Abstract
Purpose
An emerging approach in medical genetics is to identify de novo mutations in patients with severe early-onset genetic disease that are absent in population controls and in the patient’s parents. This approach, however, frequently misses post-zygotic “mosaic” mutations that are present in only a portion of the healthy parents’ cells and are transmitted to offspring.
Methods
We constructed a mosaic transmission screen for variants that have an ~50% alternative allele ratio in the proband but are significantly less than 50% in the transmitting parent. We applied it to two family-based genetic disease cohorts consisting of 9 cases of sudden unexplained death in childhood (SUDC) and 338 previously published cases of epileptic encephalopathy.
Results
The screen identified six parental-mosaic transmissions across the two cohorts. The resultant rate of ~0.02 identified transmissions per trio is far lower than that of de novo mutations. Among these transmissions were two likely disease-causing mutations: an SCN1A mutation transmitted to an SUDC proband and her sibling with Dravet syndrome, as well as an SLC6A1 mutation in a proband with epileptic encephalopathy.
Conclusion
These results highlight explicit screening for mosaic mutations as an important complement to the established approach of screening for de novo mutations.
Keywords: epilepsy, mosaic, mutation, sudden unexplained death in childhood, whole-exome sequencing
Variants in DNA sequence data are often considered ubiquitous to DNA from all cells of an individual. However, during development there is a low frequency of spontaneous mutations that occur in different cell subdivisions. Such mutations may therefore be found in some but not all of a person’s cells.1 These somatic “mosaic” mutations are similar to de novo mutations in that they have not been subject to purifying selection and are therefore more likely to be deleterious than variants inherited from unaffected parents.2,3 De novo mutations are a major component of the genetic architecture of many Mendelian and complex diseases.4–6 Mosaic mutations transmitted from a parent to a child might be enriched for pathogenicity, similar to de novo mutations identified in disease cohorts.7 Thus, a methodology to thoroughly identify mosaic transmissions in family-based sequence data is critical.
Exome sequencing is a powerful tool for studying genetic disease, targeting sequence intervals that are most likely to harbor disease-causing variants. Exome sequence data can also be used to identify mosaic mutations, as higher read depth in targeted sequence loci increases the power to confidently identify mosaic mutations that occur in a low frequency of variant-carrying reads.7
We screened for mosaic mutations among two cohorts: (i) sudden unexplained death in childhood (SUDC) and (ii) epileptic encephalopathy. The SUDC cohort included nine families in which each proband died unexpectedly in childhood and no cause of death was identified after thorough pathological and toxicological investigation.8 We suspect that SUDC patients are enriched for underlying epileptic and cardiac disorder phenotypes. An observed 24–32% of SUDC cases have a history of febrile seizures, versus 2–4% in controls.9,10 In addition, SUDC cases have been reported to have an increased mutational burden in cardiac genes compared with control exomes.8 The epileptic encephalopathy cohort of 338 proband–parent trios (Epilepsy Phenome/Genome Project and Epi4K studies) has a high portion of cases with pathogenic de novo mutations.11 Written informed consent was received from all participants or their guardians.
MATERIALS AND METHODS
Presentation of an SUDC family with mosaic transmission
One of the nine SUDC families consisted of two healthy parents with three offspring: an asymptomatic son, a daughter (SUDC proband) who died unexpectedly at age 20.8 months, and a son living with Dravet syndrome (Figure 1). Both the proband and the sibling with Dravet syndrome presented with febrile seizures. The proband was born full-term; she had normal developmental milestones and no medical problems. She had seven febrile seizures between the ages of 7 months and the morning of her death; all were generalized in onset. The first febrile seizure lasted 25 min, and subsequent ones were progressively shorter. A computerized tomography scan of the brain and routine electroencephalogram were normal. After her final febrile seizure she was prescribed amoxicillin for otitis media. That afternoon, she was placed in crib for a nap and was found 1 hour later face down and unresponsive with emesis in her nose and mouth. Emergency responders reported she was blue, apneic, and asystolic on a monitor. Resuscitation efforts were ineffective.
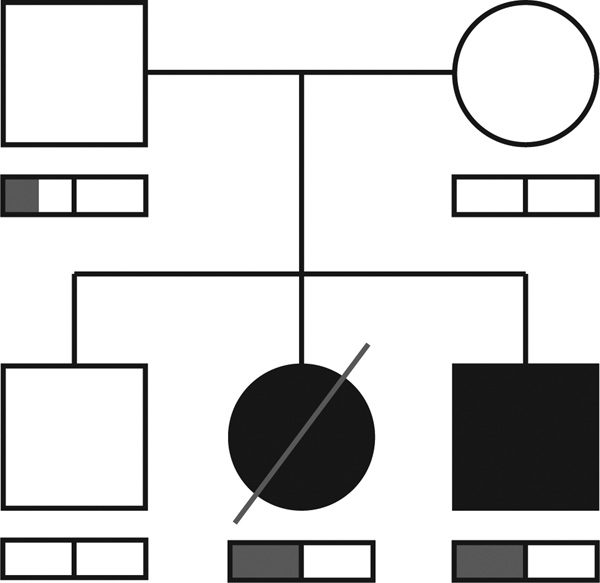
Figure 1. Transmission of a parental-mosaic SCN1A mutation in an affected family.
Two siblings were affected (proband with sudden unexplained death in childhood, sibling with Dravet syndrome). Both parents were phenotypically normal. The bar below each family-member symbol represents the portion of reads at the SCN1A variant position that carried the mutation (in grey); the black bar in the middle represents the 50% ratio expected from a heterozygote. Around 25% of reads carried the pathogenic variant in sequence data from the father.
The sibling with Dravet syndrome had his first febrile seizure at age 5 months and later had numerous febrile and afebrile seizures (status epilepticus), with focal and generalized semiologies. Results of his electrocardiogram, Holter monitor, and echocardiogram were normal. His seizure burden declined while on clobazam, valproic acid, and the ketogenic diet. His growth and development were normal at the last evaluation (age 30 months).
One hypothesis was that the proband and sibling living with Dravet syndrome shared a causal variant. Our initial screens for recessive genotypes shared by both children were negative. We then developed a screen to enable us to test this family and the other SUDC families for the transmission of potentially damaging variants from a parental mosaic origin.
Mosaic mutation transmission screen
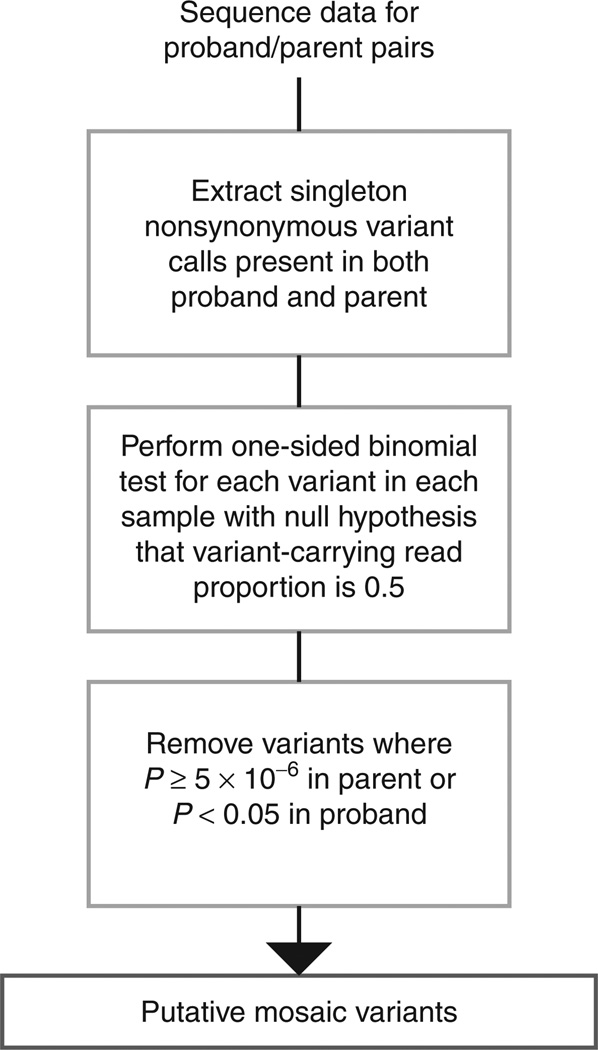
We constructed a systematic screen for identifying parental mosaic mutation transmissions in family-based sequence data. For a given parent–child pair, the screen identifies high-quality nonsynonymous variant calls that are absent among available controls of convenience (singletons), are heterozygous in the proband, and are also observed among the reads of a parent (Figure 2; Supplementary Methods online). For these variants, we then perform a binomial exact test on the mutant allele read ratio for the carrier parent and proband child to determine the probability of obtaining the observed read ratio in the parent and child, given an expected value of 0.5 for inherited heterozygous variants. We set a significance threshold of 5 × 10−6 for both the SUDC and epileptic encephalopathy screen, deriving this threshold based on a total of 13,555 total nonsynonymous singleton transmissions detected across both cohorts (0.05/13,555).
Figure 2. Workflow for the mosaic variant screen used on the sudden unexplained death in childhood and epileptic encephalopathy cohorts.
The workflow relies on extracting singleton variants of high quality from next-generation sequence data and determining which have the strongest evidence against the parent being heterozygous for the variant, while not defying the heterozygote model in the proband. For a more detailed description of the screen used, see Supplementary Methods online.
RESULTS
Mosaic variant screen in the SUDC cohort
Applying the mosaic transmission screen to the SUDC cohort identified 606 nonsynonymous singleton variant transmissions across 17 parent–child pairings. Requiring the variant allele read ratio to be significantly departed from the 0.5 expectation in the transmitting parent, we found two high-confidence inherited variants where the parent significantly defied the expected proportion of reads carrying the variant but the proband did not (Supplementary Tables S1 and S2 online).
Both of the mosaic variants in the SUDC cohort occurred in the family described earlier. This included mosaic missense variants in the SCN1A gene (NM_001165963.1:c.182T>C; Leu61Pro) and in the NTNG1 gene (NM_001113226.2.1:c.1 12T>C;Ser112Arg). SCN1A codes for Nav1.1, a voltage-gated ion channel protein that is critical to generating and propagating action potentials through the nervous and neuromuscular systems. NTNG1 codes for a protein that influences axonal guidance and signaling.12
The SCN1A variant in the SUDC proband and the sibling with Dravet syndrome has strong evidence for mosaic transmission. Sequence data from the normal father, who has no history of seizures, supports a mosaic carrier status for this variant. Of the 167 aligned paternal reads overlapping the single-nucleotide variant position, 42 (~25%) carried the variant (Supplementary Figure S1 online). Given the proportion of reads carrying the variant expected from a heterozygote, his proportion is highly unlikely by chance (P = 4.5 × 10−11, one-sided binomial exact test). By contrast, the SUDC proband and her sibling with Dravet syndrome carried the variant in 47 and 46% of reads, respectively (Supplementary Figure S1 online). The father’s mosaicism, the affected siblings’ heterozygous carrier status, and the other family members’ lack of the variant were confirmed via Sanger sequencing (Supplementary Figure S2 online). The affected amino acid is in the cytoplasmic N-terminal domain of the protein and is highly conserved in evolution. SCN1A mutations occur in 70–80% of patients with Dravet syndrome.13,14 A different mutation at this same amino acid position (Leu61Phe) was previously reported in a boy with Dravet syndrome.14
The evidence for true mosaic origin and pathogenicity for the NTNG1 variant was insufficient. Results of the mosaic variant screen suggest that the NTNG1 variant was inherited from the mother (Supplementary Figure S3 online). While the mother and SUDC proband carried the NTNG1 variant, the Sanger sequencing data do not confirm or refute the mother’s status as a mosaic (Supplementary Figure S4 online). This result, in combination with the detection of the NTNG1 variant in the healthy sibling, suggests that this variant is unlikely to contribute to the proband’s ascertained phenotype.
Mosaic variant screen in epileptic encephalopathy
We next applied the mosaic screen to a cohort of 676 Epilepsy Phenome/Genome Project parent–child pairings and identified 13,142 nonsynonymous singleton variant transmissions. Among these variants, four met the required criteria (Supplementary Tables S1 and S2 online). Only one of these four variants fell into an intolerant gene, SLC6A1 (RVIS_v2 percentile of 9.9%).15 The variant in the proband and the mosaic variant in the parent (NM_003042.3:c.1495G>A; Gly362Arg) were both confirmed with Sanger sequencing (Supplementary Figures S5 and S6 online). SLC6A1 codes for a γ-aminobutyric acid transporter, whose knockout in a mouse model leads to absence seizures.16 This gene had recurrent de novo mutation burden across a large case cohort with myoclonic-atonic seizures.17 The Gly362Arg mutation falls within the fourth extracellular loop of the γ-aminobutyric acid transporter, which has been shown to be critical for substrate binding.18 Mutagenesis of this specific site reduces transporter activity to as little as 10% as that of the wild type.18 The exceptional observation of this parental mosaic transmission, combined with the existing gene–disease association between this gene and severe infantile epilepsy, suggests that the dysfunction of this gene may contribute to the pathogenesis of epileptic encephalopathy.17
Observed rate of transmitted nonsynonymous mosaic mutations
Taking the previously described identifiable nonsynonymous mosaic singleton variants observed per cohort (two in the SUDC cohort and four in the epileptic encephalopathy cohort), we estimated the rate of identifiable nonsynonymous mosaic mutation transmission events when the mosaic mutation is identified based on variants in ectoderm (circulated blood cells and the epithelium of the mouth). Given 6 detected transmissions among 693 parent–child pairings (17 with SUDC and 676 with epileptic encephalopathy), we estimated an identifiable transmitted mosaic mutation incidence per trio of ~0.018.
DISCUSSION
Motivated by the inheritance of a parental mosaic SCN1A mutation in an SUDC proband and her brother with Dravet syndrome, we developed a general screen to identify parental mosaic transmissions in family cohorts. When we applied this screen to the nine-family SUDC cohort, as well as to an epileptic encephalopathy cohort of 338 trios, we identified 6 transmissions in 693 parent–child pairs—a rate of ~0.009 detectable transmissions per parent–child pair (0.018 per trio). In addition to the SCN1A mutation, we identified a second, possibly pathogenic SLC6A1 mutation among the epileptic encephalopathy cohort.
Our results demonstrate that this basic framework can already be used to systematically screen sequence data for putative parental mosaic mutation transmissions. Since these transmitted mosaic mutations, like de novo mutations, have not had to go through purifying selection, they are of particular interest when identified in patients ascertained for severe early-onset disorders. It is unclear how many undiagnosed genetic disorders can be explained by a parental mosaic transmission, especially given the low frequency at which we could detect such events using community-standard exome sequencing data. However, our yield suggests that a formal screen, alongside screens for de novo mutations, could become a routine part of identifying candidate pathogenic variants in clinical genetics environments.
Regarding the SUDC proband, given the SCN1A mutation and the history of febrile seizures, death probably occurred from a seizure-related mechanism. Most febrile seizures are convulsive, and convulsive seizures precede most sudden unexpected death in epilepsy, a death that is not caused by status epilepticus, trauma, drowning, or other known causes.19 Sudden unexpected death in epilepsy is common among children with Dravet syndrome.19 A similar mechanism may be relevant in other SUDC cases with febrile seizures. Without afebrile seizures, however, neither epilepsy nor sudden unexpected death in epilepsy would be diagnosed. Given the significantly elevated rate of febrile seizures among SUDC cases9,10 and the identification of cardiac channelopathy genes in other SUDC cases,8 genetic screening should be performed when possible.
Supplementary Material
Acknowledgments
The authors are grateful for the participation of the affected families and researchers in the SUDC Registry and Research Collaborative, as well as the Epilepsy Phenome/Genome Project. The authors gratefully acknowledge the contributions of the following colleagues: Dawei Wang, Ann Poduri, Mark Hannibal, Michael Ackerman, and David Tester; Sergey Kisselev, Donna Dorshorst, and Prevention Genetics. This study was supported by grants from the National Institute of Neurological Disorders and Stroke (the Epilepsy Phenome/Genome Project (NS053998); Epi4K—Sequencing, Biostatistics and Bioinformatics Core (NS077303)), as well as funding from the Lange Shaw Family Foundation and Finding a Cure for Epilepsy and Seizures. The authors also thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison (http://exac.broadinstitute.org/about).
DISCLOSURE
L.C. and O. D. are on the board of directors and scientific advisory board respectively for the SUDC Foundation.
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http://www.nature.com/gim
REFERENCES
- 1.Hall JG. Review and hypotheses: somatic mosaicism: observations related to clinical genetics. Am J Hum Genet. 1988;43:355–363. [PMC free article] [PubMed] [Google Scholar]
- 2.Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8:610–618. doi: 10.1038/nrg2146. [DOI] [PubMed] [Google Scholar]
- 3.Zhu Q, Ge D, Maia JM, et al. A genome-wide comparison of the functional properties of rare and common genetic variants in humans. Am J Hum Genet. 2011;88:458–468. doi: 10.1016/j.ajhg.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epi KC, Allen AS, et al. Epilepsy Phenome/Genome Project. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iossifov I, O’Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Acuna-Hidalgo R, Bo T, Kwint MP, et al. Post-zygotic point mutations are an underrecognized source of de novo genomic variation. Am J Hum Genet. 2015;97:67–74. doi: 10.1016/j.ajhg.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narula N, Tester DJ, Paulmichl A, Maleszewski JJ, Ackerman MJ. Post-mortem whole exome sequencing with gene-specific analysis for autopsy-negative sudden unexplained death in the young: a case series. Pediatr Cardiol. 2015;36:768–778. doi: 10.1007/s00246-014-1082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hesdorffer DC, Crandall LA, Friedman D, Devinsky O. Sudden unexplained death in childhood: a comparison of cases with and without a febrile seizure history. Epilepsia. 2015;56:1294–1300. doi: 10.1111/epi.13066. [DOI] [PubMed] [Google Scholar]
- 10.Kinney HC, Chadwick AE, Crandall LA, et al. Sudden death, febrile seizures, and hippocampal and temporal lobe maldevelopment in toddlers: a new entity. Pediatr Dev Pathol. 2009;12:455–463. doi: 10.2350/08-09-0542.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.EuroEPINOMICS-RES Consortium; Epilepsy Phenome/Genome Project; Epi4K Consortium. De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am J Hum Genet. 2014;95:360–370. doi: 10.1016/j.ajhg.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakashiba T, Ikeda T, Nishimura S, et al. Netrin-G1: a novel glycosyl phosphatidylinositol-linked mammalian netrin that is functionally divergent from classical netrins. J Neurosci. 2000;20:6540–6550. doi: 10.1523/JNEUROSCI.20-17-06540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claes L, Del-Favero J, Ceulemans B, Lagae L, Van Broeckhoven C, De Jonghe P. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am J Hum Genet. 2001;68:1327–1332. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Depienne C, Trouillard O, Saint-Martin C, et al. Spectrum of SCN1A gene mutations associated with Dravet syndrome: analysis of 333 patients. J Med Genet. 2009;46:183–191. doi: 10.1136/jmg.2008.062323. [DOI] [PubMed] [Google Scholar]
- 15.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cope DW, Di Giovanni G, Fyson SJ, et al. Enhanced tonic GABAA inhibition in typical absence epilepsy. Nat Med. 2009;15:1392–1398. doi: 10.1038/nm.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carvill GL, McMahon JM, Schneider A, et al. EuroEPINOMICS Rare Epilepsy Syndrome Myoclonic-Astatic Epilepsy & Dravet working group. Mutations in the GABA transporter SLC6A1 cause epilepsy with myoclonic-atonic seizures. Am J Hum Genet. 2015;96:808–815. doi: 10.1016/j.ajhg.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zomot E, Kanner BI. The interaction of the gamma-aminobutyric acid transporter GAT-1 with the neurotransmitter is selectively impaired by sulfhydryl modification of a conformationally sensitive cysteine residue engineered into extracellular loop IV. J Biol Chem. 2003;278:42950–42958. doi: 10.1074/jbc.M209307200. [DOI] [PubMed] [Google Scholar]
- 19.Devinsky O. Sudden, unexpected death in epilepsy. N Engl J Med. 2011;365:1801–1811. doi: 10.1056/NEJMra1010481. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.