Structural and functional characterization of the extracellular domain of the human CaSR with bound Mg2+ and a tryptophan derivative.
Keywords: Calcium Sensing Receptor, TNCA, Crystal Structure, Mg2+, L-Phe, Binding Affinity, EC50
Abstract
Ca2+-sensing receptors (CaSRs) modulate calcium and magnesium homeostasis and many (patho)physiological processes by responding to extracellular stimuli, including divalent cations and amino acids. We report the first crystal structure of the extracellular domain (ECD) of human CaSR bound with Mg2+ and a tryptophan derivative ligand at 2.1 Å. The structure reveals key determinants for cooperative activation by metal ions and aromatic amino acids. The unexpected tryptophan derivative was bound in the hinge region between two globular ECD subdomains, and represents a novel high-affinity co-agonist of CaSR. The dissection of structure-function relations by mutagenesis, biochemical, and functional studies provides insights into the molecular basis of human diseases arising from CaSR mutations. The data also provide a novel paradigm for understanding the mechanism of CaSR-mediated signaling that is likely shared by the other family C GPCR [G protein (heterotrimeric guanine nucleotide–binding protein)–coupled receptor] members and can facilitate the development of novel CaSR-based therapeutics.
INTRODUCTION
The discovery of the parathyroid Ca2+-sensing receptor (CaSR) established a new paradigm in which extracellular Ca2+ ([Ca2+]o) can act as a first messenger for regulation of diverse cellular processes, including regulating the secretion of parathyroid hormone (PTH) and modulating calcium reabsorption by the kidney, in addition to its well-known role as a second messenger (1, 2). Extracellular divalent cations, particularly [Ca2+]o and magnesium ([Mg2+]o), along with amino acids and neurotransmitters, regulate numerous cellular processes via CaSR and 14 other family C G protein (heterotrimeric guanine nucleotide–binding protein)–coupled receptors (cGPCRs), including metabotropic glutamate (mGluR) and γ-aminobutyric acid type B (GABAB) receptors (3–7). Small changes in [Ca2+]o or [Mg2+]o trigger CaSR-mediated intracellular Ca2+ signaling and activate mitogen-activated protein kinase [extracellular signal–regulated kinase (ERK) 1 or 2] (8). CaSRs play a central role in regulating [Ca2+]o and [Mg2+]o homeostasis by stimulating phospholipase C to generate inositol 1,4,5-trisphosphate, which triggers release of calcium from its intracellular calcium stores to increase the intracellular free calcium concentration ([Ca2+]i) and activate [Ca2+]i signaling (9–11), which, in turn, inhibits PTH release, stimulates calcitonin secretion, and promotes renal Ca2+ excretion (12–15). l-Amino acids, especially those with aromatic side chains, potentiate high [Ca2+]o–elicited activation of CaSR via positive heterotropic functional cooperativity (5). Like other cGPCRs, CaSR functions as a dimer (16–18), with a long (~600 amino acids) N-terminal extracellular domain (ECD) playing an important role in the receptor’s cooperative responses to its agonists (7). More than 400 mutations in CaSR cause human disorders with abnormal [Ca2+]o and [Mg2+]o homeostasis, including familial hypocalciuric hypercalcemia (FHH), neonatal severe hyperparathyroidism (NSHPT), and autosomal dominant hypocalcemia (ADH); 225 of the mutations map to the ECD, highlighting its critical role (19). To clarify the mechanism for cooperative activation of CaSR by [Ca2+]o, [Mg2+]o, and amino acids, we solved the first crystal structure of human CaSR-ECD bound with Mg2+ ions. Unexpectedly, a high-affinity tryptophan derivative was found in the crystal structure of CaSR. Further identification and characterization of the CaSR ligand (CaSRL) suggest that it plays a role in potentiating the function of CaSR.
Results and Discussion
The Venus flytrap domain of human CaSR ECD (hCaSR-ECD; residues 20 to 541) expressed in human embryonic kidney (HEK) 293S (GnTI) cells was crystallized in the presence of 200 mM Mg2+ and 10 mM Ca2+. The structure was solved by molecular replacement using the structure of mGluR2 [Protein Data Bank (PDB) ID: 4XAQ] as the search template (Fig. 1A and table S1). hCaSR-ECD contains two globular lobes with an overall structure similar to other cGPCR family members, despite a low sequence similarity between these cGPCR family members (20 to 30%) (fig. S1) (20). Both the large lobe (subdomain 1) and the small lobe (subdomain 2) are typical α/β folds where the central parallel β strands are sandwiched by α helices. hCaSR-ECD forms a homodimer in solution (fig. S2) and in the crystal structure, with both protomers in a closed conformation (Fig. 1B) similar to the equivalent closed conformation of mGluR1 bound with glutamate [root mean square deviation of 1.24 Å for C (Fig. 1C)]. In addition, the direct and extensive homodimeric subdomain 2 interactions in hCaSR-ECD are analogous to those observed in the mGluR2 dimer with a bound agonist (PDB ID: 4XAQ), strongly suggesting that hCaSR-ECD crystal structure represents an active conformation (fig. S3) (21).
Fig. 1. Crystal structure of hCaSR-ECD.
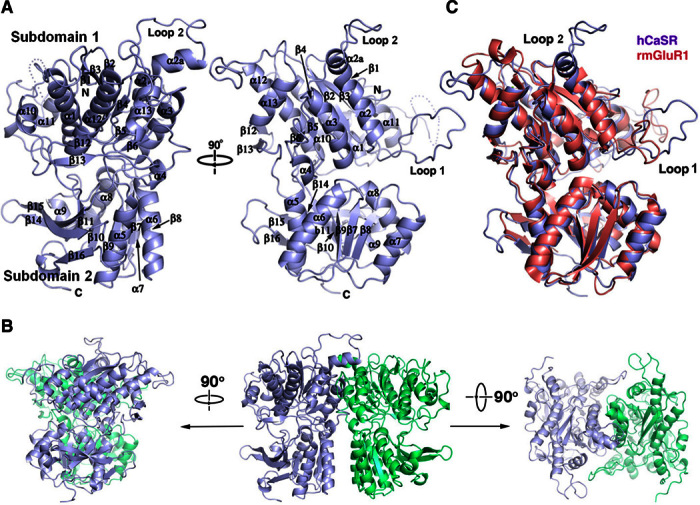
(A) Monomeric hCaSR-ECD with labeled secondary structural elements. (B) Homodimer of hCaSR-ECD. (C) Structural overlap of hCaSR-ECD with rat mGluR1 (rmGluR1) in the closed conformation (PDB ID: 1EWK).
Our data indicate that Mg2+ binds to hCaSR-ECD and elicits CaSR-mediated [Ca2+]i signaling and ERK1/2 phosphorylation in CaSR-expressing cells with a lower potency than Ca2+ (Figs. 2, A and B, and 3C and fig. S4A) (22). Similar to [Ca2+]o activation, [Mg2+]o activation is further potentiated by the known CaSR co-agonist, l-Phe (Fig. 3F) (5, 7). [Ca2+]o potentiates [Mg2+]o-stimulated intracellular response mediated by CaSR because an increase of [Ca2+]o from 0.5 to 1.5 mM results in a reduction of the median effective concentration (EC50) of [Mg2+]o from 7.2 ± 0.4 to 4.5 ± 0.3 mM for stimulation of [Ca2+]i signaling. These results suggest that there is an additive effect of both Ca2+ and Mg2+ and that they share a similar activation mechanism (Fig. 2A, fig. S5, and table S2) (22, 23). The binding of Mg2+ can be visualized by the reduction of intrinsic Trp fluorescence upon addition of Mg2+ to the purified ECD and the reduction of Tb3+-sensitized energy transfer by Mg2+ competition (fig. S6). In the crystal structure, two Mg2+ binding sites were identified at positions designated as site 1 and site 2 (Fig. 2B). Site 1 is located at the dimerization interface of subdomain 2 and the bound Mg2+ coordinates with S240 and four water molecules with an ideal geometry for a Mg2+ ion. Notably, site 1 is surrounded by highly conserved residues (E228, E231, and E241*) (* means from the other protomer) within 5 Å from an “acidic patch” composed of negatively charged residues on subdomain 2 (fig. S7). Site 2 is found on the periphery of subdomain 1, coordinated by S84 and backbone interactions with I81, L87, and L88, as well as two water molecules. An equivalent cation binding site has been observed in mGluR (20) and likely plays a structural role. To locate additional high off-rate metal binding sites, we generated Gd3+-derived crystals and identified another metal binding site (site 3) on the acidic patch in proximity to the subdomain 2 dimerization interface (Fig. 2C) and adjacent to Mg2+ binding site 1. Site 3 largely overlaps with a previously predicted Ca2+ binding site 3 (24). Mutation of site 3 coordinating residues (E228I or an E228I/E229I double mutant) reduced Ca2+/Mg2+ sensing as well as Mg2+-evoked intracellular Ca2+ mobilization. These results suggest the critical role of these metal binding sites on the acidic patch in both metal sensing (24) and regulation of CaSR function (Fig. 2G, figs. S7 and S9, and tables S3 and S4).
Fig. 2. Structural basis for Mg2+/Ca2+ modulated CaSR activities.
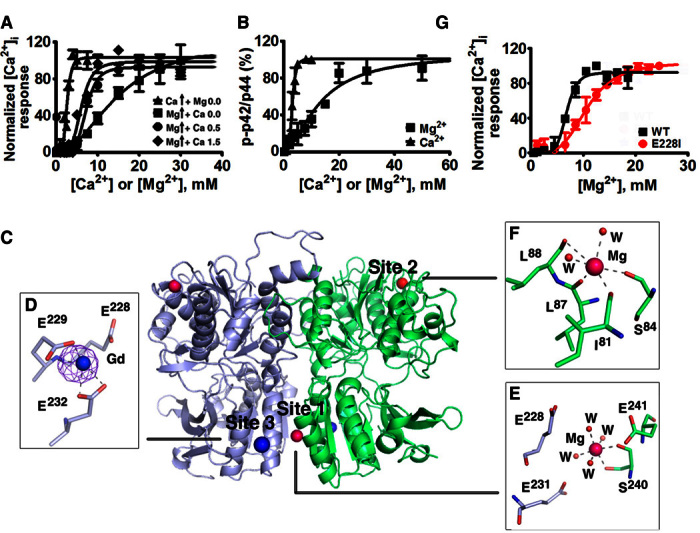
(A) CaSR-mediated [Ca2+]i responses measured by imaging of single-cell calcium oscillations with Fura-2 using HEK293 cells transfected with CaSR in the presence of various concentrations of [Ca2+]o and [Mg2+]o and fit to the Hill equation. (B) ERK1/2 activities upon stimulation by agonists were detected using Western blot and further quantified using ImageJ. The measurements were plotted against different concentrations of [Ca2+]o or [Mg2+]o and fit to the Hill equation. (C) Identified metal binding sites in the structure of hCaSR-ECD homodimer. Mg2+ and Gd3+ are depicted as hot pink and dark blue spheres, respectively. An anomalous difference map of Gd3+ (σ = 8.0) is shown in purple. W, water molecules. (D to F) Both site 1 (E) and site 3 (D) are on the “acidic patch” at the dimerization interface of subdomain 2 (fig. S7), whereas Mg2+ at site 2 in subdomain 1 (F) is primarily coordinated by the backbone carbonyl oxygen atoms. (G) Single mutations of E228I on the acidic patch significantly reduce CaSR-mediated [Ca2+]i responses in the cell population assay.
Fig. 3. Identification and characterization of a tryptophan derivative bound to hCaSR-ECD as a novel high-affinity co-agonist of CaSR.
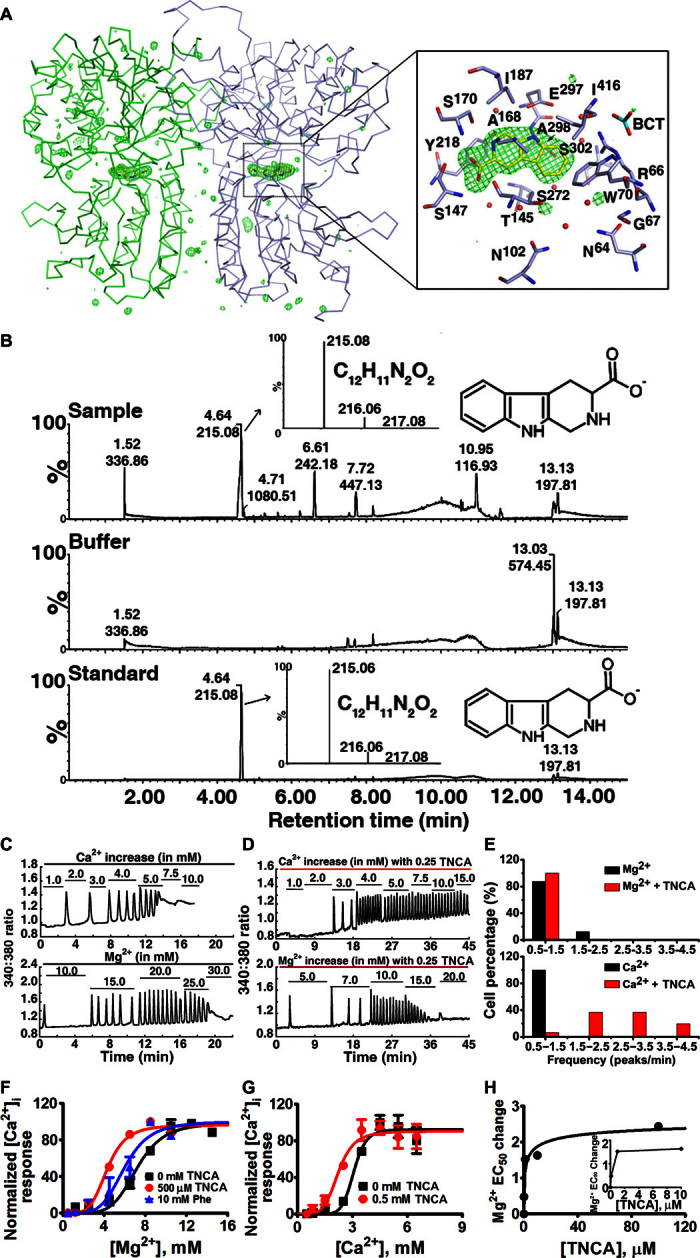
(A) Fo-Fc omit map (Fo and Fc are the observed and the calculated structure factor amplitudes, respectively) of TNCA at σ = 4.5. The protein is shown in ribbon mode, and the ligand is shown in stick mode. The residues around TNCA are labeled in the zoomed-in figure. (B) LC-ESI-MS of protein sample (top), buffer (middle), and the standard compound (bottom) in negative-ion mode. The high-resolution isotopic MS spectra of the indicated peaks are shown in the inserted figures. (C and D) A representative oscillation pattern from a single HEK293 cell stimulated with various concentrations of extracellular Ca2+ or Mg2+ in the absence (C) and presence (D) of 0.25 mM TNCA. (E) Frequency distribution of the [Ca2+]i oscillation frequency (peak/min) in HEK293 cells transfected with wild-type CaSR stimulated with metals in the presence (red bar) and absence (black bar) of TNCA. The frequency was recorded at the point when more than 50% single cells started to oscillate. Around 40 cells were analyzed and further plotted as a bar chart. (F and G) TNCA potentiates [Mg2+]o- or [Ca2+]o-evoked [Ca2+]i responses in a population assay in 5001 cells measured by Fura-2 acetoxymethyl (AM) in the absence (black square) or presence of Phe (blue triangular) or TNCA (red closed circle). (H) A maximally active concentration of 0.1 to 0.5 mM TNCA markedly reduces the EC50 for activation of [Ca2+]i signaling by [Mg2+]o in the presence of 0.5 mM [Ca2+]o. Inset: The EC50 changes of [Mg2+]o are shown over a narrow concentration range of TNCA.
Unexpectedly, an elongated planar electron density was observed in the hinge region between the two subdomains where orthosteric ligand binding is thought to occur (Fig. 3A). No naturally occurring CaSRLs or reagents that were used in sample preparation and crystallization or any currently known CaSRLs fit the density well, suggesting a novel CaSRL. High-resolution liquid chromatography–electrospray ionization–mass spectrometry (LC-ESI-MS) of the purified protein preparation (Fig. 3B) identified a species that was eluted at ~4.65 min with a mass/charge ratio (m/z) of 215.0824 in negative-ion mode. The predicted elemental formula based on the observed mass corresponds to C12H11N2O2 (calculated mass, 215.0821; mass accuracy, 1.4 ppm) (Fig. 3B and fig. S8). A search of PubChem identified a tryptophan derivative, l-1,2,3,4-tetrahydronorharman-3-carboxylic acid (TNCA) with the predicted molecular weight (Mr) and shape of the observed density. When compared to tryptophan, TNCA contains one extra carbon atom linking the amine nitrogen atom and the C2 atom of the indole ring. TNCA can be detected in various food and biological systems and is likely produced by tryptophan reacting with formaldehyde in humans (25), and is perhaps generated during production of the recombinant protein in HEK cells. Elution time, molecular weight, and MS fragmentation of synthetic TNCA matched those of the CaSRL, confirming the identity of the compound as TNCA (Fig. 3B and fig. S8). The ligand will be referred to hereafter as TNCA.
TNCA is a strong co-agonist with [Mg2+]o in activating [Ca2+]i oscillations and ERK1/2 phosphorylation (Fig. 3, D to F, and fig. S9). Similar to Trp and other amino acids, addition of exogenous TNCA alone cannot activate the receptor. However, TNCA is ~1000-fold more potent than Phe in reducing the EC50 for [Mg2+]o or [Ca2+]o activation of [Ca2+]i signaling in both wild-type and mutant CaSRs (Fig. 3, F and G), with an apparent EC50 of ≤2 μM (Fig. 3H). The apparent EC50 of TNCA was indirectly determined through the EC50 change of [Mg2+]o when incubated with different concentrations of TNCA (Fig.3, F to H). Because the bound TNCA can be partially replaced by incubation with 150 mM Phe as assessed by MS (fig. S10), TNCA and Phe likely share the similar binding site in CaSR-ECD. Together, TNCA is a novel, high-affinity co-agonist of CaSR in the activation of both [Ca2+]i signaling and ERK activity.
CaSR strongly prefers aromatic amino acid ligands, such as Phe and Trp, over negatively charged Glu, which is the ligand for mGluRs. Structural comparison of the ligand binding pocket in the hinge region between subdomains 1 and 2 of hCaSR-ECD with that of mGluR1 reveals the structural basis of ligand selectivity (fig. S11). Although the amino acid backbone of TNCA adopts a similar conformation as Glu in mGluR1 through extensive interactions with S147, A168, S170, and Y218 (S156, S186, T188, and Y236 in mGluR1) (20), hCaSR and mGluR1 recognize the side chains of their preferred ligands differently. Two positively charged residues in mGluR1 (R78 and K409) that associate with the carboxylate group of the Glu ligand are replaced in hCaSR by W70 and I416, which interact with the indole ring of TNCA. Bulky residues (Y74, W110, and R323) that limit the mobility of the Glu side chain are replaced by smaller residues in hCaSR (G67, N102, and S302). As a result, the size of the ligand binding pocket of hCaSR is significantly larger than that of mGluR1, consistent with the preference of CaSR for larger ligands. Unlike Trp, TNCA is in a fixed and presumably preferred conformation, accounting for the higher binding affinity compared to Trp.
Mapping of disease-associated mutations on the structure of hCaSR-ECD shows that the mutations are clustered in two regions: the hinge region between subdomains 1 and 2 and the dimerization interface (fig. S12) (26, 27). Indeed, our structural and functional data strongly support the pivotal roles of these two regions in CaSR function. The hinge region between subdomains 1 and 2 harbors the binding site of TNCA, supporting its role as a co-agonist of CaSR. Two other co-agonists of CaSR, Phe and Trp, likely bind in the same position (fig. S10). We did not observe metal binding at the previously proposed site 1 for Ca2+ (6, 24, 28). A close inspection of the structure reveals that the side chain of E297, a critical residue predicted for Ca2+ binding in this proposed site 1, swings away from the other residues in site 1 (S170, D190, Q193, and Y218), probably due to the extra carbon atom and the rigid structure of TNCA, ultimately resulting in its failure to capture the Ca2+ ion in combination with other site 1 residues (fig. S13). Nevertheless, the essential role of E297 in Ca2+ sensing has been supported by previous mutational studies (7, 24) and in the abrogated Mg2+ sensing of the E297I mutant (tables S3 and S4). A bicarbonate anion was also identified in the hinge region in proximity with TNCA, coordinated by the side chains of R66, R69, W70, and S417 and the backbone amide nitrogen atoms of I416 and S417 (figs. S11 and S14), potentially contributing to the known pH sensitivity of the CaSR (29).
Several lines of evidence indicate a critical role of CaSR-ECD dimerization in CaSR function (Figs. 1 and 4). First, two metal binding sites (site 1 and site 3) are identified within the acidic patch at the dimerization interface of subdomain 2 (Fig. 2 and fig. S7). A double mutant of CaSR (E228I/E229I) in site 3 showed a significantly decreased responsiveness to [Ca2+]o, and the E228I mutation also reduced activation of [Ca2+]i oscillations induced by [Mg2+]o as well as reduced Mg2+ binding, despite a similar level of membrane expression as wild-type CaSR (Fig. 2, fig. S9, and tables S3 and S4) (6). These data strongly suggest a role of metal binding at the acidic patch in metal sensing and signal transduction. Second, two loops that mediate subdomain 1 dimerization (loop 1 and loop 2; Fig. 1) are functionally important (Fig. 4A). Loop 2, following α2, is largely disordered in mGluR structures but is known to participate in two intermolecular disulfide bonds in CaSR through two conserved cysteine residues (C129 and C131) (17, 30) (Fig. 4B). The N-terminal part of loop 2 forms a short α helix (α2a) extended from α2 with a kink at N118. The α2a segments from each protomer embrace each other, likely stabilizing dimerization (Fig. 4A). Because several activating ADH mutations (L125P/F, E127G/A/K, C129Y/F/S/R, and N118K) and one inactivating FHH mutation are present on loop 2, subdomain 1 dimerization that is facilitated by loop 2 appears to be crucial in regulating the function of CaSR. Moreover, the highly conserved loop 1, which is significantly longer than the corresponding loop in mGluRs (fig. S15), reaches across the dimerization interface to a hydrophobic surface on α13*. The hydrophobic interaction, primarily mediated by P55, L51, and W458, stabilizes an extended conformation of loop 1, and a conserved positively charged patch also appears to contribute to dimerization of subdomain 1 (Fig. 4A). Notably, mutation of P55 causes FHH, indicative of a critical role of loop 1.
Fig. 4. Key determinants for the molecular basis of disease-associated mutations and regulation.
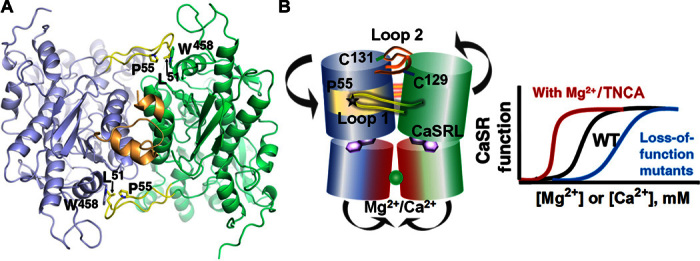
(A) Involvement of loop 1 (yellow) and loop 2 (gold) in dimerization. (B) Working model for activation occurs through a conformational change induced by ligand binding at the hinge region between subdomains 1 and 2, as well as bridging interactions provided by metal ion binding at the acidic patch at the interface between the two subdomain 2 regions of their respective protomers. Mutations at these key determinants in the ECD of CaSR cause human disorders with abnormal [Ca2+]o and [Mg2+]o homeostasis.
Figure 4B summarizes our present model for receptor activation. The presumed conformational change induced by ligand/metal binding in the hinge region between subdomains 1 and 2, with homodimerization of protomer subdomains 1 through loops 1 and 2, facilitates the approach of subdomain 2 from their respective protomers. By neutralizing the repulsive effects of the conserved and negatively charged acidic patch, metal binding would stabilize subdomain 2 interactions (fig. S15). Dimerization of subdomain 2 is also critical for the activation of mGluRs and GABAB receptors (31, 32) and therefore appears to be a common activation mechanism among cGPCRs that presumably leads to conformational changes of the transmembrane domain, through which the intracellular signal cascades are initiated. Thus, our proposed model, which is based on structural and associated biochemical and cellular studies on wild-type and mutant proteins, reveals the structural basis of agonist/co-agonist binding to CaSR and provides a framework for further studies on the mechanism of receptor activation. The discovery of an unexpected tryptophan derivative ligand (TNCA) with unusually high affinity, which potentiates the activation of CaSR by [Ca2+]o and [Mg2+]o, also opens new avenues for the development of agonists and antagonists as therapeutics for CaSR-related diseases (33).
CONCLUSION
We report here the first crystal structure of the ECD of human CaSR bound with Mg2+ and a tryptophan derivative ligand at 2.1 Å. The structure reveals important determinants for cooperative activation of the CaSR by metal ions and aromatic amino acids, including key residues in the ECD participating in Mg2+ and amino acid binding. In addition, extensive interactions between the disulfide-linked dimeric ECDs and their respective subdomains that produce a closed and likely active conformation of the Venus flytrap motifs of the two ECDs are present. The unexpected tryptophan derivative that was bound in the hinge region between two ECD subdomains represents a novel high-affinity co-agonist of CaSR that potentiates activation of the full-length CaSR expressed in HEK293 cells by Ca2+ and Mg2+. Our dissection, by mutagenesis, biochemical, and functional studies, of structure function relations provides insights into the molecular basis of human diseases arising from CaSR mutations. Finally, these data also provide a novel paradigm for understanding the mechanism of CaSR-mediated signaling that is likely shared by the other members of the family C GPCRs and can facilitate the identification and development of novel CaSR-based therapeutics.
MATERIALS AND METHODS
Purification of hCaSR-ECD secreted from HEK293S GnTI cells
hCaSR-ECD (from residues Tyr20 to Phe612) (fig. S1) was expressed in suspension culture of HEK293S GnTI cells and purified from the culture medium by Ni2+–nitrilotriacetic acid (NTA) chromatography, as previously described (34). To deglycosylate the purified protein, hCaSR-ECD was incubated with recombinant endoglycosidase F1 (Endo F1) at a 1:100 mass ratio of Endo F1 to hCaSR-ECD (35) overnight at 4°C in 10 mM tris buffer (pH 7.4). Further separation of hCaSR-ECD from Endo F1 was achieved by size exclusion chromatography (SEC) in 10 mM Hepes buffer (pH 7.3). hCaSR-ECD forms a homodimer, as determined by the elution volume observed in SEC. The electrophoretic mobility in reducing/nonreducing SDS–polyacrylamide gel electrophoresis (SDS-PAGE) indicates that intermolecular disulfide bonds contribute to dimerization (fig. S2).
Crystallization, data collection, and structure determination
The dimeric hCaSR-ECD was concentrated to 10 mg/ml and crystallized in 10% polyethylene glycol (PEG)–8000, 200 mM MgCl2, 10 mM CaCl2, and 100 mM tris-HCl (pH 7.0), using the sitting drop approach at 21°C. No crystals were formed in the absence of Ca2+ or Mg2+. The plate-shaped crystals were cryoprotected using 25% glycerol and were flash-frozen in liquid nitrogen. Dehydration by soaking the crystal in 12% PEG-8000 overnight improved the resolution from 3.5–4 to 2–3 Å. The diffraction data of the crystals were collected on the beamline of 21-ID-D at LS-CAT (Life Sciences Collaborative Access Team) in APS (Argonne Photon Source) and indexed, integrated, and scaled in HKL2000 (36). The structure was solved at 2.1 Å by molecular replacement using Auto-MR (automated molecular replacement) in PHENIX (37). The structure of chain A of mGluR2 with a bound agonist (PDB ID: 4XAQ) was used as the search template (21). The electron density map after molecular replacement was clear enough to identify the unique features of hCaSR-ECD, and iterative model building and refinement were performed using COOT (38) and Refmac5 in the CCP4 (39) suite, respectively. The restraints of TNCA (CaSRL) were generated by JLigand in COOT.
To generate the Gd3+ derivative, the native crystals were soaked with a solution containing 12% PEG-8000, 200 mM MgCl2, 10 mM CaCl2, 100 mM tris-HCl (pH 7.0), and 0.5 mM GdCl3 overnight at 21°C. The anomalous signals of a data set at 2.7 Å collected at the wavelength of 1.6985 Å were used to locate Gd3+ in the structure. The structure was solved by molecular replacement using the previously determined structure as the search template. All the figures of protein structures were generated by PyMOL version 1.3 (Schrodinger LLC).
Using local geometric restraints and electron density intensity as major criteria, Mg2+ bound at site 1 and site 2 (Fig. 2B) were unambiguously identified. Evidence also suggested a potential Mg2+ binding site in the hinge region in proximity with the bound TNCA (fig. S16). Modeling a water molecule at this position led to a B factor (26 to 32 Å2) substantially smaller than the coordinating atoms (35 to 40 Å2), suggestive of a slightly heavier atom occupying this position. Considering the negatively charged coordination sphere (A214, D216, Y218, S272, D275, and water), it was possible that this density corresponded to a bound Mg2+. Mutations such as Y218K and D275I resulted in diminished intracellular calcium responses using both the cell population and single-cell calcium imaging assays, although these mutations retained their surface expression (fig. S16B). Alternatively, it may be a highly ordered water molecule trapped in the hinge region because the distances to the coordinating oxygen atoms (2.5 to 2.8 Å) were significantly greater than typically observed Mg-O distance in biomolecules (2.1 Å) (40). Nevertheless, the low occupancy of Mg2+ due to its weak affinity may also cause a distorted geometry. At this stage, we have placed a water molecule in the model; however, given the importance of the hinge region in CaSR function and regulation, more functional and structural studies are warranted to further investigate the potential Mg2+ binding site identified here.
High-resolution LC-ESI-MS and identification of TNCA
As shown in Fig. 3A, there was an unidentified ligand (CaSRL) bound at the putative orthosteric ligand binding site of CaSR-ECD. We examined the known CaSRLs, including phenylalanine, tryptophan, glutathione, and polyamines, as well as the reagents used in sample preparation and crystallization, but none of them fit the density well. Among these initial trials, tryptophan appeared to be the best fit to the electron density of the unknown ligand, but an additional density was unaccounted for when tryptophan was used to fit the electron density. The size of the density suggested that the CaSRL contained 14 to 18 heavy atoms (C/N/O/S/P), and the absence of anomalous signal indicated that it did not contain sulfur or phosphate. Accordingly, the Mr of CaSRL must be within the range of ~180 to 250 daltons.
Considering that the CaSRL was tightly bound with hCaSR-ECD, it was conceivable that hCaSR-ECD had to be denatured to release CaSRL. To extract CaSRL, 50 μl of purified hCaSR-ECD (10 mg/ml) was mixed with 120 μl of acetonitrile and was vortexed. After high-speed centrifugation, 10 μl of the CaSRL extract was injected onto a reverse-phase ACQUITY UPLC BEH C18 column (2.1 × 100 mm, 1.7-μm particle size; Waters). Column temperature was maintained at 40°C. The flow rate was 0.3 ml/min with starting conditions at 99% solvent A (water + 0.1% formic acid) and 1% solvent B (acetonitrile). The 15-min gradient profile for elution was as follows: starting at 1% solvent B, hold for 1 min, then ramp to 98% B at 10 min, hold at 98% B to 12 min, return to 99% A/1% B at 12.01 min, and maintain until 15 min. The samples were analyzed using a Waters Xevo G2-XS QToF LC-MS interfaced to a Waters ACQUITY UPLC system. The MS settings were as follows: electrospray ionization in negative-ion mode, capillary voltage of 2.00 kV, source temperature of 100°C, desolvation temperature of 350°C, desolvation nitrogen gas flow rate of 600 liters/hour, cone voltage of 35 V, and mass range of m/z 50 to 1500 with spectra accumulated at 0.1 s per function. Three separate acquisition functions were performed to generate spectra at different collision energies (5, 25, and 60 eV) providing both nonfragmenting and fragmenting conditions. Analyses of samples by electrospray ionization in positive-ion mode were performed under the same conditions as negative-ion mode except for the collision energies (5, 20, and 40 eV). Fragmentation, formula, and abundances were analyzed with Waters MassLynx Software.
Using the above approach, we identified a species eluting at ~4.65 min, detected by MS in both positive-ion mode (m/z of 217.0990) and negative-ion mode (m/z of 215.0824), exclusively present in protein samples from several different batches but not in the sample buffer. The predicted elemental compositions based on mass are C12H13N2O2 (calculated mass = 217.0977 daltons) for positive-ion mode and C12H11N2O2 (calculated mass = 215.0824 daltons) for negative-ion mode. A thorough search in the PubChem database led to a list of candidates containing up to 200 compounds with the same Mr and formula. By manually fitting the density map with these compounds, only TNCA fit the density perfectly. Synthetic TNCA dissolved in the SEC buffer was treated in the same way as the protein samples in the LC-ESI-MS experiment, and resulted in a peak detected at the same retention time and having the same mass spectrum. In the LC-ESI-MS experiment, we also noticed a minor species that eluted at ~4.57 min (fig. S10), which was detectable only in the positive-ion mode (m/z of 215.0836) and having a predicted elemental formula of C12H11N2O2. The 2-dalton smaller Mr for this related compound suggested that it was a derivative of TNCA, likely due to a double bond formation between the backbone N and a neighboring C. Because it is also likely to be a tryptophan derivative, we cannot exclude the possibility that it binds hCaSR-ECD with high affinity. This compound may also form during extraction of TNCA from the protein sample. Nevertheless, TNCA perfectly fits the electron density at 2.1 Å, and any extra double bonds in the CaSRL structure would likely be detrimental to fitting the density.
A phenylalanine replacement experiment was carried out by mixing purified hCaSR-ECD protein (0.26 mg/ml) with phenylalanine (final concentrations are 0, 50, and 150 mM, respectively). After overnight incubation at room temperature, hCaSR-ECD in each sample was repurified with Ni2+-NTA beads. The protein samples were adjusted to the same concentration using SEC buffer and analyzed by LC-ESI-MS.
Monitoring Mg2+ binding to CaSR-ECD by fluorescence spectroscopy
The imidazole in fractions of hCaSR-ECD eluted from the Ni2+-NTA column was removed by passing the protein through desalting columns in Hepes buffer [10 mM Hepes (pH 7.2)]. The Trp fluorescence spectra of hCaSR-ECD were recorded on a QM1 fluorescence spectrophotometer (Photon Technology International) in a 1-cm-pathlength cell with a xenon short-arc lamp at ambient temperature. The emission between 300 and 400 nm was acquired during excitation at 282 nm. A solution containing 2 μM hCaSR-ECD in 10 mM Hepes (pH 7.2), 120 mM NaCl, and 10 mM KCl was gradually titrated by the addition of Ca2+ prepared in the same Hepes buffer. The binding constants of Mg2+ to CaSR-ECD were calculated by fitting the titration curve with the Hill equation. The Ca2+-Tb3+ competition experiments were performed in solutions containing 35 μM Tb3+ and 2 μM hCaSR-ECD as the starting point. MgCl2 was added to the mixture from a 1 M stock solution while maintaining a constant Tb3+ concentration in the solution. The Mg2+ binding affinity of the protein was calculated by fitting normalized fluorescence intensity data using the Hill equation
| (1) |
where ΔS is the total signal change in the equation, Kd is the apparent binding affinity, n is the Hill coefficient, and [M] is the free metal concentration.
TNCA and Mg2+ binding site mutation design
All of the full-length CaSR mutants were generated by site-directed mutagenesis on the basis of the sequence of the human CaSR in the pcDNA3.1(+) expression vector (provided by E. Brown). Site-directed mutagenesis was performed using the QuikChange kit (Stratagene) according to the manufacturer’s instructions. Briefly, a pair of complementary primers of 27 to 35 bases was designed for generating each mutant with the mutation placed at the middle of the primers. The template human CaSR in pcDNA3.1(+) was amplified using Pfu DNA polymerase (Stratagene) with these primers for 16 cycles in a polymerase chain reaction instrument (Techne). After digestion of the template DNA with Dpn I (New England Biolabs), the amplified mutant DNA was transformed into XL10-Gold Ultracompetent cells. All the DNA sequences were verified by Genewiz (www.genewiz.com).
Cell culture and transfection
Monolayer cultures of HEK293 cells were purchased from American Type Culture Collection (ATCC CRL-1573) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and high glucose (4.5 g/liter) at 37°C. Wild-type CaSR or its mutants were transfected into HEK293 cells using Lipofectamine 2000 (Life Technologies) by following the manufacturer’s instructions.
Immunostaining
Cells transfected with hCaSR-pcDNA3.1(+) were used in the immunostaining experiments, and this construct contained a FLAG tag between Asp371 and Thr372. After 48 hours post-transfection, cells were fixed with 3.7% formaldehyde for 15 min at room temperature, followed by washing with phosphate-buffered saline (PBS) three times. Mouse anti-FLAG monoclonal antibody was diluted 500 times and incubated with cells overnight at 4°C to stain the cell-surface CaSR. The cells were subsequently washed with PBS and stained with goat anti-mouse Alexa 488–conjugated secondary antibody for 1 hour at room temperature. Nuclei were stained with 4′,6-diamidino-2-phenylindole. Fluorescence was visualized using a Zeiss LSM780 confocal microscope.
Measurement of [Ca2+]i changes triggered by [Mg2+]o in single CaSR-transfected cells
Measurement of intracellular free Ca2+ was assessed as described by Huang et al. (6). Briefly, wild-type CaSR or its mutants were transiently transfected into HEK293 cells grown on coverslips and cultured for 48 hours. The cells were subsequently loaded for 15 min using 2 μM Fura-2 AM in 2 ml of physiological saline buffer (10 mM Hepes, 140 mM NaCl, 5 mM KCl, and 1.0 mM MgCl2) (pH 7.4). The coverslips were mounted on a bath chamber on the stage of a Leica DM6000 fluorescence microscope. The cells were alternately illuminated with 340- or 380-nm light, and the fluorescence at an emission wavelength of 510 nm was recorded in real time as the [Ca2+]o and/or [Mg2+]o was increased in a stepwise manner in the presence or absence of 0.25 mM TNCA in buffer (10 mM Hepes, 155 mM NaCl, 5 mM KCl, 2 mM NaH2PO4, and 0.5 mM MgCl2) (pH 7.4). The ratio of the emitted fluorescence intensities resulting (7) from excitation at both wavelengths was utilized as a surrogate for changes in [Ca2+]i and was further plotted and analyzed as a function of [Ca2+]o. All experiments were performed at room temperature. The signals from 20 to 100 single cells were recorded for each measurement. Oscillations were identified as three successive fluctuations in [Ca2+]i after the initial peak.
Determination of the effect of TNCA on Mg2+-evoked [Ca2+]i signaling by stimulation of CaSR in cell populations
Changes in [Ca2+]i elicited by [Mg2+]o in a population of cells were measured by fluorimetry as previously described (6). A cell line stably expressing CaSR (5001) was seeded on 13.5 × 20–mm coverslips and cultured in DMEM. After reaching 95% confluence, cells were washed three times using loading buffer [20 mM Hepes (pH 7.4), 125 mM NaCl, 5 mM KCl, 1.25 mM CaCl2, 1 mM MgCl2, 1 mM NaH2PO4, 1% glucose, and 1% bovine serum albumin (BSA)] and subsequently incubated with 4 μM Fura-2 and 4 μM Pluronic F127 for 20 min at 37°C to enable sufficient dye loading in the same buffer. After removing the excess Fura-2, coverslips with cells were diagonally positioned in a quartz cuvette filled with 3 ml of experimental buffer (125 mM NaCl, 5 mM KCl, 0.5 mM CaCl2, 0.5 mM MgCl2, 1% glucose, and 1% BSA). Measurements of Fura-2 fluorescence at 510 nm when excited at 340 or 380 nm were performed on a QM1 fluorescence spectrophotometer (Photon Technology International). The emission ratio of 340/380 was calculated and used to reflect the changes in [Ca2+]i when different concentrations of [Mg2+]o were applied to the cells.
To examine the coactivation of CaSR by TNCA and [Mg2+]o or [Ca2+]o, different concentrations of TNCA were placed in the experimental buffer with a fixed concentration of [Ca2+]o and varying concentrations of [Mg2+]o, or vice versa, as described in the Results section. The effects of other ligands were analyzed by comparing the changes in [Ca2+]i produced by [Mg2+]o alone or by coapplication of Mg2+ with other ligands. The EC50 of [Mg2+]o obtained during incubation with various concentrations of TNCA is compared with that observed in the presence of [Mg2+]o alone. The EC50 changes were plotted as a function of TNCA concentration, and the curve was fit to the Hill equation. The activation of CaSR by the TNCA, functioning as a co-agonist with [Mg2+]o, was indicated by the increasingly left-shifted EC50 for [Mg2+]o as the concentration of TNCA increases (table S5).
Determination of ERK1/2 phosphorylation
The 5001 cell line stably expressing hCaSR was starved in serum-free DMEM medium supplemented with 0.2% (w/v) BSA overnight, followed by washing with Hanks’ balanced salt solution (HBSS) three times and a subsequent 10-min HBSS incubation in the morning of the second day. To induce ERK1/2 phosphorylation, varying concentrations of [Mg2+]o (0 to 50 mM) or [Ca2+]o (0 to 30 mM) with or without 0.5 mM TNCA were added to cells and incubated for 10 min at 37°C. The cells were then lysed with Pierce radioimmunoprecipitation assay buffer (Thermo Scientific). Total protein concentration was measured using the Bio-Rad assay. Lysates containing 100 μg of total protein were loaded onto 4 to 20% gradient SDS-PAGE gels for separation. After electrophoresis, proteins on the gel were transferred to nitrocellulose membranes and were further analyzed by Western blotting. Anti–phospho-p44/42 ERK (1:1000 dilution) and anti-p44/42 (1:2000) polyclonal antibodies were utilized as probes to detect the phospho-ERK1/2 and total ERK1/2, respectively. A chemiluminescent detection method (AP Conjugate Substrate Kit, Bio-Rad) was applied to detect phospho-ERK1/2 and total ERK1/2. The respective bands on Western blots were evaluated by densitometry. The EC50 of [Mg2+]o- or [Ca2+]o-dependent responses were obtained by fitting the [Mg2+]o or [Ca2+]o concentration-response curves with the Hill equation (Eq. 1).
Supplementary Material
Acknowledgments
We thank R. Das and D. Cox for their help with the confocal experiments. We also thank D. A. Jones at Michigan State University for experimental instructions and thoughtful discussion of mass spectrometry experiments. We thank Y. Zhou for his critical review of this manuscript. Funding: This work was supported by NIH grants GM081749 and EB007268 (to J.J.Y.) and by NIH grant GM115373 and by a Michigan State University Startup Fund (to J.H.). This work was also supported by P41GM103390 (to K.W.M.), a Center for Diagnostics and Therapeutics fellowship (to C.Z. and R.G.), Brain and Behavior Fellowship (J.Z. and C.L.M.), and funds from the Georgia Research Alliance. Author contributions: J.J.Y., J.H., and E.M.B. formulated the project; C.Z., C.L.M., J.Z., and R.G. performed experiments in constructing plasmids, protein expression and purification, and cell assays; J.-Y.Y. and S.W. expressed the protein; T.Z. and J.H. conducted crystallographic work and structural analysis; A.S., T.Z., and J.H. performed mass spectrometry experiments; K.H. performed computational analysis; J.Z., C.L.M., C.Z., R.G., J.J.Y., J.H., E.M.B., and K.W.M. analyzed the data; J.J.Y., J.H., E.M.B., and K.W.M. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. The atomic coordinates and structure factors (accession codes 5FBK and 5FBH) have been deposited in PDB, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick (www.rcsb.org/).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/5/e1600241/DC1
table S1. Crystallographic statistics of hCaSR-ECD and hCaSR-ECD/Gd3+.
table S2. EC50 of [Mg2+]o for stimulation of [Ca2+]i signaling in the presence of different coactivators.
table S3. EC50 of [Mg2+]o for stimulation of [Ca2+]i signaling in cell population assay with or without TNCA.
table S4. EC50 of [Mg2+]o for stimulation of [Ca2+]i signaling in single cell assay with or without TNCA.
table S5. EC50 of [Mg2+]o-elicited [Ca2+]i responses in cell population assay with coapplication of various concentrations of TNCA.
fig. S1. Structure-based sequence alignment of CaSRs and mGluRs (by PROMALS3D).
fig. S2. Size exclusion chromatography of purified hCaSR-ECD.
fig. S3. Comparison of CaSR and mGluR2 structures.
fig. S4. CaSR-mediated ERK1/2 activation.
fig. S5. [Ca2+]i responses of CaSR stimulated by increasing [Mg2+]o.
fig. S6. Determining Mg2+ binding to hCaSR-ECD.
fig. S7. Metal binding at the acidic patch.
fig. S8. Identification of TNCA.
fig. S9. Determining TNCA binding capability to hCaSR-ECD.
fig. S10. Replacement of TNCA by phenylalanine.
fig. S11. Structural comparison of CaSRL binding site with that of mGluR1.
fig. S12. Disease-related mutations on CaSR ECD.
fig. S13. Structure of the proposed calcium binding site 1.
fig. S14. Identification of a bicarbonate anion near the ligand binding site.
fig. S15. A positively charged pocket for loop 1 association.
fig. S16. Identification of a potential Mg2+ binding site in the hinge region.
REFERENCES AND NOTES
- 1.Brown E. M., Gamba G., Riccardi D., Lombardi M., Butters R., Kifor O., Sun A., Hediger M. A., Lytton J., Hebert S. C., Cloning and characterization of an extracellular Ca2+-sensing receptor from bovine parathyroid. Nature 366, 575–580 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Grant M. P., Stepanchick A., Cavanaugh A., Breitwieser G. E., Agonist-driven maturation and plasma membrane insertion of calcium-sensing receptors dynamically control signal amplitude. Sci. Signal. 4, ra78 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Permyakov E. A., Kretsinger R. H., Cell signaling, beyond cytosolic calcium in eukaryotes. J. Inorg. Biochem. 103, 77–86 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Kubo Y., Miyashita T., Murata Y., Structural basis for a Ca2+−sensing function of the metabotropic glutamate receptors. Science 279, 1722–1725 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Conigrave A. D., Quinn S. J., Brown E. M., l-amino acid sensing by the extracellular Ca2+−sensing receptor. Proc. Natl. Acad. Sci. U.S.A. 97, 4814–4819 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Y., Zhou Y., Yang W., Butters R., Lee H.-W., Li S., Castiblanco A., Brown E. M., Yang J. J., Identification and dissection of Ca2+-binding sites in the extracellular domain of Ca2+–sensing receptor. J. Biol. Chem. 282, 19000–19010 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C., Huang Y., Jiang Y., Mulpuri N., Wei L., Hamelberg D., Brown E. M., Yang J. J., Identification of an l-phenylalanine binding site enhancing the cooperative responses of the calcium-sensing receptor to calcium. J. Biol. Chem. 289, 5296–5309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bapty B. W., Dai L.-j., Ritchie G., Jirik F., Canaff L., Hendy G. N., Quamme G. A., Extracellular Mg2+- and Ca2+-sensing in mouse distal convoluted tubule cells. Kidney Int. 53, 583–592 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Magno A. L., Ward B. K., Ratajczak T., The calcium-sensing receptor: A molecular perspective. Endocr. Rev. 32, 3–30 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Brown E. M., MacLeod R. J., Extracellular calcium sensing and extracellular calcium signaling. Physiol. Rev. 81, 239–297 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Chang W., Shoback D., Extracellular Ca2+-sensing receptors—an overview. Cell Calcium 35, 183–196 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Fudge N. J., Kovacs C. S., Physiological studies in heterozygous calcium sensing receptor (CaSR) gene-ablated mice confirm that the CaSR regulates calcitonin release in vivo. BMC Physiol. 4, 5 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hebert S. C., Extracellular calcium-sensing receptor: Implications for calcium and magnesium handling in the kidney. Kidney Int. 50, 2129–2139 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Ho C., Conner D. A., Pollak M. R., Ladd D. J., Kifor O., Warren H. B., Brown E. M., Seidman J. G., Seidman C. E., A mouse model of human familial hypocalciuric hypercalcemia and neonatal severe hyperparathyroidism. Nat. Genet. 11, 389–394 (1995). [DOI] [PubMed] [Google Scholar]
- 15.Hofer A. M., Brown E. M., Extracellular calcium sensing and signalling. Nat. Rev. Mol. Cell Biol. 4, 530–538 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Bai M., Structure and function of the extracellular calcium-sensing receptor (Review). Int. J. Mol. Med. 4, 115–125 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Ray K., Hauschild B. C., Steinbach P. J., Goldsmith P. K., Hauache O., Spiegel A. M., Identification of the cysteine residues in the amino-terminal extracellular domain of the human Ca2+ receptor critical for dimerization. Implications for function of monomeric Ca2+ receptor. J. Biol. Chem. 274, 27642–27650 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Suzuki Y., Moriyoshi E., Tsuchiya D., Jingami H., Negative cooperativity of glutamate binding in the dimeric metabotropic glutamate receptor subtype 1. J. Biol. Chem. 279, 35526–35534 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Pidasheva S., D’Souza-Li L., Canaff L., Cole D. E. C., Hendy G. N., CASRdb: Calcium-sensing receptor locus-specific database for mutations causing familial (benign) hypocalciuric hypercalcemia, neonatal severe hyperparathyroidism, and autosomal dominant hypocalcemia. Hum. Mutat. 24, 107–111 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Kunishima N., Shimada Y., Tsuji Y., Sato T., Yamamoto M., Kumasaka T., Nakanishi S., Jingami H., Morikawa K., Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature 407, 971–977 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Monn J. A., Prieto L., Taboada L., Pedregal C., Hao J., Reinhard M. R., Henry S. S., Goldsmith P. J., Beadle C. D., Walton L., Man T., Rudyk H., Clark B., Tupper D., Baker S. R., Lamas C., Montero C., Marcos A., Blanco J., Bures M., Clawson D. K., Atwell S., Lu F., Wang J., Russell M., Heinz B. A., Wang X., Carter J. H., Xiang C., Catlow J. T., Swanson S., Sanger H., Broad L. M., Johnson M. P., Knopp K. L., Simmons R. M. A., Johnson B. G., Shaw D. B., McKinzie D. L., Synthesis and pharmacological characterization of C4-disubstituted analogs of 1S,2S,5R,6S-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylate: Identification of a potent, selective metabotropic glutamate receptor agonist and determination of agonist-bound human mGlu2 and mGlu3 amino terminal domain structures. J. Med. Chem. 58, 1776–1794 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Quinn S. J., Thomsen A. R. B., Egbuna O., Pang J., Baxi K., Goltzman D., Pollak M., Brown E. M., CaSR-mediated interactions between calcium and magnesium homeostasis in mice. Am. J. Physiol. Endocrinol. Metab. 304, E724–E733 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrè S., Hoenderop J. G. J., Bindels R. J. M., Sensing mechanisms involved in Ca2+ and Mg2+ homeostasis. Kidney Int. 82, 1157–1166 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Huang Y., Zhou Y., Castiblanco A., Yang W., Brown E. M., Yang J. J., Multiple Ca2+-binding sites in the extracellular domain of the Ca2+-sensing receptor corresponding to cooperative Ca2+ response. Biochemistry 48, 388–398 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herraiz T., Galisteo J., Chamorro C., l-tryptophan reacts with naturally occurring and food-occurring phenolic aldehydes to give phenolic tetrahydro-β-carboline alkaloids: Activity as antioxidants and free radical scavengers. J. Agric. Food Chem. 51, 2168–2173 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Hannan F. M., Nesbit M. A., Zhang C., Cranston T., Curley A. J., Harding B., Fratter C., Rust N., Christie P. T., Turner J. J. O., Lemos M. C., Bowl M. R., Bouillon R., Brain C., Bridges N., Burren C., Connell J. M., Jung H., Marks E., McCredie D., Mughal Z., Rodda C., Tollefsen S., Brown E. M., Yang J. J., Thakker R. V., Identification of 70 calcium-sensing receptor mutations in hyper- and hypo-calcaemic patients: Evidence for clustering of extracellular domain mutations at calcium-binding sites. Hum. Mol. Genet. 21, 2768–2778 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Hannan F. M., Thakker R. V., Calcium-sensing receptor (CaSR) mutations and disorders of calcium, electrolyte and water metabolism. Best Pract. Res. Clin. Endocrinol. Metab. 27, 359–371 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Silve C., Petrel C., Leroy C., Bruel H., Mallet E., Rognan D., Ruat M., Delineating a Ca2+ binding pocket within the Venus flytrap module of the human calcium-sensing receptor. J. Biol. Chem. 280, 37917–37923 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Quinn S. J., Bai M., Brown E. M., pH sensing by the calcium-sensing receptor. J. Biol. Chem. 279, 37241–37249 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Minami H., Yoshida T., Okutsu K., Zhang Q., Inoue S., Atsuya I., Direct determination of silicon in powdered aluminium oxide by use of slurry sampling with in situ fusion graphite-furnace atomic-absorption spectrometry. Fresenius J. Anal. Chem. 370, 855–859 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Tsuchiya D., Kunishima N., Kamiya N., Jingami H., Morikawa K., Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+. Proc. Natl. Acad. Sci. U.S.A. 99, 2660–2665 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geng Y., Bush M., Mosyak L., Wang F., Fan Q. R., Structural mechanism of ligand activation in human GABAB receptor. Nature 504, 254–259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nemeth E. F., Goodman W. G., Calcimimetic and calcilytic drugs: Feats, flops, and futures. Calcif. Tissue Int. 98, 341–358 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Zhang C., Zhuo Y., Moniz H. A., Wang S., Moremen K. W., Prestegard J. H., Brown E. M., Yang J. J., Direct determination of multiple ligand interactions with the extracellular domain of the calcium-sensing receptor. J. Biol. Chem. 289, 33529–33542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng L., Forouhar F., Thieker D., Gao Z., Ramiah A., Moniz H., Xiang Y., Seetharaman J., Milaninia S., Su M., Bridger R., Veillon L., Azadi P., Kornhaber G., Wells L., Montelione G. T., Woods R. J., Tong L., Moremen K. W., Enzymatic basis for N-glycan sialylation: Structure of rat α2,6-sialyltransferase (ST6GAL1) reveals conserved and unique features for glycan sialylation. J. Biol. Chem. 288, 34680–34698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in the oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 37.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Cryst. D66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S., Overview of the CCP4 suite and current developments. Acta Cryst. D67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang W., Lee H.-W., Hellinga H., Yang J. J., Structural analysis, identification, and design of calcium-binding sites in proteins. Proteins 47, 344–356 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/5/e1600241/DC1
table S1. Crystallographic statistics of hCaSR-ECD and hCaSR-ECD/Gd3+.
table S2. EC50 of [Mg2+]o for stimulation of [Ca2+]i signaling in the presence of different coactivators.
table S3. EC50 of [Mg2+]o for stimulation of [Ca2+]i signaling in cell population assay with or without TNCA.
table S4. EC50 of [Mg2+]o for stimulation of [Ca2+]i signaling in single cell assay with or without TNCA.
table S5. EC50 of [Mg2+]o-elicited [Ca2+]i responses in cell population assay with coapplication of various concentrations of TNCA.
fig. S1. Structure-based sequence alignment of CaSRs and mGluRs (by PROMALS3D).
fig. S2. Size exclusion chromatography of purified hCaSR-ECD.
fig. S3. Comparison of CaSR and mGluR2 structures.
fig. S4. CaSR-mediated ERK1/2 activation.
fig. S5. [Ca2+]i responses of CaSR stimulated by increasing [Mg2+]o.
fig. S6. Determining Mg2+ binding to hCaSR-ECD.
fig. S7. Metal binding at the acidic patch.
fig. S8. Identification of TNCA.
fig. S9. Determining TNCA binding capability to hCaSR-ECD.
fig. S10. Replacement of TNCA by phenylalanine.
fig. S11. Structural comparison of CaSRL binding site with that of mGluR1.
fig. S12. Disease-related mutations on CaSR ECD.
fig. S13. Structure of the proposed calcium binding site 1.
fig. S14. Identification of a bicarbonate anion near the ligand binding site.
fig. S15. A positively charged pocket for loop 1 association.
fig. S16. Identification of a potential Mg2+ binding site in the hinge region.


