Abstract
Regulated expression of the Ena1 Na+-ATPase is a crucial event for adaptation to high salt and/or alkaline pH stress in the budding yeast Saccharomyces cerevisiae. ENA1 expression is under the control of diverse signaling pathways, including that mediated by the calcium-regulatable protein phosphatase calcineurin and its downstream transcription factor Crz1. We present here a quantitative study of the expression of Ena1 in response to alkalinization of the environment and we analyze the contribution of Crz1 to this response. Experimental data and mathematical models substantiate the existence of two stress-responsive Crz1-binding sites in the ENA1 promoter and estimate that the contribution of Crz1 to the early response of the ENA1 promoter is about 60%. The models suggest the existence of a second input with similar kinetics, which would be likely mediated by high pH-induced activation of the Snf1 kinase.
Introduction
Excessive intracellular concentration of sodium cations is deleterious for most eukaryotic cells and, therefore, diverse mechanisms to avoid cytosolic accumulation of this cation have been evolutionarily developed. For instance, the yeast Saccharomyces cerevisiae, often used as a model to study the mechanisms of cation homeostasis, resorts to both sequestration of sodium into organelles (mainly the vacuole) and to extrusion of this cation through transporters located at the cell membrane [1].
Extrusion of sodium is achieved by two main mechanisms. The first one is based on the H+/Na+ antiporter encoded by NHA1, which exchanges protons with Na+, Li+, and K+ cations, and has major biological significance under normal (acidic) growth conditions [2]. The second strategy is based on a P-type ATPase pump encoded by the ENA system, which becomes essential for growth particularly at pHs above neutrality. ENA genes are found exclusively in fungi, bryophyta and protozoa and ENA homologues are likely to exist in all fungal genomes [3]. In most budding yeast strains ENA genes exist as a cluster of 3 to 5 elements encoding nearly identical proteins. The relevance of Ena1 in the tolerance to increased extracellular levels of toxic monovalent cations (Na+ and Li+) or alkaline pH is documented by the extreme sensitivity of cells lacking functional ENA genes under these circumstances (see [4] and references therein). Whereas the function of a Na+-ATPase pump is evident in response to high salt, the role of such protein under high pH stress, even in the absence of elevated sodium levels in the medium, became evident only recently [5]. This report demonstrated that Ena1 is required to pump out the sodium cations introduced by the Na+/phosphate cotransporter Pho89, whose role in phosphate uptake becomes more relevant at high external pH [6,7].
Under standard growth conditions (acidic pH and absence of osmotic stress) expression of the ENA genes is minimal. However, addition of NaCl above 0.3–0.4 M or raising the external pH above 7.2 results in rapid induction of the first member of the cluster, usually denoted as ENA1 [8–11]. Work during the last twenty years has revealed the complex regulatory network that represses ENA1 under normal growth conditions and promotes rapid increase of ENA1 mRNA levels upon salt stress or high pH. This network includes the Rim101, Snf1, PKA and calcineurin pathways (see [12] and references therein). In addition to these pathways, activation of the osmosensitive Hog1 pathway becomes an important component of the response to high salt [13]. However, diverse evidences demonstrate that the Hog1 pathway does not contribute to either high pH stress tolerance nor to ENA1 induction in response to high pH stress [10,14].
Work in our laboratory [14] illustrated the relative contribution of the calcineurin, Snf1 and Rim101 pathways to the induction of the ENA1 gene in response to high pH. Although the analysis was semi-quantitative, it became evident that the calcineurin pathway was an important contributor to the alkaline responsiveness of ENA1. In budding yeast calcineurin (also known as protein phosphatase type 2B) is a heterodimeric Ser/Thr phosphatase composed of one of two possible redundant catalytic subunits (Cna1 and Cna2) plus a unique regulatory subunit (Cnb1). Calcineurin is activated by calcium and calmodulin. In response to alkaline pH stress, activation of calcineurin occurs as a consequence of the almost immediate entry of extracellular calcium through the Cch1-Mid1 calcium channel [15], and it affects the expression of a substantial number of genes [16,17]. The transcriptional effects of calcineurin activation occur mainly through the dephosphorylation of the transcription factor Crz1/Tcn1/Hal8
[18–20]. Dephosphorylated Crz1 enters the nucleus via the importin Nmd5 [21] and binds to its target promoters via specific sequences, known as CDREs (Calcineurin Dependent Response Elements). After phosphorylation, the transcription factor returns to the cytosol via the exportin Msn5 [22]. It has been proposed that regulation of ENA1 in response to calcineurin activation would involve two functional Crz1-binding regions in the ENA1 promoter at positions -813/-821 and -719/-727. The more upstream element (relative to the initiating Met codon) binds Crz1 with lesser affinity and it has been proposed to be responsible for the basal ENA1 expression [23].
Despite early reports suggesting posttranscriptional regulation of Ena1 activity [24] and the evidence of diverse components necessary for the correct localization of Ena1 at the plasma membrane [25,26], it is widely accepted that regulation of Ena1 function in response to stress occurs principally by regulation of the mRNA levels. It has been assumed that changes in ENA1 mRNA levels occur mainly in response to modulation of ENA1 promoter activity. However, recent work has showed that, after alkaline stress, the modification of mRNA levels for many genes is largely dominated by changes in mRNA stability [27]. In this work we address, in a quantitative study, the different steps relevant for the expression of Ena1 in response to high pH stress and, in particular, the contribution of the Crz1 transcription factor in this process.
Materials and Methods
Yeast strains and growth conditions
S. cerevisiae strains used in this study were all derived from BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0; obtained from EUROSCARF), which has an S288C background. Strain SP020, which expresses the Crz1 protein C-terminally tagged with the 3xHA epitope was previously described [5]. Strains carrying a Crz1 (SP017) or Ena1 (MLM001) protein C-terminally tagged with GFP epitope were constructed by transformation of the wild type strain with a cassette amplified from plasmid pFA6-GFP-kanMX6 (for Crz1) or pFA6-GFP(S65T)-HIS3MX6 (for Ena1) [28] using oligonucleotides CRZ1-C-pFA6-dir/CRZ1-C-pFA6-rev and ENA1-C-pFA6-dir/ENA1-C-pFA6-rev, respectively. The ENA1 gene was tagged similarly in the crz1::kanMX4 background to generate strain MLM002. Correct integration was verified by diagnostic PCR. Since members of the ENA cluster are highly similar, care was taken to select the transformants carrying specifically a C-terminally GFP tagged version of ENA1. To this end, two pairs of primers were used: ENA1-pFA6-comp-Cter and pFA6a_GFP_rev that amplify a region regardless the member of ENA cluster receiving the tag, and ENA1-pFA6-comp-Nter and pFA6a_GFP_rev that amplify a specific fragment for ENA1 integration. Oligonucleotides used in this work are described in S1 Table.
Strain SP039 used for nuclear colocalization studies and quantification of Crz1 in single cells by confocal microscopy was generated by crossing SP017 (BY4741 CRZ1:GFP-kanMX6) strain to BY4742 SIK1:mRFP-kanMX6 [29] and subsequent separation of tetrads by micromanipulation. The correct genotype of clones carrying both mutations was verified by diagnostic PCR analysis and analyzed for positive GFP and RFP signals by fluorescence microscopy. Strain BY4742 SIK1:mRFP-kanMX6 was a gift of H. Sychrova (Institute of Physiology, Prague). Strains employed in this work are described in Table 1.
Table 1. Strains used in this study.
| Strain | Genotype | Source /Reference |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | [67] |
| SP017 | BY4741 CRZ1-GFP-kanMX6 | This work |
| SP020 | BY4741 CRZ1-3XHA-kanMX6 | [5] |
| MLM001 | BY4741 ENA1-GFP-HIS3 | This work |
| MLM002 | BY4741 crz1:: kanMX4 ENA1-GFP-HIS3 | This work |
| SP039 | BY4741 CRZ1-GFP-kanMX6 SIK1-mRFP-kanMX6 | This work |
| SIK1-mRFP | BY4742 SIK1-mRFP-kanMX6 | [29] |
To induce alkaline pH stress response, S. cerevisiae strains were usually grown overnight to mid-exponential phase at 28°C in liquid rich medium (YPD; 10 g/l yeast extract, 20 g/l peptone and 20 g/l dextrose) and then resuspended in fresh YPD media containing 50 mM TAPS adjusted to pH 5.5 to OD600 = 0.2. Cells were grown for 4–5 hours to reach OD600 = 0.6–0.8 and treated with 1 M KOH to reach external pH 8.0. Samples (10–40 ml) were taken at the appropriate times and then either harvested by centrifugation (directly or after formaldehyde treatment for ChIP or ChIP-Seq) or, for RNA extraction, by filtration on 0.45 m GN-6 filters (Pall Co.). When subcellular localization of Crz1-GFP was inspected by either confocal microscopy or fluorescence microscopy in combination with a microfluidic device, SP039 and SP017 cells respectively, were cultivated in a low-fluorescent minimal medium lacking riboflavin and folic acid [30] containing 50 mM TAPS, and high pH stress was initiated as described above. For immunoblot analysis of Ena1-GFP, cells were grown overnight in YPD at 28°C in liquid medium and then resuspended in 250 ml of fresh YPD containing 50 mM TAPS adjusted to pH 5.5 to OD600 = 0.1. Cells were grown to reach OD600 = 0.4 and then treated with 5 M KOH.
Plasmids and recombinant DNA techniques
Escherichia coli DH5α strain was used as plasmid DNA host and grown in Luria-Bertani (LB) broth at 37°C. If necessary, E. coli cultures were supplemented with ampicillin and/or chloramphenicol to a final concentration of 100 μg/ml and 34 μg/ml, respectively. Recombinant techniques and bacterial and yeast transformations were carried out by standard methods. Plasmid pLMB127 [22], containing the triple GFP fusion to N-terminus of Crz1, was a generous gift of M. Cyert (University of Stanford). Plasmid pGEX-6P1-3HA-YPI1 was constructed as follows. The DNA fragment containing YPI1 ORF, N-terminally tagged with 3 copies of the HA (hemagglutinin) epitope, was removed from pWS-Ypi1 [31] by BglII and SalI digestion and subcloned into the BamHI/SalI site of pGEX-6P-1 to express a GST-3HA-Ypi1 fusion protein in E. coli. The proper fusion of GST to 3HA-YPI1 DNA sequence was confirmed by restriction and DNA sequencing analysis.
Expression and purification of recombinant GST-3HA-Ypi1 protein in E. coli
Purification of the fusion protein GST-3HA-Ypi1 was carried out as described in [31], with some modifications. E. coli BL21-Codon Plus (DE3)-RIL cells were transformed with pGEX-6P1-3HA-YPI1 and grown overnight at 37°C in LB medium containing ampicillin and chloramphenicol. The culture was then diluted in the same medium to grow about 4 hours at 37°C to reach OD600 0.3, isopropyl-1-β-D-thiogalactopyranoside was added to a final concentration of 0.1 mM, and culture was grown overnight at 26°C. Cells were harvested and resuspended in ice-cold lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% Triton X-100, 2 mM DTT, 0.5 mM PMSF (phenylmethylsulfonyl fluoride) and complete protease inhibitor mixture (Roche Applied Science)). Cells were ruptured using a Bioruptor Plus UCD-300 equipment (Diagenode) provided with a cooling system (4°C) for 10 cycles (high intensity; 30 s of sonication followed by 30 s pause). The fusion protein was purified by incubating the bacterial crude lysate with Glutathione Sepharose resin (ABT beads, #4B-GLU-10, 50% solution in PBS) overnight at 4°C with gentle shaking. GST-3HA-Ypi1 fusion protein was eluted with 10 mM reduced glutathione and aliquots were stored at -80°C. The amounts of the eluted fusion protein were quantified using protein gels. Three different amounts of each protein eluate were loaded on an SDS/PAGE gel alongside with serial dilutions of BSA (Sigma-Aldrich) ranging from 0.6 to 2 μg. The Coomassie Brilliant Blue stained gel was scanned and the Gel Analyzer software (http://gelanalyzer.com/) used to quantify the amount of GST-3HA-Ypi1 by comparison of the staining intensity of the bands corresponding to the fusion protein and the BSA bands.
Preparation of total protein yeast extracts
For Crz1-3HA immunodetection, whole cell protein extracts were prepared by harvesting 10 ml of yeast cultures (OD600 ~ 0.7 at t = 0), whereas for Ena1-GFP the appropriate volume was centrifuged to harvest ~12 OD600 units of cells. In both cases cells were resuspended in ice-cold lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 10% glycerol, 1 mM DTT, 2 mM PMSF and complete protease inhibitor mixture (Roche Applied Science)). Zirconia/Silica beads (0.1 g, 0.5 mm diameter; #11079105z, BioSpec) were added to the suspensions, and then cells were ruptured using FastPrep®-24 (MP Biomedicals) with potency 5.5 for ~ 2 min. The lysates were centrifuged at 500 x g for 10 min at 4°C and aliquots of the supernatants stored at -80°C.
Quantitative immunoblotting
The amounts of Crz1-3xHA and Ena1-GFP were determined by quantitative immunoblotting by comparing the Crz1 signal with GST-3HA-Ypi1 prepared and quantified as described above and the Ena1-GFP signal with recombinant GFP (rGFP) standard (#MB0752; Vector Laboratories, Inc.). Calibration standards were prepared by diluting rGFP or GST-3HA-Ypi1 into total protein extracts (described above) from the relevant strains to provide a range of the standard from 0.22 to 1.33 ng (Ypi1-3HA) and 2.5 to 10 ng (rGFP). 80 μg (for Ena1-GFP) or 70 μg (for Crz1-HA) of total protein were loaded on 10% (Ena1) or 8% (Crz1) SDS gels, electrophoresed and transferred to Immobilon®-P PVDF Transfer Membrane (Millipore). To ensure the homogeneity of the process, each complete experiment was contained in a single gel. Membranes were incubated overnight with anti-GFP (#11 814 460 001; Roche Applied Science) or with anti-HA (#11 583 816 001; clone 12CA5; Roche Applied Science) at 1:1000 dilution to detect Ena1-GFP and Crz1-3xHA, respectively. A 1:10000 dilution of horseradish peroxidase-conjugated anti-mouse antibody was used to detect primary antibodies. HA immunocomplexes were visualized by the ECL Prime Western Blotting Detection kit (GE Healthcare Life Sciences) and GFP immunocomplexes were visualized by the WESTAR Supernova kit (Cyagen). The chemiluminescence was recorded with a VersaDocTM 4000 MP imaging system (BioRad). GelAnalyzer software was used to quantify signal intensities.
Chromatin Immunoprecipitation (ChIP) and ChIP-Sequencing (ChIP-Seq) assays
ChIP experiments were performed as described previously [5]. The biological material for ChIP-Seq was prepared essentially as for ChIP with the following exceptions: i) chromatin was fragmented using a Bioruptor Plus UCD-300 equipment (Diagenode) provided with a cooling system (4°C) for 30 cycles (high intensity; 30 s of sonication followed by 60 s pause) to generate fragments of ≤ 500 bp length; ii) the supernatant was pre-cleared with protein G sepharoseTM fast flow (GE Healthcare, #17-0618-01) beads for 1 h at 4°C and then, the pre-cleared cell lysate was incubated with 1 μg of polyclonal anti-HA ChIP-grade (Abcam, #ab9110) antibody overnight at 4°C; iii) anti-HA-Crz1-DNA complexes were collected with protein-G-sepharose beads incubating for 1 hour at 4°C and iv) after reverse crosslinking, DNA was extracted using a NucleoSpin® Gel and PCR Clean-up kit (Macherey Nagel, #740609) and NTB buffer (Macherey-Nagel, #740595); v) recovered DNA (40 ng/reaction) was subjected to PCR using oligonucleotides that recognize various regions of ENA1 promoter sequences. Oligonucleotide primer sequences are given in S1 Table. ChIP libraries were prepared using the TruSeq ChIP Sample Preparation Kit (Illumina) and then subjected to paired-end deep sequencing with the MiSeq Reagent Kit v2 (300 cycle) to provide reads of around 150 nt. FASTQ files were mapped using Bowtie2 (version 2.1.0) to generate the corresponding SAM files. Mapped reads (1.9 to 3.7 million/time point) were sorted and indexed using IGV tools [32]. Subsequent analysis was performed using the SeqMonk software (Babraham Institute, http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/). The S. cerevisiae EF4 genomic data was employed. The chromosomal coordinates of the intergenic regions and information on the flanking genes were obtained from the Saccharomyces Genome Database using the YeastMine tool. This information was loaded into SeqMonk as an Annotation set and was used to generate 6303 intergenic probes. Subsequently, each of these probes were tiled in contiguous sections of 50 nt. This generated a total number of 65198 probes that were quantified by the read count quantification method, with identical reads removed and correction for total read count referred to the largest Dataset.
RNA preparation and cDNA synthesis
Total RNA was purified using the RiboPure Yeast kit (Ambion; #AM1926) following the manufacturer’s instructions. RNA quality was assessed by electrophoresis in a denaturing 1.2% agarose FlashGel RNA cassette (Lonza). RNA concentrations were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). The RNA was reverse-transcribed with the iScript cDNA synthesis kit (Bio-Rad; #170–8890) following the manufacturer’s guidelines. The reaction contains 500 ng of total RNA as a template in a reaction volume of 10 μl.
qPCR assay
Real time quantitative PCR (qPCR) was performed using the SsoAdvancedTM SYBR® Green Supermix kit (BioRad; #172–5260) in 25 μl QPCR reaction according to the manufacturer’s protocol in a 96-well plate (4titude Ltd. United Kingdom; #4ti-0720/C). The samples were amplified using a CF96TM Real-Time PCR System (BioRad). For quantification of ChIP-derived samples a calibration curve was made using plasmid pKC201 that carries the entire ENA1 promoter [11]. In all ChIP experiments, no-template controls, no-IP controls and input samples were tested in parallel for every primer set used. To determine enrichment, the 2-∆∆Ct method was used by comparing enrichment values for positive primer pairs to a negative primer pair between experimental ChIP experiments and reference (pKC201). The amount of ENA1 mRNA was quantified with a calibration curve made with plasmid pCM262-ENA1-GFP [26] containing the entire ENA1 DNA sequence. The ENA1 and ACT1 (used as a reference) specific pairs of primers (S1 Table) were used to determine the respective mRNA levels.
β-galactosidase activity assays
The wild type strain BY4741 and its derivative crz1::kanMX4 mutant strain were transformed with plasmid pKC201 [11,33], which contains the entire promoter of ENA1 fused to lacZ. Transformed yeast cells were grown to saturation under selective condition in SD media and then inoculated in YPD at A600 0.2. Growth was resumed until A600 = 0.8, then the cultures were distributed into 10 ml aliquots, centrifuged and resuspended in the appropriate media. To induce alkaline stress cells were resuspended in YPD containing 50 mM TAPS adjusted to pH 8.0 with 1 M KOH. Non-induced cells were resuspended in YPD medium. At the appropriate times 1 ml aliquots were centrifuged and β-galactosidase activity was measured as described [34].
Time-lapse fluorescence microscopy
The time-lapse movie was created by using a combination of microfluidic device and fluorescence microscopy using cells expressing Crz1-3xGFP from plasmid pLMB127. We observed that overexpression had no effect on Crz1 localization dynamics (compared with SP017 cells containing chromosomally-tagged GFP version of Crz1). During microscopy experiments cells were maintained in a fixed position in the CellASIC microfluidic flow chamber (Y04C plate) controlled by the ONIX Control System (CellASIC) and followed by fluorescence microscopy during repeated exchange of medium of pH 5.8 and pH 8.0. The microfluidic device was mounted onto a Nikon TE2000-E inverted microscope with Hamamatsu Camera Controller C-4742-80-12AG, and images were taken with an oil-immersion 40x objective. GFP excitation was detected by illumination with blue light (450–490 nm) and images were acquired at 10 s intervals, automated with Metamorph (Molecular Devices).
For quantification of cytoplasmic vs. nuclear Crz1 in a cell population before and after induction of alkaline pH stress, living cells carrying Crz1-GFP and Sik1-mRFP chromosomally-tagged versions (SP039 strain) were monitored by confocal microscopy over a time course of 20 min. The mRFP signal was used to define the position of nucleus (Sik1-mRFP is constitutively localized in the nucleus / nucleolus). Cell suspensions of strain SP039 were deposited onto a glass bottom dish and allowed to settle. Alkaline pH stress was induced by gentle removing the media placing the pipette tip at the edge of the dish and then quickly replaced by the low-fluorescent medium adjusted to pH 8.0. The Leica TCS SP2 confocal laser scanning microscope equipped with objective HCX PL APO 63x/1.40–0.60 oil was used to obtain confocal images. Images were acquired using emission interval 450–490 nm for GFP and 580–660 nm for mRFP. Pinhole was 1.25 AU. Generated GFP, mRFP, merged GFP-mRFP and bright field images of cell populations at all time points were analyzed using LAS AF Lite (Leica Microsystems) software. Bright field images were taken slightly below (~2 μm) the focal plane until a dark ring surrounds cell (useful for the Cell-ID software to find boundaries of cells, see below).
Image analysis
We performed image analysis and data processing using the open-source software packages ImageJ and Cell-ID version 1.4 as described [35]. Briefly, a time-related series of GFP, mRFP and out-of-focus bright field images acquired by confocal microscopy were converted into grey-level TIFF files of 8 bits using ImageJ software. Individual cells of regular yeast shape were found by Cell-ID software using an algorithm that calculates cell boundaries based on comparison of pixels of dark cell boundaries and bright background. The cell boundaries were then translated to the corresponding fluorescence images (from both GFP and RFP channels) and the values corresponding to either nucleus, total or background fluorescence were calculated. To calculate the nuclear fluorescence signal of Crz1-GFP in a single cell, we divided the nuclear fluorescence value by the calculated nuclear area and subtracted fluorescence background signal. The final nuclear GFP value was corrected for photobleaching effect as follows: the region of interest (ROI) of 65 individual cells was bleached with a 488 nm laser under the same conditions as the experiment was performed (i.e., scan speed, zoom, laser power, number of exposures to laser, microscope objective). Average fluorescence intensity of all ROIs for each time point was calculated and normalized to initial fluorescence (100%). The intensity drop attributable to the bleaching of a single ROI was typically 20% at the end of the experiment. Calculated average intensity loss per each time point was subtracted from the nuclear Crz1-GFP values of the alkaline pH experiment. In total approximately 600 individual cells exposed to high pH were tracked.
Other techniques
Cell volume of BY4741 cells growing in YPD supplemented with 50 mM TAPS before and 2 min after induction of alkaline pH stress was measured using a ScepterTM 2.0 Cell Counter and 40 μm-Scepter Sensors (Millipore) according to the manufacturer’s instructions.
Mathematical modeling
The mathematical models created here use systems of ordinary differential equations. Given that the mechanisms involved in the various processes are not known in detail, we used the power law formalism [36–38]. This formalism allows us to mathematically represent linear and non-linear dynamics of biological processes whose details are poorly characterized [39,40]. This formalism facilitates the systemic integration of processes into a single mathematical model, enabling the analysis and understanding of the integrated behavior in the system (see Methods and [41–44]. The power law formalism has two types of parameters: apparent rate constants and apparent kinetic orders.
We used constrained optimization to estimate parameter values for the models. The constraints are used as a means to ensure that parameter values have physically plausible values. Apparent rate constants are constrained to be larger than zero, as negative rate constants are physically impossible. Apparent kinetic orders are constrained to be larger than zero and smaller than a small integer, as described [45]. This is due to the fact that such kinetic order cannot be larger than the number of binding sites for the variable in the process of interest.
Differential equations were fitted to the experimental data as described in [45,46]. We implemented the parameter fitting procedure using the NonLinearModelFit function in Mathematica, for all parameter fitting calculations. Initial estimates for the parameter values in the fitting process were obtained as described in S1 Text.
A commonly accepted general assumption was made in the modeling of mRNA dynamics: we assumed that mRNA degradation linearly depends on mRNA abundance. This assumption has been used to model mRNA degradation in many systems, ranging from bacteria, to multicellular eukaryotes (e.g. [45,47–53], and including yeast (implicit in Eq. 6 of ref. [54] and explicit in Ref. [55]). Although simplifying, this assumption is sufficiently accurate in most cases (e. g. [45,49,51] and is consistent with many experimental measurements (e.g. [53,54]).
Results and Discussion
Time-course analysis of ENA1 promoter activity and mRNA accumulation
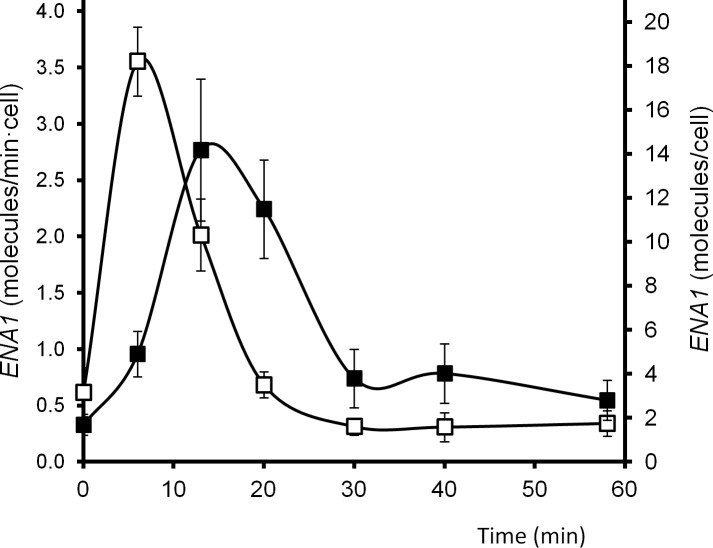
The promoter activity of ENA1 was calculated from the data recently obtained from our Genomic Run On (GRO) analysis [27] of the changes caused by sudden transition from pH 5.5 to a moderate alkalinization (pH 8.0). This specific shift was selected because it generates a vigorous ENA1 mRNA accumulation response and results only in a transient delay in proliferation with no significant loss of cell viability. In addition, we determined that this change barely affects cell volume (60 fL at pH 5.5 vs 56 fL at pH 8.0). As shown in Fig 1, transcription of ENA1 peaks after 6 min of initiation of the stress and at this moment the transcription rate is estimated in 3.5 molecules/min*cell. ENA1 mRNA levels peaks at 13 min after the stress, and reach a maximum of 14 molecules per cell. Therefore, the transition between peak transcription and maximum mRNA levels takes approximately 7 min. The comparison of the profiles of transcription rate and mRNA levels suggests that changes in mRNA stability would play a relatively minor role and that the amount of ENA1 mRNA would depend largely of the activity of the promoter.
Fig 1. Transcription rate (TR) and mRNA accumulation (RA) of ENA1 upon high pH stimulation.
The transcription rate (open squares) and mRNA amount (closed squares) for the ENA1 gene under alkaline stress was obtained from GRO experiments [27]. In order to convert the radioactive intensities into real units, the mRNA concentrations and mRNA half-lives for the most expressed genes published in (Wang et al., 2002) were compared with radioactive intensities of RA and TR for the same genes at time 0 (steady-state conditions) to obtain a conversion factor. The conversion factor was applied to the complete set of GRO data under alkaline stress. ENA1 data were extracted from this set and represented as the average ± SEM of three experiments.
Kinetics of alkaline pH-induced nuclear-cytoplasmic shuttling of Crz1
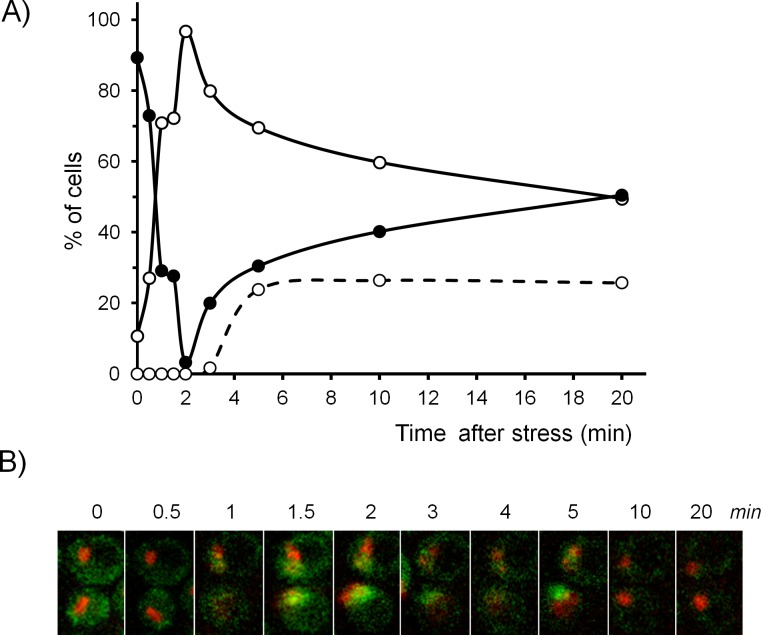
The rapid response of the ENA1 promoter is congruent with a substantial presence of the calcineurin-activated transcription factor Crz1 in the nucleus after 2.5–5 min upon alkaline stress, as it was documented earlier [17]. We therefore considered necessary to quantitatively monitor, in a time-resolved manner, the traffic between cytosol and nucleus of Crz1 upon alkalinization. To this end, we used strain SP039, which expresses chromosomally encoded version of Crz1 fused to GFP and Sik1 (a nuclear localization marker) fused to mRFP. The subcellular distribution of Crz1 in cells shifted to pH 8.0 was monitored using time-lapse confocal microscopy during 20 min. As observed in Fig 2A, an increase in the number of cells with nuclear Crz1 localization is already detected after 30 s of alkalinization, and a peak with >95% of cells show nuclear localization of the transcription factor, is reached after 2 min. Interestingly, the subsequent rapid decline in the number of cells in which Crz1 is nuclear is attenuated after 5 min of the onset of the stress due to a second wave of Crz1 returning to the nucleus that is observed in about 25% of the cells monitored (an example is shown in Fig 2B). Although in a given cell Crz1 could subsequently shift back again to the cytoplasm, the overall percentage remained constant during the rest of the experiment, with about half of the cell population showing nuclear Crz1 localization. The extremely fast nuclear entry of Crz1 shown in Fig 2A is in keeping with the almost immediate peak of intracellular calcium detected 10–20 sec after raising the pH of the medium from 5.5 to 8.1 [15] and suggests that the interaction of Crz1 with the ENA1 promoter is an important determinant in the rapid transcriptional induction observed.
Fig 2. Time-course of the subcellular distribution of Crz1 upon high pH stress.
A) Cultures of strain SP039 (CRZ1:GFP-kanMX6 SIK1:mRFP-kanMX6) were shifted from pH 5.5 to pH 8.0 and the localization of Crz1 monitored by fluorescence confocal microscopy as described in Materials and Methods. Open circles denote the percentage of cells with nuclear Crz1 localization; closed circles, cytosolic localization. The discontinuous line show cells in which Crz1 entered the nucleus, then left the nucleus and later returned to it (they are, therefore, a subset of the total nuclear Crz1 values). Sik1 signal was used as marker of constitutive nuclear (nucleolar) localization. Data is presented as percentages calculated for 90 to 160 cells examined for each time-point. B) Example of time-lapse images of SP039 cells before and after induction of alkaline stress over a 20 min time-course (as in panel A).
It is worth noting that the pattern of nuclear accumulation of Crz1 upon high pH stress differs from previously reported examples. Thus, it has been documented that significant accumulation of Crz1 in the nucleus upon exposure to calcium occurs only after 5 to 20 min [21,56–58], whereas 10 to 15 min of incubation with some toxic compounds, such as methylglyoxal or arsenite, is required to detect nuclear Crz1. For other stimuli able to activate calcineurin, such as blue light, a period of as much as 10–20 min is necessary to detect significant nuclear accumulation of Crz1 [59]. This is in contrast with the very rapid accumulation of Crz1 in the nucleus upon alkaline stress. In addition, treatment of yeast cells with 0.2 M calcium results in sporadic, unsynchronized nuclear localization bursts [57], whereas nuclear localization of Crz1 upon exposure to blue light is persistent [59]. It must be noted that the second wave of nuclear Crz1 detected in our experiments cannot be explained by the nuclear localization bursts observed upon stimulation of the cells with calcium [57], since such bursts typically lasted 2 min, whereas we observe a rather stable profile during the time of alkaline stress (see S1 Video in Supplemental Material). Therefore, alkaline stress might invoke a distinctive and unique pattern of nuclear accumulation of Crz1. It is currently unclear whether this late return of Crz1 to the nucleus is caused by a second spike of cytosolic calcium, because after the initial burst upon alkaline stress, calcium levels return to baseline values in a matter of seconds and remain unchanged for at least three minutes [60]. It is worth noting that nuclear Crz1 regained cytosolic localization very shortly (1–2 min) after extracellular pH decreased to pH 5.8 and returned quickly to the nucleus when pH was raised again to 8.0, with no signal of refractory period (see accompanying S1 Video).
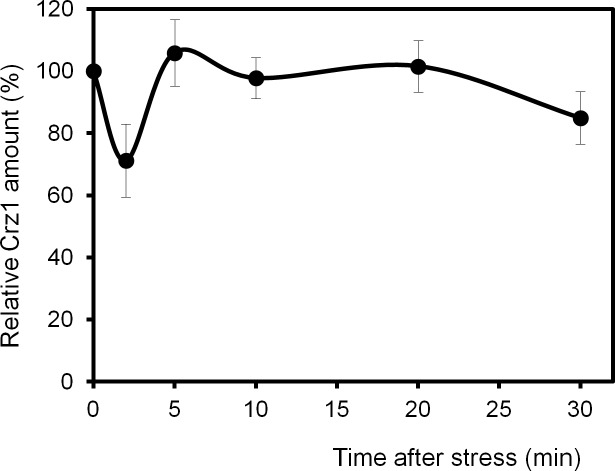
Although significant changes in CRZ1 gene expression upon cell stress have not been reported, we considered important to monitor, in addition to its subcellular localization, hypothetic changes in the cellular amounts of the transcription factor after high pH stress. This was tested by immunoblot using a strain expressing a chromosomally encoded Crz1 protein C-terminally tagged with a triple HA epitope and using known amounts of recombinant GST-3HA-Ypi1 protein as an internal standard for signal comparison. Using this method we determined a value of 186 ± 32 molecules of Crz1/cell in non-stressed cells, which is somewhat lower than that reported by most previous determinations [60]. It must be noted that the existing measurements largely differ among them (up to 20-fold). In any case, the amount of Crz1 during the first 30 min upon high pH stress did not change significantly (Fig 3), with the exception of the minute time point, 2 where a decrease of almost 30% was observed (p<0.05). However, we believe that these variations occur too fast to represent actual changes in the amounts of cellular Crz1 and we postulate that this difference, which corresponds to the time of maximum nuclear Crz1 accumulation (see Fig 2A) and massive Crz1 binding to its genomic target regions (Fig 4B), might be an artifact caused by the inability of the method used for protein extract preparation to quantitatively recover nuclear, DNA-bound Crz1.
Fig 3. Relative levels of Crz1 after switching cells to pH 8.0.
The number of Crz1 molecules/cell was calculated as described in Material and Methods and the value just prior alkalinization of the medium was set as the reference (100%). Data correspond to the mean ± SEM from 8 independent experiments.
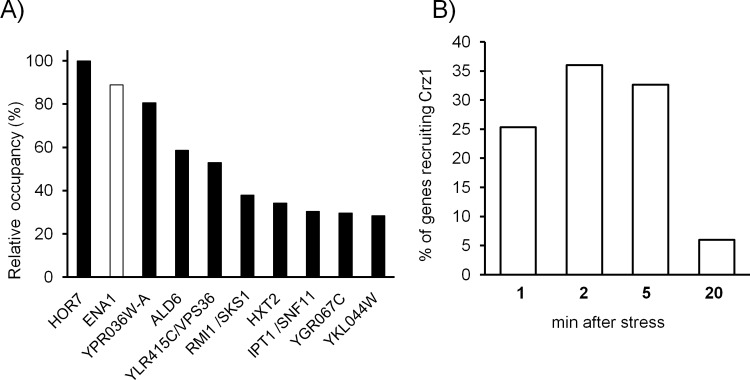
Fig 4. Genome-wide recruitment of Crz1 to gene promoters in response to high pH stress.
ChIP-seq technology was used to identify intergenic regions immunoprecitating with Crz1 and accumulating at least two-fold reads than the genome average. A) The 150 genes showing peaks of reads above the indicated threshold were ranked according the number of reads at the peak. The relative occupancy with respect the highest value (HOR7) is shown for the top ten genes in the ranking. B) The percentage of genes showing the peak of accumulation of reads at a given time after alkalinization of the medium is shown.
Analysis of Crz1 binding to the ENA1 promoter upon high pH stress
As a next step, we studied the kinetics of Crz1 binding to the ENA1 promoter after high pH stress. This was done by genome-wide examination of Crz1 binding to high pH-responsive promoters by ChIP-Seq analysis followed by specific analysis of previously identified CDREs. Samples were taken 1, 2, 5 and 20 min after shifting cells from pH 5.5 to 8.0. Accumulation of Crz1 was detected at 152 intergenic regions, of which 100 could be attributed to specific genes, whereas 52 corresponded to divergent promoters. ENA1 showed a very robust response, as it was observed to be the gene with the second highest number of reads at the promoter (Fig 4A). No reads were mapped at the ENA2 and ENA5 promoter regions, in agreement with the almost null response of these genes to alkaline stress (data not shown).
Recruitment of Crz1 was a very rapid process (Fig 4B), with over 60% of the promoters showing maximum binding within the first two minutes after alkaline pH stress. In many cases (66%), binding to the promoter decreased substantially after 20 min (as determined by the % of cases in which reads at that time were less than at any other time after the onset of stress). These observations fit well with the timing for nuclear Crz1 entry described above (see Fig 2). A detailed study of the genome-wide promoter binding of Crz1 will be published elsewhere (Roque et al. manuscript submitted).
Concerning ENA1, maximal occupancy was observed at 2 and 5 min after alkalinization, with significant increase already after one min. Interestingly, occupancy of the promoter region was still substantial (∼65% of the maximum) after 20 min, suggesting a sustained activation. This is in contrast with the very low transcription rate observed at 20 min (Fig 1), and suggests that negative regulatory components of ENA1 transcription that are removed from the promoter upon high pH stress are resuming their repressor function. This interpretation fits with the observation that the Mig2 repressor, known to act on the ENA1 promoter [14], exits the nucleus in the first few minutes upon alkalinization but recover their nuclear localization 15 min after induction of the stress [5].
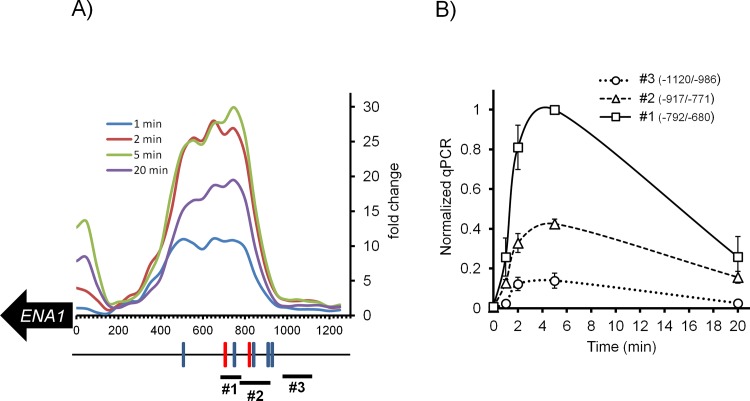
As shown in Fig 5A, several putative CDREs can be predicted in the ENA1 promoter, although the accumulation of Crz1 peaks in a region containing the two CDRE regions characterized by Serrano's laboratory [23], denoted in red in Fig 5A. Because of the close proximity of both CDREs (-813/-821 and -719/-727 relative to the ENA1 initiating Met codon) ChIP-Seq data does not provide the required resolution to evaluate the relative contribution of each element for Crz1 binding to the promoter. To solve this question we carried out ChIP experiments followed by qRT-PCR analysis using oligonucleotide pairs specifically encompassing each CDRE (denoted as #1 and #2 in Fig 5A). As shown in Fig 5B, the downstream element (-719/-727) was more effective in recruiting Crz1 (>2-fold compared with the upstream one). In both cases binding was much more prominent after 2 and 5 min of shifting cells to pH 8.0. Amplification of a region between nt -986/-1120 (#3), for which no CDRE is predicted, showed only residual accumulation of Crz1. These results confirm in vivo the previously observed in vitro binding of Crz1 to these sequences [23], although our data suggest that both CDREs are stress-responsive (see below). Recruitment to the region -493/-595, containing the putative CDRE at position -528/-537, was barely above the negative reference region (not shown), suggesting that this sequence is not functional. This perfectly fits with the existing evidence that the region between nucleotides -573/-490 of the ENA1 promoter was unable to drive transcription when cells were exposed to calcium chloride and that, while able to transcriptionally respond to high pH, this effect was insensitive to FK506 (an inhibitor of calcineurin) and not affected by mutation of CRZ1 or CNB1 [10].
Fig 5. Time-course profile of Crz1 recruitment at the ENA1 promoter following alkaline stress.
A) Cumulative counts of reads using a 50-nt window spanning the ENA1 promoter region are represented as fold-change over reads counted at time 0. The positions of CDRE deduced from computational analysis using the PWMTools website (http://ccg.vital-it.ch/pwmtools/pwmscan.php) of the Swiss Bioinformatics Institute are indicated as colored boxes on the black line. Red boxes denote the CDRE previously characterized in [23]. B) Evaluation of relative Crz1 binding to CDREs at positions -813/-821 and -719/-727 (red boxes in panel A). Chromatin immunoprecipitation using HA-tagged Crz1 was carried out and the immunoprecipitate used to amplify the -680/792; -771/-917; and -986/-1120 promoter regions (denoted as thick short lines 1, 2 and 3, respectively, in panel A). Data is presented as the mean ± SEM from 3–4 individual experiments assayed in duplicate and expressed as relative to the maximum value.
Contribution of Crz1 to alkaline pH-induced ENA1 transcription and protein accumulation
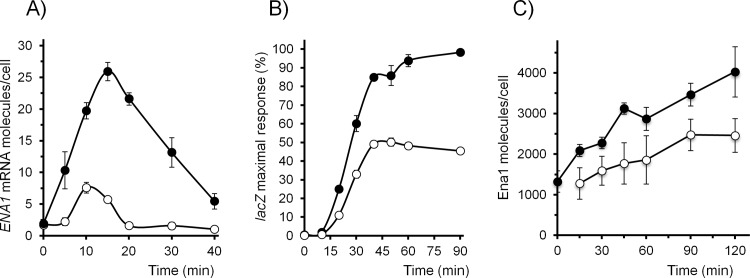
The contribution of Crz1 to the increase of ENA1 mRNA levels was investigated by qRT-PCR. As shown in Fig 5A, mRNA levels in the wild type strain peaked at 15 min with a value of approximately 25 molecules/cell. These kinetics profile of accumulation fits fairly well with that shown in Fig 1, taking into account the different technologies employed (Fig 1 data derives from DNA macroarray experiments [27]. Lack of Crz1 results in a drastic reduction of mRNA accumulation (∼ 6 molecules/cell). In addition, the peak is observed slightly earlier than in wild type cells (10 min, instead of 15). This indicates that Crz1 is not the only factor triggering the early induction of the ENA1 promoter.
We then tested the effect of the absence of Crz1 on the ENA1 promoter using a translational fusion of this promoter with the lacZ gene, encoding β-galactosidase. In contrast to the dramatic effect of the absence of Crz1 on ENA1 mRNA levels, a reduction of only about 50% of the total activity was detected (Fig 6B). We next monitored by immunoblot the total amount of Ena1 protein produced in the cell using a GFP-fused version of the protein. As shown in Fig 6C, the abundance of Ena1 in non-stressed cells was estimated in 1324 molecules/cell. This value is very close to the estimated value integrated from the existing literature for non-stressed cells [60] although it must be noted that the experimentally reported values show a very wide range (from nearly absent to about 7000 molecules/cell). Stimulation of cells with pH 8.0 caused the almost continuous accumulation of Ena1, reaching 4030 molecules/cell at the end of the experiment (120 min). Evaluation of Ena1 in the crz1 mutant background in non-stressed cells was not possible because the amount of the protein was below the sensitivity of the assay (∼500 molecules/cell). Upon induction by high pH, we observed that Ena1 accumulated at lower rate than in wild type cells, reaching a maximum (2476 molecules/cell) at 90 min.
Fig 6. Impact of the absence of Crz1 on mRNA levels and Ena1 protein accumulation after high pH stress.
A) Time-course of the accumulation of Ena1 mRNA after shifting cells to pH 8.0. Total RNA from wild type (closed circles) and crz1 (open circles) cells was extracted and mRNA levels were determined by qRT-PCR and quantified as indicated in Materials and Methods. B) The strains described above were transformed with plasmid pKC201, carrying the lacZ reporter fused to the ENA1 promoter. Cells were exposed to pH 8.0 for the indicated times, collected, and β-galactosidase activity measured. C) Strain MLM001 (CRZ1) and MLM002 (crz1), carrying in both cases C-terminally GFP-tagged version of ENA1 were shifted to pH 8.0 for the indicated times and the amount of total Ena1 quantified by immunoblot using anti-GFP antibodies and internal standards of recombinant GFP protein. Data is presented as the mean ± SEM from 3 (panel A) or 4 (panels B and C) independent experiments.
Data-driven modeling of Crz1-regulated Ena1 dynamics
The experimental results described above were integrated into a multilevel mathematical model in order to understand the differences between the dynamic behavior of Ena1 in wild type and mutant strains from 0 to 60 min after the stress challenge. We modeled the dynamic behavior of Ena1 at the mRNA and protein levels, through a set of ordinary differential equations that use the power law formalism (see Methods).
Eq 1 describes the dynamic behavior of ENA1 mRNA:
| Eq 1 |
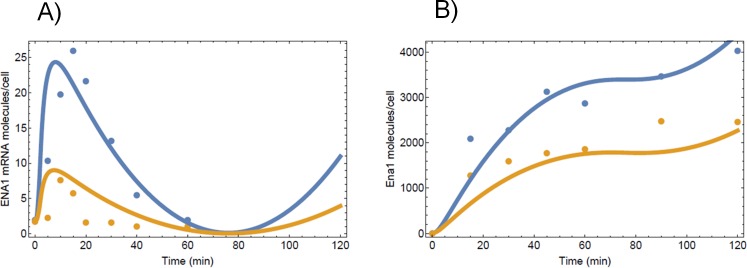
As observed in Fig 7A, there is a basal expression of Ena1 mRNA that is independent of Crz1. This is represented by the term α0(t), which we estimated using a time-dependent spline function adjusted to fit the curve corresponding to the crz1 mutant in Fig 6A. The term α1 Crz1ng1 represents Crz1 driven Ena1 mRNA synthesis. α1 represents an apparent rate constant, Crz1 represents the nuclear Crz1 concentration, and g1 represents the apparent kinetic order of nuclear Crz1 in the Ena1 mRNA synthesis. Values for α1 (= 0.00025 molecules/cell min-1) and g1 (= 2) were obtained by fitting this term to a time curve obtained by subtracting Crz1-independent Ena1 mRNA synthesis from total Ena1 mRNA synthesis (crz1 curve—WT curve in Fig 6A). Finally, the term α2 Ena1mRNA represents the degradation of Ena1 mRNA. The value for the apparent rate constant α2 was calculated after assuming that degradation linearly depends on mRNA concentrations (see Methods). Using the parameter fitting procedure described in Methods and the data from Fig 7A we estimated that α2 = 0.2 molecules/cell min-1).
Fig 7. Modeling of Ena1 mRNA and protein abundances upon high-pH stimulation.
Time-course of the Ena1 mRNA (panel A) and protein accumulation (panel B) after shifting cells to pH 8.0. Experimental data are shown as dots and the mathematical estimations, as lines (blue for wild type and yellow for mutant strains).
Eq 2 describes the dynamic behavior of the Ena1 protein:
| Eq 2 |
The term α3 mRNAg3 represents protein synthesis. We calculate the values for α3 (= 14 molecules/cell min-1) and g3(0.6) by adjusting this function to to experimental data (Fig 7B). The term α4 Ena1pr represents the degradation of the Ena1 protein. This degradation is also assumed to be proportional to the amount of protein, as it has been reported to occur in many situations [54]. The rate constant α4 (= 0.3 molecules/cell min-1) is also calculated by fitting the function to the crz1 experimental data from Fig 6C.
The parameter values given above were calculated for the wild type strain. For the crz1 mutant strain, Ena1 mRNA dynamics is represented by Eq 3
| Eq 3 |
We maintain the values calculated for the wild type α0(t) and α2 in the mutant model. The dynamic behavior of the wild type and crz1 models are given in Fig 7, where we also show the experimental dynamic behavior of the system. As observed, these models fit the experimental data very well, as shown by the Adjusted R2 (0.72 for the mRNA and 0.87 for the protein). These values indicate the percentage of variation in the experimental measurements that are justified by the models. In addition to the well adjusted R2s of our models, there is another experimental observation that supports our model. Based on the available experimental data, our parameter fitting for the multi-level model only uses data from times 0 to 60 min after stress induction. Nevertheless, Ena1 protein level measurements are available for up to 120 minutes after stress induction. When we run the model for 120 minutes, it quantitatively describes the full 120 min time course for Ena1 (Fig 7).
Model-based biological hypothesis generation
There are several hypotheses that can be inferred from this data driven model. The first hypothesis relates to the function of Crz1 in regulating ENA1 mRNA gene expression. Crz1 relocates from the cytoplasm to the nucleus during the first stage of the stress response. It then shuttles back to the cytoplasm, and subsequently part of the Crz1 population returns to the nucleus. It must be noted that correct fitting of our model to the observed dynamic behavior of the wild type and crz1 strains requires the second wave of nuclear Crz1 to be fully effective in regulating ENA1 gene expression. The second hypothesis comes from that fact that the value for the apparent kinetic order for Crz1 in the Crz1-dependent Ena1 mRNA synthesis is 2, which would be consistent with two effective Crz1 binding sites required for Ena1 mRNA synthesis. This would fit with our in vivo data (Figs 6 and 7) and previous evidence indicating in vitro binding of Crz1 at two separate promoter regions [23]. It has been shown that mutation of the downstream ENA1 CDRE element was able to largely abolish the transcriptional response to calcium and high pH stress, and there is evidence that short regions containing the ENA1–712/-733 region are able to drive transcription in response to calcium [23], high pH [10], or as a result of the deletion of the PPZ1 phosphatase gene [61]. Therefore, it seems clear that in vivo binding of Crz1 to this genomic region triggers a functional transcriptional response resulting from activation of calcineurin. In contrast, it was shown that the upstream element alone was unable to drive transcriptional response to calcium stress, leading to the proposal that this region was mostly responsible for basal expression of ENA1 [23]. However, we find significant recruitment of Crz1 to this region in response to high pH stress, accounting for about one third of the total recruitment to the promoter (Fig 5B) and our model supports a 2-site regulatory scenario. It is plausible that recruitment of Crz1 to the downstream region alone would be insufficient to activate the promoter, but would enhance the effect derived from Crz1 recruitment to the upstream region. Such synergistic effect would fit with the model proposed earlier by Mendizabal and coworkers [23].
Our third hypothesis is somewhat more complex. We observe that the time course for the stress-dependent expression of Ena1 mRNA in the crz1 mutant almost perfectly correlates with that of the wild type. This can be seen when calculating the correlation between β-galactosidase activity driven by the ENA1 promoter in wild type and crz1 strains (Spearman correlation = 0.83). This high correlation suggests that Ena1 mRNA expression might be regulated by other transcription factors that have a dynamic behavior similar to that of Crz1. If this is so, the dynamics of Crz1 in the wild type could be used as a proxy to estimate the dynamics of these other transcription factors in the crz1 mutant, allowing us to rewrite Eq 3 as:
| Eq 4 |
If we approximate f(Crz1) using the same power law formalism as above, then
| Eq 5 |
Adjusting this new function to the experimental data, we obtain α'0 = 0.00015 and g0' = 2. We note that α'0≈α1/1.7, which is the approximate average ratio between the expression of β-galactosidase from the ENA1 promoter in the wild type vs. the crz1 mutant. This suggests that the regulation of Ena1 mRNA expression by its different transcription factors during stress response might be additive. In addition, the value for g0' suggests that the number of effective binding sites for transcription factors inducing stress-dependent Ena1 mRNA expression is 2. However, as deduced from Fig 6A, this Crz1-independent event must take place very shortly after the initiation of the stress. It is known that activation of ENA1 upon high pH stress is under the direct control of three pathways: calcineurin/Crz1, Snf1 and Rim101 [14],. However, the Rim101 pathway affects ENA1 expression by repressing the expression of the Nrg1 repressor [62] which binds to the ENA1 promoter at a region located between nucleotides -561/-573 [14]. Therefore, the impact of the activation of Rim101 on ENA1 transcription can only occur at medium-long term time-points. In fact, a delayed effect for Rim101 action has been shown for the high-pH responsive PHO89 promoter [5]. In contrast, Snf1 is rapidly activated by phosphorylation in response to high pH stress [5,63]. Snf1 can functionally interact with two pairs of repressors, Nrg1/Nrg2, and Mig1/Mig2. Nrg1 has been proposed to mediate Snf1-activated function in response to some stress conditions [64–66] and very fast Mig1 and Mig2 Snf1-dependent phosphorylation, concurrent with rapid exit of these repressors from the nucleus, has been recently documented [5]. As both Nrg1 and Mig2 are known to repress the ENA1 promoter under non-stress conditions, we propose that the short-term, Crz1-independent ENA1 transcriptional activity is essentially promoted by activation of the Snf1 pathway.
Supporting Information
(DOCX)
(DOCX)
(MP4)
Acknowledgments
We acknowledge the excellent technical assistance of Montserrat Robledo and Anna Barceló (Servei de Genòmica i Bioinformàtica, UAB) for support in ChIP-Seq experiments We thank Núria Barba (Institut de Neurociències, UAB) for assistance with confocal microscopy and the microfluidics device. Thanks are given to Hana Sychrova for the SIK1:mRFP strain and Martha Cyert for plasmid pLMB127.
Data Availability
Raw data concerning Crz1 ChIP-Seq experiments are held at the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE79342 and will be available immediately after publication of the paper.
Funding Statement
This work was supported by grants BFU2011-30197-C3-01, BFU2014-54591-C2-1-P and EUI2009-04147 (SysMo2) to JA. (Ministry of Industry and Competitivity, Spain, and Fondo Europeo de Desarrollo Regional [FEDER]). JA is the recipient of an Ajut 2014SGR-4 award (Generalitat de Catalunya). DC was recipient of a predoctoral fellowship from the Spanish Ministry of Education. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Arino J, Ramos J, Sychrova H. Alkali metal cation transport and homeostasis in yeasts. Microbiol Mol Biol Rev 2010;74(1):95–120 10.1128/MMBR.00042-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banuelos MA, Sychrova H, Bleykasten-Grosshans C, Souciet JL, Potier S. The Nha1 antiporter of Saccharomyces cerevisiae mediates sodium and potassium efflux. Microbiology 1998;144 (Pt 10)2749–58 [DOI] [PubMed] [Google Scholar]
- 3.Ramos J, Arino J, Sychrova H. Alkali-metal-cation influx and efflux systems in nonconventional yeast species. FEMS Microbiol Lett 2011;317(1):1–8 10.1111/j.1574-6968.2011.02214.x [DOI] [PubMed] [Google Scholar]
- 4.Ruiz A, Arino J. Function and regulation of the Saccharomyces cerevisiae ENA sodium ATPase system. Eukaryot Cell 2007;6(12):2175–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serra-Cardona A, Petrezselyova S, Canadell D, Ramos J, Arino J. Coregulated Expression of the Na+/Phosphate Pho89 Transporter and Ena1 Na+-ATPase Allows Their Functional Coupling under High-pH Stress. Mol Cell Biol 2014;34(24):4420–4435 10.1128/MCB.01089-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez P, Persson BL. Identification, cloning and characterization of a derepressible Na+-coupled phosphate transporter in Saccharomyces cerevisiae. Mol Gen Genet 1998;258(6):628–38 [DOI] [PubMed] [Google Scholar]
- 7.Zvyagilskaya RA, Lundh F, Samyn D, Pattison-Granberg J, Mouillon JM, Popova Y et al. Characterization of the Pho89 phosphate transporter by functional hyperexpression in Saccharomyces cerevisiae. FEMS Yeast Res 2008;8(5):685–96 10.1111/j.1567-1364.2008.00408.x [DOI] [PubMed] [Google Scholar]
- 8.Haro R, Garciadeblas B, Rodriguez-Navarro A. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett 1991;291(2):189–91 [DOI] [PubMed] [Google Scholar]
- 9.Garciadeblas B, Rubio F, Quintero FJ, Banuelos MA, Haro R, Rodriguez-Navarro A. Differential expression of two genes encoding isoforms of the ATPase involved in sodium efflux in Saccharomyces cerevisiae. Mol Gen Genet 1993;236(2–3):363–68 [DOI] [PubMed] [Google Scholar]
- 10.Serrano R, Ruiz A, Bernal D, Chambers JR, Arino J. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol Microbiol 2002;46(5):1319–33 [DOI] [PubMed] [Google Scholar]
- 11.Alepuz PM, Cunningham KW, Estruch F. Glucose repression affects ion homeostasis in yeast through the regulation of the stress-activated ENA1 gene. Mol Microbiol 1997;26(1):91–98 [DOI] [PubMed] [Google Scholar]
- 12.Serra-Cardona A, Canadell D, Ariño J. Coordinate responses to alkaline pH stress in budding yeast. Microbial Cell 2015;2(6):182–96 10.15698/mic2015.06.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marquez JA, Serrano R. Multiple transduction pathways regulate the sodium-extrusion gene PMR2/ENA1 during salt stress in yeast. FEBS Lett 1996;382(1–2):89–92 [DOI] [PubMed] [Google Scholar]
- 14.Platara M, Ruiz A, Serrano R, Palomino A, Moreno F, Arino J. The Transcriptional Response of the Yeast Na+-ATPase ENA1 Gene to Alkaline Stress Involves Three Main Signaling Pathways. J Biol Chem 2006;281(48):36632–42 [DOI] [PubMed] [Google Scholar]
- 15.Viladevall L, Serrano R, Ruiz A, Domenech G, Giraldo J, Barcelo A et al. Characterization of the calcium-mediated response to alkaline stress in Saccharomyces cerevisiae. J Biol Chem 2004;279(42):43614–24 [DOI] [PubMed] [Google Scholar]
- 16.Yoshimoto H, Saltsman K, Gasch AP, Li HX, Ogawa N, Botstein D et al. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J Biol Chem 2002;277(34):31079–88 [DOI] [PubMed] [Google Scholar]
- 17.Ruiz A, Serrano R, Arino J. Direct regulation of genes involved in glucose utilization by the calcium/calcineurin pathway. J Biol Chem 2008;283(20):13923–33 10.1074/jbc.M708683200 [DOI] [PubMed] [Google Scholar]
- 18.Matheos DP, Kingsbury TJ, Ahsan US, Cunningham KW. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev 1997;11(24):3445–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendizabal I, Rios G, Mulet JM, Serrano R, de Larrinoa IF. Yeast putative transcription factors involved in salt tolerance. FEBS Lett 1998;425(2):323–28 [DOI] [PubMed] [Google Scholar]
- 20.Stathopoulos AM, Cyert MS. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev 1997;11(24):3432–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polizotto RS, Cyert MS. Calcineurin-dependent nuclear import of the transcription factor Crz1p requires Nmd5p. J Cell Biol 2001;154(5):951–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boustany LM, Cyert MS. Calcineurin-dependent regulation of Crz1p nuclear export requires Msn5p and a conserved calcineurin docking site. Genes Dev 2002;16(5):608–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendizabal I, Pascual-Ahuir A, Serrano R, de Larrinoa IF. Promoter sequences regulated by the calcineurin-activated transcription factor Crz1 in the yeast ENA1 gene. Mol Genet Genomics 2001;265(5):801–11 [DOI] [PubMed] [Google Scholar]
- 24.Wieland J, Nitsche AM, Strayle J, Steiner H, Rudolph HK. The PMR2 gene cluster encodes functionally distinct isoforms of a putative Na+ pump in the yeast plasma membrane. EMBO J 1995;14(16):3870–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wadskog I, Forsmark A, Rossi G, Konopka C, Oyen M, Goksor M et al. The yeast tumor suppressor homologue Sro7p is required for targeting of the sodium pumping ATPase to the cell surface. Mol Biol Cell 2006;17(12):4988–5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marques MC, Zamarbide-Fores S, Pedelini L, Llopis-Torregrosa V, Yenush L. A functional Rim101 complex is required for proper accumulation of the Ena1 Na+-ATPase protein in response to salt stress in Saccharomyces cerevisiae. FEMS Yeast Res 2015;15(4):fov017 10.1093/femsyr/fov017 [DOI] [PubMed] [Google Scholar]
- 27.Canadell D, Garcia-Martinez J, Alepuz P, Perez-Ortin JE, Arino J. Impact of high pH stress on yeast gene expression: A comprehensive analysis of mRNA turnover during stress responses. Biochim Biophys Acta 2015;1849(6):653–64 10.1016/j.bbagrm.2015.04.001 [DOI] [PubMed] [Google Scholar]
- 28.Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998;14(10):953–61 [DOI] [PubMed] [Google Scholar]
- 29.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS et al. Global analysis of protein localization in budding yeast. Nature 2003;425(6959):686–91 [DOI] [PubMed] [Google Scholar]
- 30.Verduyn C, Postma E, Scheffers WA, Van Dijken JP. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 1992;8(7):501–17 [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Gimeno MA, Munoz I, Arino J, Sanz P. Molecular characterization of Ypi1, a novel Saccharomyces cerevisiae type 1 protein phosphatase inhibitor. J Biol Chem 2003;278(48):47744–52 [DOI] [PubMed] [Google Scholar]
- 32.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013;14(2):178–92 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cunningham KW, Fink GR. Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol Cell Biol 1996;16(5):2226–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds A, Lundblad V, Dorris D, Keaveney M. Yeast Vectors and Assays for expression of cloned genes Current Protocols in Molecular Biology, John Wiley & Sons; 1997. 13.6.1–13.6.6 [DOI] [PubMed] [Google Scholar]
- 35.Bush A, Chernomoretz A, Yu R, Gordon A, Colman-Lerner A. Using Cell-ID 1.4 with R for microscope-based cytometry. 2012. Unit 14.18 [DOI] [PMC free article] [PubMed]
- 36.Savageau MA. Biochemical Systems Analysis: A Study of Function and Design in Molecular Biology, CreateSpace Independent Publishing Platform; 2010.
- 37.Voit EO. Computational Analysis of Biochemical Systems: A Practical Guide for Biochemists and Molecular Biologists, Cambridge University Press; 2000. [Google Scholar]
- 38.Voit EO. A First Course in Systems Biology, Garland Science; 2012. [Google Scholar]
- 39.Wall ME, Hlavacek WS, Savageau MA. Design of gene circuits: lessons from bacteria. Nat Rev Genet 2004;5(1):34–42 [DOI] [PubMed] [Google Scholar]
- 40.Savageau MA. Biochemical systems analysis. I. Some mathematical properties of the rate law for the component enzymatic reactions. J Theor Biol 1969;25(3):365–69 [DOI] [PubMed] [Google Scholar]
- 41.Salvado B, Karathia H, Chimenos AU, Vilaprinyo E, Omholt S, Sorribas A et al. Methods for and results from the study of design principles in molecular systems. Math Biosci 2011;231(1):3–18 10.1016/j.mbs.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 42.Alves R, Vilaprinyo E, Hernadez-Bermejo B, Sorribas A. Mathematical formalisms based on approximated kinetic representations for modeling genetic and metabolic pathways. Biotechnol Genet Eng Rev 2008;251–40 [DOI] [PubMed] [Google Scholar]
- 43.Vilaprinyo E, Alves R, Sorribas A. Use of physiological constraints to identify quantitative design principles for gene expression in yeast adaptation to heat shock. BMC Bioinformatics 2006;7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vilaprinyo E, Alves R, Sorribas A. Minimization of biosynthetic costs in adaptive gene expression responses of yeast to environmental changes. PLoS Comput Biol 2010;6(2):e1000674 10.1371/journal.pcbi.1000674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Comas J, Benfeitas R, Vilaprinyo E, Sorribas A, Solsona F, Farre G et al. Identification of line-specific strategies for improving carotenoid production in synthetic maize through data-driven mathematical modelling. Plant J 2016;in press [DOI] [PubMed] [Google Scholar]
- 46.Lee Y, Chen PW, Voit EO. Analysis of operating principles with S-system models. Math Biosci 2011;231(1):49–60 10.1016/j.mbs.2011.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rybakova KN, Tomaszewska A, van MS, Blom J, Westerhoff HV, Carlberg C et al. Tracing the molecular basis of transcriptional dynamics in noisy data by using an experiment-based mathematical model. Nucleic Acids Res 2015;43(1):153–61 10.1093/nar/gku1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palumbo MC, Farina L, Paci P. Kinetics effects and modeling of mRNA turnover. Wiley Interdiscip Rev RNA 2015;6(3):327–36 10.1002/wrna.1277 [DOI] [PubMed] [Google Scholar]
- 49.Mitarai N, Benjamin JA, Krishna S, Semsey S, Csiszovszki Z, Masse E et al. Dynamic features of gene expression control by small regulatory RNAs. Proc Natl Acad Sci U S A 2009;106(26):10655–59 10.1073/pnas.0901466106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie ZR, Yang HT, Liu WC, Hwang MJ. The role of microRNA in the delayed negative feedback regulation of gene expression. Biochem Biophys Res Commun 2007;358(3):722–26 [DOI] [PubMed] [Google Scholar]
- 51.Farre G, Maiam RS, Alves R, Vilaprinyo E, Sorribas A, Canela R et al. Targeted transcriptomic and metabolic profiling reveals temporal bottlenecks in the maize carotenoid pathway that may be addressed by multigene engineering. Plant J 2013;75(3):441–55 10.1111/tpj.12214 [DOI] [PubMed] [Google Scholar]
- 52.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J et al. Global quantification of mammalian gene expression control. Nature 2011;473(7347):337–42 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proc Natl Acad Sci U S A 2002;99(9):5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Sousa AR, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst 2009;5(12):1512–26 10.1039/b908315d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deneke C, Lipowsky R, Valleriani A. Complex degradation processes lead to non-exponential decay patterns and age-dependent decay rates of messenger RNA. PLoS One 2013;8(2):e55442 10.1371/journal.pone.0055442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stathopoulos-Gerontides A, Guo JJ, Cyert MS. Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes Dev 1999;13(7):798–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature 2008;455(7212):485–90 10.1038/nature07292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finn EM, DeRoo EP, Clement GW, Rao S, Kruse SE, Kokanovich KM et al. A subset of FG-nucleoporins is necessary for efficient Msn5-mediated nuclear protein export. Biochim Biophys Acta 2013;1833(5):1096–103 10.1016/j.bbamcr.2012.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bodvard K, Jorhov A, Blomberg A, Molin M, Kall M. The yeast transcription factor Crz1 is activated by light in a Ca2+/calcineurin-dependent and PKA-independent manner. PLoS One 2013;8(1):e53404 10.1371/journal.pone.0053404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang M, Herrmann CJ, Simonovic M, Szklarczyk D, von MC. Version 4.0 of PaxDb: Protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics 2015;15(18):3163–68 10.1002/pmic.201400441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruiz A, Yenush L, Arino J. Regulation of ENA1 Na(+)-ATPase gene expression by the Ppz1 protein phosphatase is mediated by the calcineurin pathway. Eukaryot Cell 2003;2(5):937–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lamb TM, Mitchell AP. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol Cell Biol 2003;23(2):677–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hong SP, Carlson M. Regulation of snf1 protein kinase in response to environmental stress. J Biol Chem 2007;282(23):16838–45 [DOI] [PubMed] [Google Scholar]
- 64.Berkey CD, Vyas VK, Carlson M. Nrg1 and nrg2 transcriptional repressors are differently regulated in response to carbon source. Eukaryot Cell 2004;3(2):311–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vyas VK, Berkey CD, Miyao T, Carlson M. Repressors Nrg1 and Nrg2 regulate a set of stress-responsive genes in Saccharomyces cerevisiae. Eukaryot Cell 2005;4(11):1882–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ye T, Elbing K, Hohmann S. The pathway by which the yeast protein kinase Snf1p controls acquisition of sodium tolerance is different from that mediating glucose regulation. Microbiology 2008;154(Pt 9):2814–26 10.1099/mic.0.2008/020149-0 [DOI] [PubMed] [Google Scholar]
- 67.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998;14(2):115–32 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(MP4)
Data Availability Statement
Raw data concerning Crz1 ChIP-Seq experiments are held at the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE79342 and will be available immediately after publication of the paper.