Abstract
Colonization of the infant gut is believed to be critically important for a healthy growth as it influences gut maturation, metabolic, immune and brain development in early life. Understanding factors that influence this process is important, since an altered colonization has been associated with a higher risk of diseases later in life. Fecal samples were collected from 108 healthy neonates in the first half year of life. The composition and functionality of the microbiota was characterized by measuring 33 different bacterial taxa by qPCR/RT qPCR, and 8 bacterial metabolites. Information regarding gender, place and mode of birth, presence of siblings or pets; feeding pattern and antibiotic use was collected by using questionnaires. Regression analysis techniques were used to study associations between microbiota parameters and confounding factors over time. Bacterial DNA was detected in most meconium samples, suggesting bacterial exposure occurs in utero. After birth, colonization by species of Bifidobacterium, Lactobacillus and Bacteroides was influenced by mode of delivery, type of feeding and presence of siblings, with differences found at species level and over time. Interestingly, infant-type bifidobacterial species such as B. breve or B. longum subsp infantis were confirmed as early colonizers apparently independent of the factors studied here, while B. animalis subsp. lactis presence was found to be dependent solely on the type of feeding, indicating that it might not be a common infant gut inhabitant. One interesting and rather unexpected confounding factor was gender. This study contributes to our understanding of the composition of the microbiota in early life and the succession process and the evolution of the microbial community as a function of time and events occurring during the first 6 months of life. Our results provide new insights that could be taken into consideration when selecting nutritional supplementation strategies to support the developing infant gut microbiome.
Introduction
Composed of 1011 to 1012 micro-organisms per gram of feces, and estimated to harbor more than 500 bacterial species, the human gastrointestinal microbiota is a large and diverse community of microorganisms and is in close cross-talk with its host [1–3]. Despite the vast number of bacteria and complexity found in the adult gut, the microbiota of the infant gut is initially a simple ecosystem which gradually undergoes successional changes until it reaches high diversity. For years, the infant gut was considered to be sterile, only becoming colonized after birth from the maternal microbiota, diet, and the environment [4–7]. However, recent findings suggest that microbial exposure may start already during gestation and colonization starts with the early settlers derived from the maternal microbiota and environment immediately from birth [8–11]. The development of the infant gut microbiota is profoundly influenced by host genotype, gestational age, antibiotic use, mode of delivery, diet and the context in which the infant is born (rural vs urban, presence of siblings, pets and other factors) [4, 5, 12–17].
The first colonizers of the infant gut are facultative anaerobic bacteria, such as Staphylococcus, Streptococcus, Enterococcus and Enterobacter spp, which by reducing the oxygen levels that initially exist, pave the way for anaerobes such as Bifidobacterium, Bacteroides and Clostridium spp. to start colonizing the gut. It is known that the type of bacteria that initially colonize the infant gut and the time frame in which this occurs, is highly dependent on the mode of delivery. Previous studies have demonstrated that strains originating from the maternal gut and vagina are transferred to the infant’s gut in case of a vaginal delivery (VD) [18–20], while infants born by cesarean section (CS) are suggested to be initially colonized by bacteria from the environment such as from maternal skin, hospital staff or other neonates [21].
The first weeks of life is a period in which the intestinal microbiota is very dynamic, with nutrition governing the developing ecosystem [15]. Breastfed infants typically have a bifidobacteria-dominated microbiota whereas formula-fed infants have a more diverse microbiota. After the introduction of solid foods, bacterial succession continues gradually diversifying with adult-like species such as Bacteroides spp. and Clostridium cluster IV and XIV. The exact age at which a stable adult-like composition is established is still unclear but it is thought to be around 3 years of age [22, 23]. However, this process continues past 3 years of age, and events occurring later in life, such as hormonal changes during puberty or changes in eating habits may also influence the microbiota composition [24, 25].
To what extend early colonization is influencing the microbiota composition in later life needs to be further elucidated. However, increasing evidence suggests that the initial colonization does influence gut maturation, immune, brain and metabolic development. Early-life events (ie., mode of delivery, type of feeding, antibiotic use) that are known to influence this process, may thus drive predisposition to diseases later in life [13, 26–31]. It is therefore critical to understand the early colonization process in great detail, including the confounding factors in early life that can be of long term importance.
This study aims to describe the dynamics of early colonization during the first six months of life and identify factors that can drive changes in the composition of the gut microbiota in early life.
Material and Methods
Subjects and sampling
The present study was an observational study performed in the area of Antwerp (Belgium) between June 2009 and July 2010. The protocol was approved by the ethics committee of the hospital network of Antwerp (ZNA), Institutional Review Board-ZNA/OCMW Antwerpen. The trial is registered in the ISRCTN register: ISRCTN66704989.
One hundred and forty-three healthy pregnant women were recruited via gynecologists. Subjects were eligible for participation when they showed a normal course of pregnancy and a good physical and mental health status as determined by the medical history and general clinical examination according to the investigators’ judgment. Exclusion criteria were 1) Birth in water; 2) Participation in another clinical trial; 3) Alcohol consumption (>7 units/week); 4) Illegal drug use. Written informed consent was obtained before inclusion in the study. Subjects that fulfilled one or more of the following criteria during the study, were excluded from further participation: 1) premature delivery (before 37 weeks of pregnancy); 2) bacterial/viral infection within 2 weeks prior to delivery; 3) congenital malformation(s) in the subjects’ infant; 4) use of immune-modulatory drugs by subject and/or subjects’ infant prior to (4 weeks) delivery and during the study; 5) use of antibiotics between 2 weeks prior to delivery and 2 weeks after delivery, for any reason except for prophylactic use (e.g. cesarean section).
Upon enrolment (about 1–2 months before the expected delivery date) information with respect to the subjects’ demographics, relevant medical history, medication and nutritional supplement use were documented. Subjects were issued with questionnaires to register information on the diet, possible allergic/gastrointestinal symptoms, and medication to be completed at several time points during the study (before delivery, immediately after delivery, at 1/2/3/4 weeks after delivery, and at 2/3/4/5/6 months after delivery).
After delivery fecal samples of the neonates were collected at several time points (first defecation, 2 days after first defecation, 1 week after birth, 1/3/6 months after birth, 1 week after weaning started).
Participants were given a pre-weighed 15 ml fecal collection tube (Sarstedt AG & Co., Numbrecht, Germany) containing 2 ml of RNAlater (Thermo Fisher Scientific Inc., Waltham, MA), and an empty fecal collection tube. They placed a spoonful of fecal sample (approximately 0.5 g) into each collection tube immediately after defecation. The samples were kept at 4°C in a cooling box with refrigerants and sent to Yakult Honsha European Research Center for Microbiology ESV (Ghent, Belgium), within 72 h of collection.
Primary treatment of fecal samples
For the following total DNA extraction and 4’,6-diamidino-2-phenylindole (DAPI) staining, fecal samples, which were collected in empty tubes, were suspended in nine volumes of phosphate-buffered saline (PBS) (Nissui Pharmaceutical Co. Ltd., Tokyo, Japan). The fecal suspension in the PBS was then stored at -30°C until DNA extraction.
Another portion of the fecal samples, also from the samples collected in the same empty fecal collection tubes, was suspended in nine volumes of 0.1 M phosphate buffer (pH 5.5) to make a 10-fold fecal suspension for organic acid analysis. Then, 0.8 ml of this fecal suspension, 0.1 ml of 10% (v/v) perchloric acid and 0.1 ml of 100 mM crotonic acid (internal standard) were mixed, and kept at 4°C overnight to remove proteins. After centrifugation at 12,000 × g for 5 min, the supernatant was filtered with 0.45 μm filter and stored at -30°C until organic acid analysis was performed.
For total RNA extraction, the fecal collection tubes, which were included 2 ml of RNAlater and fecal samples collected, were weighed, and were added fresh RNAlater to make a fecal suspension (100 mg feces/ml). Then, 0.2 ml of this fecal suspension was added to 1 ml of PBS. After centrifugation of the mixture at 12,000 × g for 5 min, the supernatant was discarded by decantation, and the pellet was stored at -80°C until RNA extraction was performed.
Determination of bacterial count by DAPI staining
Total bacterial cell counts were determined by DAPI staining, according to a method described by Matsuda et al. [32]. Briefly, 100 μl of 10-fold diluted fecal homogenate in PBS was added to 3 volumes of 4% paraformaldehyde-PBS (PFA) solution, which was then incubated at 4°C for 16 h. PFA-treated samples were smeared over a Micro slide glass (Matsunami, Osaka, Japan), stained with Vectashield mounting medium with DAPI (Vector microscope Leica DM6000, image acquisition software QFluoro, and a cooled black-and-white charge-coupled device [CCD] camera Leica DFC350FX; Leica Microsystems, Wetzlar, Germany). The obtained fluorescent particles were analyzed using Image-Pro plus software version 4.5 (Media Cybernetics, Rockville, MD), to count the fluorescent cells in each fecal samples. The bacterial count was expressed as the mean value of 24 fields for each smear sample.
DNA extraction and qPCR
DNA was extracted from the PBS-suspension mentioned above and subjected to qPCR, according to a method described by Matsuki et al. [33]. Briefly, the thawed sample was mixed with 250 μl of extraction buffer (100 mM Tris-HCl, 40 mM EDTA; pH 9.0) and 50 μl of 10% sodium dodecyl sulfate. 300 mg of glass beads (diameter, 0.1 mm) (BioSpec Products, Bartlesville, OK) and 500 μl of TE-saturated phenol (Sigma-Aldrich, St. Louis, MO) were added to the suspension, and the mixture was vortexed vigorously for 30 s using a FastPrep-24 (M.P. Biomedicals, Santa Ana, CA) at a power level of 5.0. Phenol–chloroform extractions were performed, and 250 μl of the supernatant was subjected to isopropanol precipitation. Finally, the DNA was suspended in 1 ml of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0).
To determine the population sizes and prevalence of the predominant and subdominant bacterial groups, we used primers specific for the groups and species show in Table 1. PCR amplification and detection were performed with an ABI PRISM 7900HT Sequence Detection System and SDS software (ver 2.3) (Thermo Fisher Scientific). The reaction mixture (10 μl) was composed of 10 mM Tris-HCl, pH 8.3; 50 mM KCl; 1.5 mM MgCl2; 200 μM of each dNTP; 1:75,000 dilution of SYBR Green I (Thermo Fisher Scientific); 11 ng/μl of TaqStart antibody (ClonTech, Palo Alto, CA); 0.05 U/μl of Taq DNA polymerase (Takara Bio, Shiga, Japan); 0.25 μM of each of the specific primers; and 1 μl of ×10, ×100, or ×1000 diluted template DNA. The amplification program consisted of one cycle at 94°C for 5 min; 40 cycles at 94°C for 20 s, 50°C or 55°C for 20 s (Table 1) [34], and 72°C for 50 s; and finally one cycle at 94°C for 15 s. Fluorescent products were verified at the last step of each cycle.
Table 1. 16S rRNA gene targeted primers and standard strains used for qPCR.
| Target | Primer | Sequence (5’-3’) | Annealing Temp (°C) | Reference |
|---|---|---|---|---|
| Clostridium coccoides group | g-Ccoc-F | AAATGACGGTACCTGACTAA | 55 | [34] |
| g-Ccoc-R | CTTTGAGTTTCATTCTTGCGAA | |||
| Clostridium leptum group | sg-Clept-F | GCACAAGCAGTGGAGT | 55 | [35] |
| sg-Clept-R | CTTCCTCCGTTTTGTCAA | |||
| Bacteroides fragilis group | g-Bfra-F2 | AYAGCCTTTCGAAAGRAAGAT | 50 | [34] |
| g-Bfra-R | CCAGTATCAACTGCAATTTTA | |||
| Genus Bifidobacterium | g-Bifid-F | CTCCTGGAAACGGGTGG | 55 | [34] |
| g-Bifid-R | GGTGTTCTTCCCGATATCTACA | |||
| Atopobium cluster | c-Atopo-F | GGGTTGAGAGACCGACC | 55 | [35] |
| c-Atopo-R | CGGRGCTTCTTCTGCAGG | |||
| Genus Prevotella | g-Prevo-F | CACRGTAAACGATGGATGCC | 55 | [34] |
| g-Prevo-R | GGTCGGGTTGCAGACC | |||
| Bacteroides caccae | s-Bcac122-F | ACGTATCCAACCTACCTCA | 55 | [36] |
| s-Bcac586-R | ACAACTGACTTAACAATCCG | |||
| Bacteroides eggerthii | s-Begg187-F | GTTTTTCCGCATGGTTTCAC | 55 | [36] |
| s-Begg590-R | CTTTCACAACTGACTTAAGCA | |||
| Bacteroides fragilis | s-Bfra186-R | AATGATTCCGCATGGTTTCA | 55 | [36] |
| s-Bfra592-R | CAAACTTTCACAACTGACTTAC | |||
| Bacteroides ovatus | s-Bova175-F | CCGGATAGCATACGAAYAT | 55 | [36] |
| s-Bova587-R | CACAACTGACTTAACAATCC | |||
| Bacteroides thetaiotaomicron | s-Bthe175-F | CCCGATGGTATAATCAGAC | 55 | [36] |
| s-Bthe587-R | CACAACTGACTTAACTGTCC | |||
| Bacteroides uniformis | s-Buni188-F | TTCTTCCGCATGGTAGAAC | 55 | [36] |
| s-Buni590-R | CTTTCACAACTGACTTAAGCG | |||
| Bacteroides vulgatus | s-Bvul129-F | AACCTGCCGTCTACTCTT | 55 | [36] |
| s-Bvul585-R | CAACTGACTTAAACATCCAT | |||
| Bifidobacterium adolescentis group1 | BiADOg-1a | CTCCAGTTGGATGCATGTC | 55 | [33] |
| BiADOg-1b | TCCAGTTGACCGCATGGT | |||
| BiADO-2 | CGAAGGCTTGCTCCCAGT | |||
| Bifidobacterium animalis subsp. lactis | Bflact2 | GTGGAGACACGGTTTCCC | 55 | [37] |
| Bflact5 | CACACCACACAATCCAATAC | |||
| Bifidobacterium bifidum | BiBIF-1 | CCACATGATCGCATGTGATTG | 55 | [38] |
| BiBIF-2 | CCGAAGGCTTGCTCCCAAA | |||
| Bifidobacterium breve | BiBRE-1 | CCGGATGCTCCATCACAC | 55 | [38] |
| BiBRE-2 | ACAAAGTGCCTTGCTCCCT | |||
| Bifidobacterium catenulatum group2 | BiCATg-1 | CGGATGCTCCGACTCCT | 55 | [38] |
| BiCATg-2 | CGAAGGCTTGCTCCCGAT | |||
| Bifidobacterium dentium | BiDEN-1 | ATCCCGGGGGTTCGCCT | 55 | [39] |
| BiDEN-2 | GAAGGGCTTGCTCCCGA | |||
| Bifidobacterium longum subsp. infantis | BiINF-1 | TTCCAGTTGATCGCATGGTC | 55 | [39] |
| BiINF-2 | GGAAACCCCATCTCTGGGAT | |||
| Bifidobacterium longum subsp. longum | BiLON-1 | TTCCAGTTGATCGCATGGTC | 55 | [39] |
| BiLON-2 | GGGAAGCCGTATCTCTACG |
1The B. adolescentis group consists of B. adolescentis genotypes A and B.
2The B. catenulatum group consists of B. catenulatum and B. pseudocatenulatum.
RNA extraction and RT-qPCR
Total RNA fractions were extracted from fecal samples by phenol-chloroform and RT-qPCR was performed, according to a method described by Matsuda et al. [32]. DNA’se treatment was not used because previous reports have shown that untreated and DNA’se-treated samples showed identical results, indicating that contaminating DNA does not affect RT-qPCR quantification [32].
RT-qPCR was conducted in a one-step reaction, using a QIAGEN OneStep RT-PCR Kit (QIAGEN, Venlo, The Netherlands) using 10 μl of reaction mixture containing 5 μl of template RNA and each specific primer (Table 2) at a concentration of 0.6 μM. The reaction mixture was dispensed into 384-well optical plates by using a MICROLAB STARlet Liquid Handling Workstation (Hamilton Robotics, Reno, NV). The reaction mixture was incubated at 50°C for 30 min for reverse transcription. The continuous amplification program consisted of one cycle at 95°C for 15 min and 40 or 45 cycles at 94°C for 20 s, 55°C or 60°C for 20 s, and 72°C for 50 s. Amplification and detection were performed by using an ABI PRISM® 7900HT Sequence Detection System (Thermo Fisher Scientific).
Table 2. 16S or 23S rRNA gene targeted primers and standard strains used for RT-qPCR.
| Target | Primer | Sequence (5’-3’) | PCR Cycle N | Annealing Temp (°C) | Ref |
|---|---|---|---|---|---|
| Clostridium perfringens | s-Clper-F | GGGGGTTTCAACACCTCC | 40 | 60 | [32] |
| ClPER-R | GCAAGGGATGTCAAGTGT | [40] | |||
| Family Enterobacteriaceae | En-Isu-3F1 | TGCCGTAACTTCGGGAGAAGGCA | 40 | 60 | [41] |
| En-Isu-3R1 | TCAAGGCTCAATGTTCAGTGTC | ||||
| Genus Enterococcus | g-Encoc-F | ATCAGAGGGGGATAACACTT | 40 | 55 | [32] |
| g-Encoc-R | ACTCTCATCCTTGTTCTTCTC | ||||
| Genus Staphylococcus | g-Staph-F | TTTGGGCTACACACGTGCTACAATGGACAA | 40 | 60 | [32] |
| g-Staph-R | AACAACTTTATGGGATTTGCWTGA | ||||
| Lactobacillus fermentum | LFer-1 | CCTGATTGATTTTGGTCGCCAAC | 40 | 55 | [32] |
| LFer-2 | ACGTATGAACAGTTACTCTCATACGT | ||||
| Lactococcus lactis | sg-Lclac-F | TGTAGGGAGCTATAAGTTCTCTGTA | 40 | 60 | [42] |
| sg-Lclac-R | GGCAACCTACTTYGGGTACTCCC | ||||
| Lactobacillus casei subgroup | sg-Lcas-F | ACCGCATGGTTCTTGGC | 40 | 60 | [32] |
| sg-Lcas-R | CCGACAACAGTTACTCTGCC | ||||
| Lactobacillus gasseri subgroup | sg-Lgas-F | GATGCATAGCCGAGTTGAGAGACTGAT | 40 | 60 | [32] |
| sg-Lgas-R | TAAAGGCCAGTTACTACCTCTATCC | ||||
| Lactobacillus plantarum subgroup | sg-Lpla-F | CTCTGGTATTGATTGGTGCTTGCAT | 40 | 60 | [32] |
| sg-Lpla-R | GTTCGCCACTCACTCAAATGTAAA | ||||
| Lactobacillus sakei subgroup | sg-Lsak-F | CATAAAACCTAMCACCGCATGG | 45 | 60 | [32] |
| sg-Lsak-R | TCAGTTACTATCAGATACRTTCTTCTC | ||||
| Lactobacillus reuteri subgroup | sg-Lreu-F | GAACGCAYTGGCCCAA | 45 | 60 | [32] |
| sg-Lreu-R | TCCATTGTGGCCGATCAGT | ||||
| Lactobacillus ruminis subgroup | sg-Lrum-F | CACCGAATGCTTGCAYTCACC | 40 | 60 | [32] |
| sg-Lrum-R | GCCGCGGGTCCATCCAAAA |
1Specific primers are targeting 16S rRNA genes, except for En-Isu-3F/En-Isu-3’R, which targets 23S rRNA genes
Determination of bacterial number by qPCR and RT-qPCR
The bacterial counts was estimated from the slope of the standard curve as described earlier [43], generated by using the following standard strains: Ruminococcus productus ATCC 27340T (for the Clostridium coccoides group), Faecalibacterium prausnitzii ATCC 27768T (for the Clostridium leptum group), Bacteroides vulgatus ATCC 8482T (for the Bacteroides fragilis group), Bifidobacterium longum subsp. longum ATCC 15707T (for the Genus Bifidobacterium), Collinsella aerofaciens ATCC 25986T (for the Atopobium cluster), Prevotella melaninogenica ATCC 25845T (for the Genus Prevotella), Bacteroides caccae ATCC 43185T (for Bacteroides caccae), Bacteroides eggerthii ATCC 27754T (for Bacteroides eggerthii), Bacteroides fragilis ATCC 25285T (for Bacteroides fragilis), Bacteroides ovatus ATCC 8483T (for Bacteroides ovatus), Bacteroides thetaiotaomicron ATCC 29148T (for Bacteroides thetaiotaomicron), Bacteroides uniformis ATCC 8492T (for Bacteroides uniformis), Bacteroides vulgatus ATCC 8482T (for Bacteroides vulgatus), Bifidobacterium adolescentis ATCC 15703T (for the Bifidobacterium adolescentis group), Bifidobacterium animalis subsp. lactis DSM 10140T (for Bifidobacterium animalis subsp. lactis), Bifidobacterium bifidum ATCC 29521T (for Bifidobacterium bifidum), Bifidobacterium breve ATCC 15700T (for Bifidobacterium breve), Bifidobacterium pseudocatenulatum JCM 1200T (for the Bifidobacterium catenulatum group), Bifidobacterium dentium ATCC 27534T (for Bifidobacterium dentium), Bifidobacterium longum subsp. infantis ATCC 15697T (for Bifidobacterium longum subsp. infantis), Bifidobacterium longum subsp. longum ATCC 15707T (for Bifidobacterium longum subsp. longum), Clostridium perfringens JCM 1290T (for Clostridium perfringens), Escherichia coli ATCC 11775T (for the Family Enterobacteriaceae), Enterococcus faecalis ATCC 19433T (for the Genus Enterococcus), Staphylococcus aureus ATCC 12600T (for the Genus Staphylococcus), Lactobacillus fermentum ATCC 14931T (for Lactobacillus fermentum), Lactococcus lactis subsp. lactis JCM 5805T (for the Lactococcus lactis subgroup), Lactobacillus casei ATCC 334T (for the Lactobacillus casei subgroup), Lactobacillus gasseri DSM 20243T (for the Lactobacillus gasseri subgroup), Lactobacillus plantarum ATCC 14917T (for the Lactobacillus plantarum subgroup), Lactobacillus sakei subsp. sakei JCM 1157T (for the Lactobacillus sakei subgroup), Lactobacillus reuteri JCM 1112T (for the Lactobacillus reuteri subgroup), and Lactobacillus ruminis JCM 1152T (for the Lactobacillus ruminis subgroup).
Identification and quantification of fecal organic acids
The supernatant, which was prepared as mentioned in the section entitled Primary treatment of fecal samples, was injected into a Water 432 Conductivity Detector (Waters Corporation, Milford, MA) to determine the concentrations of fecal organic acids (acetic acid, butyric acid, formic acid, iso-valeric acid, lactic acid, propionic acid, succinic acid, and valeric acid) by HPLC as previously reported [44].
Statistical Analyses
The data of bacterial counts contained proportions of non-detected values due to detection limits. The non-detected values were substituted with the detection limit divided by the square root of 2 when the percentage of non-detected samples at a time point was less than 10% [45]. When the percentage of non-detected samples at a time point was greater than 40% then the outcome was directly converted to a binary result (detected versus non-detected). The remaining outcomes that had a percentage of non-detected samples between 10% and 40% at a time point were considered for imputation. At each time point a semi-parametric density estimate was fitted to the bacterial counts and the non-detected values were replaced by imputed values from this estimated semi parametric density [46].
Analysis by random coefficients mixed model (for continuous outcome variables) and generalized linear mixed model (for binary outcome variables) were used to study associations between the outcome variables (gut microbiota and organic acids) and potential covariates. The effect of mode of delivery, type of feeding, antibiotic use, number of siblings, presence of allergens, and gender on colonization of the gut was studied. The selection of important covariates was done using likelihood ratio tests, starting from full models including all potential covariates and where possible covariate by time interaction terms. The covariate selection continued using a backward selection method. At each step, changes in the standard errors and point estimates were checked in order not to miss any confounding effects.
First defecation samples (meconium) were analyzed separately from the samples taken at other time points due to high percentages of specific assays being below detection limit in these samples. Time (in days) at which first defecation sample was taken was included as an additional covariate.
The measured gut microbiota variables and organic acids values from samples taken at 2, 7, 30 days and 3 months were analyzed in a longitudinal model that started with the assumption of a second order polynomial trajectory over time. However, during the backward selection, the time-squared terms could be deleted from the model if there was no suggestion for a curvature over time.
Since solid foods were introduced from 4 months onwards, the six months samples were analyzed separately and this variable was included as an additional covariate.
In order to check the consistency of our modeling results with the observed data, we included figures of the observed data and checked them against figures obtained from the fitted models. For the species that were not detected in at least 40% of the samples, we used the binary variable as an outcome (detected versus non-detected), permitting us to obtain the probability to be detected from the fitted models (with the correspondent p-value) which can be examined with the prevalence calculated from the observed data. For the species that were detected in at least 60% of the samples, we used the continuous variable as an outcome (log10 cell count/g of feces or μmol/g of feces for bacteria or organic acids, respectively), thus we could obtain the estimated amount from the fitted model (with the correspondent p-value) and the observed data as measured.
Results & Discussion
Study population
143 subjects were included in the study of which 34 dropped-out during the study. No longer fulfilling the inclusion and exclusion criteria during the post-delivery re-check, was considered the major reason for drop out (N = 18). The other early terminators were due to withdrawal of informed consent (N = 4), protocol violation (N = 6), lost to follow up (N = 2), and for other reasons (N = 4). 109 subjects were included in the study but data of 3 subjects was not properly collected and two subjects delivered twins, so in total fecal samples of 108 subjects were suitable to be analyzed. The study population did not have a significant difference in the numbers of boys and girls (51.9 and 48.1%, respectively), and half of the total population had at least one older sibling (48.1%). Twenty eight infants were born by CS (25.9%) and only 5 out of the 108 infants were born at home (4.6%).
Other characteristics that vary in time are summarized in Table 3. These characteristics include type of feeding (breastfeeding, mix feeding or formula feeding at the sampling time), time of introduction of solid food and antibiotic use.
Table 3. Population characteristics varying in time.
| Characteristics (Number of subjects) | Meconium1 | + 2 days2 | Day 7 | Day 30 | Day 90 | Day 180 | |
|---|---|---|---|---|---|---|---|
| Type of feeding | Exclusive breastfeeding | 100 | 76 | 89 | 79 | 59 | 10 |
| Exclusive Formula feeding | 3 | 2 | 3 | 5 | 15 | 0 | |
| Mix feeding3 | 2 | 4 | 11 | 23 | 30 | 3 | |
| Introduction of solid food4 | 0 | 0 | 0 | 3 | 3 | 88 | |
| Missing information | 3 | 26 | 5 | 1 | 1 | 7 | |
| Antibiotic use | Yes/no | 0/108 | 0/108 | 0/108 | 0/108 | 2/106 | 6/102 |
| Number of eecal samples collected per time point | 96 | 80 | 98 | 106 | 105 | 102 | |
| Nmber of infants | 108 | 108 | 108 | 108 | 108 | 108 | |
1Meconimum samples were taken at 0.77 ± 1.04 days (mean±st dev)
2 The consequent sample was taken 2 days after, which occurred at 2.88 ± 0.99 days (mean±st dev)
3Mix-feeding is defined as the combination of breastfeeding and formula feeding
4 Introduction of solid food does not exclude breastfeeding or mix-feeding.
Early colonization
It was always believed that the gut of a newborn is sterile and that the colonization process only started at birth. This concept has been challenged in the last years due to the recent discovery of bacteria or bacterial DNA in the umbilical cord blood, placenta and meconium [8–11], suggesting in utero exposure to bacteria. In this study, the first fecal sample (meconium) was collected from 96 infants (88.89%) and bacteria were detected in 74 out of the 96 meconium samples (77.08%). The most prevalent bacteria were Staphylococcus which was detected in 67.71% of the samples, followed by Enterobacteriaceae, Enterococcus, Lactobacillus and Bifidobacterium. The later showed the highest cell count number in meconium (7.69±1.13 log10 cells/g of feces). Interestingly, bacteria were detected in the meconium of infants born by C-section (as described later in this article), suggesting that colonization by certain bacteria might start before birth. Acetic acid was the organic acid the most often detected (63% of the meconium samples) with a mean amount of 9.12 ±8.74 μmol/g of feces.
These results are in agreement with a recent study reporting evidence of the presence of the same spectrum of bacteria in 66% of meconium samples collected within 24 hours after birth [47].
In that study, Hansen et al., concluded that there is limited bacterial colonization at birth and it occurs rapidly thereafter. In our study we defined meconium as the first fecal sample after birth but collection was not restricted to the first 24 hours of birth, the mean time at which they were collected was at 0.77 days (± 1.04 days). Since time was included in our model as a variable, our results showed that delaying the collection of meconium samples by one day increased the chance of detecting the following bacteria: Bifidobacterium; Bacteroides fragilis group; Enterococcus; Staphylococcus; L. gasseri subgroup; and B. adolescentis by 3.1 (p = 0.003); 2.35 (p = 0.01); 10.69 (p<0.001); 2.26 (p = 0.016), 3.01 (p = 0.018); and 3.9 (p = 0.008) fold respectively. This could be explained by the growth of microorganisms that were already present before delivery as recently suggested [8–11] or by the presence of those microorganisms entering the ecosystem from the environment after birth.
After 2 days, the prevalence and count of the different bacterial groups followed a temporal and dynamic change during the first 6 months of life as illustrated in S1 Table.
The microbiota composition evolves over time which is the result of the combined impact of different events (i.e. different type of feeding, mode of delivery, and antibiotic use) and will be discussed below. Interestingly, for some species no suggestion for a statistically significant influence of any of the covariates emerged except for time. Examples are the increased probability to detect B. uniformis during the first 3 months of life (longitudinal model, p = 0.003), the decreased probability to detect L. fermentum (six months samples; p = 0.049), the increased probability to detect B. animalis subsp. lactis (six months samples; p = 0.0046 and the decreased in Staphylococcus counts from 3 to 6 months (six months samples; p = 0.009). These findings may suggest that these bacteria are transient in the infant gut but may play a key role in maturing the immune system in the first months of life.
Influence of mode of delivery on the infant gut microbiota during the first 6 months of life
The mode of delivery is often described as a key factor in selecting the early colonizers of the infant gut. Infants born by VD are exposed to a vast number of bacteria when passing through the birth canal, whereas CS-born infants lack the direct contact with the maternal microbiota, and are exposed to a more sterile environment at birth. It has been suggested that the aberrant gut microbiota composition observed in early life in infants born by CS may explain why CS-born infants are at a higher risk for developing allergy and obesity later in life [26, 29, 48–51].
In this study (in which 25.9% infants were born by CS) mode of delivery was indeed found to be one of the factors that had a strong impact on the early microbiota composition. Infants born by VD had significantly higher bacterial counts in the meconium (as measured by DAPI) than those born by CS (8.47 vs. 7.76 log10 cells/g of feces, first defecation sample, p = 0.024), which may have resulted from the transfer of bacteria during birth. This difference in total bacteria count was observed during the whole study period, although cell counts increased faster over time in CS-born infants (longitudinal model, p = 0.004). In addition, CS-born infants had a delayed colonization of several bacterial groups or species, and this was still the case for some species at 6 months of age as illustrated in Table 4.
Table 4. Prevalence of different bacterial groups in infants born by vaginal delivery or C-section.
| Prevalence (%) | Vaginal Delivery (N = 80) | Cesarean section (N = 28) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mec1 | D22 | D7 | D30 | D90 | D180 | Mec1 | D22 | D7 | D30 | D90 | D180 | |
| Bifidobacteria | 16.7 | 82.3 | 90.7 | 94.9 | 97.4 | 97.4 | 16.7 | 22.2 | 69.6 | 75 | 100 | 100 |
| Bacteroides fragilis group | 12.5 | 80.7 | 68 | 74.4 | 77.9 | 82.9 | 4.2 | 0 | 0 | 3.6 | 32.1 | 57.7 |
| Enterococcus | 21.3 | 62.9 | 72.6 | 72.2 | 89.7 | 97.4 | 12 | 61.1 | 75 | 92.9 | 96.5 | 100 |
| Clostridium perfringens | 5.3 | 11.3 | 21.9 | 24.1 | 33.3 | 36.4 | 0 | 16.7 | 50 | 53.6 | 35.7 | 34.6 |
| Atopobium | 1.4 | 29 | 29.3 | 39.7 | 54.5 | 71.4 | 0 | 0 | 0 | 17.9 | 64.3 | 69.2 |
| B. fragilis | 2.8 | 20.9 | 12.5 | 12.8 | 22.1 | 34.2 | 0 | 0 | 0 | 3.6 | 14.3 | 26.9 |
| B. caccae | 1.4 | 17.7 | 5.3 | 10.3 | 18.2 | 23.7 | 0 | 0 | 0 | 3.6 | 10.7 | 11.5 |
| B. vulgatus | 11.11 | 67.7 | 56 | 58.9 | 66.2 | 65.8 | 0 | 0 | 0 | 3.6 | 14.3 | 38.5 |
| B. ovatus | 1.4 | 14.5 | 17.3 | 25.6 | 24.7 | 30.3 | 0 | 0 | 0 | 0 | 7.1 | 7.7 |
| B. uniformis | 0 | 22.6 | 14.7 | 21.8 | 31.2 | 40.8 | 0 | 0 | 0 | 3.6 | 14.3 | 15.4 |
| B. catenulatum group | 2.8 | 29 | 28 | 29.5 | 37.7 | 55.3 | 0 | 0 | 0 | 7.1 | 28.6 | 53.9 |
| B. longum | 11.1 | 62.9 | 69.3 | 71.8 | 77.92 | 78.9 | 0 | 0 | 8.7 | 39.3 | 71.4 | 96.2 |
| B. bifidum | 12.5 | 51.6 | 45.3 | 50 | 66.2 | 77.6 | 8.3 | 11.1 | 4.3 | 21.4 | 60.7 | 69.2 |
| L. gasseri subgroup | 17.3 | 70.1 | 39.7 | 49.3 | 39.7 | 35.1 | 8 | 11.11 | 33.3 | 46.4 | 50 | 26.9 |
| L. reuteri subgroup | 20 | 20.9 | 17.8 | 18.9 | 23.1 | 22.1 | 4 | 5.5 | 8.3 | 21.4 | 21.4 | 19.2 |
Table representing only those bacterial groups that were significantly dependent on mode of delivery; Prevalence presented as % of infants in which the bacteria were detected
1The meconium sample was taken at 0.77 ± 1.04 days (mean±st dev)
2 The consequent sample was taken 2 days after, which occurred at 2.88 ± 0.99 days (mean±st dev)
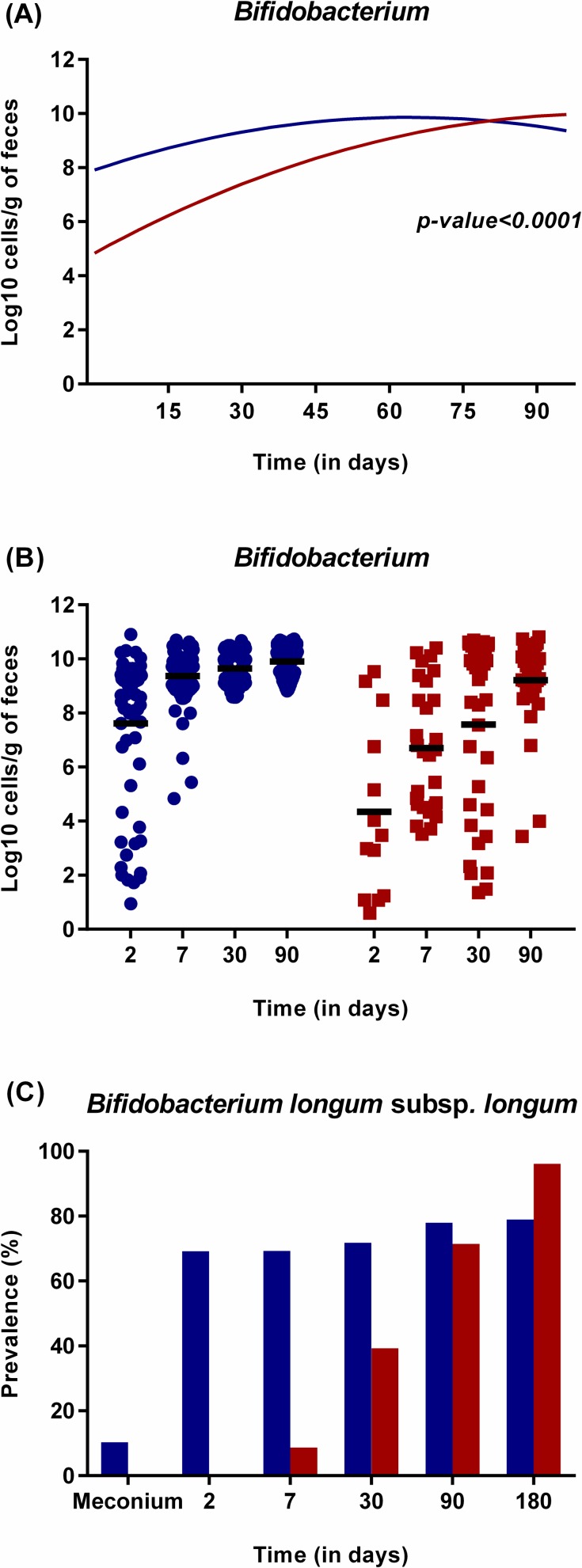
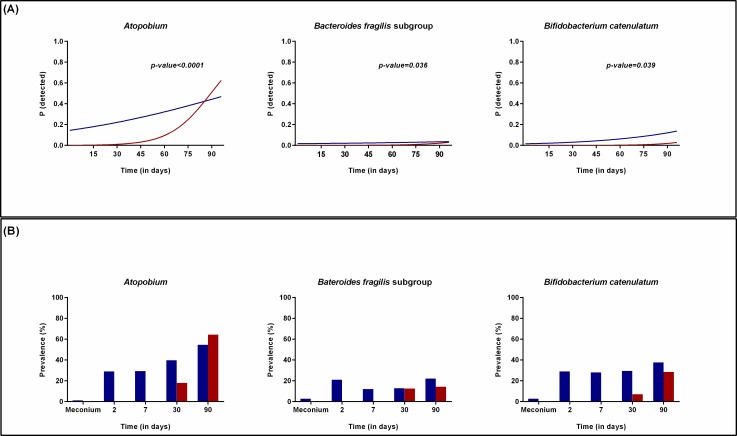
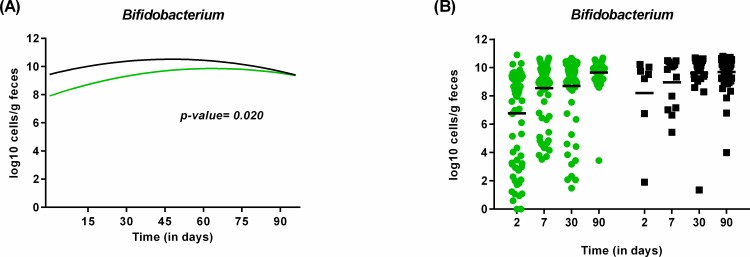
The delayed colonization in CS-born infants was demonstrated by the significantly lower probability to detect Bacteroides fragilis group (longitudinal model, p = 0.003), B. ovatus (longitudinal model, p = 0.006), B. vulgatus (longitudinal model, p<0.001), B. uniformis (longitudinal model, p = 0.001), B. caccae (longitudinal model, p = 0.028) (Original data represented in Fig 1), and B. longum subsp. longum (longitudinal model, p<0.001) (Original data represented in Fig 2) during the first three months of life. During this period, bifidobacterial counts were significantly higher in VD-born infants, although it increased rapidly in the CS group (longitudinal model, p<0.001) (Fig 2A). In addition, the probability to detect Atopobium cluster (longitudinal model, p<0.001), B. fragilis subgroup (longitudinal model, p = 0.036) and B. catenulatum group (longitudinal model, p = 0.039) in VD-infants was higher than in CS-born infants and followed a different trajectory over time (Fig 3A).
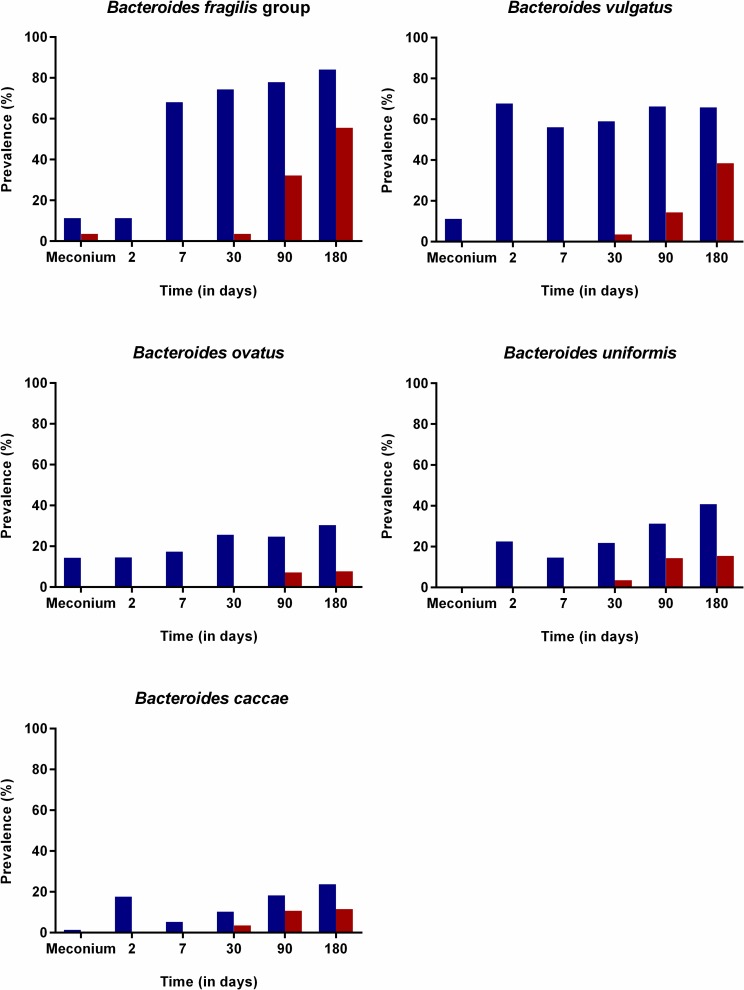
Fig 1. Effect of mode of delivery on the colonization by Bacteroides species.
Prevalence of different Bacteroides species in fecal samples of infants born by caesarean section (red) or vaginal delivery (blue) at birth, 2, 7, 30, 90 and 180 days of life, using the original data.
Fig 2. Effect of mode of delivery on the colonization by Bifidobacterium.
Bifidobacterium count (in log10 cell/g of feces) in infants born by caesarean section (red) compared to those born by vaginal delivery (blue) over time as calculated by the statistical model (A) or as represented from original data (B). (C) Prevalence of B. longum in infants born by CS (red) or vaginal delivery (blue) at birth, 2, 7, 30, 90 and 180 days of life. The graph is generated using the raw data.
Fig 3. Effect of mode of delivery on the probability to detect different bacteria.
(A) Probability to detect Atopobium cluster, B. fragilis group and B. catenulatum group in infants born by VD (blue) compared to those born by CS (red) over time as calculated by the statistical model. (B) Prevalence of Atopobium cluster, B. fragilis group and B. catenulatum group in in infants born by VD (blue) compared to those born by CS (red) over time as represented by the original data.
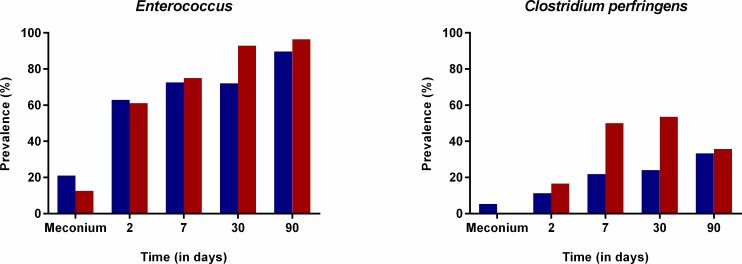
As previously described, the lack of bacterial transmission during CS delivery or more likely; the exposure to sterile hospital surfaces or the skin of medical personnel/parents, seems to favor the colonization and growth of facultative aerobes and skin related bacteria such as Staphylococcus, Propionibacterium and Clostridium species [52, 53]. In our study, the probability to detect Enterococcus (longitudinal model, p = 0.008) and C. perfringens (longitudinal model, p = 0.013) was significantly higher in infants born by CS compared to those born by VD (Original data represented in Fig 4).
Fig 4. Effect of mode of delivery on the colonization by Enterococcus and Clostridium perfringens.
Prevalence of Enterococcus and Clostridium perfringens in fecal samples of infants born by caesarean section (red) or vaginal delivery (blue) at birth, 2, 7, 30, 90 and 180 days of life, using the original data.
There is no consensus about the timing at which the differences observed between the microbiota composition of VD and CS-born infants may disappear, if at all. Whereas some authors have reported that there are no differences after 1 month, others reported differences even at 7 years of age [52]. In our study most differences disappeared at 6 months of age, except for the higher probability to detect B. fragilis-group, B. uniformis, B. vulgatus and B. ovatus (six months samples, p = 0.01, p = 0.002, p = 0.007, p = 0.008, respectively) in VD-born than in CS-born infants (Original data represented in Fig 1) for which longer study periods than the 6 months studied here would be required.
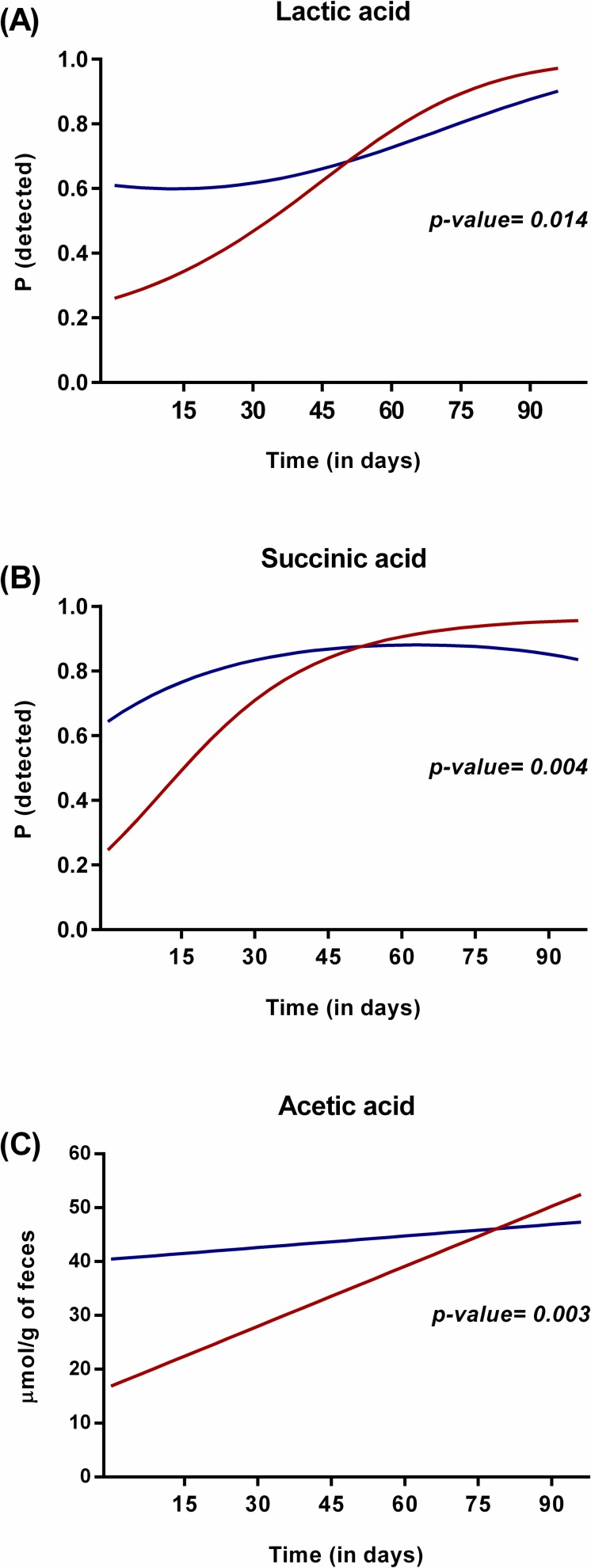
The differences observed in the microbiota composition of infants born by CS or VD were also in line with differences in organic acids detected in the fecal samples. Significant differences were found for the detection of succinic acid (longitudinal model, p = 0.004) and lactic acid (longitudinal model p = 0.014), and for the amount of acetic acid (longitudinal model p = 0.003) between the two groups (Fig 5). When analyzing the data at each time point separately, succinic and lactic acid were most often detected in infants born by VD only at day 2 (p<0.001 and p = 0.026, respectively) while the amount of acetic acid was significant until 1 week (p = 0.04). The pattern of organic acids described reflects the colonization pattern with high levels of bifidobacteria in these two groups of infants (VD vs CS). The same trajectory over time was observed for the Bifidobacterium count and the acetic acid concentration, one of the major metabolites produced by Bifidobacterium as a result of the metabolism of carbohydrates (Figs 2A and 5C).
Fig 5. Effect of mode of delivery on the organic acid profile.
Probability to detect organic acids (acetic, lactic and succinic acid) in infants born by caesarean section (red) compared to those born by vaginal delivery (blue) over time as calculated by the statistical model.
Our results are in agreement with previous studies reporting a delayed colonization by Atopobium cluster, Bifidobacterium and Bacteroides species in infants born by CS, whereas they are more often colonized by Enterococcus and Clostridium species [12, 15, 54–57]. However, we could not confirm the higher prevalence of Staphylococcus in CS-born infants previously described by Dominguez-Bello et al. [53], which might be explained by the geographical differences of the study populations (Europe and USA).
Our data confirmed that the mode of delivery influences the early colonization and development of the infant gut microbiota, and showed that differences in species composition can still be found until at least 6 months of age.
Impact of type of feeding on the differences observed as a function of mode of delivery
It is often suggested that the differences observed in the microbiota composition of infants born by CS are also due to a different type of feeding. It is even argued that breastfeeding is more difficult after a CS delivery [58, 59], and therefore, that infants born by CS are often not breastfed, or that they start breastfeeding later than infants born by VD. The results we have reported so far were found to be dependent only on the mode of delivery, however, in this study we also found that the presence of the L. reuteri subgroup, the L. gasseri subgroup and B. bifidum was dependent on the interaction between mode of delivery and the type of feeding (longitudinal model; p = 0.003, p = 0.03, p = 0.009, respectively) (Table 5).
Table 5. Impact of mode of delivery and type of feeding1.
| Prevalence | Vaginal delivery | Caesarean section | |||
|---|---|---|---|---|---|
| Species | Time | Exclusive BF % (N) | Mix Feeding % (N) | Exclusive BF % (N) | Mix Feeding % (N) |
| B. bifidum | Meconium2 | 11.26 (71) | 25 (4) | 8.33 (25) | 0 (1) |
| Day 23 | 51.72 (58) | 50 (4) | 5.58 (17) | 50 (2) | |
| Day 7 | 47.69 (65) | 37.5 (8) | 5.58 (17) | 0 (6) | |
| Day 30 | 51.66 (60) | 41.18 (17) | 11.76 (17) | 36.36 (11) | |
| Day 90 | 70.45 (44) | 60.60(33) | 46.15 (13) | 73.33 (15) | |
| L. gasseri | Meconium2 | 18.3 (71) | 0 (4) | 8 (25) | 0 (1) |
| Day 23 | 55.17 (58) | 25 (4) | 11.76(17) | 0 (2) | |
| Day 7 | 41.64 (65) | 25 (8) | 33.33 (18) | 33.33 (6) | |
| Day 30 | 47.54 (61) | 41.18 (17) | 47.06 (17) | 45.45 (11) | |
| Day 90 | 40 (45) | 40.63 (32) | 76.92 (13) | 23.08(13) | |
| L. reuteri | Meconium2 | 8.45 (71) | 0 (4) | 4 (25) | 0 (1) |
| Day 23 | 22.41 (58) | 0 (4) | 5.58 (17) | 0 (2) | |
| Day 7 | 16.92 (65) | 25 (8) | 0 (18) | 33.33 (6) | |
| Day 30 | 18.03 (61) | 23.53 (17) | 5.88 (17) | 45.45 (11) | |
| Day 90 | 11.11 (45) | 40.63 (32) | 15.38 (13) | 15.38 (13) | |
1Original data
2The meconium sample was taken at 0.77 ± 1.04 days (mean±st dev)
3 The consequent sample was taken 2 days after, which occurred at 2.88 ± 0.99 days (mean±st dev)
N (Number of infants born by VD or CS and being exclusive or mix-fed). % (proportion of infants in which the specific species was detected)
Infants born by VD who were exclusively breastfed were the ones most often colonized by B. bifidum (longitudinal model, p = 0.009) and L. gasseri subgroup (longitudinal model, p = 0.029), compared to those exposed to formula feeding (mixed feeding) or born by CS.
Numbers of B. bifidum were higher in infants born by VD during the first week of life. However, infants born by CS, who were exclusively breast fed, reached a level comparable to that observed in VD infants earlier than CS-born infants that were exposed to formula (mixed feeding). Similarly, members of the L. gasseri subgroup were more frequently detected in exclusively breastfed infants. Interestingly, at 3 months, the highest prevalence of both L. gasseri subgroup and B. bifidum was found in infants born by cesarean section. All together, these results suggest that vaginal delivery in combination with breastfeeding favor the colonization by B. bifidum and L. gasseri subgroup in the infant gut, and that breastfeeding may compensate in time in those infants born by CS by favoring the colonization of these species. Both bacterial species have been previously found in human milk which may explain the differences observed [60, 61]. On the contrary, infants born by CS and exposed to formula feeding were most often colonized by L. reuteri subgroup, whereas those born by CS but exclusively breastfed were the least often colonized by this species (Table 5). Our results suggest that exposure to formula feeding seems to favor the colonization of this species which may be the result of the use of formula supplemented with probiotic strains belonging to this species.
To our knowledge this is the first time that an interaction between mode of delivery and type of feeding is reported. However, the number of infants born by CS was low and the findings need to be confirmed in future trials.
Influence of the type of feeding on the infant gut microbiota during the first 6 months of life
The feeding pattern varied along the study and it is described in Table 3. Most of the infants were exclusively breastfed in the first month of life (73.15%), and thus only a small proportion received infant formula either exclusively or in combination with breastfeeding.
At 3 months of age, 54.63% of the infants were still exclusively breastfed, 13.88% were exclusively formula fed, 27.77% received mix feeding, while 2.77% of the infants had received solid foods. At 6 months of age, 9.26% of infants were exclusively breastfed and 2.94% received mix feeding but did not receive solid foods, whereas 81.48% of the infants had received solid foods even though they might continue receiving breast milk and/or infant formula and are considered within the group of weaned infants (Table 3).
Due to the low numbers of exclusively formula fed infants, the data from these infants and the ones receiving mix-feeding were regarded as one group. The differences reported are found when comparing exclusive breastfeeding to mix feeding (partially breastfed or exclusively formula fed).
During the first 3 months of life, exclusively breastfed infants had lower total bacterial counts (by DAPI staining) than those receiving other type of feeding (longitudinal model, p = 0.027), but had higher counts of Staphylococcus (longitudinal model, p = 0.037). Staphylococcus has been shown to be transferred from breast milk to the infant gut and is known to be one of the most dominant bacteria found in the infant gut [62]. However, as discussed earlier, it is not yet clear whether it is a permanent or transient colonizer or whether they play a key in role in the infant gut [62].
Infants exposed to formula feeding (either exclusively or in combination with breastfeeding) were more often colonized by Enterococcus (longitudinal model, p<0.001), C. coccoides group (longitudinal model, p = 0.002), Atopobium cluster (longitudinal model, p = 0.002), B. vulgatus (longitudinal model, p = 0.027) and B. longum subsp. longum (longitudinal model, p = 0.033) (Original data represented in S1 Fig).
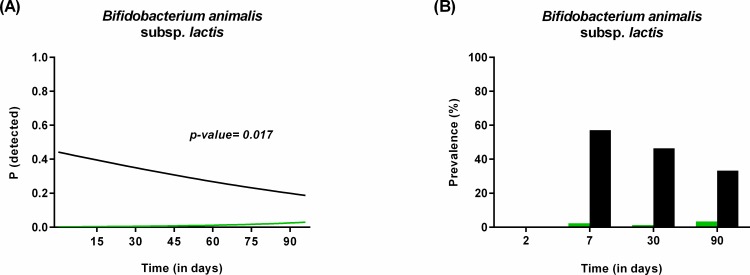
The colonization of Bifidobacterium, C. leptum group, C. perfringens, L. casei subgroup and B. animalis subsp. lactis was also influenced by type of feeding, but the differences followed a different trajectory over time. Contrary to previous studies and much to our surprise [63–66], exclusively breastfed infants had lower counts of Bifidobacterium, especially at earlier time points (longitudinal model, p = 0.02; Fig 6A). The high bifidobacterial count found in the mix-fed group can however be explained by the inclusion of a few exclusively formula-fed infants that received a formula supplemented with a probiotic strain (B. animalis subsp. lactis). Indeed, infants exposed to formula feeding were most often colonized by B. animalis subsp. lactis (Fig 7A), whereas this species was always below the detection limit in exclusively breastfed infants. This finding, together with the fact that the probability to detect B. animalis subsp. lactis in mix-fed infants decreased over time (longitudinal model, p = 0.017; Fig 7A), shows that this species is not a common inhabitant of the gut of healthy breast-fed infants.
Fig 6. Influence of type of feeding on the colonization by Bifidobacterium during the first 3 months of life.
Bifidobacterium count (in log10 cell/g of feces) in exclusively breastfed infants (green) compared to those receiving mix-feeding (black) over time as calculated by the statistical model (A) or as represented from the original data (B).
Fig 7. Influence of type of feeding on the presence of B. animalis subsp. lactis during the first 3 months of life.
(A) Probability to detect B. animalis in exclusively breastfed infants (green) compared to those receiving mix-feeding (black) as calculated by the statistical model. (B) Prevalence of B. animalis in infants receiving mix-feeding (black) or exclusively breast fed (green) at birth, 2, 7, 30 and 90 days of life as represented by the original data.
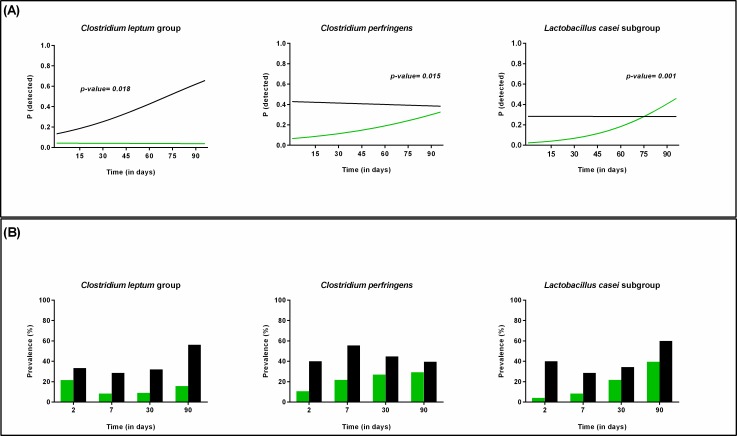
Exclusively breastfed infants were less often colonized by C. perfringens (longitudinal model, p = 0.015) and L. casei subgroup (longitudinal model, p = 0.001), but the probability to detect these bacteria increased more rapidly during the first three months of life in this group of infants than in the group exposed to infant formula (Fig 8). On the other hand, infants exposed to formula feeding were most often colonized by C. leptum group, and its prevalence increased over time (longitudinal model, p = 0.018; Fig 8).
Fig 8. Effect of type of feeding on the colonization pattern of different bacteria during the first 3 months of life.
(A) Probability to detect C. leptum group, C. perfringens, and L. casei subgroup in infants receiving exclusive breastfeeding (green) compared to those receiving mix-feeding (black) over time as calculated by the statistical model. (B) Prevalence of C. leptum group, C. perfringens, and L. casei subgroup in infants receiving mix-feeding (black) or exclusive breast feeding (green) over time as represented by the original data.
After 3 months, a considerable number of infants ceased breastfeeding (45%) and/or received solid food. In our study, we investigated the effect of these two factors on the microbiota composition of the fecal sample taken at 6 months.
Diet in the first 3 months of life still has an impact on the microbiota composition at 6 months. Exclusive breastfeeding for the first 3 months in contrast to mix-feeding was associated with a higher count of Bifidobacterium (10.07 log10 cells/g feces vs 9.49 log10 cells/g feces; six months samples, p = 0.037) and higher prevalence of L. casei subgroup (9.5 times more prevalent, six months samples, p = 0.019) and B. adolescentis group (3 times more prevalent, six months samples, p = 0.044) at 6 months (Original data represented in S2A Fig). The probability to detect L. gasseri subgroup was also higher in those infants exclusively breastfed for 3 months, although it decreased over time (six months samples, p = 0.017) (Data represented in S3 Fig). The predominance of lactic acid producers described above is also accompanied by a higher probability to detect lactic acid (almost 3 times higher; six months samples, p = 0.049), and lower probability to detect butyric acid (6.4 times lower; six months samples, p = 0.007) (Original data represented in S2B Fig) in those infants exclusively breastfed for 3 months, which may indicate a less complex ecosystem. These results suggest that exclusive breastfeeding in the first three months of life may have a long lasting effect on the colonization process.
Similarly, the introduction of solid foods is associated with the replacement of the early colonizers by a more complex microbiota, which is reflected by a change in the organic acid profile, with lower chance to detect succinic acid (4 times lower, six months samples, p = 0.05) and lactic acid (8 times lower, six months samples, p = 0.015), and a higher chance to detect butyric acid (10 times higher, six months samples, p = 0.05) compared to those infants not receiving solid foods. Indeed, the introduction of solid foods was associated with a higher prevalence of Atopobium cluster (six months samples, p = 0.027) and some butyrate-producing bacteria like the C. coccoides group (six months samples, p = 0.002) (Original data represented in S4 Fig). The increased prevalence of the latter group after the introduction of solid foods has been previously reported, and can be explained by the introduction of complex carbohydrates through the diet that are easily metabolized by these type of bacteria in the infant gut [57, 67, 68]. Another effect of the early introduction of solid food was the decreased amount of early colonizers such as Enterobacteriaceae (7.7 log10 cells/g of feces vs. 8.1 log10 cells/g of feces; six months samples, p = 0.04) and Staphylococcus (5.06 log10 cells/g of feces vs 5.6 log10 cells/g of feces; six months samples, p = 0.024), which were significantly lower in those infants receiving solid foods than in those not receiving solid foods, as previously described [67]. Unlike other authors, we did not find a decrease in the prevalence of infant-type bifidobacterial species after the introduction of solid foods. Surprisingly, the prevalence of B. longum subsp. longum was even 9.4 times higher (six months samples, p = 0.002). This discrepancy may be due to different ages of the children in the different studies or to the influence that exclusive breastfeeding may have on this process. We investigated the effect of early introduction of solid foods (before 6 months) whereas Bergtrom et al, looked at infants between 9 and 36 months of age [67]. It has been recently described that cessation of breast-feeding, rather than introduction of solid food, drives further maturation into adult like microbiota [15], and that breast milk may provide the gut microbiome with a greater plasticity that eases the transition into solid foods [69].
Influence of antibiotic use on the infant gut microbiota during the first 6 months of life
Antibiotic use can cause an ecological disruption that might be even more pronounced when administered in early life, a period of critical development. Indeed, antibiotic exposure in early life is associated with numerous diseases later in life [4]. Antibiotic use was very low throughout the study; only 6 infants received antibiotics, which all occurred after 3 months. Infants receiving antibiotics had slightly lower total bacterial count (as measured by DAPI) (9.97 log10 cells/g of feces vs 10.44 log10 cells/g of feces, six months samples, p<0.001), lower Bifidobacterium (8.5 log10 cells/g of feces vs 9.7 log10 cells/g of feces, six months samples, p = 0.003), and Staphylococcus (4.9 log10 cells/g of feces vs. 5.7 log10 cells/g of feces, six months samples, p = 0.043). In addition, the probability to detect B. longum subsp. longum at 6 months was reduced by 7.4 times in case of antibiotic treatment (six months samples, p = 0.007).
The reduced colonization by bifidobacteria and staphylococci after antibiotic treatment is in line with previous findings [70–72]. We could not confirm the increase of Enterococcus or Enterobacteria following antibiotic treatment previously reported by others [54, 72], most probably due to the smaller sample size in our study.
Other factors
Besides mode of delivery, mode of feeding, and antibiotic use, other factors associated with early exposure to microbes, such as farm exposure, place of birth, presence of siblings or pets are suggested to influence the early colonization process. For example, it has been previously described that infants born at hospital are more often colonized by C. difficile [13, 73]. In this study only 4.6% infants were born at home, and even though our results suggest that place of birth influences the colonization pattern of B. breve (higher probability to be detected when born at home; longitudinal model, p = 0.024), this should be further confirmed in larger studies.
Early exposure to microbes is suggested to decrease the risk of allergies later in life; and these associations are commonly attributed as “hygiene hypothesis” [74–76]. Although the mechanism of such a protective effect remains to be unraveled; it seems that contact with bacteria in early life plays a pivotal role in triggering the immune response and tolerance induction [14, 77]. Up to now, little evidence is available showing how the contact with a less sterile environment in early life, such as living in a rural environment, keeping indoor pets or having older siblings, differentially impact the gut microbiota composition.
A) Presence of older siblings or household pets
Exposure to pets has been associated with an over-representation of Clostridium species, Veillonella, Peptostreptococcaceace and Coprococcus and an under-representation of Bifidobacterium, or specifically B. breve, in the infant gut [78]. However, in our study a negative association was found only between exposure to pets and the colonization by the L. reuteri subgroup. The probability to detect L. reuteri subgroup was significantly higher in those infants not exposed to households pets (N = 90) (longitudinal model, p = 0.006) and those that were firstborns (N = 56) (longitudinal model, p<0.001). The latest was still significant at 6 months (six months samples, p = 0.013) (35.85% present in firstborns vs 6.15% if there were older siblings in the family as represented by the original data in S5 Fig). Our result suggests that colonization by bacteria belonging to L. reuteri subgroup is favored by a more sterile environment and exposure to formula feeding as discussed before, in which the exposure to microbes is limited. However, the relevance of this finding deserves further confirmation since the association between colonization by L. reuteri and increased allergy risk has not been established before, and in fact supplementation by a specific L. reuteri strain seems to increase the immune-regulatory capacity during infancy [79].
Besides the results on the L. reuteri subgroup described above, more correlations were found between prevalence of specific bacteria and the presence/absence of older siblings. Firstborn infants were 2.5 times more often colonized by B. dentium than those having older siblings (longitudinal model, p = 0.035), which was still significant at 6 months (six months samples, p = 0.003). On the contrary, infants with older siblings were 9 times more often colonized with B. catenulatum group (longitudinal model, p = 0.015) during the first 3 months of life, and the difference was still significant at 6 months (six months samples, p = 0.004) (Original data represented in S6 Fig).
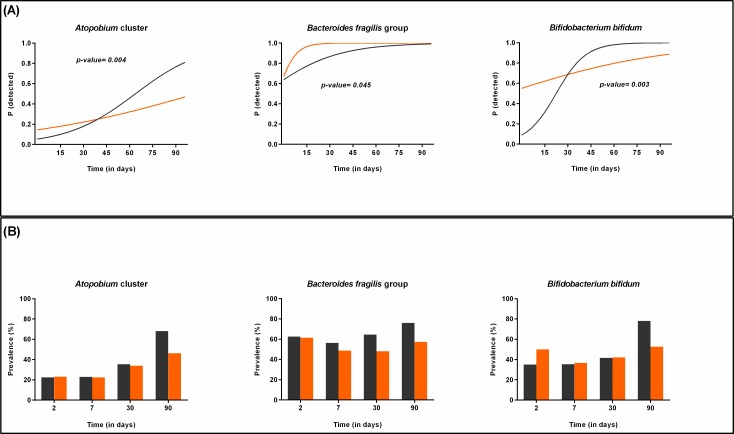
Infants having older siblings were more often colonized by B. fragilis group (longitudinal model, p = 0.045) during the first 3 months of life and by B. fragilis subgroup at 6 months (six months samples, p = 0.041). In addition, the probability to detect Atopobium and B. bifidum in those infants, increased faster throughout the first three months of life when compared to firstborns (longitudinal model, p = 0.004 and p = 0.003, respectively) (Fig 9A). This difference was still significant for B. bifidum at 6 months of age (six months samples, p = 0.005) (84% vs 67.31% detected in firstborns as represented by the original data).
Fig 9. Bacterial groups or species influenced by the presence/absence of older siblings (I).
(A) Probability to detect Atopobium cluster, B. fragilis group and B. bifidum in firstborn infants (orange) compared to those having older siblings (grey) during the first 3 months of life as calculated by the statistical model. (B) Prevalence of Atopobium cluster, B. fragilis group and B. bifidum in firstborn infants (orange) compared to those having older siblings (grey) during the first 3 months of life as represented by the original data.
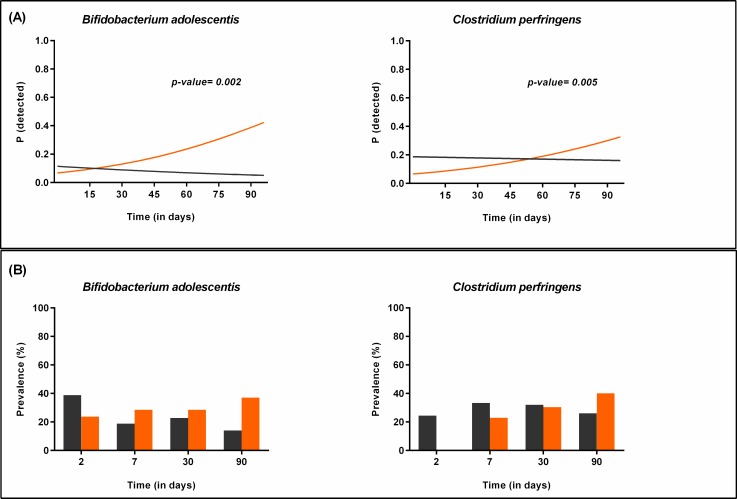
On the other hand, the probability to detect B. adolescentis group and C. perfringens increased faster in firstborn infants during the first three months of life, when compared to those infants having older siblings (longitudinal model, p = 0.002 and p = 0.005, respectively; Fig 10A).
Fig 10. Bacterial groups or species influenced by the presence/absence of older siblings (II).
(A) Probability to detect B. adolescentis and C. perfringens in firstborn infants (orange) compared to those having older siblings (grey) during the first 3 months of life as calculated by the statistical model. (B) Prevalence of B. adolescentis and C. perfringens in firstborn infants (orange) compared to those having older siblings (grey) during the first 3 months of life as represented by the original data.
Several studies have reported that sibling number is associated with microbiota composition; first born infants are thought to be most often colonized by enterobacteria and clostridia, whereas the colonization by bifidobacteria, Bacteroides and Lactobacillus increase with an increasing number of siblings [13, 57, 80]. A positive correlation between numbers of older siblings and richness and bacterial diversity within the phyla of Firmicutes and Bacteroidetes at 18 months of age has been described recently [81].
Although we have not confirmed all previous findings, our results show that sibling number influences the microbiota composition at 6 months of age and describes differences at the species level. Whereas a reduced colonization by bifidobacteria has been reported before in first born infants [13, 57], we have shown that this is true for B. bifidum and B. catenulatum group, but not for B. dentium or B. adolescentis group, which were more often detected in first-born infants. Similarly, the previously reported reduced colonization by Lactobacillus in firstborn infants, seems not to be true for L. reuteri subgroup, which was most often detected in firstborn infants than in infants with older siblings even at 6 months of age. To our knowledge, this is the first time that Atopobium cluster has been shown to be dependent on number of siblings.
Our results provide new supporting evidence to the hygiene hypothesis, since the exposure to a less sterile environment (number of siblings) seems to favor the colonization by the type of bacteria that are reported to be absent in allergic infants in early life (B. fragilis, B. bifidum and B. adolescentis), suggesting their role in the development of the immune system [82–85].
C. Gender
We included 56 boys (51.9%) and 52 girls (48.1%) in this study. At birth, boys had higher total bacterial count (as measured by DAPI) than girls (first defecation samples, p = 0.029). Girls were six times more frequently colonized by L. ruminis subgroup (first defecation samples, p = 0.003) at birth, 4 times by L. gasseri subgroup (longitudinal model, p<0.001) and 3 times by L. reuteri subgroup (longitudinal model, p<0.001) from 2 days to 3 months after birth.
To our knowledge, little is known about the influence of gender on the microbiota composition in early life. So far, only one study has reported gender differences in adults, showing a larger colonization by Bacteroides in males than in females [86]. Although puberty is thought to influence the microbiota composition due to hormonal changes, data supporting this idea is lacking in humans. Recently, it was shown through animal models that the gut microbiota of mice differs in males and females after puberty and that this trend is reversed by male castration [87–89]. Results from these animal models might point toward the difference in microbiota composition between male and female, as an explanation for a predisposition to certain immune diseases that are known to be gender-dependent (ie., diabetes type 1 in females or autism in males) [90].
Interestingly, we have found that Lactobacillus, a dominant genus in the vaginal microbiota and known to be regulated by female estrogens [91, 92], colonizes the gut of girls more often than boys in early life. Further research is needed to confirm this finding and elucidate whether microbiota and hormones can work together to influence gender bias in different diseases risk, and more importantly, whether changes in the gut microbiota in early life could modulate this susceptibility.
Conclusions
To our knowledge this is one of the few studies using a statistical model to elucidate the complex process of early life colonization connecting longitudinal data and the different factors influencing this dynamic process. However, whereas currently 16S sequencing is often used to study the composition and diversity of complex microbial ecosystems, we chose to use a qPCR targeted approach, which allowed us to follow quantitatively the colonization of key taxa at species level.
This study contributes to understand not only the composition of the microbiota in early life, but also the succession process and the evolution of the microbial community as a function of time and important events or ecological drivers occurring the first 6 months of life.
Our results confirm that mode of delivery and type of feeding are important factors influencing this process, providing more detailed information about the timing of this influence and a deeper level of bacterial identification beyond genus. Additionally, this study reveals that other factors, which may have not yet been sufficiently studied or overlooked in the past; including presence of siblings, pets, and gender also influence this process. Even though the difference may be viewed as subtle, we need to further understand the implications of the differential effects observed and the association with potential health outcomes in the future. As expected, known early colonizers such as Bifidobacterium and/or specific Bifidobacterium species, are shown to be greatly influenced by most factors investigated in our study. Surprisingly, some of the known infant type bifidobacterial species, such B. breve and B. longum subsp. infantis were confirmed in this study to be early colonizers but were hardly dependent on the covariates studied. On the contrary, different Lactobacillus species are shown to be influenced by several covariates even though they are not normally dominant in the infant gut. In addition, our results also point out that Bacteroides is a key bacterial genus in the early colonization process, which is influenced by most factors included in the model to a larger extend than Bifidobacterium, which may indicate the need to consider strains belonging to this genus as potential probiotic interventions in infant nutrition in the future.
Our results also suggest that there is at least a certain degree of bacterial exposure in utero since 77.08% of the meconium samples contained bacterial DNA.
One needs to keep in mind that these results are generated using data driven covariate selection and therefore further studies are warranted to confirm these findings.
In summary, this study provides new insights that need to be taken into consideration when selecting the most relevant nutritional supplementation strategies to establish the microbiota composition in early life and potentially influence health later in life. According to our results these strategies may need to differ depending on the timing of supplementation, the environment in which the infant lives and maybe even whether it is a boy or a girl, which seems to point out to the need to develop personalized infant nutrition in the future.
Supporting Information
Prevalence of different bacterial groups significantly dependent on the type of feeding; at 2,7, 30 and 90 days of life. Graph representing original data in exclusive breastfed (green) or mix-fed infants (black).
(TIF)
(A) Prevalence of L. casei subgroup and B. adolescentis at 6 months in infants that were exclusive breastfed for the first 3 months of life (green) or exposed to mix-feeding (black) as represented by the original data. (B) Prevalence of different organic acids at 6 months in infants that were exclusive breastfed for the first 3 months of life (black) or exposed to mix-feeding (grey) as represented by the original data.
(TIF)
(A) Prevalence of L. gasseri subgroup from 3 to 6 months in infants that were exclusive breastfed for the first 3 months of life (green) or exposed to mix-feeding (black) as calculated by the statistical model. (B) Prevalence of L. gasseri subgroup at 3 and 6 months in infants that were exclusive breastfed for the first 3 months of life (green) or exposed to mix-feeding (black) as represented by the original data.
(TIF)
Effect of the introduction of solid foods on the prevalence of C.coccoides group and Atopobium cluster in the fecal sample taken at 6 months of age.
(TIF)
(A) Prevalence of L. reuteri subgroup in infants not exposed to households’ pets (orange) or having pets at home (grey) over time. (B).Prevalence of L. reuteri subgroup in first-born infants (orange) and those having older siblings (grey) over time.
(TIF)
Prevalence of L. reuteri subgroup, B. dentium and B. catenulatum group in firstborn infants (orange) and those having siblings (grey) over time,
(TIF)
(PDF)
Acknowledgments
We thank to all midwives involved in subject recruitment and families who kindly participate in this study
Data Availability
All relevant data are within the paper and its Supporting Information files. In case of need, data are from the Neonates study and authors may be contacted at rocio.martin@danone.com.
Funding Statement
The study was funded by Yakult and Nutricia Research. The following authors are paid by Nutricia Research: RM, KBA, MR, SS, ACY, JK, whereas HM, HK, TS, EI, KO, AK are paid by Yakult. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–20. [DOI] [PubMed] [Google Scholar]
- 2.Blaut M, Clavel T. Metabolic diversity of the intestinal microbiota: implications for health and disease. The Journal of nutrition. 2007;137(3 Suppl 2):751S–5S. [DOI] [PubMed] [Google Scholar]
- 3.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: health and disease. Frontiers in immunology. 2014;5:427 Epub 2014/09/25. 10.3389/fimmu.2014.00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wopereis H, Oozeer R, Knipping K, Belzer C, Knol J. The first thousand days—intestinal microbiology of early life: establishing a symbiosis. Pediatric allergy and immunology: official publication of the European Society of Pediatric Allergy and Immunology. 2014;25(5):428–38. Epub 2014/06/06. [DOI] [PubMed] [Google Scholar]
- 6.Avershina E, Storro O, Oien T, Johnsen R, Pope P, Rudi K. Major faecal microbiota shifts in composition and diversity with age in a geographically restricted cohort of mothers and their children. FEMS microbiology ecology. 2014;87(1):280–90. Epub 2013/10/12. 10.1111/1574-6941.12223 [DOI] [PubMed] [Google Scholar]
- 7.Munyaka PM, Khafipour E, Ghia JE. External influence of early childhood establishment of gut microbiota and subsequent health implications. Frontiers in pediatrics. 2014;2:109 Epub 2014/10/28. 10.3389/fped.2014.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jimenez E, Marin ML, Martin R, Odriozola JM, Olivares M, Xaus J, et al. Is meconium from healthy newborns actually sterile? Research in microbiology. 2008;159(3):187–93. Epub 2008/02/19. 10.1016/j.resmic.2007.12.007 [DOI] [PubMed] [Google Scholar]
- 9.Satokari R, Gronroos T, Laitinen K, Salminen S, Isolauri E. Bifidobacterium and Lactobacillus DNA in the human placenta. Letters in applied microbiology. 2009;48(1):8–12. Epub 2008/11/21. 10.1111/j.1472-765X.2008.02475.x [DOI] [PubMed] [Google Scholar]
- 10.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Science translational medicine. 2014;6(237):237ra65 Epub 2014/05/23. 10.1126/scitranslmed.3008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jimenez E, Fernandez L, Marin ML, Martin R, Odriozola JM, Nueno-Palop C, et al. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Current microbiology. 2005;51(4):270–4. Epub 2005/09/28. [DOI] [PubMed] [Google Scholar]
- 12.Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ: Canadian Medical Association journal = journal de l'Association medicale canadienne. 2013;185(5):385–94. Epub 2013/02/13. 10.1503/cmaj.121189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penders J, Gerhold K, Stobberingh EE, Thijs C, Zimmermann K, Lau S, et al. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. The Journal of allergy and clinical immunology. 2013;132(3):601–7 e8. Epub 2013/08/01. 10.1016/j.jaci.2013.05.043 [DOI] [PubMed] [Google Scholar]
- 14.Penders J, Gerhold K, Thijs C, Zimmermann K, Wahn U, Lau S, et al. New insights into the hygiene hypothesis in allergic diseases: mediation of sibling and birth mode effects by the gut microbiota. Gut microbes. 2014;5(2):239–44. Epub 2014/03/19. 10.4161/gmic.27905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host & Microbe. 2015;17(6):852. [DOI] [PubMed] [Google Scholar]
- 16.Laursen M, Zachariassen G, Bahl M, Bergström A, Høst A, Michaelsen K, et al. Having older siblings is associated with gut microbiota development during early childhood. BMC Microbiol. 2015;August(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nermes M, Endo A, Aarnio J, Salminen S, Isolauri E. Furry pets modulate gut microbiota composition in infants at risk for allergic disease. Journal of Allergy and Clinical Immunology. [DOI] [PubMed] [Google Scholar]
- 18.Makino H, Kushiro A, Ishikawa E, Muylaert D, Kubota H, Sakai T, et al. Transmission of intestinal Bifidobacterium longum subsp. longum strains from mother to infant, determined by multilocus sequencing typing and amplified fragment length polymorphism. Applied and environmental microbiology. 2011;77(19):6788–93. 10.1128/AEM.05346-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makino H, Kushiro A, Ishikawa E, Kubota H, Gawad A, Sakai T, et al. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant's microbiota. PloS one. 2013;8(11):e78331 Epub 2013/11/19. 10.1371/journal.pone.0078331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, Ferrario C, et al. Exploring Vertical Transmission of Bifidobacteria from Mother to Child. Applied and environmental microbiology. 2015;81(20):7078–87. Epub 2015/08/02. 10.1128/AEM.02037-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biasucci G, Benenati B, Morelli L, Bessi E, Boehm G. Cesarean delivery may affect the early biodiversity of intestinal bacteria. The Journal of nutrition. 2008;138(9):1796S–800S. Epub 2008/08/22. [DOI] [PubMed] [Google Scholar]
- 22.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proceedings of the National Academy of Sciences of the United States of America. 2011;108 Suppl 1:4578–85. Epub 2010/07/30. 10.1073/pnas.1000081107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–7. Epub 2012/06/16. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agans R, Rigsbee L, Kenche H, Michail S, Khamis HJ, Paliy O. Distal gut microbiota of adolescent children is different from that of adults. FEMS microbiology ecology. 2011;77(2):404–12. Epub 2011/05/05. 10.1111/j.1574-6941.2011.01120.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollister EB, Riehle K, Luna RA, Weidler EM, Rubio-Gonzales M, Mistretta TA, et al. Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome. 2015;3:36 Epub 2015/08/27. 10.1186/s40168-015-0101-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darmasseelane K, Hyde MJ, Santhakumaran S, Gale C, Modi N. Mode of delivery and offspring body mass index, overweight and obesity in adult life: a systematic review and meta-analysis. PloS one. 2014;9(2):e87896 Epub 2014/03/04. 10.1371/journal.pone.0087896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fooladi AA, Khani S, Hosseini HM, Mousavi SF, Aghdam EM, Nourani MR. Impact of altered early infant gut microbiota following breastfeeding and delivery mode on allergic diseases. Inflammation & allergy drug targets. 2013;12(6):410–8. Epub 2013/12/07. [DOI] [PubMed] [Google Scholar]
- 28.Mueller NT, Whyatt R, Hoepner L, Oberfield S, Dominguez-Bello MG, Widen EM, et al. Prenatal exposure to antibiotics, cesarean section and risk of childhood obesity. Int J Obes (Lond). 2015;39(4):665–70. Epub 2014/10/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sevelsted A, Stokholm J, Bonnelykke K, Bisgaard H. Cesarean section and chronic immune disorders. Pediatrics. 2015;135(1):e92–8. Epub 2014/12/03. 10.1542/peds.2014-0596 [DOI] [PubMed] [Google Scholar]
- 30.Storro O, Avershina E, Rudi K. Diversity of intestinal microbiota in infancy and the risk of allergic disease in childhood. Current opinion in allergy and clinical immunology. 2013;13(3):257–62. Epub 2013/04/04. 10.1097/ACI.0b013e328360968b [DOI] [PubMed] [Google Scholar]
- 31.Kostic AD, Gevers D, Siljander H, Vatanen T, Hyotylainen T, Hamalainen AM, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17(2):260–73. Epub 2015/02/11. 10.1016/j.chom.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuda K, Tsuji H, Asahara T, Matsumoto K, Takada T, Nomoto K. Establishment of an analytical system for the human fecal microbiota, based on reverse transcription-quantitative PCR targeting of multicopy rRNA molecules. Applied and environmental microbiology. 2009;75(7):1961–9. 10.1128/AEM.01843-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuki T, Watanabe K, Fujimoto J, Kado Y, Takada T, Matsumoto K, et al. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Applied and environmental microbiology. 2004;70(1):167–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuki T, Watanabe K, Fujimoto J, Miyamoto Y, Takada T, Matsumoto K, et al. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Applied and environmental microbiology. 2002;68(11):5445–51. Epub 2002/10/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuki T, Watanabe K, Fujimoto J, Takada T, Tanaka R. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Applied and environmental microbiology. 2004;70(12):7220–8. Epub 2004/12/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takaishi H, Matsuki T, Nakazawa A, Takada T, Kado S, Asahara T, et al. Imbalance in intestinal microflora constitution could be involved in the pathogenesis of inflammatory bowel disease. International journal of medical microbiology: IJMM. 2008;298(5–6):463–72. Epub 2007/09/28. [DOI] [PubMed] [Google Scholar]
- 37.Ventura M, Reniero R, Zink R. Specific identification and targeted characterization of Bifidobacterium lactis from different environmental isolates by a combined multiplex-PCR approach. Applied and environmental microbiology. 2001;67(6):2760–5. Epub 2001/05/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuki T, Watanabe K, Tanaka R, Oyaizu H. Rapid identification of human intestinal bifidobacteria by 16S rRNA-targeted species- and group-specific primers. FEMS microbiology letters. 1998;167(2):113–21. Epub 1998/11/11. [DOI] [PubMed] [Google Scholar]
- 39.Matsuki T, Watanabe K, Tanaka R, Fukuda M, Oyaizu H. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Applied and environmental microbiology. 1999;65(10):4506–12. Epub 1999/10/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kikuchi E, Miyamoto Y, Narushima S, Itoh K. Design of species-specific primers to identify 13 species of Clostridium harbored in human intestinal tracts. Microbiology and immunology. 2002;46(5):353–8. Epub 2002/07/26. [DOI] [PubMed] [Google Scholar]
- 41.Matsuda K, Tsuji H, Asahara T, Kado Y, Nomoto K. Sensitive quantitative detection of commensal bacteria by rRNA-targeted reverse transcription-PCR. Applied and environmental microbiology. 2007;73(1):32–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kubota H, Tsuji H, Matsuda K, Kurakawa T, Asahara T, Nomoto K. Detection of human intestinal catalase-negative, Gram-positive cocci by rRNA-targeted reverse transcription-PCR. Applied and environmental microbiology. 2010;76(16):5440–51. Epub 2010/06/29. 10.1128/AEM.03132-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ririe KM, Rasmussen RP, Wittwer CT. Product Differentiation by Analysis of DNA Melting Curves during the Polymerase Chain Reaction. Analytical Biochemistry. 1997;245(2):154–60. [DOI] [PubMed] [Google Scholar]
- 44.Chonan O, Matsumoto K, Watanuki M. Effect of galactooligosaccharides on calcium absorption and preventing bone loss in ovariectomized rats. Bioscience, biotechnology, and biochemistry. 1995;59(2):236–9. Epub 1995/02/01. [DOI] [PubMed] [Google Scholar]
- 45.Lubin JH, Colt JS, Camann D, Davis S, Cerhan JR, Severson RK, et al. Epidemiologic evaluation of measurement data in the presence of detection limits. Environmental health perspectives. 2004;112(17):1691–6. Epub 2004/12/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lambert P, Eilers PH. Bayesian proportional hazards model with time-varying regression coefficients: a penalized Poisson regression approach. Statistics in medicine. 2005;24(24):3977–89. Epub 2005/12/02. [DOI] [PubMed] [Google Scholar]
- 47.Hansen R, Scott KP, Khan S, Martin JC, Berry SH, Stevenson M, et al. First-Pass Meconium Samples from Healthy Term Vaginally-Delivered Neonates: An Analysis of the Microbiota. PloS one. 2015;10(7):e0133320 Epub 2015/07/29. 10.1371/journal.pone.0133320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huh SY, Rifas-Shiman SL, Zera CA, Edwards JW, Oken E, Weiss ST, et al. Delivery by caesarean section and risk of obesity in preschool age children: a prospective cohort study. Archives of disease in childhood. 2012;97(7):610–6. Epub 2012/05/25. 10.1136/archdischild-2011-301141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li HT, Zhou YB, Liu JM. Cesarean section might moderately increase offspring obesity risk. The American journal of clinical nutrition. 2012;96(1):215–6; author reply 6. Epub 2012/06/22. 10.3945/ajcn.112.038760 [DOI] [PubMed] [Google Scholar]
- 50.Bager P, Wohlfahrt J, Westergaard T. Caesarean delivery and risk of atopy and allergic disease: meta-analyses. Clinical and experimental allergy: journal of the British Society for Allergy and Clinical Immunology. 2008;38(4):634–42. Epub 2008/02/13. [DOI] [PubMed] [Google Scholar]
- 51.Thavagnanam S, Fleming J, Bromley A, Shields MD, Cardwell CR. A meta-analysis of the association between Caesarean section and childhood asthma. Clinical and experimental allergy: journal of the British Society for Allergy and Clinical Immunology. 2008;38(4):629–33. Epub 2008/03/21. [DOI] [PubMed] [Google Scholar]
- 52.Salminen S, Gibson GR, McCartney AL, Isolauri E. Influence of mode of delivery on gut microbiota composition in seven year old children. Gut. 2004;53(9):1388–9. Epub 2004/08/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(26):11971–5. Epub 2010/06/23. 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R, et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. Journal of pediatric gastroenterology and nutrition. 2010;51(1):77–84. Epub 2010/05/19. 10.1097/MPG.0b013e3181d1b11e [DOI] [PubMed] [Google Scholar]
- 55.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014;63(4):559–66. Epub 2013/08/09. 10.1136/gutjnl-2012-303249 [DOI] [PubMed] [Google Scholar]
- 56.Tsuji H, Oozeer R, Matsuda K, Matsuki T, Ohta T, Nomoto K, et al. Molecular monitoring of the development of intestinal microbiota in Japanese infants. Beneficial microbes. 2012;3(2):113–25. Epub 2012/06/12. 10.3920/BM2011.0038 [DOI] [PubMed] [Google Scholar]
- 57.Yap GC, Chee KK, Hong PY, Lay C, Satria CD, Sumadiono, et al. Evaluation of stool microbiota signatures in two cohorts of Asian (Singapore and Indonesia) newborns at risk of atopy. BMC Microbiol. 2011;11:193 Epub 2011/08/31. 10.1186/1471-2180-11-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rowe-Murray HJ, Fisher JR. Baby friendly hospital practices: cesarean section is a persistent barrier to early initiation of breastfeeding. Birth. 2002;29(2):124–31. Epub 2002/05/10. [DOI] [PubMed] [Google Scholar]
- 59.Zanardo V, Svegliado G, Cavallin F, Giustardi A, Cosmi E, Litta P, et al. Elective cesarean delivery: does it have a negative effect on breastfeeding? Birth. 2010;37(4):275–9. Epub 2010/11/19. 10.1111/j.1523-536X.2010.00421.x [DOI] [PubMed] [Google Scholar]
- 60.Martin R, Jimenez E, Heilig H, Fernandez L, Marin ML, Zoetendal EG, et al. Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Applied and environmental microbiology. 2009;75(4):965–9. Epub 2008/12/18. 10.1128/AEM.02063-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin R, Langa S, Reviriego C, Jiminez E, Marin ML, Xaus J, et al. Human milk is a source of lactic acid bacteria for the infant gut. The Journal of pediatrics. 2003;143(6):754–8. Epub 2003/12/06. [DOI] [PubMed] [Google Scholar]
- 62.Jimenez E, Delgado S, Maldonado A, Arroyo R, Albujar M, Garcia N, et al. Staphylococcus epidermidis: a differential trait of the fecal microbiota of breast-fed infants. BMC Microbiol. 2008;8:143 Epub 2008/09/12. 10.1186/1471-2180-8-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fan W, Huo G, Li X, Yang L, Duan C. Impact of diet in shaping gut microbiota revealed by a comparative study in infants during the six months of life. Journal of microbiology and biotechnology. 2014;24(2):133–43. Epub 2013/10/31. [DOI] [PubMed] [Google Scholar]
- 64.Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. Journal of pediatric gastroenterology and nutrition. 2000;30(1):61–7. Epub 2000/01/12. [DOI] [PubMed] [Google Scholar]
- 65.Lee SA, Lim JY, Kim BS, Cho SJ, Kim NY, Kim OB, et al. Comparison of the gut microbiota profile in breast-fed and formula-fed Korean infants using pyrosequencing. Nutrition research and practice. 2015;9(3):242–8. Epub 2015/06/11. 10.4162/nrp.2015.9.3.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology. 2010;156(Pt 11):3329–41. Epub 2010/09/25. 10.1099/mic.0.043224-0 [DOI] [PubMed] [Google Scholar]
- 67.Bergstrom A, Skov TH, Bahl MI, Roager HM, Christensen LB, Ejlerskov KT, et al. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Applied and environmental microbiology. 2014;80(9):2889–900. Epub 2014/03/04. 10.1128/AEM.00342-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fallani M, Amarri S, Uusijarvi A, Adam R, Khanna S, Aguilera M, et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology. 2011;157(Pt 5):1385–92. Epub 2011/02/19. 10.1099/mic.0.042143-0 [DOI] [PubMed] [Google Scholar]
- 69.Thompson AL, Monteagudo-Mera A, Cadenas MB, Lampl ML, Azcarate-Peril MA. Milk- and solid-feeding practices and daycare attendance are associated with differences in bacterial diversity, predominant communities, and metabolic and immune function of the infant gut microbiome. Frontiers in cellular and infection microbiology. 2015;5:3 Epub 2015/02/24. 10.3389/fcimb.2015.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrobial agents and chemotherapy. 2012;56(11):5811–20. Epub 2012/09/06. 10.1128/AAC.00789-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lindberg E, Adlerberth I, Matricardi P, Bonanno C, Tripodi S, Panetta V, et al. Effect of lifestyle factors on Staphylococcus aureus gut colonization in Swedish and Italian infants. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2011;17(8):1209–15. Epub 2010/11/16. [DOI] [PubMed] [Google Scholar]
- 72.Tanaka S, Kobayashi T, Songjinda P, Tateyama A, Tsubouchi M, Kiyohara C, et al. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS immunology and medical microbiology. 2009;56(1):80–7. Epub 2009/04/24. 10.1111/j.1574-695X.2009.00553.x [DOI] [PubMed] [Google Scholar]
- 73.van Nimwegen FA, Penders J, Stobberingh EE, Postma DS, Koppelman GH, Kerkhof M, et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. The Journal of allergy and clinical immunology. 2011;128(5):948–55 e1-3. Epub 2011/08/30. 10.1016/j.jaci.2011.07.027 [DOI] [PubMed] [Google Scholar]
- 74.Strachan DP . Family size, infection and atopy: the first decade of the "hygiene hypothesis". Thorax. 2000;55 Suppl 1:S2–10. Epub 2000/08/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karmaus W, Botezan C. Does a higher number of siblings protect against the development of allergy and asthma? A review. Journal of epidemiology and community health. 2002;56(3):209–17. Epub 2002/02/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.von Mutius E, Vercelli D. Farm living: effects on childhood asthma and allergy. Nature reviews Immunology. 2010;10(12):861–8. Epub 2010/11/10. 10.1038/nri2871 [DOI] [PubMed] [Google Scholar]
- 77.Nermes M, Niinivirta K, Nylund L, Laitinen K, Matomaki J, Salminen S, et al. Perinatal pet exposure, faecal microbiota, and wheezy bronchitis: is there a connection? ISRN allergy. 2013;2013:827934 Epub 2013/06/01. 10.1155/2013/827934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Sears MR, et al. Infant gut microbiota and the hygiene hypothesis of allergic disease: impact of household pets and siblings on microbiota composition and diversity. Allergy, asthma, and clinical immunology: official journal of the Canadian Society of Allergy and Clinical Immunology. 2013;9(1):15. Epub 2013/04/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Forsberg A, Abrahamsson TR, Bjorksten B, Jenmalm MC. Pre- and post-natal Lactobacillus reuteri supplementation decreases allergen responsiveness in infancy. Clinical and experimental allergy: journal of the British Society for Allergy and Clinical Immunology. 2013;43(4):434–42. Epub 2013/03/23. [DOI] [PubMed] [Google Scholar]
- 80.Adlerberth I, Strachan DP, Matricardi PM, Ahrne S, Orfei L, Aberg N, et al. Gut microbiota and development of atopic eczema in 3 European birth cohorts. The Journal of allergy and clinical immunology. 2007;120(2):343–50. Epub 2007/07/03. [DOI] [PubMed] [Google Scholar]
- 81.Laursen MF, Zachariassen G, Bahl MI, Bergstrom A, Host A, Michaelsen KF, et al. Having older siblings is associated with gut microbiota development during early childhood. BMC Microbiol. 2015;15:154 Epub 2015/08/02. 10.1186/s12866-015-0477-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Suzuki S, Shimojo N, Tajiri Y, Kumemura M, Kohno Y. Differences in the composition of intestinal Bifidobacterium species and the development of allergic diseases in infants in rural Japan. Clinical and experimental allergy: journal of the British Society for Allergy and Clinical Immunology. 2007;37(4):506–11. Epub 2007/04/14. [DOI] [PubMed] [Google Scholar]
- 83.Ouwehand AC, Isolauri E, He F, Hashimoto H, Benno Y, Salminen S. Differences in Bifidobacterium flora composition in allergic and healthy infants. The Journal of allergy and clinical immunology. 2001;108(1):144–5. Epub 2001/07/12. [DOI] [PubMed] [Google Scholar]
- 84.Lahtinen SJ, Boyle RJ, Kivivuori S, Oppedisano F, Smith KR, Robins-Browne R, et al. Prenatal probiotic administration can influence Bifidobacterium microbiota development in infants at high risk of allergy. The Journal of allergy and clinical immunology. 2009;123(2):499–501. Epub 2009/01/13. 10.1016/j.jaci.2008.11.034 [DOI] [PubMed] [Google Scholar]
- 85.Johansson MA, Sjogren YM, Persson JO, Nilsson C, Sverremark-Ekstrom E. Early colonization with a group of Lactobacilli decreases the risk for allergy at five years of age despite allergic heredity. PloS one. 2011;6(8):e23031 Epub 2011/08/11. 10.1371/journal.pone.0023031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Applied and environmental microbiology. 2006;72(2):1027–33. Epub 2006/02/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339(6123):1084–8. Epub 2013/01/19. 10.1126/science.1233521 [DOI] [PubMed] [Google Scholar]
- 88.Markle JG, Frank DN, Adeli K, von Bergen M, Danska JS. Microbiome manipulation modifies sex-specific risk for autoimmunity. Gut microbes. 2014;5(4):485–93. Epub 2014/07/10. 10.4161/gmic.29795 [DOI] [PubMed] [Google Scholar]
- 89.Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39(2):400–12. Epub 2013/08/27. 10.1016/j.immuni.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Nikita L, et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome. 2014;2(1):4 Epub 2014/02/04. 10.1186/2049-2618-2-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Bieda J, et al. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome. 2014;2:18 Epub 2014/07/06. 10.1186/2049-2618-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pelzer ES, Allan JA, Theodoropoulos C, Ross T, Beagley KW, Knox CL. Hormone-dependent bacterial growth, persistence and biofilm formation—a pilot study investigating human follicular fluid collected during IVF cycles. PloS one. 2012;7(12):e49965 Epub 2012/12/12. 10.1371/journal.pone.0049965 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Prevalence of different bacterial groups significantly dependent on the type of feeding; at 2,7, 30 and 90 days of life. Graph representing original data in exclusive breastfed (green) or mix-fed infants (black).
(TIF)
(A) Prevalence of L. casei subgroup and B. adolescentis at 6 months in infants that were exclusive breastfed for the first 3 months of life (green) or exposed to mix-feeding (black) as represented by the original data. (B) Prevalence of different organic acids at 6 months in infants that were exclusive breastfed for the first 3 months of life (black) or exposed to mix-feeding (grey) as represented by the original data.
(TIF)
(A) Prevalence of L. gasseri subgroup from 3 to 6 months in infants that were exclusive breastfed for the first 3 months of life (green) or exposed to mix-feeding (black) as calculated by the statistical model. (B) Prevalence of L. gasseri subgroup at 3 and 6 months in infants that were exclusive breastfed for the first 3 months of life (green) or exposed to mix-feeding (black) as represented by the original data.
(TIF)
Effect of the introduction of solid foods on the prevalence of C.coccoides group and Atopobium cluster in the fecal sample taken at 6 months of age.
(TIF)
(A) Prevalence of L. reuteri subgroup in infants not exposed to households’ pets (orange) or having pets at home (grey) over time. (B).Prevalence of L. reuteri subgroup in first-born infants (orange) and those having older siblings (grey) over time.
(TIF)
Prevalence of L. reuteri subgroup, B. dentium and B. catenulatum group in firstborn infants (orange) and those having siblings (grey) over time,
(TIF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. In case of need, data are from the Neonates study and authors may be contacted at rocio.martin@danone.com.