Abstract
The release of Wolbachia infected mosquitoes is likely to form a key component of disease control strategies in the near future. We investigated the potential of using near-infrared spectroscopy (NIRS) to simultaneously detect and identify two strains of Wolbachia pipientis (wMelPop and wMel) in male and female laboratory-reared Aedes aegypti mosquitoes. Our aim is to find faster, cheaper alternatives for monitoring those releases than the molecular diagnostic techniques that are currently in use. Our findings indicate that NIRS can differentiate females and males infected with wMelPop from uninfected wild type samples with an accuracy of 96% (N = 299) and 87.5% (N = 377), respectively. Similarly, females and males infected with wMel were differentiated from uninfected wild type samples with accuracies of 92% (N = 352) and 89% (N = 444). NIRS could differentiate wMelPop and wMel transinfected females with an accuracy of 96.6% (N = 442) and males with an accuracy of 84.5% (N = 443). This non-destructive technique is faster than the standard polymerase chain reaction diagnostic techniques. After the purchase of a NIRS spectrometer, the technique requires little sample processing and does not consume any reagents.
Author Summary
Near infrared spectroscopy (NIRS) is a technique that measures specific frequencies of light absorbed by C-H, O-H, S-H and N-H functional groups. Mosquito samples are grouped based upon absorption differences between their chemical properties. In this study, we used NIRS to differentiate 1) Aedes aegypti infected with either of the two strains of intracellular bacterium Wolbachia (wMel and wMelPop) from wild type Ae. aegypti and 2) Aedes aegypti infected with wMel from those infected with wMelPoP. NIRS facilitated the differentiation of wMel and wMelPop from wild type samples and samples infected with either of the Wolbachia infected strains with high prediction accuracies over their lifespan. Predictive models were derived from initial calibration data sets and validated against independent cohorts. Prediction accuracies were high (82–98%) regardless of the cohort mosquitoes were sampled from. The results show that NIRS may have real potential as an alternative method for monitoring Wolbachia incidence in mosquitoes. A rapid, simple and cost-effective surveillance tool suitable for resource-poor areas and large urban release programs would be of great utility for evaluating Wolbachia-based interventions. The models developed during this study require further validation using field collections.
Introduction
The mosquito Aedes aegypti is the primary vector of the four human dengue, chikungunya and Zika viruses. Options for developing effective vaccines or chemotherapeutics are limited [1] and vector control remains fundamental for the prevention of disease transmission. One of the most promising strategies exploits the maternally-transmitted intracellular bacteria, Wolbachia pipientis. Wolbachia are naturally found in up to 60% of all insect species and are propagated through insect populations via reproductive manipulations. In some insect species such as mosquitoes [2,3], tsetse flies[4] and Liriomyza trifolii [5] a phenomenon called cytoplasmic incompatibility (CI) mediated by Wolbachia, modifies the sperm of infected males such that crosses with uninfected females do not produce viable offsprings. The effect is rescued in crosses with infected females, leading to a reproductive advantage that favors Wolbachia transmission through the population. CI and phenotypes associated with Wolbachia infection can interrupt mosquito-borne pathogen transmission cycles. Transinfection of the Wolbachia-free dengue vector Ae. aegypti with the virulent Wolbachia strain wMelPop was associated with a 50% reduction in mosquito life span and 100% CI [2]. Furthermore, Wolbachia infection interferes with subsequent infections by a diverse range of pathogens, including dengue and chikungunya viruses, Plasmodium gallinaceum, and filarial worms [6–8]. The mechanisms of pathogen interference are yet to be fully defined. Wolbachia infection can stimulate the host insect immune system, leading to the "immune priming" a hypothesis of pathogen interference [7–11]. However, this hypothesis does not hold in all situations [12,13]. Alternatively, competition for nutrient resources required by Wolbachia and pathogens, particularly cholesterol, appears to be a major factor contributing to dengue virus (DENV) blocking by Wolbachia [14,15].
To date, two W. pipientis strains have been successfully transinfected into Ae. aegypti; the virulent strain wMelPop [2] and the relatively benign strain wMel [16]. In the first demonstration of the capacity of Wolbachia to reduce the risk of dengue transmission under field settings, wMel infected Ae. aegypti were released into suburbs of Cairns, Australia, harboring endemic populations of the vector. Subsequent monitoring indicated the successful invasion and stable establishment of the bacteria in the Ae. aegypti population [17]. Reduced DENV vector competence of field-collected Ae. aegypti subsequent to that invasion has been demonstrated [18] and a program of field evaluation studies and pilot releases is now occurring globally[19,20]. Future rearing and release programs will need careful monitoring to track the establishment, patterns and stability of Wolbachia invasions.
Currently, molecular techniques, such as polymerase chain reaction (PCR), are used to detect Wolbachia infections in mosquitoes [21–23] but there is concern that their cost and technical complexity are not amenable to the evaluation of large-scale, programmatic interventions in the developing world [24,25].
Near-infrared spectroscopy (NIRS) is a non-destructive and almost instantaneous technique that allows high throughput differentiation of biological samples. Mosquito samples are grouped based upon absorption differences in the NIR region that result from differences in the composition and concentration of organic molecules. The technique does not require reagents and involves minimal sample processing (up to 15 seconds per sample). Collection of NIR spectra from a mosquito requires only a single, 3 second interrogation. Once a calibration model has been developed and validated, prediction of independent samples takes a few seconds. Parameters such as age and species identity can be predicted from the same spectrum. It has been estimated that as an age grading and species identification tool, NIRS is 35 times cheaper and over 16 times faster than PCR and conventional microscopy techniques [26].
Previously, NIRS has been used to differentiate the morphologically identical African malaria vectors; Anopheles gambiae and Anopheles arabiensis with an accuracy of 80–90%. NIRS has also been used to categorise individuals within these species into age categories (> or < 7 d old); that are more relevant to the probability of Plasmodium infection. NIRS grouped mosquitoes into these categories with an accuracy of 78–90% respectively [26–33].
In this study we examined the potential of using NIRS as a high throughput technique for detecting the presence or absence of Wolbachia in laboratory-reared Ae. aegypti mosquitoes. NIRS has previously been successfully applied to discriminate species and strains of bacteria [34–36] and recently to detect Wolbachia in fruit flies [37]. This study represents the first use of NIRS to detect the presence or absence of Wolbachia in male and female Ae. aegypti.
Materials and Methods
Rearing and collection of mosquitoes
We compared wild type Ae. aegypti (from a colony established from Wolbachia-free material collected from Cairns in Jan 2015), Ae. aegypti transinfected with wMel (from a colony established from Wolbachia release suburbs in Cairns April 2015) and the PGYP1 strain of Ae. aegypti transinfected with wMelPop and subsequently out crossed to wild type Ae. aegypti from Cairns (supplied courtesy of the Eliminate Dengue group, Monash). All strains were reared at the insectary of QIMR Berghofer Medical Research Institute, Australia, in separate rooms under identical conditions; 27°C, 70% humidity, 12:12 hr day:night lighting. Larvae were fed on Tetramin tropical flakes (Tetra Melle, Germany). Pupae were transferred into cages measuring 40 × 40 × 30 cm for adult emergence. Adults were fed on 10% sugar solution daily and blood fed on a human volunteer for 15 min every 7 d according to human research ethics protocol (QIMR HREC980).
Each of the three Ae. aegypti mosquito strains (wMel transinfected, wMelPop transinfected and wild type) were represented by two separate cohorts: one was used to develop calibration and validate models and the other was used to test the models. Adults from the calibration cohort were collected at 1, 5, 10, 15, 19 and 20 d post emergence. This was to ensure that the calibration model was applicable across a wide range of age groups. For the validation set, adults were collected 5 d post emergence. Samples were stored in RNAlater for 2–4 weeks before scanning by a NIR spectrometer [28]. PCR of the wsp gene was used to confirm presence or absence of Wolbachia-infection in Ae. aegypti colonies [23]. The wMel and wMelPop colonies used in these experiments were 100% Wolbachia infected due to the PCR amplification of the wsp gene from 100% of tested specimens (N = 39 and 40, respectively) whereas the wild type Ae. aegypti colony was completely uninfected (N = 47).
Scanning of mosquitoes
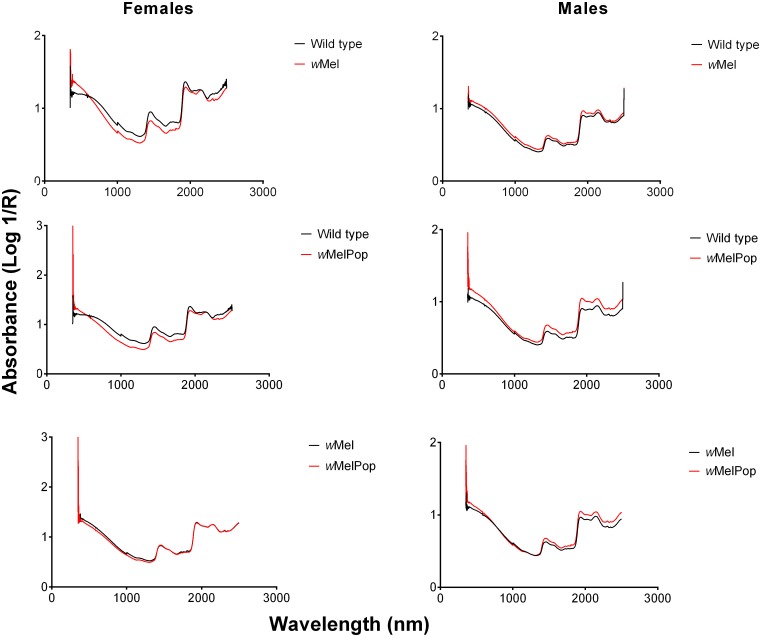
All mosquitoes collected were transferred to the Ifakara Health Institute, Tanzania for scanning using a LabSpec 5000 NIR spectrometer (ASD Inc, Boulder, CO) according to established protocols [27]. Prior to scanning, residual RNAlater was removed by blotting mosquito specimens with a filter paper. Examples of average spectra collected from heads and thoraces of individual mosquitoes are shown in Fig 1.
Fig 1. Examples of average spectra collected from heads and thoraces of Wolbachia infected and wild type male and female Ae. aegypti mosquitoes.
Data analysis
Model development and calibration was conducted as previously published [27]. In brief, spectra between 500–2350 nm were analyzed using partial least square (PLS) regression in GRAMS Plus/IQ software (Thermo Galactic, Salem, NH). Calibrations were developed to differentiate: 1) wMelPop infected females from wild type females, 2) wMel infected females from wild type females 3) wMelPop infected males from wild type males, 4) wMel infected males from wild type males 5) wMelPop infected females from wMel infected females and 6) wMelPop infected males from wMel infected males. For each data set, at least 144 mosquitoes were used to develop calibration models.
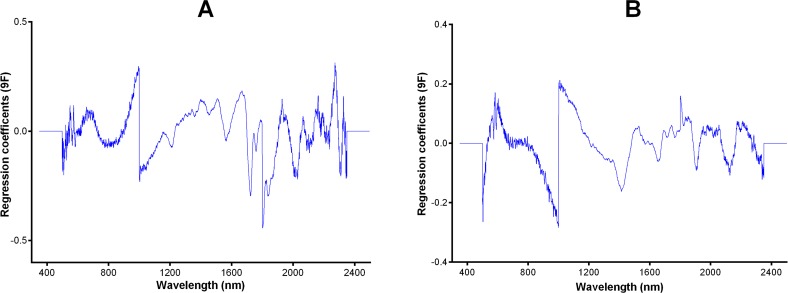
In the PLS model, a value of “1” was assigned to wild type samples whereas a value of “2” was assigned to Wolbachia infected mosquitoes. Similarly, to differentiate male and female mosquitoes infected with wMel from those infected with wMelPop, a value of “1” was assigned to wMel and a value of “2” was assigned to wMelPop infected mosquitoes. A value of 1.5 was considered the threshold for correct or incorrect classification. For example, all wild type samples predicted above the 1.5 cut-off point were considered misclassified and vice versa. These calibration models were then applied to independent data sets (mosquitoes from cohort 1 that were not used to develop the model and mosquitoes from cohort 2 to test whether they could predict: 1) the presence or absence of wMel or wMelPop in female and male Ae. aegypti mosquitoes, and 2) to differentiate between wMelPop and wMel infections. Regression coefficient plots used to differentiate wMelPop from wMel infected female Ae. aegypti and wMel infected from wild type Ae. aegypti using 9 factors in the PLS model are shown in Fig 2.
Fig 2. Regression coefficient plots used to predict the presence or absence of infection.
They are based on 9 PLS regression factors. The plots show peaks that influenced the differentiation of female wMelPop from female wMel (panel A) and female wMel from female wild type Ae. aegypti (panel B).
A logistic regression analysis was undertaken in SPSS version 22.0 (SPSS Inc. Chicago) to determine the effects of age, sex, cohort and infection type on the prediction accuracy of NIRS. Backward stepwise procedure was used to control for potential confounding factors between covariates (removal criteria, P > 0.10; re-entry criteria, P ≤ 0.05). Non-linear relationships were examined through scatter plots. Results are expressed as adjusted odds ratios and 95% confidence intervals. Where quoted, statistical significance is at the conventional P < 0.05 level.
Results and Discussion
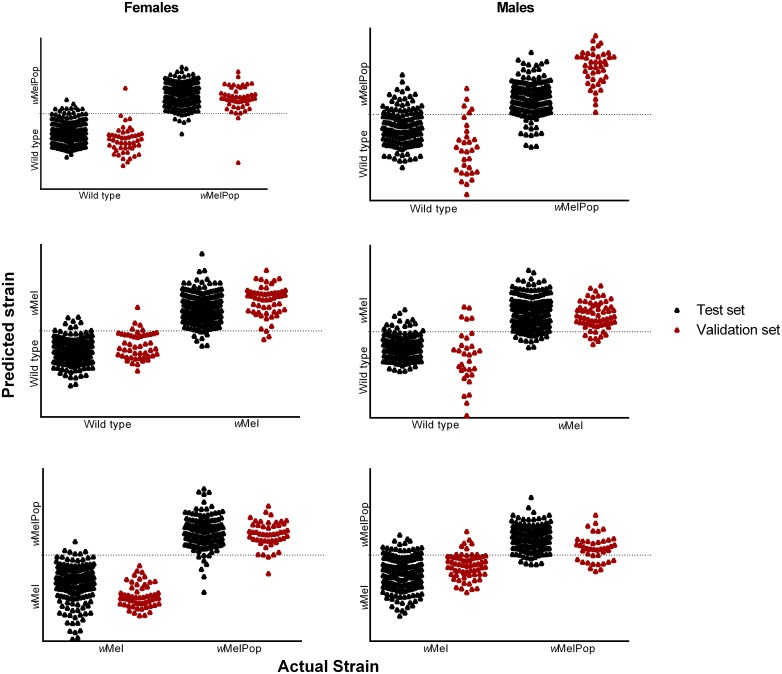
We demonstrated that NIRS has considerable potential as a high throughput tool for differentiating Wolbachia infected and uninfected mosquitoes. Using PLS models developed from a cohort of mosquitoes of various ages, NIRS could detect the presence or absence of specific Wolbachia types in independent mosquito collections. Regardless of their age category and the specific cohort, females and males transinfected with wMelPop were differentiated from wild types with an average accuracy of 96% (N = 299) and 87.5% (N = 377) respectively. Similarly, NIRS differentiated between wMel and wild type females with an average accuracy of 92% (N = 352) whereas infected males were differentiated from uninfected males with an accuracy of 88.5% (N = 444). NIRS was also able to categorise different strains of Wolbachia; female and male Ae. aegypti mosquitoes infected with wMelPop could be differentiated from wMel infected mosquitoes with an accuracy of 96.6% (N = 442) and 84.5% (N = 443), respectively. The overall accuracy for each cohort is presented in Table 1 and Fig 3.
Table 1. Percentage accuracy of Wolbachia detection using cross validation and prediction analyses.
| Infection type | % Accuracy [N] Cross validation1 | % Accuracy [N] Validation set2 | % Accuracy [N] Test set3 |
|---|---|---|---|
| wMelPop ♀/ wild type ♀ | 92 [259] | 97 [200] | 95 [99] |
| wMel ♀/ wild type ♀ | 89 [256] | 91 [248] | 93 [104] |
| wMelPop ♂/ wild type ♂ | 85 [144] | 82 [301] | 93 [76] |
| wMel ♂/ wild type ♂ | 91 [160] | 89 [346] | 88 [98] |
| wMelPop ♀/ wMel ♀ | 95 [200] | 95 [337] | 98 [105] |
| wMelPop ♂/ wMel ♂ | 89 [177] | 90 [335] | 79 [108] |
1 Accuracy of mosquitoes used to develop calibration models
2 Accuracy of cohort 1 mosquitoes that were used to validate calibration models
3 Accuracy of cohort 2 mosquitoes that were used to test calibration models
Fig 3. NIRS differentiation of Wolbachia infected and wild type male and female Ae. aegypti mosquitoes using validation and test samples.
The dotted line indicates the classification cut off point as predicted by the NIRS.
Logistic regression analysis indicated that age (P < 0.001), the interaction term between age and infection type (P < 0.001), sex (P = 0.046) and infection type (P < 0.001) significantly affected the prediction accuracy of NIRS. Odds of an accurate prediction of the presence or absence of an infection increased 1.089 fold (95% CI: 1.039–1.142) from younger (1 d old) to older (20 d old) mosquitoes. Compared to wild type Ae. aegypti, the odds of NIRS accurately predicting the presence of wMelPop changes 0.919 fold (95% CI: 0.865–0.976) and 0.862 fold (95% CI: 0.815–0.911) for wMel from younger to older age groups. NIRS was 1.647 times (95% CI: 1.008–2.691) more accurate at predicting infection in females than males. NIRS was 4.321 times (95% CI: 2.447–7.629) more accurate in predicting wMel infection, and 2.318 times (95% CI: 1.303–4.123) more accurate in predicting the presence of wMelPop relative to wild type mosquitoes.
We conducted this study under the assumption that there was a difference in the cellular constituents of Wolbachia transinfected and wild type mosquitoes. It was also hypothesized that those differences would be reflected in the heads and thoraces of the mosquitoes hence producing unique NIR spectra that could be used for their identification.
Spectral regions between 700–2350 nm have previously been reported to be specific for bacteria differentiation and these regions have been successful at detecting and identifying different species of bacteria in an isolated system [34,36] as well as for differentiation of different strains of bacteria [38] using NIRS. We have also shown in this study that bands at the1400-2350 nm spectral regions were responsible for differentiating wild type from Wolbachia infected Ae. aegypti and sharp bands at 2000–2350 nm might have been responsible for differentiating Ae. aegypti transinfected with different strains of Wolbachia. In particular, bands at 2000-2300nm spectral band region suggest aliphatic C-H and methylene stretching might have largely contributed to the differentiation of wMel from wMelPop infected mosquitoes (Fig 2A) whereas sharp bands between 1400-1700nm related to C-H first overtone of amide band combinations and aromatic groups largely contributed to differentiation of Wolbachia infected from uninfected mosquitoes [39] (Fig 2B).
Infections with wMelPop, and to a lesser extent, wMel can achieve high densities in a broad range of mosquito tissues [16]. Recent metabolic analyses demonstrate that Wolbachia utilize and modulate cellular levels of nutrients including cholesterol [14], amino acids [15], proteins carbohydrates [40] and glycogen [41]. It is possible that NIRS differentiates wild type from Wolbachia infected mosquitoes through spectral signals generated from qualitative and quantitative differences in these nutrients. Alternatively, differences in bacteria density may also differentiate wMel and wMelPop. There is very limited genomic variation between wMelPop and wMel, with the exception of a large inversion, triplication of copy number of a 19 kb region [42,43] and insertion of an IS5 element into gene WD1310 [44]. The triplication was lost during subsequent cell passaging and mosquito infection to generate the wMelPop PGYP strain used in these experiments. Currently, quantitative PCR (qPCR) of a region of the IS5 element is used to differentiate between wMel and wMelPop strains [45]. The ability to distinguish wMel or wMelPop using our rapid and non-destructive technique could introduce large time and cost savings over conventional PCR techniques.
The calibration model was developed using a range of adult ages (age is known to affect the absorbed and reflected spectra in mosquitoes) successfully predicted the Wolbachia type in single age cohorts (Table 2). NIRS predictions were generally more accurate for female mosquitoes than for males. The proportion of female individuals that were misclassified was 3–7% whereas the proportion of misclassified males was 7–12% and this difference was significant (P = 0.046). Stronger NIRS signals from females may be due to higher Wolbachia densities in females relative to males. This has been observed for Wolbachia strains wAlbA and wAlbB within Aedes albopictus [46] and may reflect higher Wolbachia densities in mosquito ovaries [16,47]. When differentiating wMel or wMelPop infected females from wild type females, 1 d old female mosquitoes contributed to the highest misclassification (Table 2). This might be explained by the presence of under-developed ovaries in 1 d old mosquitoes relative to older mosquitoes [47].
Table 2. Percentage accuracy with which pairs were differentiated at various ages (1–20 days), and the specific identification accuracy for the individual components of those pairs.
| Infection type | N | 1d | 5d | 10d | 15d | 19d | 20d | Specific [N] for first member of pair | Specific [N] for second member of pair |
|---|---|---|---|---|---|---|---|---|---|
| wMelPop ♀ / wild type ♀ | 299 | 87 | 98 | 100 | 97 | 100 | 96 | 95 [151] | 97 [148] |
| wMel ♀ / wild type ♀ | 352 | 85 | 86 | 92 | 95 | 84 | 96 | 93 [208] | 87 [144] |
| wMelPop ♂ / wild type ♂ | 377 | 70 | 87 | 87 | 89 | 80 | 92 | 89 [197] | 79 [180] |
| wMel ♂ / wild type ♂ | 444 | 90 | 93 | 100 | 81 | 76 | 95 | 90 [262] | 89 [182] |
| wMelPop ♀ / wMel♀ | 442 | 95 | 99 | 98 | 95 | 92 | 97 | 95 [199] | 97 [243] |
| wMelPop ♂ / wMel ♂ | 443 | 98 | 97 | 98 | 75 | 79 | 90 | 89 [197] | 86 [246] |
Wolbachia-mediated disease control strategies may take a number of different forms in the future. The currently established model is a replacement strategy in which a relatively benign strain such as wMel is driven into wild mosquitoes through unidirectional CI. The mosquito population that develop from these interactions are Wolbachia-infected, fit and stable but less capable of transmitting virus [17].
The wMelPop strain of Wolbachia is known for its virulent nature. Cells infected with wMelPop have been reported to cause increased cell apoptosis and reduced survival in adult mosquitoes [2]. This affects fitness of Ae. aegypti and prevents the establishment of infections in field populations, limiting its current usefulness for biological control. However, its virulence may be exploited by a proposal to first drive wMelPop into a target wild type population and then “crash” that population through the poor fitness of the resulting phenotype [48].
Another possibility is to use unidirectional CI to implement variations on the sterile insect technique (SIT). In one scenario, male-only releases of Wolbachia-infected mosquitoes could be used to overwhelm mating interactions with wild type females, a strategy referred to as the incompatible insect technique (IIT) [49,50]. Unlike conventional SIT, the object of IIT for Aedes species would be to suppress local mosquito populations below the threshold required for effective disease transmission. In that scenario, it is crucial that no Wolbachia-infected females are released (which could lead to population replacement instead of population suppression). Therefore, a proposed strategy is to combine conventional SIT with IIT, which would remove the risk of vector population replacement [50,51]. Proof-of-concept studies have shown the viability of combining irradiation and Wolbachia-based approaches [51–54]. Depending on the virus-blocking capability of the introduced Wolbachia strain, this combined approach might also have the additional benefit of eliminating the risk of virus transmission by those released females. Alternatively, bidirectional CI where no mating with Wolbachia infected mosquitoes are viable could also be used to overwhelm and replace the local population [55]. In all of these instances it will be essential to monitor the infection status of the release colonies and the stability and spread of infection population suppression strategies. NIRS may present a rapid means of making these assessments.
Our examination of over 900 male and female wild type Ae. aegypti mosquitoes or Ae. aegypti transinfected with either wMel or wMelPop demonstrated that NIRS has potential as a high throughput tool to identify Wolbachia infection across a range of mosquito age. NIRS offers demonstrable improvements over technically demanding, expensive and time consuming molecular assays. PCR techniques are suited to limited surveillance using small or pooled samples that may not represent the true spatial or temporal heterogeneity of extensive field populations. The high throughput PCR assay recently described by Lee and colleagues for detecting Wolbachia in dengue mosquitoes requires costly DNA extraction kits and PCR reagents [22,23]. Unpublished analysis of cost using a local supplier (Qiagen Pty LTD, Victoria, Australia) on 09/07/2015 demonstrated that a single sample processed using this high throughput PCR-based technique would cost approximately 6 USD after the initial outlay for a real time PCR machine with an option for high resolution melt analysis. Comparatively, NIRS can collect a diagnostic spectrum from an individual mosquito within 3 seconds.The NIRS data that result from a single interrogation can be used to predict more than one characteristic in each individual e.g. age and species in the An. gambiae complex [26–28]. No reagents or sample-specific preparations are needed, hundreds of individual mosquitoes can be scanned in a day and results can be analysed and reported upon immediately. After the initial cost of the spectrometer (Ca. 60,000 USD), only minimal labor related costs are required to run the NIRS technique. The >80% accuracy observed in this study is sufficient to rapidly assess success or failure of Wolbachia based control interventions such as those targeting dengue and other emerging infectious diseases such as Zika. If NIRS can also be used to predict the age of dengue and Zika vectors, it would define many key factors associated with vectorial capacity and the evaluation of disease transmission risk. The use of NIRS to measure a number of key disease vector characteristics has now been demonstrated in several studies [26–28]. NIRS promises to improve the speed and capacity of program evaluations and risk mapping exercises, especially in remote regions with little infra-structure or in dense urban environments with very high surveillance costs.
Acknowledgments
We thank Jill Ulrich and Jonathan Darbro of the Mosquito Control Laboratory, QIMR Berghofer for establishing wMel infected and wild type colonies at QIMR Berghofer. wMelPop was provided to us courtesy of the Eliminate Dengue group, Monash University, Australia. We thank Nikita Lysenko for his technical assistance in Tanzania.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by Grand Challenges Canada Stars for Global Health funded by the Government of Canada (grant 0439-001) awarded to MTSL and the Mosquito and Arbovirus Research Committee, Queensland, Australia. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, et al. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai school children: a randomised, controlled phase 2b trial. Lancet. 2012;380(9853):1559–1567. Epub 2012/09/15. 10.1016/s0140-6736(12)61428-7 . [DOI] [PubMed] [Google Scholar]
- 2.McMeniman CJ, Lane RV, Cass BN, Fong AWC, Sidhu M, Wang Y-F, et al. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science. 2009;323(5910):141–144. [DOI] [PubMed] [Google Scholar]
- 3.Sinkins SP. Wolbachia and cytoplasmic incompatibility in mosquitoes. Insect Biochem Mol Biol. 2004;34(7):723–729. [DOI] [PubMed] [Google Scholar]
- 4.Alam U, Medlock J, Brelsfoard C, Pais R, Lohs C, Balmand S, et al. Wolbachia symbiont infections induce strong cytoplasmic incompatibility in the tsetse fly Glossina morsitans. PLoS Pathog. 2011;7(12):e1002415 10.1371/journal.ppat.1002415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tagami Y, Doi M, Sugiyama K, Tatara A, Saito T. Wolbachia-induced cytoplasmic incompatibility in Liriomyza trifolii and its possible use as a tool in insect pest control. Biological Control. 2006;38(2):205–209. [Google Scholar]
- 6.Hedges LM, Brownlie JC, O'Neill SL, Johnson KN. Wolbachia and virus protection in insects. Science. 2008;322(5902):702–702. 10.1126/science.1162418 [DOI] [PubMed] [Google Scholar]
- 7.Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, et al. A Wolbachia symbiont in Aedes aegypti limits infection with Dengue, Chikungunya, and Plasmodium. Cell. 2009;139(7):1268–1278. 10.1016/j.cell.2009.11.042 [DOI] [PubMed] [Google Scholar]
- 8.Kambris Z, Cook PE, Phuc HK, Sinkins SP. Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science. 2009;326(5949):134–136. 10.1126/science.1177531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kambris Z, Blagborough AM, Pinto SB, Blagrove MS, Godfray HCJ, Sinden RE, et al. Wolbachia stimulates immune gene expression and inhibits Plasmodium development in Anopheles gambiae. PLoS Pathog. 2010;6(10):e1001143–e1001143. 10.1371/journal.ppat.1001143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan X, Zhou G, Wu J, Bian G, Lu P, Raikhel AS, et al. Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc Nat Acad Sci. 2012;109(1):E23–E31. 10.1073/pnas.1116932108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bian G, Xu Y, Lu P, Xie Y, Xi Z. The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS pathog. 2010;6(4):e1000833 10.1371/journal.ppat.1000833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frentiu FD, Robinson J, Young PR, McGraw EA, O’neill SL. Wolbachia-mediated resistance to dengue virus infection and death at the cellular level. PloS One. 2010;5(10):e13398 10.1371/journal.pone.0013398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osborne SE, Leong YS, O’Neill SL, Johnson KN. Variation in antiviral protection mediated by different Wolbachia strains in Drosophila simulans. PLoS Pathog. 2009;5(11):e1000656 10.1371/journal.ppat.1000656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caragata EP, Rances E, Hedges LM, Gofton AW, Johnson KN, O'Neill SL, et al. Dietary cholesterol modulates pathogen blocking by Wolbachia. PLoS Pathog. 2013;9(6):e1003459 10.1371/journal.ppat.1003459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caragata EP, Rancès E, O’Neill SL, McGraw EA. Competition for amino acids between Wolbachia and the mosquito host, Aedes aegypti. Microbial Ecol. 2014;67(1):205–218. [DOI] [PubMed] [Google Scholar]
- 16.Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476(7361):450–453. 10.1038/nature10355 [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann AA, Montgomery BL, Popovici J, Iturbe-Ormaetxe I, Johnson PH, Muzzi F, et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature. 2011;476(7361):454–457. 10.1038/nature10356 [DOI] [PubMed] [Google Scholar]
- 18.Frentiu FD, Zakir T, Walker T, Popovici J, Pyke AT, van den Hurk A, et al. Limited dengue virus replication in field-collected Aedes aegypti mosquitoes infected with Wolbachia. PLoS Negl Trop Dis. 2014;8(2):e2688 10.1371/journal.pntd.0002688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dutra HLC, dos Santos LMB, Caragata EP, Silva JBL, Villela DAM, Maciel-de-Freitas R, et al. From Lab to Field: The influence of urban landscapes on the invasive potential of Wolbachia in Brazilian Aedes aegypti mosquitoes. PLoS Negl Trop Dis. 2015;9(4): e0003689 10.1371/journal.pntd.0003689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeffery JA, Yen NT, Nam VS, Hoffmann AA, Kay BH, Ryan PA. Characterizing the Aedes aegypti population in a Vietnamese village in preparation for a Wolbachia-based mosquito control strategy to eliminate dengue. PLoS Negl Trop Dis. 2009;3(11):e552 10.1371/journal.pntd.0000552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Neill SL, Giordano R, Colbert A, Karr TL, Robertson HM. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Nat Acad Sci. 1992;89(7):2699–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SF, White VL, Weeks AR, Hoffmann AA, Endersby NM. High-throughput PCR assays to monitor Wolbachia infection in the dengue mosquito (Aedes aegypti) and Drosophila simulans. Appl Environ Microb. 2012;78(13):4740–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou W, Rousset F, O'Neill S. Phylogeny and PCR–based classification of Wolbachia strains using wsp gene sequences. P R Soc London B: Biol Sci. 1998;265(1395):509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muniaraj M, Paramasivan R, Sunish I, Arunachalam N, Mariappan T, Leo S, et al. Detection of Wolbachia endobacteria in Culex quinquefasciatus by Gimenez staining and confirmation by PCR. Journal of vector borne diseases. 2012;49(4):258–261. [PubMed] [Google Scholar]
- 25.da Silva Gonçalves D, Cassimiro APA, de Oliveira CD, Rodrigues NB, Moreira LA. Wolbachia detection in insects through LAMP: loop mediated isothermal amplification. Parasite Vector. 2014;7(1):1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sikulu M, Killeen G, Hugo L, Ryan P, Dowell K, Wirtz R, et al. Near-infrared spectroscopy as a complementary age grading and species identification tool for African malaria vectors. Parasite Vector. 2010;3(1):49 10.1186/1756-3305-3-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayagaya VS, Michel K, Benedict MQ, Killeen GF, Wirtz RA, Ferguson HM, et al. Non-destructive determination of age and species of Anopheles gambiae s.l. Using near-infrared spectroscopy. Am J Trop Med Hyg. 2009;81(4):622–630. 10.4269/ajtmh.2009.09-0192 [DOI] [PubMed] [Google Scholar]
- 28.Sikulu M, Dowell K, Hugo L, Wirtz R, Michel K, Peiris K, et al. Evaluating RNAlater as a preservative for using near-infrared spectroscopy to predict Anopheles gambiae age and species. Malar J. 2011;10(1):186 10.1186/1475-2875-10-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sikulu MT, Majambere S, Khatib BO, Ali AS, Hugo LE, Dowell FE. Using a Near-infrared spectrometer to estimate the age of Anopheles mosquitoes exposed to pyrethroids. PLoS One. 2014;9(3):e90657 10.1371/journal.pone.0090657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ntamatungiro A, Mayagaya V, Rieben S, Moore S, Dowell F, Maia M. The influence of physiological status on age prediction of Anopheles arabiensis using near infra-red spectroscopy. Parasite Vector. 2013;6(1):298 10.1186/1756-3305-6-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayagaya VS, Ntamatungiro AJ, Moore SJ, Wirtz RA, Dowell FE, Maia MF. Evaluating preservation methods for identifying Anopheles gambiae ss and Anopheles arabiensis complex mosquitoes species using near infra-red spectroscopy. Parasite Vector. 2015;8(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sikulu M. Non-destructive near infrared spectroscopy for simultaneous prediction of age and species of two major African malaria vectors: An. gambiae and An. arabiensis. NIR news. 2014;25(5):4–6. [Google Scholar]
- 33.Dowell FE, Noutcha AEM, Michel K. The effect of preservation methods on predicting mosquito age by near Infrared spectroscopy. Am J Trop Med Hyg. 2011;85(6):1093–1096. 10.4269/ajtmh.2011.11-0438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexandrakis D, Downey G, Scannell AG. Detection and identification of bacteria in an isolated system with near-infrared spectroscopy and multivariate analysis. J Agr Food Chem. 2008;56(10):3431–3437. [DOI] [PubMed] [Google Scholar]
- 35.Alexandrakis D, Downey G, Scannell AM. Detection and identification of selected bacteria, inoculated on chicken breast, using near infrared spectroscopy and chemometrics. Sens Instrum Food Qual Saf 2011;5(2):57–62. 10.1007/s11694-011-9111-y [DOI] [Google Scholar]
- 36.Cámara-Martos F, Zurera-Cosano G, Moreno-Rojas R, García-Gimeno RM, Pérez-Rodríguez F. Identification and quantification of lactic acid bacteria in a water-based matrix with near-infrared spectroscopy and multivariate regression modeling. Food Anal Method. 2012;5(1):19–28. [Google Scholar]
- 37.Aw WC, Dowell FE, Ballard JWO. Using near-infrared spectroscopy to resolve the species, gender, age, and the presence of Wolbachia infection in laboratory-reared Drosophila. G3: Genes Genom Genet. 2012;2(9):1057–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez-Saona LE, Khambaty FM, Fry FS, Calvey EM. Rapid Detection and Identification of Bacterial Strains By Fourier Transform Near-Infrared Spectroscopy. J Agric Food Chem. 2001;49(2):574–579. 10.1021/jf000776j [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Sowa MG, Ahmed MK, Mantsch HH. Photoacoustic near-infrared investigation of homo-polypeptides. J Phys Chem. 1994;98(17):4748–4755. 10.1021/j100068a043 [DOI] [Google Scholar]
- 40.Ponton F, Wilson K, Holmes A, Raubenheimer D, Robinson KL, Simpson SJ. Macronutrients mediate the functional relationship between Drosophila and Wolbachia. P R Soc London B: Biol Sci. 2015;282(1800):20142029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.da Rocha Fernandes M, Martins R, Costa EP, Pacidônio EC, de Abreu LA, da Silva Vaz I Jr, et al. The modulation of the symbiont/host interaction between Wolbachia pipientis and Aedes fluviatilis embryos by glycogen metabolism. PloS One. 2014;9(9):e109857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woolfit M, Iturbe-Ormaetxe I, Brownlie JC, Walker T, Riegler M, Seleznev A, et al. Genomic evolution of the pathogenic Wolbachia strain, wMelPop. Genom Bio Evol. 2013;5(11):2189–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun LV, Riegler M, O'Neill SL. Development of a physical and genetic map of the virulent Wolbachia strain wMelPop. J Bacteriol. 2003;185(24):7077–7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riegler M, Sidhu M, Miller WJ, O’Neill SL. Evidence for a global Wolbachia replacement in Drosophila melanogaster. Curr Biol. 2005;15(15):1428–1433. [DOI] [PubMed] [Google Scholar]
- 45.Yeap HL (2014) Phenotypic and genotypic analysis for quality control of Wolbachia-infected Aedes aegypti in a dengue biocontrol program. Minerva access: The University of Melbourne. 213 p.
- 46.Wiwatanaratanabutr I, Kittayapong P. Effects of crowding and temperature on Wolbachia infection density among life cycle stages of Aedes albopictus. J Invert Pathol. 2009;102(3):220–224. [DOI] [PubMed] [Google Scholar]
- 47.Genty L-M, Bouchon D, Raimond M, Bertaux J. Wolbachia infect ovaries in the course of their maturation: last minute passengers and priority travellers? PloS one. 2014;9(4):e94577 10.1371/journal.pone.0094577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritchie SA, Townsend M, Paton CJ, Callahan AG, Hoffmann AA. Application of wMelPop Wolbachia strain to crash local populations of Aedes aegypti. PLoS Negl Trop Dis. 2015;9(7):e0003930 10.1371/journal.pntd.0003930 PMC4512704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bourtzis K, Dobson SL, Xi Z, Rasgon JL, Calvitti M, Moreira LA, et al. Harnessing mosquito–Wolbachia symbiosis for vector and disease control. Acta tropica. 2014;132:S150–S163. 10.1016/j.actatropica.2013.11.004 [DOI] [PubMed] [Google Scholar]
- 50.Bourtzis K, Lees RS, Hendrichs J, Vreysen MJ. More than one rabbit out of the hat: Radiation, transgenic and symbiont-based approaches for sustainable management of mosquito and tsetse fly populations. Acta tropica. 2016. [DOI] [PubMed] [Google Scholar]
- 51.Zhang D, Lees RS, Xi Z, Gilles JR, Bourtzis K. Combining the sterile insect technique with Wolbachia-based approaches: II-A safer approach to Aedes albopictus population suppression programmes, designed to minimize the consequences of inadvertent female release. PloS One. 2015;10(8):e0135194 10.1371/journal.pone.0135194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brelsfoard CL, St Clair W, Dobson SL. Integration of irradiation with cytoplasmic incompatibility to facilitate a lymphatic filariasis vector elimination approach. Parasite Vectors. 2009;2(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang D, Lees RS, Xi Z, Bourtzis K, Gilles JR. Combining the sterile insect technique with the incompatible insect technique: III-robust mating competitiveness of irradiated triple Wolbachia-infected Aedes albopictus Males under semi-field conditions. PloS One. 2016;11(3):e0151864 10.1371/journal.pone.0151864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang D, Zheng X, Xi Z, Bourtzis K, Gilles JR. Combining the sterile insect technique with the incompatible insect technique: I-Impact of Wolbachia infection on the fitness of triple-and double-infected strains of Aedes albopictus. PloS One. 2015;10(4):e0121126 10.1371/journal.pone.0121126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blagrove MS, Arias-Goeta C, Failloux A-B, Sinkins SP. Wolbachia strain wMel induces cytoplasmic incompatibility and blocks dengue transmission in Aedes albopictus. Proc Nat Acad Sci. 2012;109(1):255–260. 10.1073/pnas.1112021108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.