Abstract
GenomeC wide gene expression quantitative trait loci (eQTL) mapping have been focused on single nucleotide polymorphisms and have helped interpret findings from diseases mapping studies. The functional effect of structure variants, especially short insertions and deletions (indel) has not been well investigated. Here we imputed 1,380,133 indels based on the latest 1000 Genomes Project panel into 3 eQTL datasets from multiple tissues. Imputation of indels increased 9.9% power and identified indel specific eQTLs for 325 genes. We found introns and vicinities of UTRs were more enriched of indel eQTLs and 3.6 (singleC tissue)C 9.2%(multiC tissue) of previous identified eSNPs were taggers of eindels. Functional analyses identified epigenetics marks, gene ontology categories and disease GWAS loci affected by SNPs and indels eQTLs showing tissueC consistent or tissueC specific effects. This study provides new insights into the underlying genetic architecture of gene expression across tissues and new resource to interpret function of diseases and traits associated structure variants.
Introduction
In the past decade, most common single nucleotide polymorphism (SNP) with allele frequency >5% have been identified and genomeC wide association studies (GWAS) have been focusing on these common variants. As of February 2014,1785 studies have detected disease susceptibility loci at genomeC wide significant level1 (www.genome.gov/GWAStudies). However, discovery has only explained a modest portion of disease risk2. The undetected variants could be due to common SNPs but without sufficiently large effect, structure variants such as short insertion and deletion (indel), or low frequency SNP not covered by genotyping platforms or imputation3,4 based on previous releases of the HapMap5 and the 1000 Genomes pilot projects6.
The latest release (phase 1) of the 1000 Genomes Project haplotypes consists of 39.7 million genetic markers including 1.4 million indels. By applying genotype imputation techniques3,4 on this high quality reference panel from the 1000 Genomes Project (1000G)7, we can assess the genetic effect of indels as well as low frequency SNPs on disease phenotypes and gene expression. Indels are the second most abundant category of genetic variants and are widely distributed in the human genome. Comparing to SNPs, it is still unknown whether this type of structural variant has a larger causal effect on traits of interest, or serves as better tags of the causal genetic variants. A recent study based on 179 sequenced samples from the 1000 Genomes Project has shown that indels are generally subject to stronger purifying selection than SNPs and they are enriched in associations with gene expression8. Imputation of these newly identified genetic variants into existing genomeC wide association studies may help identifying novel disease loci not discovered by previous genotyping platforms and imputation. But it is not known how much unidentified disease heritability is due to indels and to what extent previously identified disease associated SNPs are due to linkage disequilibrium with indels of bigger impact on disease phenotype.
InterC individual variation in gene expression levels has a significant heritable component9C,13, and studies have mapped individual genetic variants associated with gene expression levels, known as expression quantitative trait locus (eQTL), in diverse cell types9,14C,20. Large scale gene expression data, which provide complex traits with full spectrum of heritability and genetic architecture, is ideal for evaluation of the power of association study using imputation of the newly identified indels. This information will be useful to the research community as to what should be expected from the imputation of indels and guide the design of genotyping platforms for next generation association studies.
Functional annotations generated from eQTL mapping, most of which available to the public, is an important resource to interpret variants of human genome21. It is well known that eQTLs can be a useful tool to characterize the function of a diseaseC associated variant and point to the underlying biological pathways22C,48.With the available 1000G indel reference panel, existing genomeC wide association studies are doing imputation on these indels. Once diseaseC associated indels are identified, their functional interpretation will become essential. We expect that indel eQTL will be a useful tool to characterize the findings of GWAS based on indels, either by imputation or genotyping experiments.
TissueC specific effects of small insertions and deletions on gene expression have not been examined before. Whether the tissue specificity of eQTL effects shows different patterns in SNPs and structure variants is unknown. In this study, we used 1000G imputed indels from 718 samples of multiple tissue types to answer the above questions and discussed their implication for disease mapping studies.
Results
Indel imputation
We collected tissue gene expression data from 3 studies (1) gene expression in lymphoblastoid cell lines from the MRCA family panel of 206 siblings of British descent13. A total of 368 children were genotyped using the Illumina Sentrix HumanHap300 BeadChip (ILMN300K) and the Illumina Sentrix HumanC 1 Genotyping BeadChip (ILMN100K); (2) gene expression in peripheral blood mononuclear cells (PBMC) from 47 Germany eczema nuclear families49. A total of 240 individuals (107 children, 133 parents) were genotyped using Affy500K and Affy 6.0 SNP array; (3) Normal skin tissues of 57 unrelated healthy controls and unaffected skin of 53 patients from a Psoriasis GWAS50. A total of 110 individuals were genotyped with Perlegen 400K array. Gene expression was measured using the Affymetrix HGC U133 Plus 2.0 GeneChip. After quality control on genotypes and expression, 376,877 SNPs from the LCL expression dataset, 687,364 from the PBMC expression dataset, 433,964 from the skin expression dataset, as well as 51,190 gene expression probe sets remain for downstream analysis (see Methods for details).
A total of 39.7 million genetic markers including 1.4 million indels from the phase 1 release of 1000 Genomes Project7 were imputed3 into these three datasets, separately. Among these variants, 814,715 indels and 10,129,531 SNPs have high quality score (imputation R2 >0.3 in all 3 studies). Across the entire allele frequency spectrum, indel imputation quality was generally comparable to that of SNPs but showed a slightly smaller fraction with extremely high imputation score (R2>0.95, for common variants, Supplementary Fig. S1). Imputation quality for both SNPs and indels were similar across studies based on different genotype platforms (Pearson correlation between quality score (MACH Rsq) from PBMC and skin datasets were 0.727 (SNP) and 0.811 (indel); 0.696 (SNP) and 0.786 (indel) between PBMC and LCL datasets; 0.736 (SNP) and 0.814 (indel) between LCL and skin datasets). All downstream analyses were based on SNPs and indels with imputation quality R2>0.3 across all 3 studies.
eQTL metat analysis
Within each individual study, we tested for association between gene expression and imputed SNPs and indels using MERLIN package51,52 accounting for family relatedness and including sex and expression principal components in the model53. The number of expression principal components was chosen to maximize the number of transcription probes that can be mapped by a variant within 1Mb of the probe set with FDR<5%. Results from individual studies were then combined using weighted z-score meta-analysis with sample size and imputation R2 as weights. We corrected for multiple testing using the Benjamini-Hochberg false discovery rate54,55 (FDR) accounting for all probe set-variant pairs ((814715 indel + 10129531 SNP)*51190 probe set). At FDR < 5%, a total of 5,898 unique genes (corresponding to 10,364 unique probe sets) were mapped by both SNP and indel; 325 unique genes (428 unique probe sets) can only be mapped by indels and 3,186 unique genes (6,663 unique probe sets) can only be mapped by SNPs (Figure 1A). Of the 9,409 genes mapped by SNPs or indels or both, 3,024 (32.1%) genes have indel as the most significant eQTL. We summarized our results based on the eQTLs that passed the > 5% FDR threshold. For genes that have both significant SNP and indel, the heritability explained by the eQTL is apparently larger than the genes mapped by only SNP or indel (Figure 1B).
Figure 1. Meta-analysis result for association between gene expression and imputed indels and SNPs.

A: Venn diagram for unique number of genes only mapped by SNP or indel or both.
B: Density curve of heritability of gene expression (H2) explained by the top SNP and indel. Red: H2 of the top indel for the 5898 genes in panel A. Green: H2 of the top indel for the 325 genes in panel A. Blue: H2 of the top SNP for the 5898 genes in panel A. Purple: H2 of the top SNP for the 3168 genes in panel A.
C: Percent of probes mapped by eQTL (<5% FDR) by total narrow sense heritability for 51,190 transcription probes. Probes were categorized by total narrow sense heritability previously estimated based on the MRCA family panel. The red bar shows the percent of probes mapped by eQTL using the MRCA LCL data alone with imputation of the 1000G pilot releases. The blue bar shows the percent of probes mapped using imputation of 1000G phase 1 variants in MRCA dataset excluding probes with indel as the top eQTL. The desh box on top of the blue bar indicates the percent of mapped probes before excluding probes with indel as top eQTL. The green bar shows the percent of probes mapped using imputation of 1000G phase 1 variants in MRCA dataset. The purple bar shows the percent of probes mapped using meta-analysis across three tissues excluding the probes with indel as the top associated variants. The orange bar shows the percent of probes mapped using meta-analysis across three tissues. The numbers on top of each group of bars are the number of probes in each heritability category.
Previous studies have shown that the power to detect eQTL increases with total additive heritability of gene expression estimated from pedigree data13. In Figure 1C, we compared the power to detect association (FDR<5%) across different strategies. The overall heritability is the narrow sense heritability estimated based on previous study using the MRCA family panel53. Within each heritability bin, the red bar shows the percent of expression probes mapped by using the MRCA LCL data alone with imputation of the 1000G pilot release (8 million SNPs). Using the same MRCA sample but with imputation of the 1000G phase1 SNPs and indels (green bars), we observed that 9.90% more probes were mapped at the same FDR (green bars vs. red bars). By combining results from multiple tissues, we observed that 41.9% more probe sets can be mapped by at least one genetic variant (orange bars vs. red bars).
Disease or trait associated genetic variants identified from genomeC wide association studies were usually not the causal variants but markers in linkage disequilibrium with the causal variants. Almost all genomeC wide association studies used SNPs as genetic markers to tag underlying causal genetic variants of the disease or trait of interest. By contracting the results based on different imputation strategies, we estimated how many previously identified associations might be tagging the association between the trait and indel. The blue bar in Figure 1C shows the percent of probes mapped by 1000G phase1 SNPs after removing the probes with indel as the top eQTL. Comparing the red and the blue bars we found that 3.62% of previously identified SNP QTL (eSNP) in LCL were likely to be tagging the indel eQTL (eindel) of the same gene in LCL. The dash bar on top of the blue bar shows the percent of probes mapped by 1000G phase1 SNPs before removing the probes with indel as the top eQTL, indicating a small gain of power by using the phase 1 SNPs. After combining multiple tissues to increase sample size, this difference increased to 9.20% (purple vs. orange) indicating that indels are more likely to be the causal genetic variants for these genes.
Comparison of SNP and indel eQTLs
To better understand the eQTLs due to SNP and indel effect, we sought to characterize the difference in indel eQTL and SNP eQTL by effect size, allele frequency spectrum and genomic distribution. We first tested whether indel were enriched with eQTL by randomly selecting 100,000 SNP and 100,000 indel with high imputation quality (R2>0.3) from the 1000 Genomes reference. There were 7,987 eSNPs and 11,386 eindels in the selected set of SNPs and indels. The enrichment of eQTLs in indels was significantly more than among SNPs (ChiC squared test for homogeneity, pC value < 2.2*10C 16).
Of the 5,898 genes mapped by both SNPs and indel that passed the 5% FDR threshold, the effect size (H2 explained by the variant) of eindel was similar to eSNP (mean difference=C 0.007, Standard deviation=0.048, Figure 2A).
Figure 2. Comparison of eSNP and eindel.
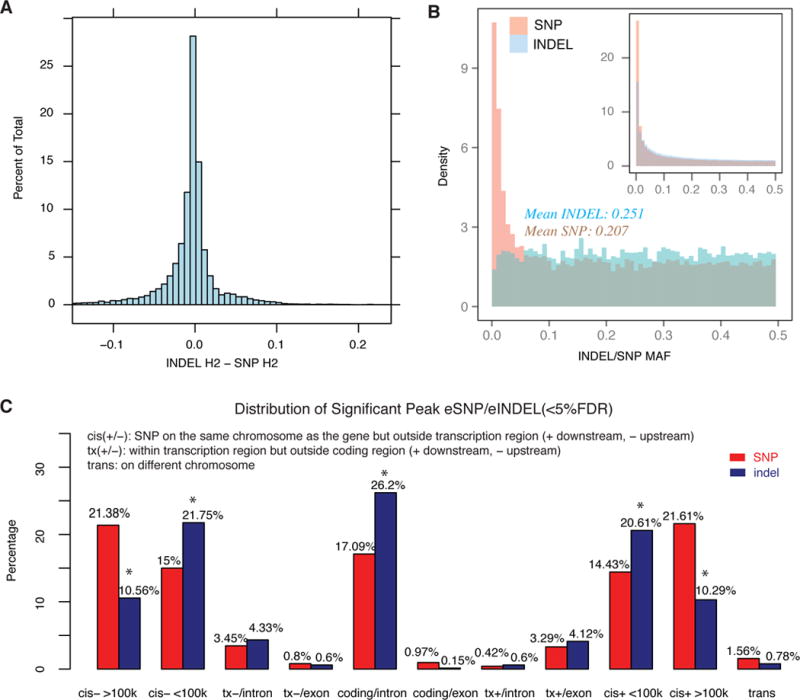
A: Histogram of the effect size (heritability explained by the individual eSNP or eindel) difference between eindel and eSNP.
B: Histogram of minor allele frequency for eindel (blue) and eSNP (red). Top right small histogram shows minor allele frequency distribution for all indel (blue) and SNP (red) with good imputation quality (Rsquare>0.3).
C: Genomic distribution of peak eQTLs (cis or trans) related to associated gene. For a particular SNP-probe pair or indel-probe pair, we divided the genome into 11 regions related to the gene: upstream TSS >100kb (cis->100k), upstream TSS <100kb (cis-<100k), intron between TSS and translation start sites (tx-/intron), exon between TSS and translation start sites (tx-/exon), intron in translation region(coding/intron), exon in translation region (coding/exon), intron between translation stop sites and TES (tx+/intron), exon between translation stop sites and TES (tx+/exon), downstream of TES <100kb (cis+ <100k), the downstream TES >100kb (cis+ >100k) and on different chromosomes (trans). We then assigned the peak eSNP or eindel into one of these categories, respectively for SNP and indel, and reported the percentage of total peak eSNP or eindel fell into each category. A star on the top of bar indicates significant difference between eSNP and eindel (p<10−4).
Among all eSNPs and eindels that passed 5% FDR, allele frequency of eindels were higher than eSNPs (mean MAF of eindel=0.25, mean MAF of eSNP=0.21, Figure 2B, main panel). This difference is not due to the distribution of SNPs and indels with good imputation quality, where most low frequency indels were not eQTL (Figure 2B, small panel).
Next we examined the genomic distribution of eSNPs and eindels by focusing on the peak eQTL (either SNP or indel, cis or trans) for each probe set with at least one significant eQTL (5% FDR) and divided the genome into 11 regions: upstream TSS >100kb (cis->100k), upstream TSS <100kb (cis-<100k), intron between TSS and translation start sites (tx-/intron), exon between TSS and translation start sites (tx-/exon), intron in translation region(coding/intron), exon in translation region (coding/exon), intron between translation stop sites and TES (tx+/intron), exon between translation stop sites and TES (tx+/exon), downstream of TES <100kb (cis+ <100k), the downstream TES >100kb (cis+ >100k) and on different chromosomes (trans). We found that eindels were significantly enriched in intron in translation region, upstream 5′ UTR (cis-<100K) and downstream 3′UTR (cis+ <100K) of the associated gene, and were depleted in exon, distal cis effect (cis->100K and cis+ >100K, Figure 2C). This pattern did not change after restricting to common SNPs and indels. (Supplementary Fig. S2 panel A) and eindels seem to show larger effect size and more significant evidence (larger LOD score) than eSNPs in these three regions (Supplementary Fig. S2 panel B and C). Except for the well known depletion of indel in exons7, the enrichment and depletion in other regions cannot be explained by genomic distribution of available SNPs and indels with high imputation quality (Supplementary Table 1). We hypothesized that eindel are more likely to be causal eQTLs in intron in translation region and regions close to 5′ UTR and 3′UTR, but this remains to be confirmed by experiments.
Tissue specific eQTLs
For a particular SNPC probe set pair, the eQTL effect may present (coded as 1) in a particular tissue or not (coded as 0). For 3 tissues we studied, this resulted in 8 possible scenarios (from not being an eQTL in any tissues 000 to being eQTL in all 3 tissues 111, corresponding to the order of LCL, PBMC and SKIN). We investigated tissue-specific eQTL effects for each SNP-probe set pair by estimating the posterior probability of each of the 8 possible scenarios (see methods for detail). We denoted P111 as the posterior probability that the eQTL effect presented in all 3 tissues and Ptissue specific eQTL (tse) = 1-P000-P111 as the probability that eQTL effect presented in at least one but not all tissues. We found that for cis eQTLs (defined as SNP/indel and probe set are within 1Mb of each other), the mean of Ptse across all 930,775 SNP-probe set pairs from meta-analysis (<5% FDR) were 0.448 (s.d. 0.369), the mean of P111 were 0.552 (s.d. 0.369) and the mean of P000 was 6.15*10−5. This suggests that cis eQTLs are more likely to be shared between tissues but still many were tissue-specific. On the contrary, the mean PTSE for trans eQTLs (defined as SNP and probe set being either on different chromosomes or >500Kb apart) was 0.806 (s.d. 0.121), and the mean of P111 was 0.192 (s.d. 0.121) (Supplementary Fig. S3 A–D). Results for eindels were similar (Supplementary Fig. S3 E–H). This clearly showed that trans eQTLs are much less likely to be shared between tissues.
Genes with shared genetic regulators across tissues might have different functions than genes with tissue-specific regulators. Characterizing gene pathways and functional groups by tissue-sharing of genetic regulators would help understanding the underlying regulation of such pathways and help prioritizing genetic studies using related tissues. We used Gene Ontology (GO) biological process to characterize the functions of genes based on annotation information downloaded from the manufactures website. For each scenario of eQTL sharing (P000, …, P111) both Z-score and permutation-based P-values were used to assess significance. For each GO category, we focused on the peak eQTL on the same chromosome of the probes belonging to genes in this category. We then asked whether eQTLs for this GO category showed significant high level of posterior probability for each of the 8 sharing scenarios, respectively. Accounting for 199 GO terms (number of genes>20) and a 5% family-wise false-positive rate, the Bonferroni correction gave a significant P-value threshold of 2.5 × 10−4 (Supplemental table 2). We found that genetic regulators for genes involved in “translation”(p< 1.62 × 10−5), “oxidation-reduction” (p<1.65 × 10−4), and “proton transport” (p<2.89 × 10−4) were likely shared between all three tissues (P111). Genes involved in “response to protein stimulus” (p<6.57 × 10−7) were likely sharing similar genetic regulation in PBMC and skin (P011). Genes involved in “oxidation-reduction” (p<2.29 × 10−7) and “metabolic process” (p<4.18 × 10−6) were likely sharing similar genetic regulation in LCL and skin (P101). “tRNA processing” (p<6.30 × 10−4) were marginal significantly shared between LCL and PBMC (P110) (Supplementary Table 2).
Potential epigenetic driving factors for eQTLs
Genetic regulatory variants may affect gene expression level by different functional mechanisms. To identify the potential functional role of eQTLs, functional elements predicted by using ENCODE data were downloaded from UCSC genome browser (hg19)56. We pooled the annotation data from the tracks for skin(NHEK) and blood(GM12878) for mapping and analysis of chromatin state dynamics of peak eQTLs (either eSNPs or eindels in cis or trans) identified from this study. This strategy has been shown to be a powerful way to interpret the function of eQTLs and generate specific hypothesis for gene expression regulation, as demonstrated in the Crohns disease and PEGER4 gene eQTL example57.
We divided the peak eSNPs and eindels into three groups according to minor allele frequency: MAF<0.05, 0.05≤MAF<0.10 and 0.10≤MAF≤0.50, and examined the overlap with each available functional element. Across all three MAF groups, we found that both types of eQTLs (eSNP and eindel) were significantly enriched (pvalue < 10−4by permutation test comparing to MAF-matched and distance-to-gene-matched SNPs and indels chosen from 1000Genomes Project) in functional elements, including Active Promoter, Weak Promoter, Transcriptional Elongation and Weak Transcribed (Figure 3 B–D). They were both significantly depleted in Heterochromatin marks (Figure 3, A). Both types of eQTLs showed non-significant overlap with functional elements for Inactive/poised Promoter, Insulator, Polycomb-repressed and common Copy Number Variation (Figure 3 B–D), and were marginally significant for rare CNVs (p=0.0012 for both eSNP and eindel). For Strong Enhancer and Weak/poised Enhancer, common eQTLs were enriched in these two functional elements while rare eQTLs were marginally significantly enriched (p ranges from 0.0011 to 0.0015). Finally, for Transcriptional Transition, both common and low frequent eindels were enriched but only common eSNPs (MAF>5%) were enriched in this functional element (rare eSNP pvalue=0.58). To compare between indels and SNPs, we found that transcriptional elongation and weak transcribed were significant different between indels and SNPs (P value < 1 × 103 4). Active Promoter was significantly different in common eSNPs (MAF>5%) (P value < 1 × 103 4). Transcriptional transition was significantly different in rare eSNPs (MAF<10%) (P value < 1 × 103 4). P value was calculated using simulation by randomly selected SNPs or indels from the same MAF category and genome within +/C 500Kb of any genes. These functional annotations for individual peak eQTLs that passed the 5% FDR threshold are available in supplementary Table 3. Association results (effect size, LOD score, pvalue, etc) for these eQTLs were also provided in this table.
Figure 3. Distribution of peak eQTLs in functional regulator elements based on ENCODE data.
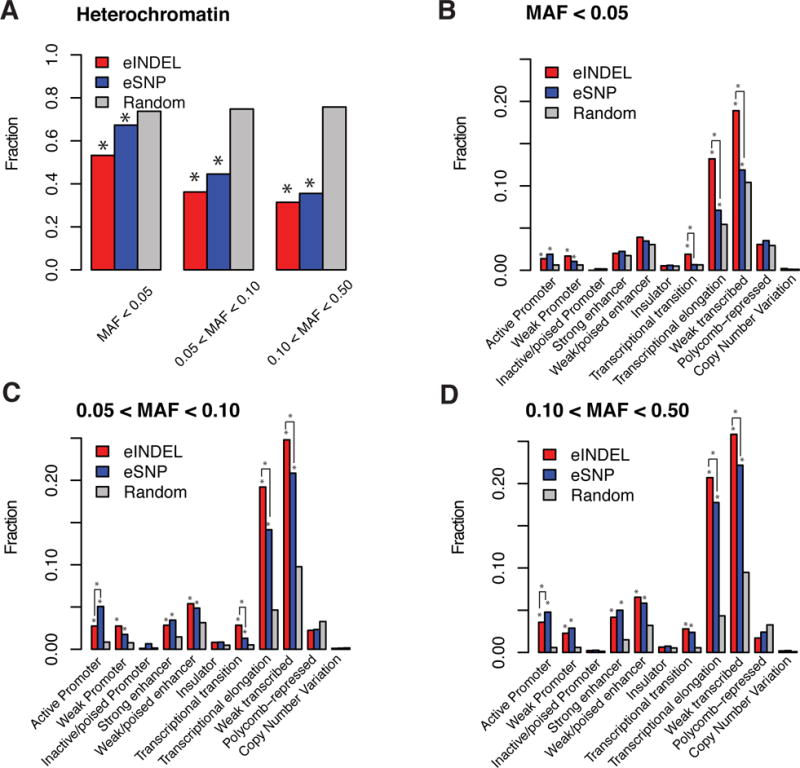
A: The fraction of eindel (red bars) and eindel (blue bars) within Heterochromatin marks (Ernst et al 2011 Nature) by different ranges of minor allele frequency (MAF < 0.05, 0.05 ≤ MAF < 0.1, 0.1 ≤ MAF ≤ 0.5). A star on the top of bar indicates significant depletion (p<10−4, Bonferroni correction for 12*3 categories is 0.05/36=1.39*10−3)
B,C,D: The fraction of eindel (red bars) and eindel (blue bars) in functional elements (Ernst et al 2011 Nature) by different ranges of minor allele frequency (B: MAF < 0.05, C: 0.05 ≤ MAF < 0.1, D: 0.1 ≤ MAF ≤ 0.5) compared with randomly selected SNPs from the same MAF category and genome within +/−500Kb of any genes (grey color). Statistically different between eindels and eSNPs are also compared. A star on the top of bar indicates significant enrichment (p<10−4, Bonferroni correction for 12*3 categories is 0.05/36=1.39*10−3). A star with solid lines connecting the SNP and indel bars indicates significant difference between the SNP and indel categories (p<10−4).
Comparison of eQTLs with known GWAS loci
SNP eQTLs have been widely used to characterize the function of a diseaseC associated variant and point to the underlying biological pathways22C,48. The newly identified eSNPs and in particular eindels may continue to help interpret function of GWAS loci that could not be explained before. We examined the disease and trait associated loci from the NHGRI GWAS category (downloaded from http://www.genome.gov/GWAStudies/ on March 15, 2013). Considering the diseases and traits with more than 10 reported genes (Supplemental table 4) and all cis and trans eQTLs that passed the 5% FDR threshold, we found that top diseases or traits enriching genes with significant eSNPs include mean corpuscular hemoglobin concentration, response to amphetamines and red blood cell count, while top diseases or traits enriching genes with significant eindelS include tonometry, IgE levels, mean corpuscular hemoglobin concentratio, celiac disease and rheumatoid arthritis. We also examined tissue specific eQTLs among GWAS variants and found that tonometry was among the top list with ≥50% trait associated genes were regulated by tissue specific eindelS, while temperament (bipolar disorder), adiponectin levels and glycated hemoglobin levels were among the top list with ≥60% disease or trait associated genes regulated by tissue specific eSNPs.
Discussion
This is the first study to impute short insertion and deletion (indel) genomeC wide in eQTL mapping study, which provided unique opportunities to answer several important questions. Our results suggested that imputation of indels can increase power of genomeC wide association studies for complex traits by about 10%. Although this was an estimate based on gene expression traits with complex genetic architecture and full spectrum of heritability, the power gain for particular disease or traits would vary by their specific genetic background. After the completion of the 1000 Genomes Project, many more high quality indels will be available for imputation and the reference panel will be increased from the current 1092 subjects to 2500 subjects. We expected that the power gain by imputation of indel will be even larger.
Our results also suggested that a substantial fraction of previous identified disease and trait associated SNPs were markers in linkage disequilibrium of indels with larger effect. Imputation of indels into GWAS would help fine map the causal variants that were tagged by previous studies. As seen in our study, this fraction of SNPs tagged by indel would increase as power increases. In our case, it increases from 3.62% to 9.20% as sample size increases from 368 to 718.
Previous studies and this study has shown that SNP eQTLs were enriched in intron and regions closed to UTRs, our study is the first time to show that indel eQTLs were even more enriched in these three regions (defined as intron in translation region, 100kb upstream of 5′ UTR and 100kb of downstream of 3′ UTR). We hypothesized that this is because indel is more likely to be causal regulator as they are more likely to destruct splice sites and promoter regions but further experiments are required to validate these hypotheses.
Finally this study showed that cis eQTLs were more likely to be shared across tissues, while trans eQTLs were more likely to be tissue specific. This is consistent with previous findings10. Indel eQTLs and SNP eQTLs showed similar pattern for tissue specificity for cis and trans. It suggested that the tissue differentiated genetic regulation is not related to the size of the genetic variants.
All significant SNP and indel eQTLs identified from this study are freely accessible to the public. We expect that it will be an important resource for GWAS to interpret function of genetic variants for complex diseases and traits, particular for structure variants.
Methods
Data resources
This study includes data from three former studies: (1) MRCA contained 206 siblings of British descent13. A total of 368 children were genotyped using the Illumina Sentrix HumanHap300 BeadChip (ILMN300K) or the Illumina Sentrix HumanC 1 Genotyping BeadChip (ILMN100K) or both. Global gene expression in lymphoblastoid cell lines (LCLs) was measured using Affymetrix HGC U133 Plus 2.0 GeneChip (including 54,675 transcript probes). (2) 240 individuals from 47 Germany eczama families were genotyped with Affy500K and Affy 6.0 SNP array. The gene expression level of their peripheral blood mononuclear cells was evaluated with Affymetrix U133 Plus 2.0 GeneChip. (3) Normal skin tissues of 110 subjects from a former Psoriasis GWAS were genotyped with Perlegen 400K array and the RNA expression level was evaluated with the same Affymetrix GeneChip. 3,423 probe set which can be mapped to multiple genome position (based on HG U133 Plus_2 annotations file, release 34) and 62 Affymetrix control probes were removed in our analysis.
SNP quality control and imputation
SNPs were excluded from further analysis with the following criteria: (1) The SNP has more than 2 alleles; (2) The SNP is not presented in the 1000 Genomes Project phase 1 release; (3) The SNPs were genotyped in less than 95% samples; (4) The Hardy-Weinbergtestis significant with P-value<10−6; (5) The minor allele frequency of the SNP <0.01. SNPs and indels from the 1000 Genomes Project phase 1 release (2012-03-14 haplotypes) were imputed using MINIMAC58. A total of 814,715 indels and 10,129,531 SNPs had high quality score (Rsquare > 0.3 in three studies).
Meta analysis
We used a weighted-z score in meta-analysis. To account for different imputation qualities and sample sizes in the three studies, we used a weighted scheme: and here the weight is a combination of sample size and imputation quality: , where is as defined in Li et al 20103. To control for multiple testing, a cut-off of FDR<0.05 accounting for all SNP/indel-probe set pairs were used, corresponding to p-value < 2.58*10−6.
Inference of tissue specific effect
Following Wakefield 200759, we used y to denote the observed data and H1 the alternative hypothesis, then for the quantitative trait: y = xTγ + Zθ, where β = (γ,θ) are parameters corresponding to the effects of covariates x and SNP z. According to Wakefield 200759, when calculating Asymptotic Bayesian factor (ABF), we only need to consider the sampling distribution of the MLE: and the prior for θ ~.N(0, W), which gives:
| (1) |
An advantage of using ABF is that the calculation only involves and its standard error from individual studies. And from the above we can calculate ABF1, ABF2 and ABF3 for the three tissues, respectively. We defined all possible scenarios for tissue sharing as in table 1. Then the posterior probability of each scenario can be calculated.
Table 1.
Definition of all different tissue specific eQTLs.
| Case | LCL | PBMC | SKIN | Prob |
|---|---|---|---|---|
| C1 | 0 | 0 | 0 | P(000) |
| C2 | 1 | 0 | 0 | P(100) |
| C3 | 0 | 1 | 0 | P(010) |
| C4 | 0 | 0 | 1 | P(001) |
| C5 | 1 | 1 | 0 | P(110) |
| C6 | 1 | 0 | 1 | P(101) |
| C7 | 0 | 1 | 1 | P(011) |
| C8 | 1 | 1 | 1 | P(111) |
Note: Here we use “1” to indicate the eQTL is present in the corresponding tissue, and “0” to indicate eQTL is absent in that tissue.
We defined probability of being tissue-specific eQTL (Ptse) as:
| (2) |
Here P111 is the posterior probability of sharing eQTLs across all three tissues, while P000 is the probability that the eQTL does not occur in any tissue. Bayesian factor for each scenario in table 1 can be computed based on Bayesian factors from individual studies, for example BFc1 is given in formula.
| (3) |
Then the posterior probability of P111 is calculated as:
assuming equal prior probabilities πi = 1/8, i=000 to 111. The posterior probability of the other 7 scenarios can be computed in a similar way.
GO enrichment analysis
The Affymetrix expression probes were grouped into GO categories using annotation information downloaded from the manufacturer’s website. For each GO category, we focused on the peak eQTL (FDR<5%) on the same chromosome of the probes belonging to genes in this category and calculated the mean of P(000), …, P(111) of these eQTLs, denoted as Gi for the ith GO category for each of the 8 scenarios, respectively. The Z-score for the ith GO category is , where μ is the overall mean of P(000), …, P(111) for all probes annotated to the 9,409 genes, respectively, σ is the corresponding standard deviation, and ni is the number of probes for the ith GO category that mapped by eQTL. The P-value is computed by comparing the Z-score with a standard normal distribution for one sided test.
We also computed the permutation based P-value by shuffling the correspondence between probes and GO categories while maintaining the same number of genes for each GO term. From 10,000 permutations, we counted how many times (Mi) the Gi based on permutated data were larger than Gi based on observed data. The permutation based P-value is Mi/10000.
Genomic distribution of SNP and indel eQTLs
Genome annotation was obtained from the UCSC genome browser (hg19). To examine the distribution of the physical location of cis and trans eSNPs and eindels, we focused on peak eQTLs (cis or trans) for each probe set. For a particular SNP-probe pair or indel-probe pair, we divided the genome into 11 regions related to the gene: upstream TSS >100kb (cis->100k), upstream TSS <100kb (cis-<100k), intron between TSS and translation start sites (tx-/intron), exon between TSS and translation start sites (tx-/exon), intron in translation region(coding/intron), exon in translation region (coding/exon), intron between translation stop sites and TES (tx+/intron), exon between translation stop sites and TES (tx+/exon), downstream of TES <100kb (cis+ <100k), the downstream TES >100kb (cis+ >100k) and on different chromosomes (trans). We then assigned the peak eSNP or eindel into one of these categories, respectively for SNP and indel, and reported the percentage of peak eSNP or eindel fell into each category.
Distribution of SNP and indel eQTLs related to epigenetic factors
Functional elements predicted by using ENCODE data were downloaded from UCSC genome browser (hg19)56. We pooled the data from the skin(NHEK) and blood(GM12878) tracks for mapping and analysis of chromatin state dynamics. SNPs and indels located within these functional elements were considered as related to epigenetic factors. For this analysis we focused on peak eQTLs (either eSNPs or eindels in cis or trans). For each minor allele frequency category (MAF ≤ 0.05, 0.05 < MAF ≤ 0.1, 0.1 < MAF ≤ 0.5), we calculated the fraction of eSNPs or eindels fell into each functional element category. The significance of enrichment was determined by randomly selecting the same number of SNPs or indels from the same MAF category and computed the fraction of random SNPs or indels fell into these functional regulatory regions. We counted how many times (M) the fraction of random SNP or indel were larger than the fraction based on eSNPs or eindels. Enrichment P-value (Figure 3, B–D) was calculated as M/10,000 for 10,000 permutations. Depletion P-value (Figure 3, A) was calculated as 1−M/10,000.
Overlapping with known GWAS reported genes
GWAS results were obtained from the NHGRI GWAS database (downloaded from http://www.genome.gov/admin/gwascatalog.txt on January 10, 2014). GWAS reported genes were obtained from the “Reported Gene” column in gwascatalog.txt file. For each disease or trait, we calculated the percent of reported genes associated with at least one SNP or indel, respectively. We also calculated the percent of reported genes associated with eSNP or eindel showed tissue specific effects (Ptse>0.5)
Supplementary Material
Genomic location was based on UCSC genome browser. The first two columns showed the percentage of SNPs or indels with imputation Rsquare>0.3 fell into each category. The last column “genome” shown the percentage of each type of region covered the whole genome. Intergenic regions were further divided into two parts at the last two rows: < or > 100Kb of the closest UTR.
P(000) to P(111) are average posterior probabilities of each sharing scenario. Pvalue_z is pvalue calculated based on z-score method. Pvalue_permutation is pvalue calculated based on permutation test. Count is the total number of genes belongs to the GO category.
For each probe, the peak associated SNP or indel are listed. Association results includes effect allele label, effect size, standard error of effect size, heritability explained by the associated SNP or indel, LOD score and pvalue, z-score of the metaanalysis combining all three tissues. If the SNP or indel is located in functional elements (Ernst et al 2011 Nature), it is indicated as “Y”. Otherwise, it is indicated as “N”.
Gene Count is the number of genes reported for the disease or trait in NHGRI GWAS database.
PercentIndelRegulatedGenes is the percentage of GWAS reported genes regulated by indel.
PercentSNPRegulatedGenes is the percentage of GWAS reported gene regulated by SNP.
PercentINDELTissueSpec is the percentage of GWAS reported genes regulated by indel with Ptse>0.5.
PercentSNPTissueSpec is the percentage of GWAS reported genes regulated by SNP with Ptse>0.5.
Figure S1. Histogram of imputation quality for indels and SNPs.
SNPs and indels were categorized based on MAF (A: <0.05, B: 0.05–0.10 and C: 0.10–0.50).
Figure S2. Genomic distribution of peak eQTLs (cis or trans) related to associated gene.
A: Genomic distribution of peak eQTLs (cis or trans) related to associated gene. Similar to Figure 2 panel C but restricted to common SNPs and indels (MAF>5%)
B: Boxplots of heritability explained by peak eQTLs (SNPs or indels) by genomic location related to the associated gene. Include all common and rare SNPs and indels.
C: Boxplots of LOD score of peak eQTLs (SNPs or indels) by genomic location related to the associated gene. Include all common and rare SNPs and indels.
Figure S3. Histogram of the posterior probability of tissue specific (Ptse) and tissue consistent (P111) eQTL categorized by eSNP and eindel in cis (variant and probe<1Mb) and trans.
Footnotes
Online resources
On our website (http://eqtl.rc.fas.harvard.edu/indeleQTL/), we provided flat tables (csv files) for all eQTL results with meta-analysis FDR<0.05.
References
- 1.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature Genetics. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 5.The International HapMap Consortium. The International HapMap Project. Nature. 2005;437:1299–320. [Google Scholar]
- 6.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Consortium TGP. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montgomery SB, et al. The origin, evolution, and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome Res. 2013;23:749–61. doi: 10.1101/gr.148718.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–24. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price AL, et al. Single-tissue and cross-tissue heritability of gene expression via identity-by-descent in related or unrelated individuals. PLoS Genet. 2011;7:e1001317. doi: 10.1371/journal.pgen.1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grundberg E, et al. Mapping cis-and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44:1084–9. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morley M, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–7. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon AL, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 14.Dimas AS, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–50. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fairfax BP, et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012;44:502–10. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nica AC, et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schadt EE, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeller T, et al. Genetics and beyond–the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stranger BE, et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang L, et al. A cross-platform analysis of 14,177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome Res. 2013 doi: 10.1101/gr.142521.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–94. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas G, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 23.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohns disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 25.Libioulle C, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genetics. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi Q, Liang L, Doria A, Hu FB, Qi L. Genetic predisposition to dyslipidemia and type 2 diabetes risk in two prospective cohorts. Diabetes. 2012;61:745–52. doi: 10.2337/db11-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi Q, et al. Genome-wide association analysis identifies TYW3/CRYZ and NDST4 loci associated with circulating resistin levels. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu X, et al. A genome-wide association study identifies two new risk loci for Graves disease. Nat Genet. 2011;43:897–901. doi: 10.1038/ng.898. [DOI] [PubMed] [Google Scholar]
- 29.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–8. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heid IM, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–60. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu YH, et al. An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility Loci for osteoporosis-related traits. PLoS Genet. 2010;6:e1000977. doi: 10.1371/journal.pgen.1000977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M, et al. Integrating pathway analysis and genetics of gene expression for genome-wide association study of basal cell carcinoma. Hum Genet. 2012;131:615–23. doi: 10.1007/s00439-011-1107-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu C, et al. Genome-wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene-environment interactions. Nat Genet. 2012 doi: 10.1038/ng.2411. [DOI] [PubMed] [Google Scholar]
- 35.van der Harst P, et al. 75 genetic loci influencing the human red blood cell. Nature. 2012 doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weedon MN, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hom G, et al. Association of Systemic Lupus Erythematosus with C8orf13–BLK and ITGAM–ITGAX. New England Journal of Medicine. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 39.Levy D, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satake W, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinsons disease. Nat Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 42.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ober C, et al. Effect of Variation in CHI3L1 on Serum YKL-40 Level, Risk of Asthma, and Lung Function. New England Journal of Medicine. 2008;358:1682–1691. doi: 10.1056/NEJMoa0708801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heazlewood CK, et al. Aberrant Mucin Assembly in Mice Causes Endoplasmic Reticulum Stress and Spontaneous Inflammation Resembling Ulcerative Colitis. PLoS Med. 2008;5:e54. doi: 10.1371/journal.pmed.0050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silverberg MS, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216–220. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahn J, et al. Variation in KLK genes, prostate-specific antigen and risk of prostate cancer. Nat Genet. 2008;40:1032–4. doi: 10.1038/ng0908-1032. author reply 1035–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunt KA, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas G, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 49.Esparza-Gordillo J, et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat Genet. 2009;41:596–601. doi: 10.1038/ng.347. [DOI] [PubMed] [Google Scholar]
- 50.Ding J, et al. Gene expression in skin and lymphoblastoid cells: Refined statistical method reveals extensive overlap in cis-eQTL signals. Am J Hum Genet. 2010;87:779–89. doi: 10.1016/j.ajhg.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nature Genetics. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 52.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. American Journal of Human Genetics. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang L, et al. A cross-platform analysis of 14,177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome research. 2013;23:716–26. doi: 10.1101/gr.142521.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benjamini Y, Yekutieli D. The control of the False Discovery Rate in multiple testing under dependency. Annals of Statistics. 2001;29:1165–1188. [Google Scholar]
- 55.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journ al of the Royal Statistical Society (B) 1995;57:289–300. [Google Scholar]
- 56.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–9. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet81. 2007:208–27. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic location was based on UCSC genome browser. The first two columns showed the percentage of SNPs or indels with imputation Rsquare>0.3 fell into each category. The last column “genome” shown the percentage of each type of region covered the whole genome. Intergenic regions were further divided into two parts at the last two rows: < or > 100Kb of the closest UTR.
P(000) to P(111) are average posterior probabilities of each sharing scenario. Pvalue_z is pvalue calculated based on z-score method. Pvalue_permutation is pvalue calculated based on permutation test. Count is the total number of genes belongs to the GO category.
For each probe, the peak associated SNP or indel are listed. Association results includes effect allele label, effect size, standard error of effect size, heritability explained by the associated SNP or indel, LOD score and pvalue, z-score of the metaanalysis combining all three tissues. If the SNP or indel is located in functional elements (Ernst et al 2011 Nature), it is indicated as “Y”. Otherwise, it is indicated as “N”.
Gene Count is the number of genes reported for the disease or trait in NHGRI GWAS database.
PercentIndelRegulatedGenes is the percentage of GWAS reported genes regulated by indel.
PercentSNPRegulatedGenes is the percentage of GWAS reported gene regulated by SNP.
PercentINDELTissueSpec is the percentage of GWAS reported genes regulated by indel with Ptse>0.5.
PercentSNPTissueSpec is the percentage of GWAS reported genes regulated by SNP with Ptse>0.5.
Figure S1. Histogram of imputation quality for indels and SNPs.
SNPs and indels were categorized based on MAF (A: <0.05, B: 0.05–0.10 and C: 0.10–0.50).
Figure S2. Genomic distribution of peak eQTLs (cis or trans) related to associated gene.
A: Genomic distribution of peak eQTLs (cis or trans) related to associated gene. Similar to Figure 2 panel C but restricted to common SNPs and indels (MAF>5%)
B: Boxplots of heritability explained by peak eQTLs (SNPs or indels) by genomic location related to the associated gene. Include all common and rare SNPs and indels.
C: Boxplots of LOD score of peak eQTLs (SNPs or indels) by genomic location related to the associated gene. Include all common and rare SNPs and indels.
Figure S3. Histogram of the posterior probability of tissue specific (Ptse) and tissue consistent (P111) eQTL categorized by eSNP and eindel in cis (variant and probe<1Mb) and trans.


