Abstract
The objective of this study was to identify genomic regions that are associated with meat quality traits in the Nellore breed. Nellore steers were finished in feedlots and slaughtered at a commercial slaughterhouse. This analysis included 1,822 phenotypic records of tenderness and 1,873 marbling records. After quality control, 1,630 animals genotyped for tenderness, 1,633 animals genotyped for marbling, and 369,722 SNPs remained. The results are reported as the proportion of variance explained by windows of 150 adjacent SNPs. Only windows with largest effects were considered. The genomic regions were located on chromosomes 5, 15, 16 and 25 for marbling and on chromosomes 5, 7, 10, 14 and 21 for tenderness. These windows explained 3,89% and 3,80% of the additive genetic variance for marbling and tenderness, respectively. The genes associated with the traits are related to growth, muscle development and lipid metabolism. The study of these genes in Nellore cattle is the first step in the identification of causal mutations that will contribute to the genetic evaluation of the breed.
Introduction
Brazil is one of the world’s largest exporter of beef and possesses the second largest herd [1]. Zebu animals (Bos taurus indicus) account for 80% of this herd and the Nellore breed corresponds to 90% of these animals [2]. Studies have shown that the quality of Bos taurus indicus meat is inferior when compared to the meat of Bos taurus taurus [3,4]. Meat quality is a generic term used to describe consumer perceptions of meat [5]. In this respect, tenderness is one of the most important attributes that determines the quality of the product and consumer acceptance [6]. Another attribute, defined by sensory panel testing, which is involved in the appreciation of meat quality, is intramuscular fat [7]. This attribute affects the flavor, juiciness and chewing of meat. These traits are important to guarantee the stability and market expansion of Brazil as a beef exporter.
The application of traditional selection to these traits is difficult, since they are expensive and difficult to measure because it requires the slaughter of the animals. Genome-wide association studies (GWAS) allow the identification of single-nucleotide polymorphisms (SNPs) associated with large-effect genes that influence these traits, providing a better biological understanding of the trait and a list of candidate genes for fine mapping.
At first, classical methods were used for GWAS, which were based on testing one marker at a time. However, using those methods increases the chance of false-positives due to multicolinearity [8]. Furthermore, it is difficulty to test the significance of each marker considering the existence of thousands of SNPs [9]. Currently, different methods of modeling SNPs simultaneously have been used. In the majority of these methods, the analyses are carried out in multiple steps, which first require traditional genetic evaluation using an animal model, followed by the formation of pseudo-phenotypes and, finally, the estimation of the SNP effects [10]. In addition, those methods generally require that phenotype and genotype information are available for all animals. Recently, a method that uses pedigree, phenotype and genotype data in a single step (ssGBLUP) was proposed [11]. This method can be easily implemented and allows including in the analysis more animals with phenotypes than genotypes.
Some association studies of SNPs with meat quality traits in taurine cattle have identified different regions associated with marbling, which are located on chromosomes 2, 3, 7, 8, 10, 17, 22 and 27 and with meat tenderness, which are located on chromosomes 7, 8, 9, 10 and 29 [12–15]. Genes that affect meat tenderness, such as calpain (CAPN1) and calpastatin (CAST), are located on chromosomes 29 and 7, respectively [16,17]. The SNPs CAPN4751, CAPN4753, UOGCAST and WSUCAST, which are associated with tenderness in Bos taurus taurus, are also polymorphic in Nellore cattle [18]. In GWAS of Nellore animals, [19] identified SNPs for tenderness located near CAPN1, CAPN2, CAPN5 and CAST.
In previous studies, it is unclear whether these genes contribute to the additive genetic variance of these traits in Zebu cattle. Therefore, the objective of this study was to perform GWAS in order to identify genomic regions that are associated with meat quality traits in the Nellore breed.
Materials and Methods
Data
Tenderness and marbling were the meat quality traits used in this study. Data of male Nellore animals born between 2008 and 2010 were collected among herds located in different regions of Brazil, which belong to three breeding programs (DeltaGen, Paint, and Nelore Qualitas). The animals were finished in feedlots for approximately 90 days and slaughtered at an average age of 731 ± 81 days.
The approval of the ethics committee of the Faculty of Agrarian Sciences and Veterinary of Sao Paulo State (FCAV-UNESP) was not necessary, because the slaughter of animals was done in commercial slaughterhouses (JBS S/A, Minerva S.A and Marfrig Alimentos S.A), located in several regions of Brazil. Since such slaughterhouses have animal welfare department, staffed by professionals trained by WAG (World Animal Protection), ensuring that the animals are killed humanely, by use of captive bolt pistol in the stunning process.
The carcasses were maintained in a cold chamber for 24 to 48 h post-mortem and longissimus dorsi muscle (tenderloin) samples were detached with bone between the 12th and 13th rib of the left half-carcasses. All samples were frozen and none of them was aged. Animals from the same farm and year of birth were slaughtered in the same slaughterhouse and slaughter date.
Longissimus dorsi samples measuring 2.54 cm in thickness were obtained for analysis of tenderness. The procedure standardized and proposed by [20] was adopted, which consists of the measurement of shear force using a mechanical Salter Warner-Bratzler Shear Force device. The degree of marbling was scored on a scale from 1 to 10 according to the method of [21], where 1 = practically absent; 2 = traces; 3 = slight; 4 = small; 5 = modest; 6 = moderate; 7 = slightly abundant; 8 = moderately abundant; 9 = abundant, and 10 = very abundant.
There were a total of 7,436 animals in the relationship matrix. Data from 1,875 animals with tenderness and marbling records were available. Measures that were 3.5 standard deviations above or below the mean of the contemporary group (year, farm and management group at yearling) were excluded from the analyses. Additionally, contemporary groups containing fewer than three animals were eliminated. Thus, there were records of 1,822 animals with tenderness measures and of 1,873 animals with marbling measures (Table 1). The heritabilities were estimated using Bayesian approach with BLUPF90 family of programs [22]. The model included fixed effects of contemporary groups (year, farm of birth and management group at yearling) and age at slaughter and days between slaughter and physicochemical analysis of meat as covariates (linear effect).
Table 1. Descriptive statistics and heritability (h2) estimates of meat quality traits in Nellore cattle.
| Traits | N | Mean | SD | Med | h2 | SE |
|---|---|---|---|---|---|---|
| Tenderness (kg) | 1,822 | 5.17 | 1.37 | 5.04 | 0.12 | 0.07 |
| Marbling | 1,873 | - | - | 2.80 | 0.10 | 0.07 |
N: number of observations; SD: standard deviation; Med: Median; h2: heritability; SE: Standard error
The animals were genotyped using a high-density panel containing 777,962 SNPs (Illumina High-Density Bovine BeadChip). The criteria for exclusion of SNPs were: non-autosomal SNPs, SNPs at the same position, with a minor allele frequency ≤ 0.02, a p value for Hardy-Weinberg equilibrium ≤ 10−5, GenCall score ≤ 0.70, call rate ≤ 0.98, and r² (correlation between SNPs) > 0.995 with adjacent SNPs within a window of 100 SNPs. Samples with a call rate ≤ 0.92 and without a valid phenotype were also excluded. After quality control, 1,630 animals genotyped for tenderness, 1,633 genotyped for marbling, and 369,722 SNPs remained.
GWAS
The ssGWAS method proposed by [9] was used for GWAS. A single-trait model was considered for the studied traits:
where y is the vector of phenotypic observations; X is an incidence matrix relating the phenotypes to the fixed effects; b is the vector of fixed effects, including the contemporary group (year, farm and management group at yearling) and slaughter age and days between slaughter and physicochemical analysis of meat as covariates (linear effect); Z is an incidence matrix relating the animal to the phenotype; a is the vector of effects of the animals, and e is the vector of residual effects.
The variances of a and e can be written as:
where σ2a is the direct additive genetic variance; σ2e is the residual variance; H is a matrix that combines pedigree and genomic information as proposed by [10], and I is an identity matrix. The inverse of matrix H is:
where A is the pedigree matrix for all animals; A22 is the relationship matrix for genotyped animals, and G is the genomic relationship matrix, which was calculated as described by [23].
The effects of the SNPs (ȗ) were obtained using the equation described by [9]:
where ȗ is the vector of SNP effects; λ is the variance ratio calculated according to [23]; ȃg is the animal effect of genotyped animals; Z is a matrix that relates the genotypes of each locus; D is a diagonal matrix of the weights of SNP variances. For this study the weights of SNP was not used, so D = I (identity matrix). G is the genomic relationship matrix.
The analyses were carried out using the BLUPF90 family of programs [22]. The results of GWAS are reported as the proportion of variance explained by a window of 150 adjacent SNPs. The MapViewer tool of the bovine genome, available at NCBI (http://www.ncbi.nlm.nih.gov/projects/mapview/map_search.cgi?taxid=9913&build=103.1), was used to identify the genes. The DAVID software [24] was used to describe the genes.
Results and Discussion
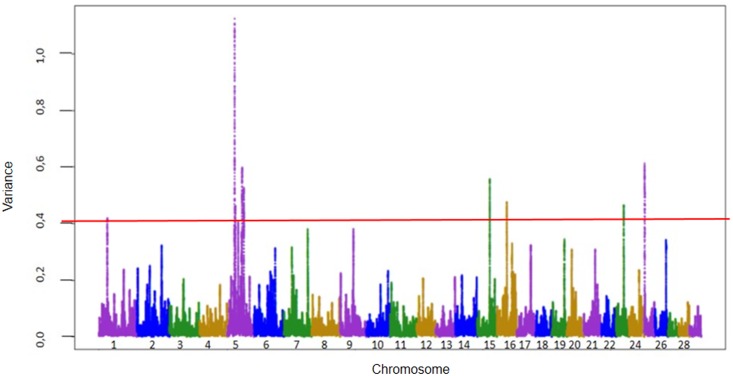
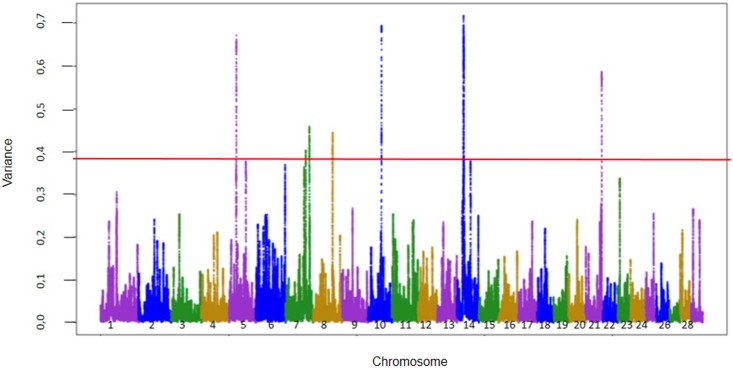
Figs 1 and 2 show the Manhattan plot with the percentages of additive genetic variance explained by windows of 150 adjacent SNPs for marbling and tenderness, respectively. Only genes inside windows with the highest percentage of additive genetic variance were presented. Others windows to complete top 10 ranking windows for genetic variance, as described by [25] are in S1 and S2 Tables and shows the chromosome, location, identification of genes and proportion of genetic variance for traits of marbling and tenderness. General information about all results of ssGWAS for both traits are in S1 File.
Fig 1. Manhattan plot of additive genetic variance explained by windows of 150 adjacent SNPs for Marbling.
Fig 2. Manhattan plot of additive genetic variance explained by windows of 150 adjacent SNPs for Tenderness.
The windows are located on chromosomes 5, 15, 16 and 25 for marbling and on chromosomes 5, 7, 10, 14 and 21 for tenderness and explained 3.89% and 3.80% of the additive genetic variance for marbling and tenderness, respectively.
Describing the results obtained from the genes for marbling and tenderness (Tables 2 and 3), it was observed that these traits have a similar window (31349253bp– 32177734bp) in the chromosome 5. For marbling, this window is the one that explains the highest percentage of additive genetic variance (1.12%) and for tenderness it is the third largest effect window (0.67%). This is an interesting result to be analyzed since the traits are moderately correlated [26,27]. Therewith, it is possible identify SNPs associated the candidate genes that exert a pleiotropic effect on meat quality traits and contributes to the genetic evaluation of both traits. Using the DAVID software, ten genes that act as olfactory receptors of the same window were identified (LOC617415, LOC507383, LOC504567, LOC523258, LOC509817, LOC781968, LOC515887, LOC506992, LOC617388, LOC781446). This cluster of genes participates in the change of GDP (guanosine diphosphate) to GTP (guanosine triphosphate), which are regulators of G proteins. GDP and GTP can be used as energy source and are responsible for transferring energy within the cell [28]. Another explanation for the effect of olfactory receptors on meat traits is their action in the duodenum, where they promote the absorption of fatty acids and thus increase the accumulation of fat as demonstrated in mice [29]. Furthermore, olfactory receptors are known to act on adipose tissue and adipocyte differentiation, increasing fat accumulation [30]. Some studies also suggest that olfactory receptors are involved in food intake because of their known action on olfactory neurons, increasing the search for food and food intake [31], and consequently weight gain and fat accumulation. Analysis of copy number variations for polyunsaturated fatty acids in the same databank also identified olfactory receptors [32]. This shows the strong influence of this group of genes on meat traits in Nellore cattle. Genes play multiple roles in metabolism. In this respect, olfactory receptors were first described to be involved in the perception of odors, but it cannot be ruled out that they exert other functions in the organism as mentioned above. Thus, specific studies demonstrating the true role of this group of genes in lipid metabolism are needed.
Table 2. Chromosome (Chr), location, identification of genes and proportion of variance (VAR) explained by windows with largest effects on marblinga.
| Chr | Location (bp) | Genes | VAR (%) |
|---|---|---|---|
| 5 | 31349253–32177734 | LALBA, LOC104972406, LOC784038 | 1.12409 |
| LOC526630, LOC614370, LOC515887 | |||
| LOC506992, LOC781446, LOC509508 | |||
| LOC781517, LOC507383, LOC781595 | |||
| OR8S1, LOC504567, LOC781756, ZNF641 | |||
| LOC781840, LOC101907855, LOC617415, | |||
| C5H12orf54, LOC509817, LOC781968, H1FNT | |||
| LOC617388, LOC617380, LOC518129 | |||
| LOC100300010, LOC523258, LOC100848234 | |||
| 25 | 2252680–2891421 | PRSS33, TCEB2, FLYWCH2, NAA60, CASP16 | 0.61217 |
| FLYWCH1, KREMEN2, PAQR4, ZNF200 | |||
| PKMYT1, CLDN9, CLDN6, ZNF597, TIGD7 | |||
| TNFRSF12A, HCFC1R1, THOC6, ZNF174 | |||
| CCDC64B, MMP25, IL32, MEFV, ZNF75A | |||
| LOC100139916, LOC100141258, ZNF263, | |||
| ZSCAN10, ZNF205, ZNF213, C25H16orf90 | |||
| 5 | 68620356–69478777 | CHST11, LOC101903254, LOC104972484 | 0.59773 |
| SLC41A2, C5H12orf45, ALDH1L2, APPL2, | |||
| KIAA1033, C5H12orf75, LOC104972485 | |||
| 15 | 54775627–55624618 | CHRDL2, LOC100139754, RNF169, GDPD5 | 0.55671 |
| LOC100848131, XRRA1, NEU3, KLHL35 | |||
| LOC101904814, LOC507022, OR2AT4 | |||
| SPCS2, LOC512627, LOC790343, MIR326 | |||
| SLCO2B1, TPBGL, ARRB1, MAP6, RPS3 | |||
| LOC104974272, SERPINH1, LOC101905131 | |||
| 5 | 76214455–77325534 | ELFN2, MFNG, CARD10, USP18, ALG10, SYT10 | 0.52581 |
| 16 | 43778493–44690610 | CASZ1, LOC101906968, PEX14, TRNAG-CCC | 0.47581 |
| DFFA, CORT, APITD1, PGD, CLSTN1, RBP7 | |||
| LOC101904547, TRNAR-CCG, CTNNBIP1 | |||
| KIF1B, LOC101904590, LOC104974439, PIK3CD | |||
| LOC104974446, UBE4B, LOC101905440 | |||
| LOC104974441, LZIC, LOC104974440, NMNAT1 |
aNCBI Symbol (Assembly UMD3.1, annotation release 103).
Table 3. Chromosome (Chr), location, identification of genes and proportion of variance (VAR) explained by windows with largest effects on tendernessa.
| Chr | Location (bp) | Genes | VAR (%) |
|---|---|---|---|
| 14 | 23048646–23659905 | ST18, LOC101906592, FAM150A, LYPLA1 | 0.71742 |
| RB1CC1, LOC104974017, TCEA1 | |||
| NPBWR1, OPRK1, ATP6V1H, RGS20 | |||
| 10 | 59864007–60992988 | TRPM7, LOC101906880, USP50, SLC27A2 | 0.69363 |
| USP8, LOC101906954, GABPB1, ATP8B4 | |||
| HDC, LOC100848857, LOC104973174 | |||
| LOC101907420, DTWD1, FAM227B | |||
| 5 | 31349253–32177734 | LALBA, LOC104972406 H1FNT, LOC617415 | 0.67148 |
| LOC526630, LOC614370, LOC515887 | |||
| LOC506992, LOC781446, LOC509508 | |||
| LOC781517, LOC507383, LOC781595 | |||
| OR8S1, LOC504567, LOC781756, LOC100848234, ZNF641 | |||
| LOC781840, LOC101907855, LOC784038 | |||
| C5H12orf54, LOC509817, LOC781968 | |||
| LOC617388, LOC617380, LOC518129 | |||
| LOC100300010, LOC523258 | |||
| 14 | 24336953–25030555 | XKR4, TRNAT-AGU, TMEM68, LOC104974019 | 0.66924 |
| TGS1, LYN, RPS20, MOS, PLAG1 | |||
| 21 | 65833836–66414612 | BCL11B, SETD3, CCNK, HHIPL1, EML1 | 0.58616 |
| CCDC85C, LOC101906687, CYP46A1 | |||
| 7 | 98394404–98767591 | CAST, LOC104968992, ERAP1 | 0.45783 |
aNCBI Symbol (Assembly UMD3.1, annotation release 103).
For marbling, the genes described in Table 2 are related to carbohydrate and lipid metabolism. This fact agrees with the physiology of interspersed fat deposition in muscle, since lipids are derived from the consumption of fats or from the excess intake of carbohydrates that are processed and stored in the form of fat. The TCEB2 gene was detected by proteomic analysis of hepatic fat droplets, which showed that high production of this protein protects against diabetes in mice [33]. The TIGD7 gene has been associated with body mass index and plasma glucose levels in humans [34]. The APPL2 gene has been shown to be associated with obesity in humans [35] and to play a role in the inhibition of glucose uptake in skeletal muscle [36]. The NEU3 gene promotes the accumulation of triglycerides in mice, which may facilitate the deposition of body fat [37]. The MFNG gene is responsible for causing bile duct abnormalities in mice, which can compromise bile flow and the consequent emulsification of fat for absorption [38]. The list of genes for marbling was compiled using the DAVID software. The CHST11, LALBA and PGD genes form a cluster and are involved in the chemical reactions resulting in the formation of carbohydrates.
The genes associated with marbling in the present study are not the same as those reported in other GWAS for the same trait in Bos taurus taurus [39–41]. In study with the same breed, [42], the authors also reported that the regions that most influenced the trait did not coincide with those of the present study.
For meat tenderness, genes related to growth and muscle development were identified (PLAG1, SLC27A2, RB1CC1, HDC, LYPLA1, XKR4 and TMEM68). The PLAG1 gene has been associated with meat tenderness in cattle [43]. This gene also is associated with carcass traits in a study on the same animals [44], in addition to other studies, with different breeds [39,45–48]. This gene has also been associated with growth traits, feed intake and reproductive and andrological traits in different breeds of the same species [39,43,49–54]. This gene has an important pleiotropic effect and can be an excellent candidate for a large-effect gene because it acts on different traits. This fact can be explained by the function of the gene, which is a transcription factor for the growth hormone IGF-2 [55].
The SLC27A2 gene has also been associated with meat tenderness in cattle [56], among other carcass traits [57]. The RB1CC1 gene is involved in muscle development and possesses non-synonymous SNPs that are associated with pectoral muscle size in broiler chickens [58]. Furthermore, the gene acts on skeletal muscle of chicken under thermal stress [59]. Since this gene acts through temperature variations in the living animal, its activity may continue postmortem. A decline in temperature is known to affect meat tenderness. Thus, different variants of the gene may act in the carcass through postmortem temperature variation, resulting in more or less tender meat.
The HDC gene is also expressed in skeletal muscle where it is involved in the processing of histamine, but its function is unknown [60]. The SETD3 gene participates in the differentiation of muscle cells [61].
The LYPLA1, XKR4 and TMEM68 genes have been associated with feed intake and growth in cattle [62]. The XKR4 gene has been associated with fat thickness in cattle [41,63] and the LYN and TGS1 genes with carcass phenotypes also in cattle [41]. Furthermore, in another study with same population as in this one [64], XKR4, TMEM68, TGS1 and LYN genes were associated to birth weight. All genes mentioned above, are located in chromosome 14 and were associated with several traits besides tenderness, suggesting a pleiotropic effect. These results corroborate those describing positive genetic correlation estimates of tenderness with growth traits [65] and with fat thickness [66].
The calpastatin (CAST) is an important gene associated with tenderness, which is known to inhibit calpain (CAPN1). These genes (CAST and CAPN1) are considered responsible for the process of meat tenderization during the postmortem period [67]. CAPN1 determines an increase in final meat tenderness, while CAST acts in the opposite manner, inhibiting this tenderizing process. In the present study, the window where CAST was located explained 0.46% of the additive genetic variance in tenderness. The CAST gene has been associated with meat tenderness in GWAS of Australian cattle [68]. However, studying approximately 500 Nellore animals, [19] found no association between the CAST gene and tenderness. The authors identified 56 genes for different tenderness measures (24 h, 7 and 14 days after slaughter), but these genes only explained a small percentage of the additive genetic variance (less than 0.2%).
Different genes have been reported in other studies conducted on the same breed and traits [19,42], indicating the existence of a genetic difference between populations analyzed within breeds. Several factors may be responsible for these differences, such as variations in allele frequency, linkage disequilibrium, coverage of the SNP chip in the breed, method and number of animals, since the studies on Nellore animals cited above used few animals when compared to the present study.
Important genomic regions associated with meat quality traits were identified in the present study, providing a better biological understanding of tenderness and marbling in Bos taurus indicus. The identification of these genes in cattle using genomic tools is the first step in the search for causal mutations that exert great influence on traits of economic interest and can contribute in the future to genetic evaluations.
Supporting Information
(XLS)
aNCBI Symbol (Assembly UMD3.1, annotation release 103).
(DOCX)
aNCBI Symbol (Assembly UMD3.1, annotation release 103).
(DOCX)
Data Availability
All relevant data are within the paper.
Funding Statement
The authors would like to thank Fapesp (Fundação de Amparo à Pesquisa do Estado de São Paulo <http://www.fapesp.br/>) for the first author’s scholarship (N° 2012/21969-7) and for funding this study (N° 2009/16118-5).
References
- 1.United States Departament of Agriculture—USDA. Livestock and Poultry: World Markets and Trade. Foreign Agricultural Service; 2014. 27p. [Google Scholar]
- 2.ABIEC. Associação Brasileira das Indústrias Exportadoras de Carne. ABIEC. [updated 2015 Aug 01; cited 2015 Sep 10]. Available: http://www.abiec.com.br/3_rebanho.asp. Accessed 10 September 2015.
- 3.O’Connor SF, Tatum JD, Wulf DM, Green RD, Smith GC. Genetic Effects on Beef Tenderness in Bos indicus Composite and Bos taurus Cattle. J. Anim. Sci. 1997; 75:1822–1830. [DOI] [PubMed] [Google Scholar]
- 4.Bressan MC, Rodrigues EC, Rossato LV, Ramos EM, da Gama LT. Physicochemical properties of meat from Bos taurus and Bos indicus. R. Bras. Zootec. 2011; 40(6):1250–1259. [Google Scholar]
- 5.Maltin C, Balcerzak D, Tilley R, Delday M. Determinants of meat quality: tenderness. Proceedings of the Nutrition Society. 2003; 62:337–347. [DOI] [PubMed] [Google Scholar]
- 6.Thu N T D. Meat quality: understanding of meat tenderness and Influence of fat content on meat flavor. Science & Technology Development. 2006; 9(12). [Google Scholar]
- 7.Williams JL. Genetic Controlo f Meat Quality Traits In: Toldrá F, editor. Meat Biotechnology. New York: Springer Science+Business Media; 2008. p. 21–60. [Google Scholar]
- 8.Hirschhorn JN. Daly MJ. Genome-wide association studies for common diseases and complex traits. Nature. 2005; 6. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, Misztal I, Aguilar I. Legarra A, Muir WM. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet. Res. Camb. 2012; 94:73–83. 10.1017/S0016672312000274 [DOI] [PubMed] [Google Scholar]
- 10.Aguilar I, Misztal I, Johnson DL, Legarra A, Tsuruta S, Lawlor TJ. Hot topic: a unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 2010; 93:743–752. 10.3168/jds.2009-2730 [DOI] [PubMed] [Google Scholar]
- 11.Misztal I, Legarra A, Aguilar I. Computing procedures for genetic evaluation including phenotypic, full pedigree, and genomic information. J. DairySci. 2009; 92:4648–4655. [DOI] [PubMed] [Google Scholar]
- 12.Casas E, Keele JW, Shackelford SD, Koohmaraie M, Sonstegard TS, Smith TP, et al. Association of the muscle hypertrophy locus with carcass traits in beef cattle. J Anim Sci. 1998;76:468–473. [DOI] [PubMed] [Google Scholar]
- 13.Casas E, Shackelford SD, Keele JW, Stone RT, Kappes SM, Koohmaraie M. Quantitative trait loci affecting growth and carcass composition of cattle segregating alternate forms of myostatin. J AnimSci. 2000; 78:560–569. [DOI] [PubMed] [Google Scholar]
- 14.Casas E, Stone RT, Keele JW, Shackelford SD, Kappes SM, Koohmaraie MA. Comprehensive search for quantitative trait loci affecting growth, and carcass composition of cattle segregating alternative forms of the myostatin gene. J Anim Sci. 2001;79:854–860. [DOI] [PubMed] [Google Scholar]
- 15.Davis GP, Moore SS, Drinkwater RD, Shorthose WR, Loxton ID, Barendse W, et al. QTL for meat tenderness in the M. longissimus lumborum of cattle. Anim Genet. 2007;39:40–45. [DOI] [PubMed] [Google Scholar]
- 16.Smith TPL, Casas E, Rexroad CE III, Kappes SM, Keele JW. Bovine CAPN1 maps to a region of BTA29 containing a quantitative trait locus for meat tenderness. J. Anim. Sci. 2000;78:2589–2594. [DOI] [PubMed] [Google Scholar]
- 17.Cheong HS, Yoon DH, Park BL, Kim LH, Bae JS, Namgoong S, et al. A single nucleotide polymorphism in CAPN1 associated with marbling score in Korean cattle. BMC Genet. 2008;9(33). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinto LFB, Ferraz JBS, Meirelles FV, Eler JP, Rezende FM, Carvalho ME, et al. Association of SNPs on CAPN1 and CAST genes with tenderness in Nellore cattle. Genet. Mol. Res. 2010; 9(3):1431–1442. [DOI] [PubMed] [Google Scholar]
- 19.Tizioto PC, Decker JE, Taylor JF, Schnabel RD, Mudadu MA, Silva FL, et al. Genome scan for meat quality traits in Nelore beef cattle. Physiol. Genomics. 2013;45:1012–1020. 10.1152/physiolgenomics.00066.2013 [DOI] [PubMed] [Google Scholar]
- 20.Wheeler TL, Koomaraie M, Shackelford SD. Standardized warner-bratzler shear force procedures for meat tenderness measurementClay Center: Roman L. Hruska U. S. MARC. USDA; 1995. 7p. [Google Scholar]
- 21.United States Departament of Agriculture. U.S. Standards for Grades of Feeder Cattle. Washington: USDA; 2000. 4p. [Google Scholar]
- 22.Misztal I. BLUPF90—a flexible mixed model program in Fortran 90. 2012. [updated 2015 Aug 05; cited 2015 Sep 17]. Available: http://nce.ads.uga.edu/wiki/lib/exe/fetch.php?media=blupf90.pdf. Accessed 17 September 2015.
- 23.VanRaden PM, Van Tassell CP, Wiggans GR, Sonstegard TS, Schnabel R D, Taylor JF, et al. Invited review: Reliability of genomic predictions for North American Holstein bulls. J. Dairy Sci. 2009; 92:16–24. 10.3168/jds.2008-1514 [DOI] [PubMed] [Google Scholar]
- 24.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009; 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Misztal I, Aguilar I, Legarra A, Fernando RL, Vitezica Z, et al. Genome-wide association mapping including phenotypes from relatives without genotypes in a single-step (ssGWAS) for 6-week body weight in broiler chickens. Front. Genet 2014. 5(134):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wheeler TL, Cundiff LV, Koch RM. Effect of Marbling Degree on Beef Palatability in Bos taurus and Bos indicus Cattle. J. Anim. Sci. 1994;72:3145–3151. [DOI] [PubMed] [Google Scholar]
- 27.O’Connor SF, Tatum JD, Wulf DM, Green RD, Smith GC. Genetic Effects on Beef Tenderness in Bos indicus Composite and Bos taurus Cattle. J. Anim. Sci. 1997;75:1822–1830. [DOI] [PubMed] [Google Scholar]
- 28.Russell PJ, Wolfe SL, Hertz PE, Starr C. Biology: The Dynamic Science Brooks Cole. 1th ed; 2007. 400p. [Google Scholar]
- 29.Primeaux SD, Braymer HD, Bray GA. High Fat Diet Differentially Regulates the Expressionof Olfactory Receptors in the Duodenum of Obesity-Proneand Obesity-Resistant Rats. Dig Dis Sci. 2013;58:72–76. 10.1007/s10620-012-2421-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Von der Heyde S, Fromm-Dornieden C, Salinas-Riester G, Beissbarth T, Baumgartner BG. Dynamics of mRNA and polysomal abundance inearly 3T3-L1 adipogenesis. BMC Genomics. 2014;15:381 10.1186/1471-2164-15-381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loch D, Heidel C, Breer H, Strotmann J. Adiponectin Enhances the Responsiveness of the Olfactory System. Plos One. 2013;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Lemos MVA, Camargo GMF, Pereira ASC, Baldi F. Association between copy number variations in the Nellore cattle genome and polyunsaturated fatty acid profile. Proceedings of XXIV Congreso de la Asociación Latino americana de Producción Animal; 2015. November 9–13; Puerto Varas—Chile.
- 33.Baumier C, Kaiser D, Heeren J, Scheja L, John C, Weise C, et al. Caloric restriction and intermittent fasting alter hepatic lipid droplet proteome and diacyglycerol species and prevent diabetes in NZO mice. Biochimica et Biophysica Acta. 2015; 1851:566–576. 10.1016/j.bbalip.2015.01.013 [DOI] [PubMed] [Google Scholar]
- 34.Berisha SZ, Serre D, Schauer P, Kashyap SR, Smith JD. Changes in Whole Blood Gene Expression in Obese Subjects with Type 2 Diabetes Following Bariatric Surgery: a Pilot Study. PlosOne. 2011;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Z, Michal JJ, Chen J, Daniels TF, Kunej T, Garcia MD, et al. Discovery of novel genetic networks associated with 19 economically important traits in beef cattle. International Journal of Biological Sciences. 2009;5(6):528–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng KKY, Zhu W, Chen B, Wang Y, Wu D, Sweeney G, et al. The adaptor protein APPL2 inhibits insulin-stimulated glucose uptake by interacting with TBC1D1 in skeletal muscle. 2014;63:3748–3757. [DOI] [PubMed] [Google Scholar]
- 37.Yoshizumia S, Suzukia S, Hiraia M, Hinokioa Y, Yamadaa T, Yamadaa T, et al. Increased hepatic expression of ganglioside-specific sialidase, NEU3, improves insulin sensitivity and glucose tolerance in mice. Metabolism Clinical and Experimental 56 2007:420–429. [DOI] [PubMed] [Google Scholar]
- 38.Ryan MJ, Bales C, Nelson A, Gonzalez DM, Underkoffler L, Segalov M, et al. Bile Duct Proliferation in Jag1/Fringe Heterozygous Mice Identifies Candidate Modifiers of the Alagille Syndrome Hepatic Phenotype. Hepatology. 2008. [DOI] [PubMed] [Google Scholar]
- 39.Saatchi M, Schnabel RD, Taylor JF, Garrick DJ. Large-effect pleiotropic or closely linked QTL segregate within and across ten US cattle breeds. BMC Genomics. 2014;15(442). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barendse W. Haplotype Analysis Improved Evidence for Candidate Genes for Intramuscular Fat Percentage from a Genome Wide Association Study of Cattle. PLoSONE. 2001;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramayo-Caldas Y, Fortes MRS, Hudson NJ, Porto-Neto LR, Bolormaa S, Barendse W, et al. A marker-derived gene network reveals the regulatory role of PPARGC1A, HNF4G, and FOXP3 in intramuscular fat deposition of beef cattle. J Anim Sci. 2014, 92:2832–2845. 10.2527/jas.2013-7484 [DOI] [PubMed] [Google Scholar]
- 42.Cesar ASM, Regitano LCA, Mourão GB, Tullio RR, Lanna DPD, Nassu RT. Genome-wide association study for intramuscular fat deposition and composition in Nellore cattle. BMC Geneticsl.2014;15:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fortes MRS, Kemper K, Sasazaki S, Reverter A, Pryce JE, Barendse W, et al. Evidence for pleiotropism and recent selection in the PLAG1 region in Australian Beef cattle. Animal Genetics. 2013; 44:636–647. 10.1111/age.12075 [DOI] [PubMed] [Google Scholar]
- 44.Fernandes Júnior GA. Seleção genômica para características de carcaça em bovinos da raça Nelore. [dissertation]: Universidade Estadual Paulista; 2015.
- 45.Sharma A, Dang CG, Kim KS, Kim JJ, Lee HK, Kim HC, et al. Validation of genetic polymorphisms on BTA14 associated with carcass trait in a commercial Hanwoo population. Animal Genetics. 2014. 10.1111/age.12204 [DOI] [PubMed] [Google Scholar]
- 46.Hoshiba H, Setoguchi K, Watanabe T, Kinoshita A, Mizoshita K, Sugimoto Y, et al. Comparison of the effects explained by variations in the bovine PLAG1 and NCAPG genes on daily body weight gain, linear skeletal measurements and carcass traits in Japanese Black steers from a progeny testing program. Animal Science Journal. 2013;84:529–534. 10.1111/asj.12033 [DOI] [PubMed] [Google Scholar]
- 47.Nishimura S, Watanabe T, Mizoshita K, Tatsuda K, Fujita T, Watanabe N, et al. Genome-wide association study identified three major QTL for carcass weight including the PLAG1-CHCHD7 QTN for stature in Japanese Black cattle. BMC Genetics. 2012;13:40 10.1186/1471-2156-13-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee SH, Choi BH, Lim D, Gondro C, Cho YM, Dang CG, et al. Genome-Wide Association Study Identifies Major Loci for Carcass Weight on BTA14 in Hanwoo (Korean Cattle). PlosOne. 2013;8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Littlejohn M, Grala T, Sanders K, Walker C, Waghorn G, Macdonald K, et al. Genetic variation in PLAG1 associates with early life body weight and peripubertal weight and growth in Bos Taurus. Animal Genetics. 2011;43:591–594. 10.1111/j.1365-2052.2011.02293.x [DOI] [PubMed] [Google Scholar]
- 50.Riley DG, Welsh TH Jr, Gill CA, Hulsman LL, Herring AD, Riggs PK, et al. Whole genome association of SNP with newborn calf cannon bone length. Livestock Science. 2013;155:186–196. [Google Scholar]
- 51.Utsunomiya YT, do Carmo AS, Carvalheiro R, Neves HHR, Matos MC, Zavarez LB, et al. Genome-wide association study for birth weight in Nellore cattle points to previously described orthologous genes affecting human and bovine height. BMC Genetics. 2013;14(52). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fortes MRS, Reverter A, Kelly M, McCulloch R, Lehnert SA. Genome-wide association study for inhibin, luteinizing hormone, insulin-like growth factor 1, testicular size and semen traits in bovine species. Andrology. 2013; 1:644–650. 10.1111/j.2047-2927.2013.00101.x [DOI] [PubMed] [Google Scholar]
- 53.Utsunomiya YT, Carmo AS, Neves HHR, Carvalheiro R, Matos MC, Zavarez LB, et al. Genome-Wide Mapping of Loci Explaining Variance in Scrotal Circumference in Nellore Cattle. PlosOne. 2014;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Camargo GMF, Porto-Neto LR, Kelly MJ, Bunch RJ, McWilliam SM, Tonhati H, et al. Non-synonymous mutations mapped to chromosome X associated with andrological and growth traits in beef cattle. BMC Genomics. 2015;16(384). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voz ML, Mathys J, Hensen K, Pendeville H, Valckenborgh IV, Huffel CV, et al. Microarray screening for target genes of the proto-oncogene PLAG1. Oncogene. 2004; 23:179–191. [DOI] [PubMed] [Google Scholar]
- 56.Jiang Z, Michal JJ, Chen J, Daniels TF, Kunej T, Garcia MD et al. Discovery of novel genetic networks associated with 19 economically important traits in beef cattle. Int. J. Biol. Sci. 2009; 5(6):528–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L, Michal JJ, O'Fallon JV, Pan Z, Gaskins CT, Reeves JJ, et al. Quantitative Genomics of 30 Complex Phenotypes in Wagyu x Angus F1 Progeny. Int. J. Biol. Sci. 2012; 8(6):838–858. 10.7150/ijbs.4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Godoy TF, Moreira GCM, Boschiero C, Gheyas AA, Gasparin G, Paduan, et al. SNP and INDEL detection in a QTL region on chicken chromosome 2 associated with muscle deposition. Animal Genetics. 2015;46(2):158–163. 10.1111/age.12271 [DOI] [PubMed] [Google Scholar]
- 59.Luo QB, Song XY, Ji CL, Zhang XQ, Zhang DX. Exploring the molecular mechanism of acute heat stress exposure in broiler chickens using gene expression profiling. Gene. 2014;546:200–205. 10.1016/j.gene.2014.06.017 [DOI] [PubMed] [Google Scholar]
- 60.Everaert I, De Naeyer H, Taes Y, Derave W. Gene expression of carnosine-related enzymes and transporters in skeletal muscle. Eur J ApplPhysiol. 2013;113:1169–1179. [DOI] [PubMed] [Google Scholar]
- 61.Eom GH, Kim K, Kim JH, Kim J, Kim J, Kee HJ, et al. Histone Methyltransferase SETD3 Regulates Muscle Differentiation. The journal of Biological Chemistry. 2011;286(40):34733–34742. 10.1074/jbc.M110.203307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lindholm-Perry AK, Kuehn LA, Smith TPL, Ferrell CL, Jenkins TG, Freetly HC, et al. A region on BTA14 that includes the positional candidate genes LYPLA1, XKR4 and TMEM68 is associated with feed intake and growth phenotypes in cattle. Animal Genetics. 2011;43:216–219. 10.1111/j.1365-2052.2011.02232.x [DOI] [PubMed] [Google Scholar]
- 63.Porto Neto LR, Bunch RJ, Harrison BE, Barendse W. Variation in the XKR4 gene was significantly associated with subcutaneous rump fat thickness in indicine and composite cattle. Animal Genetics. 2012;43:785–789. 10.1111/j.1365-2052.2012.02330.x [DOI] [PubMed] [Google Scholar]
- 64.Terakado APN. Utilizações de informações genômicas para o melhoramento genético de características de crescimento em bovinos da raça Nelore. [dissertation]: Universidade Estadual Paulista; 2015.
- 65.Tonussi RL, Espigolan R, Gordo DGM, Magalhães AFB, Venturini GC, Baldi F, et al. Genetic association of growth traits with carcass and meat traits in Nellore cattle. Genet. Mol. Res. 2015; 14(4):18713–18719. 10.4238/2015.December.28.20 [DOI] [PubMed] [Google Scholar]
- 66.De Castro LM, Magnabosco CU, Sainz RD, De Faria CU, Lopes FB. Quantitative genetic analysis for meat tenderness trait in Polled Nellore cattle. Rev. Ciênc. Agron. 2014;45(2):393–402. [Google Scholar]
- 67.Koohmaraie M. Biochemical factors regulating the toughening and tenderization process of meat. Meat Science.1996;43:193–201. [DOI] [PubMed] [Google Scholar]
- 68.Bolormaa S, Porto Neto LR, Zhang YD, Bunch RJ, Harrison BE, Goddard ME, et al. A genome-wide association study of meat and carcass traits in Australian cattle. J Anim Sci. 2011;89:2297–2309. 10.2527/jas.2010-3138 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
aNCBI Symbol (Assembly UMD3.1, annotation release 103).
(DOCX)
aNCBI Symbol (Assembly UMD3.1, annotation release 103).
(DOCX)
Data Availability Statement
All relevant data are within the paper.