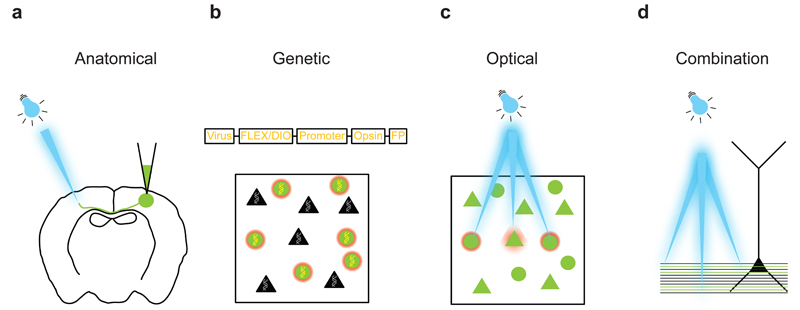
Figure 1. Intersectional strategies for targeting optogenetic manipulation.
(a) Physical delivery of virus to a given anatomical location can exploit or uncover circuit connectivity patterns either by making use of axonal projections or by using viruses that are able to cross one or more synapses. (b) Addressing cell types can be performed if the cell type of interest has a known genetic identity. (c) Directing the illumination source to a given set of cells or even individual neurons and processes is useful when the targets of interest are separated in space relative to the spatial resolution of the technique employed. (d) These three strategies can be combined as shown in this example in which axons of a particular cell class projecting to a subcellular domain of a neuron are photostimulated at different distances from the neuron.
1. Targeting optogenetic probes to the ‘right’ neurons
The brain is composed of a large variety of morphologically and functionally different neurons that can be grouped into ‘cell types’ or ‘cell classes’. The cell type concept makes more sense in some organisms and brain regions, and less so in others. A number of these neurons have their own defined function in the circuit, and therefore it is common sense to define the single neuron as the functional unit. In the mammalian retina, most neurons with a defined morphology and function exist in multiple copies occupying nodes of a spatial mosaic that covers the retina. Here the functional unit is often considered to be a mosaic of cells with the same properties, referred to as cell type. In this review, we use ‘cell type’ to refer to a population of neurons that cannot practically be divided into smaller units, and ‘cell class’ to refer to a population of neurons that is defined by some common property but which can be further divided into smaller populations.
A key advantage of optogenetics compared to electrical stimulation is that, in principle, the ‘right’ neurons as opposed to a random set of neurons can be manipulated. The ‘right’ neurons could be a cell type, such as a single retinal ganglion cell mosaic; it could be a cell class, such as parvalbumin-expressing neurons in a given brain area; ‘right’ could also represent a functionally defined cell type, such as neurons in visual cortex responding to a particular stimulus orientation; and finally, ‘right’ could also mean subcellular localization, for example, the axon terminals in a given region. Targeting the ‘right’ neurons is still a largely unsolved problem, especially in species, such as non-human primates, where genetic manipulations are often not feasible. Targeting optogenetic probes is not only important for research but also for the possible therapeutic use of optogenetics. In this review we describe various approaches for targeting optogenetics probes, focusing on using viruses alone, or in combination with the use of transgenic animals4.
Viruses as “lego” machines for optogenetic targeting
Viruses are especially useful for optogenetic targeting since they are small (roughly 20-200 nanometers) compared to neurons, they can be injected at any time into any brain region, and they can lead to high levels of expression of optogenetic tools. Viruses can be regarded as small machines containing modules with specific functions that can be modified. Many viruses incorporate only a few proteins that confer essential properties. Within a given viral family these proteins exist as many variants, and this diversity can be further increased by synthetic approaches. For example, the virus used most frequently for targeting, the adeno associated virus (AAV), has a coat protein that exists in 100 different variants in nature - and millions more can be made by DNA synthesis or mutagenesis5. A particular coat protein can confer a useful property, such as an affinity for a neuronal class or a preference to enter via axon terminals. By mutating that protein or providing a variant of that coat protein from a related virus the viral property can be changed e.g. changing the entry site from axons to soma or dendrites. Not only can variants of a given protein be exchanged within a viral family, but proteins can also be exchanged across highly different viruses. For example the vesicular stomatitis virus G coat-protein is often used in other viruses such as lentiviruses which enable efficient cellular entry6. Furthermore, combinations of different viruses can be used to enhance versatility. Rabies virus, for example, can be helped to cross one synapse with an engineered AAV or Herpes virus7–9. None of these viruses are used are in their wild-type form; rather, they are assembled element-by-element.
We therefore consider the thousands of viruses made by nature and the many variants made by researchers as a “Legoland” for neuroscientists performing optogenetics experiments, or other experiments where precise gene targeting is needed. Once a new and useful property of a viral component is published, this component can be tested in any virus. Indeed the way viruses are made is highly modular: the different properties are stored in different plasmids and by mixing these plasmids and adding them to cells, the virus is self-assembled. This modular nature of viruses facilitates innovation, providing new solutions to previously intractable problems.
Virus properties relevant for optogene targeting include the concentration at which it can be produced; whether it is an RNA or a DNA virus; whether it is replication competent or incompetent; lipid-coated or not; its physical size; and its packaging capability. The concentration of the virus is an often-overlooked variable: it can vary over many log units (106-1013/ml) and it can decrease significantly if the virus is handled improperly. Replication competent viruses are toxic to varying degrees, but if long-term stimulation is required replication incompetent viruses are needed. Our experience is that lipid coated viruses, such as rabies, lenti, VSV and herpes viruses do not penetrate well into tissues and therefore infection occurs mostly along the “needle track”. The best penetration is achieved with small, non-lipid coated viruses such as AAVs. The injection volume, injection speed, and the affinity of viruses for the surface of neighboring cells can influence access to cells further away from the injection site. Larger injection volumes deliver more viral particles, but also can result in tissue damage. Slow injection speed may help to distribute viruses better; however it is not clear if the speed or the time before needle withdrawal is the more important variable. Early needle withdrawal could result in distributing the virus along the needle track before the particles have the chance to diffuse into the tissue. High virus affinity for non-target cells could significantly decrease target cell gene expression10. Finally, packaging capabilities vary widely among viruses, which represents a serious limitation for the more ambitious experiments with large genetic payloads.
Targeting viruses to different types or classes of neurons can be based on the genetic identity of these neurons (e.g. expression of parvalbumin4), their specific circuit connectivity (e.g. neurons presynaptic to a simple cell in visual cortex), or a combination of the two3.
Virus targeting based on genetic identity
The morphology and function of different cell types is to a large extent defined by the pattern of genes they express. Past work has utilized the fact that some classes of neurons uniquely express particular signature genes - for instance, a large class of fast-spiking interneurons expresses parvalbumin - as a genetic handle that can be used to drive expression of various molecular tools exclusively in these cells. Some of these molecular tools, such as site-specific recombinases (for example Cre or Flp) can be used to drive the expression of optogenetic probes from viruses infecting these cells4. Such conditional viruses can be made from DNA viruses such as AAV11 or Herpes viruses12. Cell type specific expression from RNA viruses such as rabies requires an additional component, such as a helper AAV7. Specificity of targeting could be increased using intersectional strategies13 for example to express Cre and Flp in different but overlapping cell classes and make the virus expression to be conditional on both Cre and Flp. The main drawback of the conditional virus approach is that it requires expression of a site-specific recombinase, typically using a transgenic animal. The generation of a transgenic animal for a target neuronal type is both time consuming and unpredictable and currently only feasible in a few model organisms, while viral expression requires an additional injection.
It would therefore be highly desirable to be able to target viruses directly to cell types of wild-type animals in a variety of species by using promoter elements. The most suitable virus for this purpose would be AAV due to the lack of observable toxicity and its long term, often mouse-lifetime long, expression14. There are promoters that drive expression of AAVs in many cell types but it is difficult to find one that restricts high transgene expression to one cell type. Screening AAVs for cell type-specific expression with random or guessed, synthetic promoter elements would be highly valuable both for basic research, since once a specific and strong promoter for a cell type is found it can be used in combination with any tool, and also for translational research and medicine, since specific and safe applications of optogenetic probes in primates and humans may require cell type/class targeting15,16.
Targeting based on circuit connectivity
In many cases, targeting based on genetic identity is not possible; however, some cell types can be thought of as having a “connectivity signature” that defines them. This signature could be a specific long-range axonal projection17, as well as specific local circuit connectivity to other neurons18. Where it exists, a connectivity signature combined with viruses specialized to either infect neurons at specific locations or to infect them via their synaptic connections (transsynaptic infection) can be used for selectively targeting optogene expression (Fig. 2). Targeting based on connectivity can be performed in any species where a particular virus is able to infect neurons.
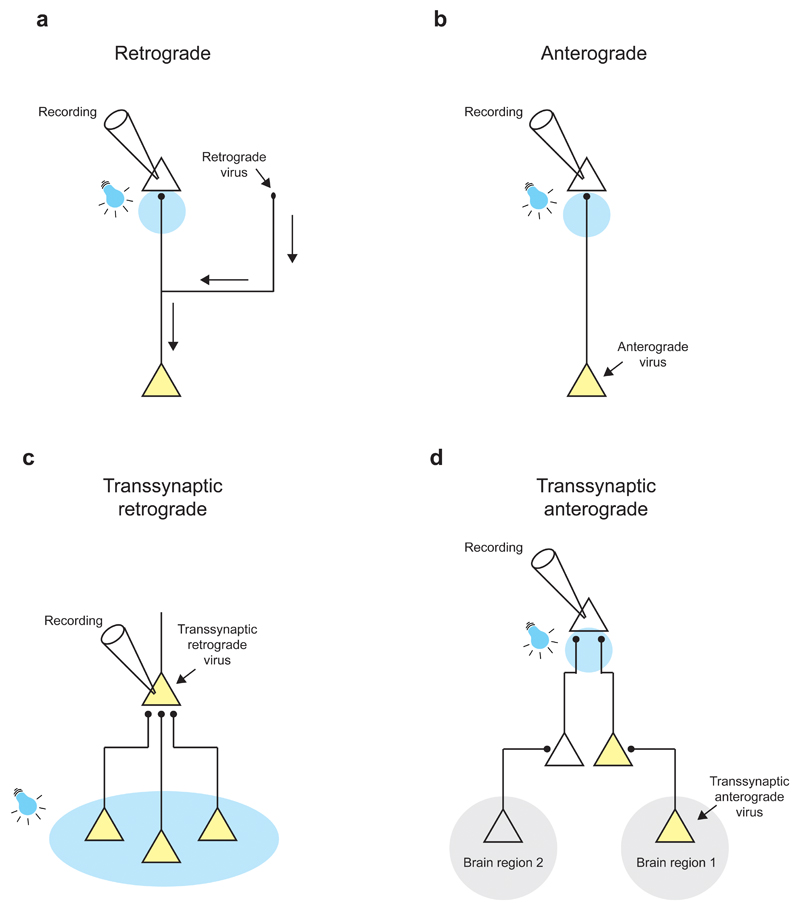
Figure 2. Viral targeting of optogenetic tools using knowledge of circuit connectivity.
Schematic illustration of different strategies for targeting optogenetic tools to specific cell types based on their connectivity pattern. Neurons expressing an optogenetic tool are indicated in yellow; arrows next to cellular processes indicate the direction of viral spread; and the location of light stimulation is shown in blue. (a) Use of a retrograde virus with targeted virus injection to an axon projection region. (b) Use of an anterograde virus with targeted virus injection to the somatic region. (c) Use of a transsynaptic retrograde virus starting from virus introduction (or infection) of a single postsynaptic cell which leads to optogene expression in monosynaptically connected presynaptic partners. (d) Use of a transsynaptic anterograde virus starting from virus injection in a given brain region to cause optogene expression in synaptically connected downstream neurons.
Labeling based on axonal projection
Retrograde
Cells that project to a brain region can be targeted using pressure injection of viruses that are able to infect neurons at the axon terminals, such as some variants of herpes19, adeno20, rabies21, VSV22, lenti23 and adeno-associated viruses24. These viruses either naturally have the ability to enter axons or they are ‘recoated’ with a protein that allows them to do so. The soma of the target cell should be far away from the injection site to ensure that light used for optogenetic stimulation does not excite all the locally infected cells (Fig. 2a). A problem inherent to this approach is that the injection can cause damage exactly at the location where postsynaptic cells of interest reside. However, if the target cell also sends axon collaterals to another brain region then this area could be used to initiate infection without damaging or infecting neurons in the intended postsynaptic zone. Rabies and herpes based retrograde labeling methods, while suitable for short-term studies over days, are too toxic for studies where long-term expression is needed. Among the viruses mentioned above, lenti- and adeno-associated viruses (LVs and AAVs) are the least toxic; however the efficiency of currently used retrograde LV and AAV variants is low, requiring identification of more efficient LV and AAV coats for axonal entry.
Anterograde
An important use of optogenetics is the mapping of inputs, arriving from different brain areas, to different spatial positions on a given target neuron25. This can be achieved by viral delivery of the optogenetic probe to the cell bodies or dendrites of projection neurons. Once the probe is anterogradely transferred to the axon terminals of infected neurons, close to the target neuron, it can be focally stimulated by light (Fig. 2b). By systematically mapping regions around the target neuron the spatial distribution of synaptic inputs from a given brain region can be reconstructed25. AAVs are excellent tools for anterograde delivery; however currently used AAVs are not exclusively anterograde and further development of non-toxic, exclusively anterograde vectors is needed.
Transsynaptic or transneuronal labeling
Retrograde
Optogenetic probes expressed using monosynaptic retrograde viral tracers, such as rabies virus26, could serve as important tools for proving putative connectivity between the virus-marked post and presynaptic cells21,22,27. A particularly attractive strategy is single-cell electroporation of a postsynaptic neuron and the subsequent initiation of a retrograde virus from only the electroporated neuron28. Light stimulation of the tracer labeled cells and simultaneous electrical or optical recording from the electroporated cell could prove functional connectivity between these cells (Fig. 2c). A limitation of this approach is that the electroporated cell also expresses the optogenetic tool and therefore is directly stimulated with unfocused light. This can be solved by a combination of pharmacology, to compare stimulation before and after the application of synaptic blockers, and 3D-patterned light stimulation as discussed below. A transneuronal approach for optogene expression which has likely little toxicity, is the use of Cre recombinase fused to wheat germ agglutinin that can be combined with conditional optogene expressing viruses29,30. This approach is well suited to perform long-term studies and for studying optogenetically manipulated behavior.
Anterograde
Axonal projection-based mapping of synaptic inputs can be extended by using a monosynaptic anterograde virus22 two synapses away from the target neuron (Fig. 2d). This is useful when the brain area, one synapse away, contains different types of neurons which receive input from different brain regions. In many experiments targeting is performed with the combined use of the knowledge of circuit connectivity and genetic identity29. Transsynaptic tracing from Cre-expressing neurons7,31 as well as the combination of axon projection-based retrograde labeling with labeling based on genetic identity are powerful ways of increasing the specificity of targeting.
Despite the large number of available viral vectors as well as the possibility of combining different viruses to target the desired cell types, viral targeting is not yet robust and simple: replication competent viruses are toxic to various degrees, growing different types of viruses in the lab requires specific safety conditions and expertise, and it often takes a long time until the targeting is optimized. These considerations highlight the urgent need for the development of non-toxic versions of purely anterograde and retrograde as well as monosynaptic tracers. Furthermore, it would be highly desirable to create vector distribution centers for neurotropic viruses where all targeting vectors are available, where experts can produce viral kits for particular experiments and advise new users.
Long-term optogene expression
Major questions in neuroscience address the circuit basis of the formation, maintenance and elimination of synaptic connection over long time periods. Addressing these using optogenetic methods requires low toxicity and long-term stability of optogenetic probe expression. Expression with lower toxicity can be achieved using mouse genetics, AAVs, electroporation or the combination of these three. However achieving stable expression is a key limitation. Expression via AAVs or in utero electroporated plasmids increases over several days or weeks and it is unclear when equilibrium is achieved. This is a particular concern since high-level, long-term expression has been shown to cause abnormal axonal morphology32. Furthermore, AAVs form a deposit in the target tissue following injection, which could lead to continued infection over time and a slow shift in optogene expression. This long-term increase in copy number is likely not a problem with electroporation but for both delivery methods the number of optogene copies could vary considerably from cell to cell. The most stable method for long-term expression is using transgenic animals where the changes across cells of the same type are uniform.
Single-cell targeting of optogene expression
An elegant way to precisely target optogenetic probes to individual neurons is to use single-cell electroporation42,44 (Fig. 3). This involves using 2-photon microscopy to target a plasmid-filled patch pipette to individual neurons in vivo and using electrical pulses33,34 to deliver the plasmid to the cell under visual control. Neurons can be targeted this way based on their somatodendritic morphology (using ‘shadowimaging’33), their genetic identity (using GFP expression as a marker) or their functional properties (such as tuned responses to sensory stimuli) for subsequent optogenetic activation34. Since up to a few dozen neurons, in any arbitrary spatial arrangement, can be electroporated using this approach, this therefore allows targeted optogene expression in a precisely defined ensemble of neurons, enabling tests of the relationships between neuron number, their identity, and their spatial arrangement on circuit processing.
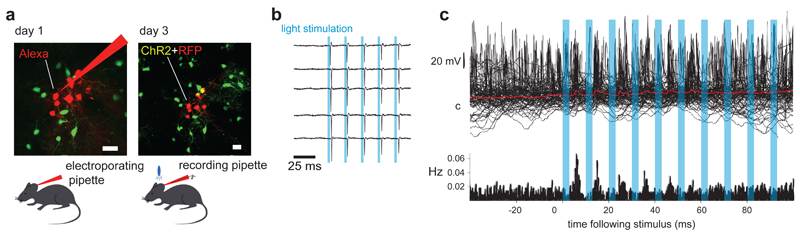
Figure 3. Targeting optogene expression using single-cell electroporation.
a, Left panel: Two-photon Z-stack projection of mouse cortex during successive single-cell electroporation; the transgenic mouse expresses GFP in GAD67-positive cortical inhibitory neurons (green cells). The red neurons have been electroporated with Alexa Fluor 594 (red cells) and plasmid DNA encoding RFP and ChR2. Right panel: 48 hours later the electroporated cells express RFP (red) suggesting that ChR2 is also expressed. b Cell-attached recording from a red cell confirms that blue light stimulation using an LED drives the neuron to fire an action potential precisely and reliably. c Whole-cell recording from a green cell (inhibitory, not electroporated) shows an increased firing probability following a light stimulus. The average membrane potential shows a depolarizing transient 5 ms after the onset of light stimulus, suggesting a direct synaptic connection from some of the electroporated neurons onto this cell. Unpublished data from M. London, L. Beeren and M. Hausser.
Activity-dependent expression of optogenes
Ultimately, it would be extremely useful to target expression of optogenetic probes to neurons not only based on genetic identity, but also based on activity patterns. This would open up many exciting experimental avenues, enabling functionally defined neuronal ensembles – rather than simply genetically defined populations – to be targeted for manipulation. For example, this approach would allow the reactivation of only the subset of neurons that had been active during a recent behavioral episode, such as during learning, allowing the minimal ensemble required for reactivating the behavior to be defined. At present, the options for implementing such a strategy remain limited. This is primarily due to the lack of known promoters that are unambiguously and specifically linked to spiking activity in neurons. Initial efforts in this direction have been made using a promoter for c-fos, an immediate early gene which has been shown to be switched on by neural activity35, to drive ChR2 expression in neurons activated during a memory task36. However, the precise relationship between spiking activity and the resulting ChR2 expression in this system remains unclear. Moreover, since c-fos expression, like that of other immediate early genes such as arc and zif268, has a timescale of hours, it lacks the temporal precision to uniquely label ensembles active on a millisecond timescale during behavior, leading to problems with background and specificity (though these can be ameliorated to some extent using a combinatorial approach, such as with the tetracycline system for gene regulation35). An alternative approach would be the use of light-sensitive promoter systems37 to label cells with optogenetic tools. For example cells that were activated during a specific behavior and were observed via two-photon calcium imaging could be forced to express optogenetic inhibitors, provided that the two-photon scanning required for the calcium imaging does not activate the promoter. In a subsequent experiment these cells could be optogenetically inhibited during the same behavior. Ultimately, it may be possible to find an appropriate promoter which is precisely temporally ‘gatable’, and yields a linear relationship between spiking and optogene expression, allowing for well-calibrated re-activation (or inactivation) of functionally defined neural ensembles.
2. Targeted light delivery
Once the challenge of targeting optogenetic probes to the ‘right’ neurons is overcome, the next challenge is to deliver light to those neurons. Ultimately, if light targeting is sufficiently precise and rapid to allow selective activation of individual cells, this automatically relaxes the constraints for genetic targeting (since all cells could then express the opsins, and only the ‘right’ neurons activated). However, we are still relatively far from this goal. Though an advanced treatment of optics is beyond the scope of this review, we describe below some considerations necessary for performing a successful optogenetics experiment. There are many options for delivering the required amount of light to a desired location, but careful consideration of the scattering nature of light in biological tissue requires control experiments to confirm correct delivery. Getting enough light to the right place depends on specific experimental goals, but the key factors to consider are the wavelength, intensity, and scattering of the light in the model system being used, in addition to the optical delivery system.
Selecting an animal model
Optogenetics approaches have been applied to animal models ranging from c. elegans and zebrafish to rodents and primates. The optical access afforded by transparent animals is obviously advantageous for light-based approaches to activation while mammals, whose nervous system tissue is much more difficult to access, are key species for modeling computations in the human brain. Much work using optogenetics has focused on rodent models, although recent work in primates shows promise. The first report of optogenetic excitation in primates38 was followed by additional functionality such as inhibition and step-function capability39. The optimization of optogenetics approaches in primates40, which present their own experimental challenges such as larger brains and the need for long-term chronic installations, has recently yielded breakthroughs, with the appearance of the first reports of behavioral responses in primates to excitatory41,42 and inhibitory43 optogenetics.
Selecting an opsin
The next step is to consider which opsin to use,which depends on the experiment. Careful consideration of the polarity of manipulation (exciting, inhibiting, or bidirectionally manipulating activity44–46), the timecourse of the manipulation (involving millisecond control of spiking47, or a more prolonged or subtle modulation48) and the selected wavelength of light (e.g. using longer wavelengths for deeper penetration, or differential wavelengths when using two opsins simultaneously). As the rapidly growing range of opsins with different properties has been described extensively in various recent reviews3,49 a comprehensive summary is not presented here.
The expression level of the chosen opsin is a key issue to consider, as viruses and promoters can often drive production of opsin molecules to extremely high levels. Overexpression can be useful in overcoming low conductances per molecule but high-level long-term expression can lead to toxicity32. In addition, driving all opsin-expressing membranes at once does not mimic physiological activity, and particular care should be taken when interpreting the results of such experiments, for example when all neurons of a particular cell type are driven synchronously. Finally, action potentials evoked optogenetically by illuminating the axon terminals can have different kinetics from spontaneous action potentials, resulting in differences in neurotransmitter release50.
Selecting a light source
Practically, the first crucial piece of equipment is the light source. This can be a mercury or xenon bulb, a light-emitting diode (LED), a continuous-wave laser, or an ultrafast pulsed laser (for two-photon excitation, see below). Mercury and xenon bulbs produce a wide spectrum of light that must be subsequently bandpass filtered for the desired wavelength. Bulbs produce the highest power output across the spectrum, but need to be replaced often (200 - 2000 hours) and disposed of appropriately. LEDs last much longer (10,000 - 100,000 hours), do not produce as much heat as bulbs, are generally inexpensive, and can generate a specific wavelength or a wide spectrum of light. Both bulbs and LEDs emit light over a wide angular area, which can make coupling into a fiber or microscope inefficient. Nevertheless, both sources, if installed correctly, have sufficient power for optogenetic activation. Laser light sources produce coherent light, which means the photons emitted are in phase with each other, a necessary property for generating holographic patterns (see SLM below) which also aids coupling efficiency into fibers. Lasers capable of yielding two-photon excitation emit ‘ultrafast’ pulses of light tens to hundreds of femtoseconds long.
Given the range of intensity of the various light sources, care should be taken to ensure the appropriate amount of light is delivered to the sample. Too few photons will result in insufficient activation of opsin molecules in the sample. Too much light can result in phototoxicity and photobleaching, or even activate neurons directly51. In addition, optogenetic tools exhibit desensitization to light over the course of seconds49, a process noted in the initial characterization of channelrhodopsin-2 (ref. 52). Ultimately, only simultaneous electrophysiological recordings can confirm directly that enough photons impinge on the opsin molecules to drive sufficient current throughout an experiment. Such a calibration experiment is crucial, particularly when prolonged or repeated photostimulation is necessary.
Delivering light from source to sample
Transmitting the light from the source to the sample is the next practical consideration in designing an optogenetics experiment. This depends on whether the experiment is performed in vitro or in vivo. An in vitro experiment, e.g. electrophysiological recordings in acute slices during optogenetic manipulation, typically involves using a microscope onto which the light source can be coupled. The excitation light on many fluorescence microscopes can be repurposed for optogenetic stimulation once the appropriate wavelength and light intensity parameters are chosen. Alternatively, an LED or laser light source can be installed on the fluorescence excitation port of such a microscope. Light sources can also be mounted remotely to the microscope and the light delivered via a fiber or liquid light guide. An advantage of in vitro preparations is not only the stability of the sample, enabling higher resolution optical manipulations, but also the ability to leverage opportunities for optical access provided by the microscope. In addition to fluorescence ports, for example, light can be focused through the condenser or tube lens using one of the various targeting strategies (see Table 1).
Table 1. Comparison of light targeting strategies.
| Targeted light strategy | Number of neurons addressed | Pros | Cons | Biological questions addressed | Representative references |
|---|---|---|---|---|---|
| 1P full field | 100 - 1000 | Many neurons activated simultaneously, high temporal resolution | Low spatial resolution using viral transfection | Circuit analysis of cell types | 47, 52 |
| 1P full field + sparse labeling | 1 - 100 | High spatial and temporal resolution; can identify cells individually | Only suitable for low numbers of neurons | Single to many-neuron computation | 34 |
| 1P fiber | 100 - 1000 | Can be used in freely moving animals | Low spatial resolution | Effect of cell types on behavior | 104 |
| 1P directed beam | 10 - 100 | Spatial resolution ~50 µm | Cannot activate individual neurons | Mapping anatomical features of cell types and projections | 25, 105 |
| 1P DMD | 100 - 1000 | Commercially available | Low spatial resolution | Effect of activation of cell types in spatial patterns | 71, 72, 103, 106 |
| 1P SLM | 100 - 1000 | Holographic patterns enable photostimulation in three dimensions | Low spatial resolution | Effect of activation of cell types in spatial patterns | 107, 108 |
| 2P directed beam | 1 | Single cell spatial resolution | Only one neuron at a time | Mapping inputs from individual neurons | 76, 83, 84, 109 |
| 2P SLM | ~50 | High-resolution holographic patterns can activate multiple individual neurons | Low temporal resolution | Manipulation of neural coding at the individual neuron level | 78, 84 |
| 2P temporal focusing | 1 - 10 | High spatial and temporal resolution: can activate multiple individual neurons | Few neurons at a time given high laser power required for each neuron | Manipulation of neural coding at the individual neuron level | 77, 78 |
| 2P AOD | 1 - ? | High spatial and temporal resolution: can activate multiple neurons sequentially over very short intervals | Untested | Manipulation of neural coding at the individual neuron level | None |
In vivo experiments can also be performed with a biological microscope if the animal is headfixed, though the presence of the animal obviously blocks optical access through many standard entry points. This situation is alleviated, however, in the case of transparent animals such as zebrafish. For freely moving behaviors in rodents or primates, fiber illumination can be used, which requires a stationary light source coupled to a flexible fiber that terminates in a mount on the animal’s skull53,54. An alternative is an LED mounted directly on the animal, which can be controlled wirelessly55. Head-mounted miniature microscopes also offer the potential to deliver patterns of light stimulation for one56 or two photon excitation57, but the usefulness of these devices has yet to be extended to optogenetics.
The impact of light scattering
The effect of light scattering must be considered when attempting to deliver light in biological tissue. Scattering is the process by which photons are deflected from their path of travel. A turbid medium, such as biological tissue, is highly scattering in an anisotropic manner due its dense and mixed composition. The mean scattering length, or distance a photon travels before being scattered, is on the order of 100 µm in biological tissue for visible light, and slightly higher for infrared light. This means that the distribution of light inside a specimen will not match the distribution of light observed when viewing the output of a light source or fiber outside a specimen. Simulations taking into account optogenetic as well as optical properties indicate that under certain experimental conditions action potentials may be initiated as far away as 1 mm from the fiber source58. The distribution of light in the sample is difficult to obtain directly, but can be estimated with simulations or calibrated directly by making simultaneous recordings.
The importance of simultaneous readout of activity
Given the difficulties associated with targeting opsins to a particular cell type, strong and stable opsin expression, and adequate delivery of light, the reliability of optogenetic manipulation in a given neuron cannot be guaranteed. It is therefore essential to have some form of readout of the activity of the neurons being manipulated. The best way to achieve this is by combining optogenetic manipulation with electrophysiological recording, which offers the highest fidelity measurement59. A range of combinations are currently available, including simply inserting a tetrode together with a fiber; combination with patch-clamp recording60; up to the development of sophisticated optrodes54,61,62. All of these approaches suffer from the following problems. First, given that any electrical recording method is subject to photoelectric effects (such as the Becquerel effect), electrical artefacts arising from light stimulation are almost inevitable53. Second, electrophysiological approaches are limited to recording from only a few, and up to hundreds of neurons; and it is challenging to target particular cell types. As a consequence, assaying activity in a targeted way across the entire optogenetically manipulated population can be difficult.
These problems could be circumvented by an all-optical approach, in which the use of calcium sensors in combination with optogenetic probes in the same cells is used to assay activity from the same neurons that are being optogenetically manipulated. Such an approach, however, faces difficult challenges, such as high expression of both sensors and activators in the same neurons, clean wavelength discrimination and detection for stimulation and imaging, cellular resolution, and adaption in vivo. Using channelrhodopsin-2 simultaneously with voltage sensitive dye imaging has provided coarse anatomical mapping at the resolution of brain regions63. Simultaneous one-photon photostimulation and imaging has been performed in worms to direct stimulation patterns while simultaneously performing calcium imaging of GCaMP at low intensity to avoid inadvertent photostimulation64. Simultaneous one-photon photostimulation and imaging has been performed with fiberoptics in vivo enabling manipulation and imaging on a finer scale of approximately hundreds of neurons65. Simultaneous one-photon activation of sparsely labelled interneurons and two-photon calcium imaging could theoretically provide single-cell resolution for both activation and imaging, by disregarding any imaging data collected during the photostimulation, but results have thus far been heterogeneous66. One-photon photostimulation of inputs to dendrites imaged with two-photon calcium imaging has enabled the dissection of subcellular circuitry67. Expressing both an activator and a genetically encoded calcium indicator in one construct enabled direct measurement of spectral crosstalk, highlighting the usefulness of a novel red indicator (RCaMP) in combination with low-light-sensitive variants of channelrhodopsin-2 to activate and record in separate populations in c. elegans68.
Patterned illumination
Both in vitro and in vivo experiments can make use of full-field illumination, in which light is delivered homogenously over a given spatial area, or patterned illumination, in which light is structured spatially to illuminate particular areas of interest. The simplest form of patterned illumination is to direct a diffraction-limited spot of light to the region of interest either by moving the sample69 or by using a pair of mirrors to direct the beam70 (Fig. 4a). To generate more complex patterns, multiple beams of light can be independently directed using a spatial light modulator (SLM) to generate spatial patterns of light, for example by using a digital micromirror device (DMD)64 (Fig. 4b). This optically simple method is restricted by its low power efficiency, as a great deal of light is lost, however this is often not an issue given the high intensity light sources available. DLP projectors incorporating DMDs can be installed to deliver illumination via a standard microscope condenser71, and have even been programmed to track movement in C. elegans72,73. A third alternative for producing patterns of light is to use holographic projection, often achieved using liquid crystal on silicon SLMs (LCoS-SLM) to create holographic patterns under a microscope objective74 (Fig. 4c). The holographic approach has the advantage that less light is wasted compared to a direct projection approach because the light is reshaped into a pattern. In any patterned illumination experiment, careful controls must be performed to ensure that scattering does not cause unacceptable distortion of the desired pattern at the intended location.
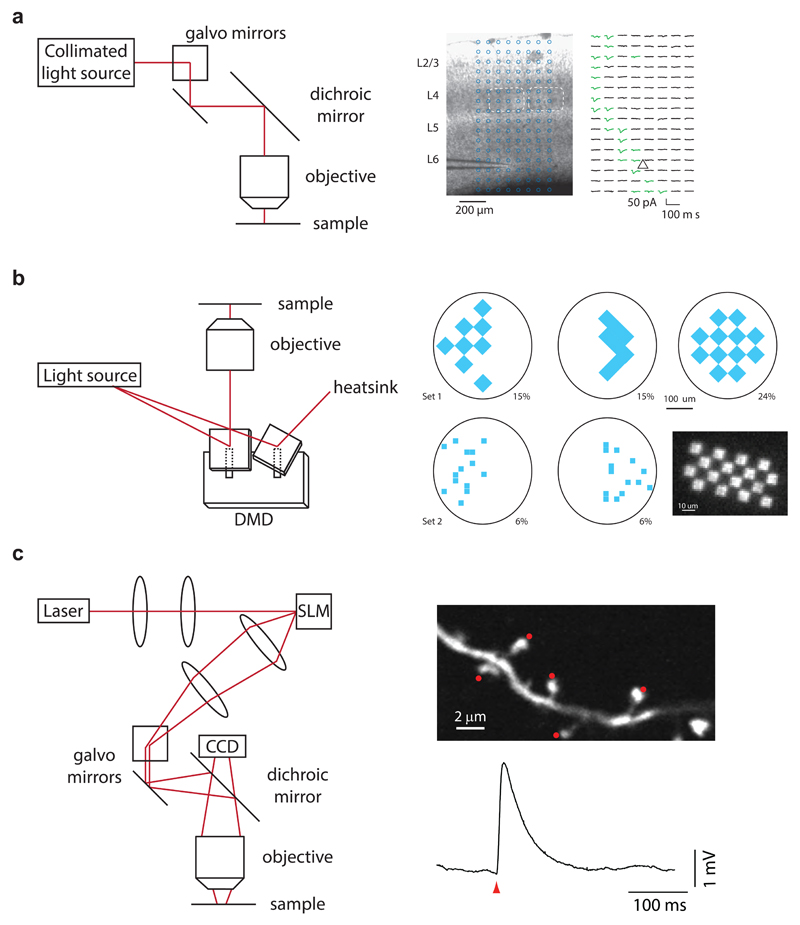
Figure 4. Patterned illumination strategies.
(a) Left, pointing a single beam with galvanometer mirrors is the most straightforward implementation of directing a focused beam of light onto different locations within a sample. Right, this approach is particularly useful for mapping studies101 in which independent activation of small, localized subsets of labeled neurons or axons is desired for readout by downstream neurons. (b) Left, pointing multiple beams with a digital micromirror device102. Right, this enables more complex patterns of activation across large areas of tissue, which has proven useful in studies of retinal circuitry71 and zebrafish behavior103. (c) Left, creating holographic patterns with a spatial light modulator combines the power of generating multiple individual beamlets with high efficiency in directing power into those beamlets. Right, this enables multi-site activation78, 84 when combined with two-photon excitation (see Fig. 5).
Two-photon excitation
The patterned illumination strategies discussed above rely on one-photon excitation, which excites any opsins in the cone of light above and below the focal plane. Two-photon excitation, on the other hand, provides high spatial resolution in both the axial and lateral dimensions by requiring two photons to be absorbed simultaneously, which only occurs within the extremely confined focal volume75 (Fig. 5a). Combining two-photon microscopy with optogenetics is difficult due to this small illumination volume. While the number of opsin molecules must be expressed at levels low enough for the neurons to remain healthy, sufficient numbers of them must be activated to generate the desired current. Sufficiency depends on the current generated per opsin molecule (which depends on its two-photon absorption cross-section and its conductance); expression level (i.e. number of opsin molecules per membrane area); temporal kinetics of the opsin (mainly the off-time); and the particular spatiotemporal illumination strategy.
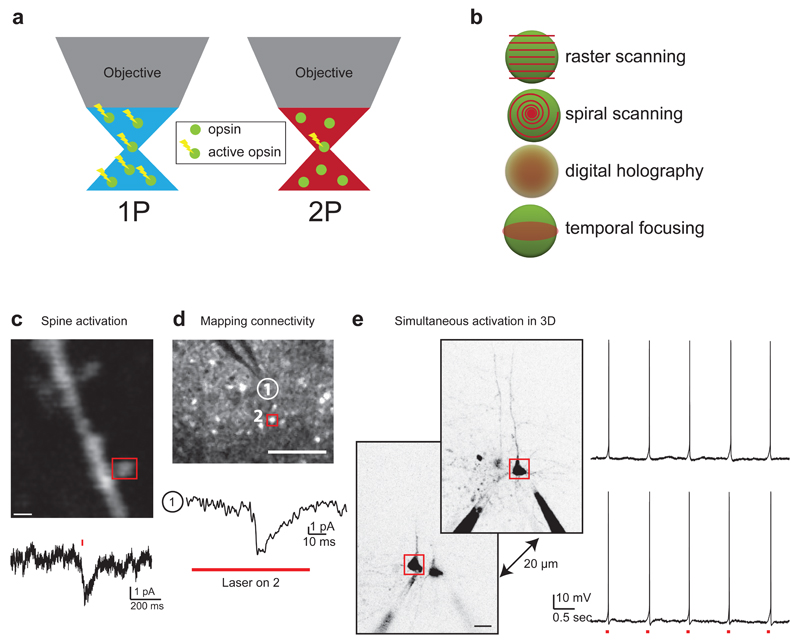
Figure 5. One-photon versus two-photon activation strategies: from spines to circuits.
(a) In one-photon excitation (left), all opsin molecules illuminated above and below the focal plane of interest are excited. Alternatively, in two-photon excitation (right), generally only opsin molecules in the focal plane are excited (but see Ref. 76), leading to optical sectioning that allows activation to be restricted to the particular neurons of interest. (b) Spatiotemporal patterns for illuminating neurons with two-photon beams require different power budgets and yield different spatial and temporal resolutions (see Table 1). (c) Two-photon point stimulation of a dendritic spine on a neuron expressing C1V1 (top panel, scale bar 1 um) generates current detectable at the soma (bottom trace). (d) Two-photon raster-scanning of neuron 2 (top panel, red box) during electrophysiological recording from neuron 1 (white circle, top panel and bottom trace) indicates that neuron 2 is monosynaptically connected to neuron 1 (scale bar 100 µm). (e) Simultaneous action potential generation in two neurons in 3D using a spatial light modulator to generate separate laser beamlets over each neuron (scale bar 20 µm). Data in panels c,d,e adapted from Ref. 84.
Optimization of these parameters can lead to successful action potential generation in neurons in acute slices and in vivo. In the initial work on two-photon excitation of channelrhodopsin76, the most commonly used variant, ChR2(H134R), was shown to absorb two photons effectively. Scanning the somata of highly expressing cultured neurons in a spiral pattern for 32 milliseconds resulted in efficient spatiotemporal integration of photostimulated current leading to reliable action potential generation (Fig. 5b). Subsequent work showed action potential generation via two-photon excitation in acute brain slices using temporal focusing to create a disk-shaped illumination pattern. This enabled simultaneous stimulation of many opsin molecules in neuronal somata with very short stimulation times (≤ 5ms)77. Combining temporal focusing with an SLM allowed structuring the two-photon illumination to match the shape of neuronal somata, further enabling the activation of more than one selected neuron simultaneously78. Such methods to shape the illumination to the soma require relatively high power on sample (70 to >100mW) to obtain sufficient power density over the extended surface area, and calculations indicate that these ‘parallel’ excitation methods may require upwards of 20 times as much power as ‘serial’ scanning methods79. This may be a problem for in vivo applications, where light scattering is severe80, although adaptive optics can be used to increase two-photon excitation deep in the tissue81. Recent work using temporal focusing (Fig. 5b) has shown robustness against scattering, enabling action potential production >200um deep in tissue82. If simultaneous stimulation of multiple neurons in vivo is the goal, improving the power per neuron ratio is required.
The recent introduction of C1V1, a red-shifted opsin with several variants, has addressed this issue directly. Expression of this opsin enables action potential generation in highly-expressing neurons via conventional two-photon raster scanning, as performed during standard two-photon imaging83 (Fig. 5b). One of the C1V1 variants, C1V1t, has off kinetics approximately twice as slow as ChR2(H134R), easing the constraints on the integration of photostimulated current. With this variant, relatively short illumination times (10-15ms) leads to robust action potential generation with only 20mW of laser power on sample. The superior spatial resolution of two-photon microscopy allows zooming in to even finer levels of detail of neuronal function, at the level of subcellular compartments. Two-photon excitation pinpointed to dendrites and individual dendritic spines also generates reliable optogenetic excitation (Fig. 5c)83,84. This technique enables the mapping of monosynaptic connections from individual neurons to electrophysiologically recorded neurons (Fig. 5d)84. Alternatively, an SLM can be used to split the laser beam into individual beamlets, mediating the activation of multiple selected neurons in three dimensions (Fig. 5e)84. The reduced power budget implies that more selected neurons can be activated, albeit with less temporal precision.
An alternative approach for two-photon activation in rapid spatial patterns could involve the use of acousto-optical deflectors (AODs), which allow for dramatically increased speed in directing light versus conventional galvo-based systems85–88. Given the high two-photon absorption cross-section of channelrhodopsin-2, it seems the optimal excitation strategy would be to minimize scan time76, though an upper limit to how quickly illumination can be performed has been found in raster-scanning applications83,84. Temporal focusing strategies77,78 can alleviate this issue by illuminating the entire neuronal soma simultaneously, but if multiple neurons are to be stimulated, AODs should be particularly helpful given their ability to redirect a laser beam within microseconds85–88, compared to conventional galvo-based systems which take ~100 microseconds. AODs could perhaps even be used to excite individual neurons more efficiently than current strategies (given the high power of an individual beam and the speed with which it can be refocused), particularly if new opsins with different kinetic properties become available.
Tradeoffs between various light targeting strategies
The tradeoffs between the number of cells activated, to what level of activity, at a given resolution are important to consider when determining which targeted light strategy is appropriate for a given experiment. Table 1 provides a comparison of the various approaches that are currently available.
3. Applications of successful optogenetic targeting
The genetic and optical targeting strategies described above are under active development across hundreds of laboratories worldwide, and should yield significant improvements over the next decade. Moreover, the various strategies can be used in concert, potentially dramatically enhancing the power of the optogenetic approach. In order to spur further development and provide a yardstick for progress, it is useful to consider what ‘dream experiments’ may become possible using these more sophisticated optogenetic strategies – which we call ‘targeted optogenetics’ – and what fundamental questions in neuroscience they can be used to answer.
Probing neural identity
The question of what defines neuronal identity continues to be an issue in neuroscience89. While traditional definitions of identity on the basis of morphological features (dendritic and axonal shape, projection patterns) have recently been complemented by electrophysiological and genetic ‘fingerprinting’90,91, rigorously defining cell types remains a difficult challenge. The ability to target optogenetic probes to precisely defined populations should allow the anatomical, genetic, and physiological definitions of identity to be combined in unprecedented ways. In particular, it may be possible to identify the precise functional role of a particular cell type during behaviour – or to reveal further subdivisions in a defined population – by activation or inactivation of that cell class. For example, restricting expression of ChR2 in a particular cell class (identified based on projection target, or genetic identity) allows these neurons to be ‘tagged’ and identified by optogenetic activation during conventional electrophysiological or optrode recordings92. Such an approach has been used to identify and distinguish cortical interneurons from pyramidal cells92–94, GABAergic and dopaminergic neurons in the ventral tegmental area95, and striatal interneurons and projection neurons96. Experimental strategies such as these should allow for greater security of cell type identification during in vivo experiments, and ultimately may also lead to richer definitions of neuronal identity.
How many neurons are enough?
Recent experiments have suggested that the activity of only few neurons97 – or perhaps only a single neuron98 – may be enough to change network activity sufficiently to influence behaviour. The relationship between the number of active neurons and behavioural readout remains unknown, and would put fundamental constraints on the design of neural circuits and their sensitivity to perturbations. Being able to target expression of optogenetic probes to defined numbers of neurons, and/or being able to activate (or inactivate) precise numbers of neurons using a targeted optical approach, should enable quantification of this relationship for different cell types during behaviour. This would provide fundamental information about the sparseness of representations in neural circuits. It may also identify if there are particular types of neurons (defined by their anatomical, genetic or functional identity – see above) or even single neurons that are unusually influential in regulating the activity of their local circuits and ultimately behaviour.
Cracking the neural code
The nature of the neural code has long been a fundamental problem in neuroscience. Given that behaviour can engage thousands of neurons in intricate patterns on millisecond timescales, probing the nature of the code presents a formidable challenge to the optogenetic approach, which in its conventional incarnation only permits synchronous activation of neural populations. Cracking the neural code – in other words, determining which spatiotemporal patterns of activity in which genetically defined sets of neurons causally drive behaviour – will require ‘playing in’ spatiotemporal patterns of activity into the circuit with the same temporal and spatial precision as the ‘real’ physiological patterns. Therefore, it will be necessary to combine optical readout of activity using activity indicators (e.g. for voltage or calcium), followed by optogenetic manipulation of the same neurons, both with high temporal and spatial resolution, ideally in a large three-dimensional volume encompassing the entire engaged circuit (Fig. 6). Such an experiment is currently not possible given the combined constraints of opsin properties and optical hardware, although the recent advances in patterned illumination described above suggest that this on the horizon. Moreover, since behaviour engages activity differentially across different populations of neurons within the same circuit – at the minimum, excitatory and inhibitory neurons – it will be necessary to use a multi-colour approach for selective manipulation of the different populations. Once these problems are overcome, however, it should be possible to test which neural codes – for example, involving different levels of temporal precision – in which neurons are required to drive specific behaviours. Similar experiments (in combination with the use of activity-dependent opsin expression) may be used to probe which activity patterns drive memory storage and retrieval. The interplay between experiment and theory is expected to play a crucial role for answering these questions, not only because theoretical approaches are extremely useful for refining design and interpretation of optogenetic experiments58,99, but also because theories can provide experimentally testable predictions100.
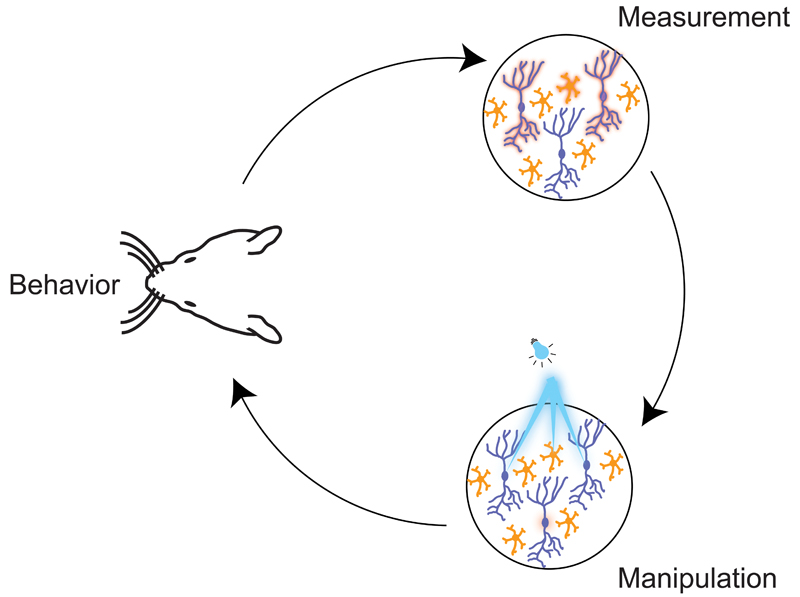
Figure 6. Using targeted optogenetics to enable ‘dream experiments’.
A schematic illustration of how ‘targeted optogenetics’ can be used to probe the neural code in a cortical circuit. The figure highlights the close interplay that is necessary between behavioural experiments, optical readout of patterns of activity, and replay of the same patterns in the ‘right’ neurons using optogenetics. Targeted optogenetics allows the precision of temporal patterns, and the precise membership of the neuronal ensemble, to be tested directly to investigate their importance for the neural code driving the behaviour.
Acknowledgements
We are grateful to Beverley Clark, Benjamin Judkewitz, Darcy Peterka, Arnd Roth, Tatsuo Sato, Spencer Smith, Dara Sosulski, Christian Wilms, Keisuke Yonehara, Rafael Yuste, and Feng Zhang for helpful discussions and for comments on the manuscript. We apologize to our colleagues whose work could not be cited owing to space constraints. This work was supported by an EMBO Long-Term Fellowship (AP) and by grants from the Friedrich Miescher Institute, Alcon, ERC, and the European Union (BR) and from the Wellcome Trust, ERC, and Gatsby Charitable Foundation (MH).
References
- 1.Miesenbock G. The optogenetic catechism. Science. 2009;326:395–399. doi: 10.1126/science.1174520. [DOI] [PubMed] [Google Scholar]
- 2.Bamann C, Nagel G, Bamberg E. Microbial rhodopsins in the spotlight. Curr Opin Neurobiol. 2010;20:610–616. doi: 10.1016/j.conb.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Madisen L, et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat Neurosci. 2012;15:793–802. doi: 10.1038/nn.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kienle E, et al. Engineering and evolution of synthetic adeno-associated virus (AAV) gene therapy vectors via DNA family shuffling. Journal of visualized experiments : JoVE. 2012 doi: 10.3791/3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cronin J, Zhang XY, Reiser J. Altering the tropism of lentiviral vectors through pseudotyping. Current gene therapy. 2005;5:387–398. doi: 10.2174/1566523054546224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wall NR, Wickersham IR, Cetin A, De La Parra M, Callaway EM. Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. PNAS. 2010;107:21848–21853. doi: 10.1073/pnas.1011756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tripodi M, Stepien AE, Arber S. Motor antagonism exposed by spatial segregation and timing of neurogenesis. Nature. 2011;479:61–66. doi: 10.1038/nature10538. [DOI] [PubMed] [Google Scholar]
- 9.Yonehara K, et al. Spatially asymmetric reorganization of inhibition establishes a motion-sensitive circuit. Nature. 2011;469:407–410. doi: 10.1038/nature09711. [DOI] [PubMed] [Google Scholar]
- 10.Calame M, et al. Retinal degeneration progression changes lentiviral vector cell targeting in the retina. PloS one. 2011;6:e23782. doi: 10.1371/journal.pone.0023782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farrow K, et al. Ambient Illumination Toggles a Neuronal Circuit Switch in the Retina and Visual Perception at Cone Threshold. Neuron. 2013 doi: 10.1016/j.neuron.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Dymecki SM, Ray RS, Kim JC. Mapping cell fate and function using recombinase-based intersectional strategies. Methods in enzymology. 2010;477:183–213. doi: 10.1016/S0076-6879(10)77011-7. [DOI] [PubMed] [Google Scholar]
- 14.Busskamp V, et al. Genetic reactivation of cone photoreceptors restores visual responses in retinitis pigmentosa. Science. 2010;329:413–417. doi: 10.1126/science.1190897. [DOI] [PubMed] [Google Scholar]
- 15.Busskamp V, Picaud S, Sahel JA, Roska B. Optogenetic therapy for retinitis pigmentosa. Gene therapy. 2012;19:169–175. doi: 10.1038/gt.2011.155. [DOI] [PubMed] [Google Scholar]
- 16.Tye KM, Deisseroth K. Optogenetic investigation of neural circuits underlying brain disease in animal models. Nat Rev Neurosci. 2012;13:251–266. doi: 10.1038/nrn3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arlotta P, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 18.Fried SI, Munch TA, Werblin FS. Mechanisms and circuitry underlying directional selectivity in the retina. Nature. 2002;420:411–414. doi: 10.1038/nature01179. [DOI] [PubMed] [Google Scholar]
- 19.Neve RL. Overview of gene delivery into cells using HSV-1-based vectors. Current protocols in neuroscience / editorial board, Jacqueline N. Crawley … [et al. 2012;Chapter 4:Unit 4 12. doi: 10.1002/0471142301.ns0412s61. [DOI] [PubMed] [Google Scholar]
- 20.Soudais C, Laplace-Builhe C, Kissa K, Kremer EJ. Preferential transduction of neurons by canine adenovirus vectors and their efficient retrograde transport in vivo. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2001;15:2283–2285. doi: 10.1096/fj.01-0321fje. [DOI] [PubMed] [Google Scholar]
- 21.Wickersham IR, et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beier KT, et al. Anterograde or retrograde transsynaptic labeling of CNS neurons with vesicular stomatitis virus vectors. PNAS. 2011;108:15414–15419. doi: 10.1073/pnas.1110854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato S, et al. A lentiviral strategy for highly efficient retrograde gene transfer by pseudotyping with fusion envelope glycoprotein. Human gene therapy. 2011;22:197–206. doi: 10.1089/hum.2009.179. [DOI] [PubMed] [Google Scholar]
- 24.Kaspar BK, et al. Targeted retrograde gene delivery for neuronal protection. Molecular therapy : the journal of the American Society of Gene Therapy. 2002;5:50–56. doi: 10.1006/mthe.2001.0520. [DOI] [PubMed] [Google Scholar]
- 25.Petreanu L, Mao T, Sternson SM, Svoboda K. The subcellular organization of neocortical excitatory connections. Nature. 2009;457:1142–1145. doi: 10.1038/nature07709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osakada F, et al. New rabies virus variants for monitoring and manipulating activity and gene expression in defined neural circuits. Neuron. 2011;71:617–631. doi: 10.1016/j.neuron.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beier KT, et al. Transsynaptic tracing with vesicular stomatitis virus reveals novel retinal circuitry. J Neurosci. 2013;33:35–51. doi: 10.1523/JNEUROSCI.0245-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshel JH, Mori T, Nielsen KJ, Callaway EM. Targeting single neuronal networks for gene expression and cell labeling in vivo. Neuron. 2010;67:562–574. doi: 10.1016/j.neuron.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gradinaru V, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu W, Sudhof TC. A neural circuit for memory specificity and generalization. Science. 2013;339:1290–1295. doi: 10.1126/science.1229534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lo L, Anderson DJ. A Cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron. 2011;72:938–950. doi: 10.1016/j.neuron.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyashita T, Shao YR, Chung J, Pourzia O, Feldman DE. Long-term channelrhodopsin-2 (ChR2) expression can induce abnormal axonal morphology and targeting in cerebral cortex. Frontiers in neural circuits. 2013;7:8. doi: 10.3389/fncir.2013.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitamura K, Judkewitz B, Kano M, Denk W, Hausser M. Targeted patch-clamp recordings and single-cell electroporation of unlabeled neurons in vivo. Nature methods. 2008;5:61–67. doi: 10.1038/nmeth1150. [DOI] [PubMed] [Google Scholar]
- 34.Judkewitz B, Rizzi M, Kitamura K, Hausser M. Targeted single-cell electroporation of mammalian neurons in vivo. Nature protocols. 2009;4:862–869. doi: 10.1038/nprot.2009.56. [DOI] [PubMed] [Google Scholar]
- 35.Reijmers L, Mayford M. Genetic control of active neural circuits. Frontiers in molecular neuroscience. 2009;2:27. doi: 10.3389/neuro.02.027.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, et al. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nature methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 38.Han X, et al. Millisecond-timescale optical control of neural dynamics in the nonhuman primate brain. Neuron. 2009;62:191–198. doi: 10.1016/j.neuron.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diester I, et al. An optogenetic toolbox designed for primates. Nat Neurosci. 2011;14:387–397. doi: 10.1038/nn.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han X, et al. A high-light sensitivity optical neural silencer: development and application to optogenetic control of non-human primate cortex. Frontiers in systems neuroscience. 2011;5:18. doi: 10.3389/fnsys.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerits A, et al. Optogenetically induced behavioral and functional network changes in primates. Curr Biol. 2012;22:1722–1726. doi: 10.1016/j.cub.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jazayeri M, Lindbloom-Brown Z, Horwitz GD. Saccadic eye movements evoked by optogenetic activation of primate V1. Nat Neurosci. 2012;15:1368–1370. doi: 10.1038/nn.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cavanaugh J, et al. Optogenetic inactivation modifies monkey visuomotor behavior. Neuron. 2012;76:901–907. doi: 10.1016/j.neuron.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han X, Boyden ES. Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PloS one. 2007;2:e299. doi: 10.1371/journal.pone.0000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kleinlogel S, et al. A gene-fusion strategy for stoichiometric and co-localized expression of light-gated membrane proteins. Nature methods. 2011;8:1083–1088. doi: 10.1038/nmeth.1766. [DOI] [PubMed] [Google Scholar]
- 46.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 47.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 48.Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nat Neurosci. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 49.Mattis J, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nature methods. 2012;9:159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schoenenberger P, Scharer YP, Oertner TG. Channelrhodopsin as a tool to investigate synaptic transmission and plasticity. Experimental physiology. 2011;96:34–39. doi: 10.1113/expphysiol.2009.051219. [DOI] [PubMed] [Google Scholar]
- 51.Hirase H, Nikolenko V, Goldberg JH, Yuste R. Multiphoton stimulation of neurons. J Neurobiol. 2002;51:237–247. doi: 10.1002/neu.10056. [DOI] [PubMed] [Google Scholar]
- 52.Nagel G, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. PNAS. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cardin JA, et al. Targeted optogenetic stimulation and recording of neurons in vivo using cell-type-specific expression of Channelrhodopsin-2. Nature protocols. 2010;5:247–254. doi: 10.1038/nprot.2009.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LeChasseur Y, et al. A microprobe for parallel optical and electrical recordings from single neurons in vivo. Nature methods. 2011;8:319–325. doi: 10.1038/nmeth.1572. [DOI] [PubMed] [Google Scholar]
- 55.Iwai Y, Honda S, Ozeki H, Hashimoto M, Hirase H. A simple head-mountable LED device for chronic stimulation of optogenetic molecules in freely moving mice. Neuroscience research. 2011;70:124–127. doi: 10.1016/j.neures.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 56.Flusberg BA, et al. High-speed, miniaturized fluorescence microscopy in freely moving mice. Nature methods. 2008;5:935–938. doi: 10.1038/nmeth.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Helmchen F, Fee MS, Tank DW, Denk W. A miniature head-mounted two-photon microscope. high-resolution brain imaging in freely moving animals. Neuron. 2001;31:903–912. doi: 10.1016/s0896-6273(01)00421-4. [DOI] [PubMed] [Google Scholar]
- 58.Foutz TJ, Arlow RL, McIntyre CC. Theoretical principles underlying optical stimulation of a channelrhodopsin-2 positive pyramidal neuron. J Neurophysiol. 2012;107:3235–3245. doi: 10.1152/jn.00501.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scanziani M, Hausser M. Electrophysiology in the age of light. Nature. 2009;461:930–939. doi: 10.1038/nature08540. [DOI] [PubMed] [Google Scholar]
- 60.Lee SH, et al. Activation of specific interneurons improves V1 feature selectivity and visual perception. Nature. 2012;488:379–383. doi: 10.1038/nature11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anikeeva P, et al. Optetrode: a multichannel readout for optogenetic control in freely moving mice. Nat Neurosci. 2012;15:163–170. doi: 10.1038/nn.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Royer S, et al. Multi-array silicon probes with integrated optical fibers: light-assisted perturbation and recording of local neural circuits in the behaving animal. Eur J Neurosci. 2010;31:2279–2291. doi: 10.1111/j.1460-9568.2010.07250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lim DH, et al. In vivo Large-Scale Cortical Mapping Using Channelrhodopsin-2 Stimulation in Transgenic Mice Reveals Asymmetric and Reciprocal Relationships between Cortical Areas. Frontiers in neural circuits. 2012;6:11. doi: 10.3389/fncir.2012.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo ZV, Hart AC, Ramanathan S. Optical interrogation of neural circuits in Caenorhabditis elegans. Nature methods. 2009;6:891–896. doi: 10.1038/nmeth.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stroh A, et al. Making waves: initiation and propagation of corticothalamic ca(2+) waves in vivo. Neuron. 2013;77:1136–1150. doi: 10.1016/j.neuron.2013.01.031. [DOI] [PubMed] [Google Scholar]
- 66.Wilson NR, Runyan CA, Wang FL, Sur M. Division and subtraction by distinct cortical inhibitory networks in vivo. Nature. 2012;488:343–348. doi: 10.1038/nature11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Little JP, Carter AG. Subcellular synaptic connectivity of layer 2 pyramidal neurons in the medial prefrontal cortex. J Neurosci. 2012;32:12808–12819. doi: 10.1523/JNEUROSCI.1616-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akerboom J, et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Frontiers in molecular neuroscience. 2013;6:2. doi: 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Callaway EM, Katz LC. Photostimulation using caged glutamate reveals functional circuitry in living brain slices. PNAS. 1993;90:7661–7665. doi: 10.1073/pnas.90.16.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shepherd GM, Pologruto TA, Svoboda K. Circuit analysis of experience-dependent plasticity in the developing rat barrel cortex. Neuron. 2003;38:277–289. doi: 10.1016/s0896-6273(03)00152-1. [DOI] [PubMed] [Google Scholar]
- 71.Munch TA, et al. Approach sensitivity in the retina processed by a multifunctional neural circuit. Nat Neurosci. 2009;12:1308–1316. doi: 10.1038/nn.2389. [DOI] [PubMed] [Google Scholar]
- 72.Leifer AM, Fang-Yen C, Gershow M, Alkema MJ, Samuel AD. Optogenetic manipulation of neural activity in freely moving Caenorhabditis elegans. Nature methods. 2011;8:147–152. doi: 10.1038/nmeth.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stirman JN, et al. Real-time multimodal optical control of neurons and muscles in freely behaving Caenorhabditis elegans. Nature methods. 2011;8:153–158. doi: 10.1038/nmeth.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nikolenko V, et al. SLM Microscopy: Scanless Two-Photon Imaging and Photostimulation with Spatial Light Modulators. Frontiers in neural circuits. 2008;2:5. doi: 10.3389/neuro.04.005.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 76.Rickgauer JP, Tank DW. Two-photon excitation of channelrhodopsin-2 at saturation. PNAS. 2009;106:15025–15030. doi: 10.1073/pnas.0907084106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Andrasfalvy BK, Zemelman BV, Tang J, Vaziri A. Two-photon single-cell optogenetic control of neuronal activity by sculpted light. PNAS. 2010;107:11981–11986. doi: 10.1073/pnas.1006620107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Papagiakoumou E, et al. Scanless two-photon excitation of channelrhodopsin-2. Nature methods. 2010;7:848–854. doi: 10.1038/nmeth.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peron S, Svoboda K. From cudgel to scalpel: toward precise neural control with optogenetics. Nature methods. 2011;8:30–34. doi: 10.1038/nmeth.f.325. [DOI] [PubMed] [Google Scholar]
- 80.Theer P, Denk W. On the fundamental imaging-depth limit in two-photon microscopy. Journal of the Optical Society of America. A, Optics, image science, and vision. 2006;23:3139–3149. doi: 10.1364/josaa.23.003139. [DOI] [PubMed] [Google Scholar]
- 81.Ji N, Sato TR, Betzig E. Characterization and adaptive optical correction of aberrations during in vivo imaging in the mouse cortex. PNAS. 2012;109:22–27. doi: 10.1073/pnas.1109202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Papagiakoumou E, et al. Functional patterned multiphoton excitation deep inside scattering tissue. Nat Photonics. 2013;7:274–278. [Google Scholar]
- 83.Prakash R, et al. Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nature methods. 2012;9:1171–1179. doi: 10.1038/nmeth.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Packer AM, et al. Two-photon optogenetics of dendritic spines and neural circuits. Nature methods. 2012;9:1202–1205. doi: 10.1038/nmeth.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Duemani Reddy G, Kelleher K, Fink R, Saggau P. Three-dimensional random access multiphoton microscopy for functional imaging of neuronal activity. Nat Neurosci. 2008;11:713–720. doi: 10.1038/nn.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Katona G, et al. Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nature methods. 2012;9:201–208. doi: 10.1038/nmeth.1851. [DOI] [PubMed] [Google Scholar]
- 87.Kirkby PA, Srinivas Nadella KM, Silver RA. A compact Acousto-Optic Lens for 2D and 3D femtosecond based 2-photon microscopy. Optics express. 2010;18:13721–13745. doi: 10.1364/OE.18.013720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grewe BF, Langer D, Kasper H, Kampa BM, Helmchen F. High-speed in vivo calcium imaging reveals neuronal network activity with near-millisecond precision. Nature methods. 2010;7:399–405. doi: 10.1038/nmeth.1453. [DOI] [PubMed] [Google Scholar]
- 89.Defelipe J, et al. New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat Rev Neurosci. 2013 doi: 10.1038/nrn3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Toledo-Rodriguez M, et al. Correlation maps allow neuronal electrical properties to be predicted from single-cell gene expression profiles in rat neocortex. Cereb Cortex. 2004;14:1310–1327. doi: 10.1093/cercor/bhh092. [DOI] [PubMed] [Google Scholar]
- 91.Siegert S, et al. Transcriptional code and disease map for adult retinal cell types. Nat Neurosci. 2012;15:487–495. S481–482. doi: 10.1038/nn.3032. [DOI] [PubMed] [Google Scholar]
- 92.Lima SQ, Hromadka T, Znamenskiy P, Zador AM. PINP: a new method of tagging neuronal populations for identification during in vivo electrophysiological recording. PloS one. 2009;4:e6099. doi: 10.1371/journal.pone.0006099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Royer S, et al. Control of timing, rate and bursts of hippocampal place cells by dendritic and somatic inhibition. Nat Neurosci. 2012;15:769–775. doi: 10.1038/nn.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cardin JA, et al. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 2009;459:663–667. doi: 10.1038/nature08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cohen JY, Haesler S, Vong L, Lowell BB, Uchida N. Neuron-type-specific signals for reward and punishment in the ventral tegmental area. Nature. 2012;482:85–88. doi: 10.1038/nature10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kravitz AV, et al. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature. 2010;466:622–626. doi: 10.1038/nature09159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huber D, et al. Sparse optical microstimulation in barrel cortex drives learned behaviour in freely moving mice. Nature. 2008;451:61–64. doi: 10.1038/nature06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Houweling AR, Brecht M. Behavioural report of single neuron stimulation in somatosensory cortex. Nature. 2008;451:65–68. doi: 10.1038/nature06447. [DOI] [PubMed] [Google Scholar]
- 99.Grossman N, et al. The spatial pattern of light determines the kinetics and modulates backpropagation of optogenetic action potentials. J Comput Neurosci. 2012 doi: 10.1007/s10827-012-0431-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.London M, Roth A, Beeren L, Hausser M, Latham PE. Sensitivity to perturbations in vivo implies high noise and suggests rate coding in cortex. Nature. 2010;466:123–127. doi: 10.1038/nature09086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat Neurosci. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- 102.Jerome J, Foehring RC, Armstrong WE, Spain WJ, Heck DH. Parallel optical control of spatiotemporal neuronal spike activity using high-speed digital light processing. Frontiers in systems neuroscience. 2011;5:70. doi: 10.3389/fnsys.2011.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Blumhagen F, et al. Neuronal filtering of multiplexed odour representations. Nature. 2011;479:493–498. doi: 10.1038/nature10633. [DOI] [PubMed] [Google Scholar]
- 104.Zhang F, et al. Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nature protocols. 2010;5:439–456. doi: 10.1038/nprot.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang H, et al. High-speed mapping of synaptic connectivity using photostimulation in Channelrhodopsin-2 transgenic mice. PNAS. 2007;104:8143–8148. doi: 10.1073/pnas.0700384104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhu P, Fajardo O, Shum J, Zhang Scharer YP, Friedrich RW. High-resolution optical control of spatiotemporal neuronal activity patterns in zebrafish using a digital micromirror device. Nature protocols. 2012;7:1410–1425. doi: 10.1038/nprot.2012.072. [DOI] [PubMed] [Google Scholar]
- 107.Nikolenko V, Peterka DS, Yuste R. A portable laser photostimulation and imaging microscope. Journal of neural engineering. 2010;7:045001. doi: 10.1088/1741-2560/7/4/045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reutsky-Gefen I, et al. Holographic optogenetic stimulation of patterned neuronal activity for vision restoration. Nature communications. 2013;4:1509. doi: 10.1038/ncomms2500. [DOI] [PubMed] [Google Scholar]
- 109.Zhu P, et al. Optogenetic Dissection of Neuronal Circuits in Zebrafish using Viral Gene Transfer and the Tet System. Frontiers in neural circuits. 2009;3:21. doi: 10.3389/neuro.04.021.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]