Abstract
Expansion microscopy is a recently introduced technique in which fluorophores on fixed specimens are linked to a swellable polymer that is physically expanded to enable super-resolution microscopy with ordinary microscopes. We have developed and characterized new methods for linking fluorophores to the polymer that now enable expansion microscopy with conventional fluorescently-labeled antibodies and fluorescent proteins. Our methods simplify the procedure, expand the palette of compatible labels, and will aid in rapid dissemination of the technique.
Boyden and coworkers recently introduced Expansion Microscopy (ExM) as a super-resolution microscopy technique that uses physical expansion of fixed specimens to allow features closer than the diffraction limit of light (~250 nm) to become resolvable in the expanded specimen.1 Unlike other super-resolution techniques which rely on specialized instruments,2,3 ExM is compatible with standard microscopes (e.g., widefield, confocal, etc.) and is poised to make a significant impact based on its accessibility and on its strong performance in thick specimens.
In the impressive initial report on ExM, imaging with ~65 nm resolution was demonstrated in cultured cells and in brain tissue using a procedure entailing: staining of a specimen with polymer-linkable probes, growth of a swellable polymer within the specimen which links to the probes, protease digestion of the specimen, and expansion of the polymer through dialysis.1 The polymer-linkable probes consisted of antibodies labeled with doubly-modified DNA oligonucleotides containing a fluorophore and a methacryloyl group designed to become covalently incorporated into the polymer. As these DNA-labeled antibodies are custom-made and require a 1–2 day multi-step protocol to prepare with expensive reagents, we sought to develop methods which would allow ExM to use standard fluorophore-labeled secondary antibodies lacking DNA. We refer to these antibodies as conventional secondary antibodies, and to their use as conventional immunostaining. We also extended our approach to allow the direct use of intrinsic fluorescent protein signal in ExM.
We initially reasoned that conventional fluorescently-labeled antibodies could potentially be used in ExM if a sufficient number of linkages could be formed between the antibodies and hydrogel so that protease-digested antibody fragments would remain linked to the hydrogel (Fig. 1). Indeed, we found that 60 min treatment of a fixed and conventionally immunostained cultured cells with a 25 mM solution of the amine-reactive small molecule MA-NHS (methacrylic acid N-hydroxy succinimidyl ester) conferred excellent retention of fluorescent signal after digestion and expansion (Fig. 2 a–d). Omission of the MA-NHS treatment resulted in distorted images with poor retention of fluorescence (Supplementary Fig. 1). MA-NHS was chosen here due to its resemblance to the methacryloyl group originally used in the DNA-labeled antibody probes; similar reactive groups are also established for linking of peptides or proteins to hydrogels.4
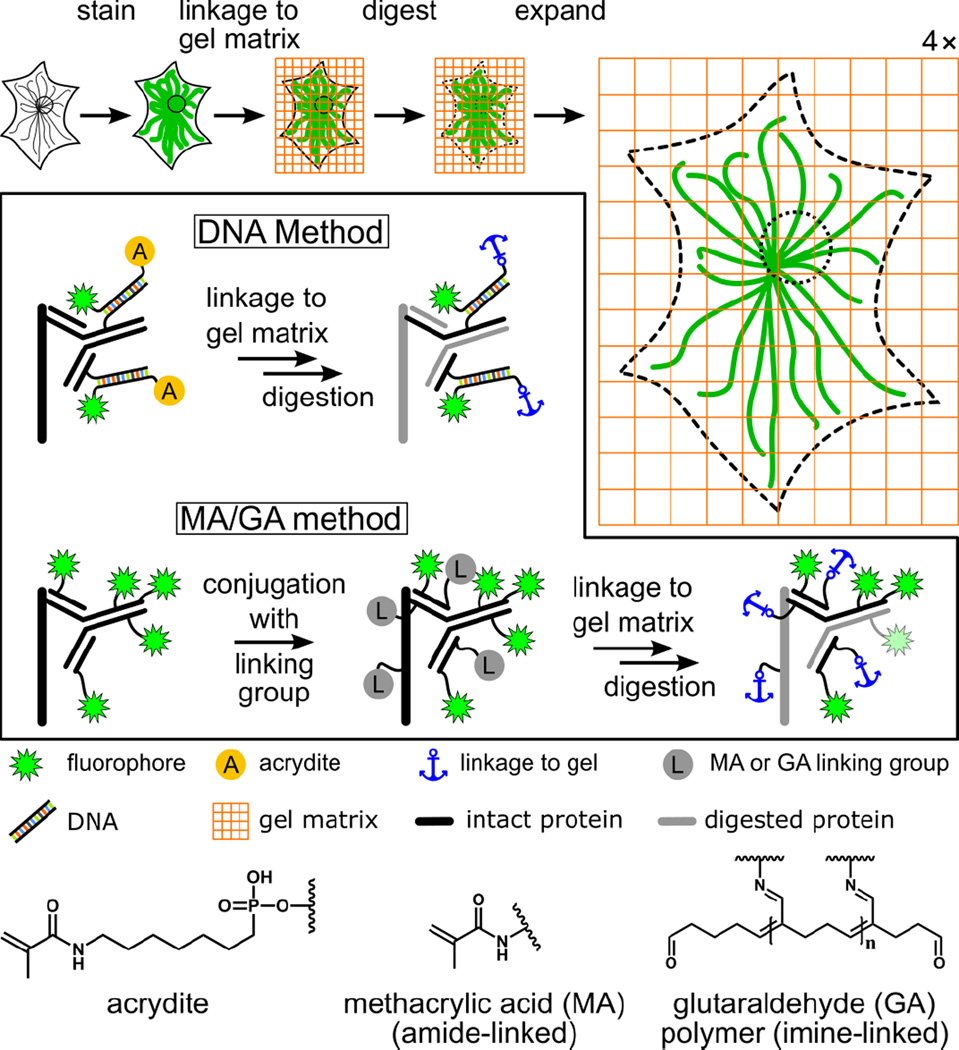
Figure 1.
Schematic illustration of expansion microscopy and label retention strategies. The boxed region highlights the difference between the original DNA method1 and the post-stain linker-group functionalization method (“MA/GA method”) presented in this work. In the DNA method, the specimen is immunostained with a custom-prepared antibody bearing doubly-modified DNA linked to a fluorophore and an acrydite moiety (A). In contrast, with the MA/GA method, methacrylic acid N-hydroxy succinimidyl ester (MA-NHS) or glutaraldehyde (GA) are used to label the entire sample with polymer-linking groups after conventional immunostaining with fluorophore-labeled antibodies (only secondary antibodies are shown). For both methods, the next steps are gelation, digestion with a protease, and expansion through dialysis into deionized water. The acrydite (A), MA, and GA groups allow formation of a linkage to the hydrogel. Dyes are retained through a connection to antibody fragments that also contain a linkage to the gel. Fluorescent proteins are also retained using the MA/GA method through a similar method but are not shown here for the sake of clarity.
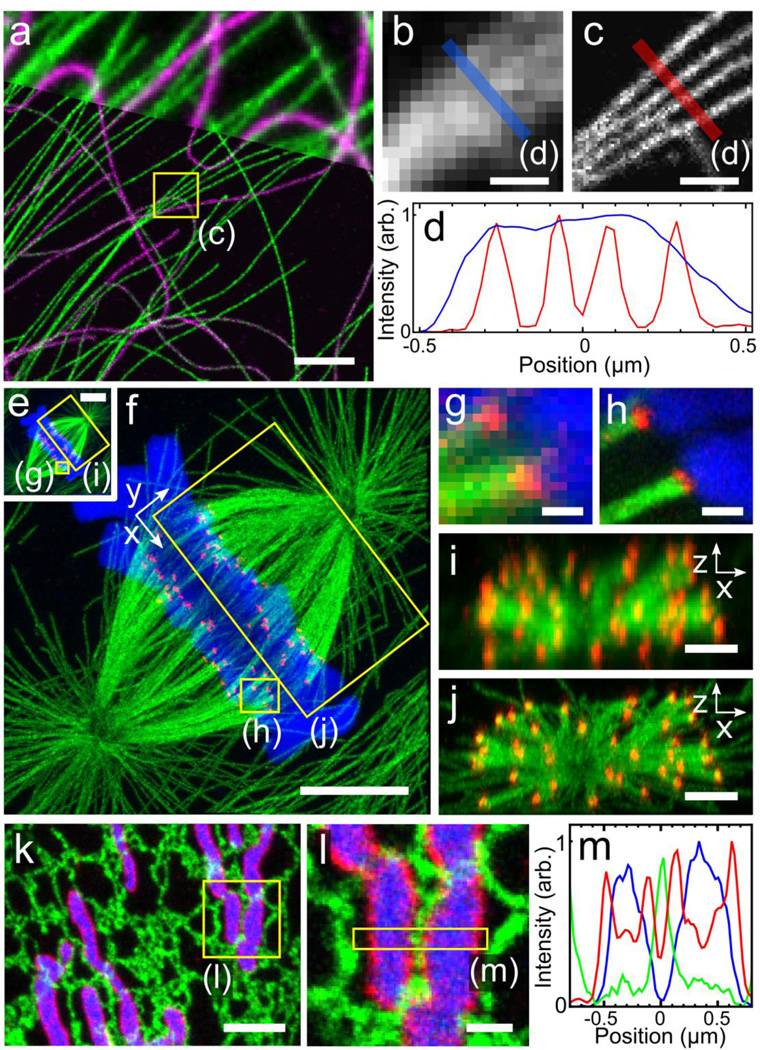
Figure 2.
Confocal fluorescence images of expanded cultured cells. (a) BS-C-1 cell immunostained for tyrosinated tubulin (green) and detyrosinated tubulin (magenta) using conventional secondary antibodies and partially overlaid with corresponding pre-expansion image (top). Specimen was treated with MA-NHS after immunostain. Zoom-in of boxed region in a showing corresponding pre-expansion (b) and post-expansion (c) images of tyrosinated tubulin signal along with corresponding line profiles (d). Pre-expansion (e) and post-expansion (f) expansion images of a dividing PtK1 cell immunostained for tubulin (green) and the kinetochore protein HEC1 (red) using conventional secondary antibodies and also stained for DNA (blue) using TO-PRO-3. Specimen was treated with GA after immunostain. (g–h) Zoom-in of microtubule-kinetochore attachments from boxed regions in e and f. End-on views of boxed regions in e, f before (i) and after (j) expansion (DNA channel omitted for clarity). (k) Maximum intensity projection of a fixed BS-C-1 cell expressing the endoplasmic reticulum (ER) tag Sec61Β-GFP (green) and the inner mitochondrial membrane tag mito-DsRed (blue) and immunostained against the outer mitochondrial membrane protein TOM20 using a conventional secondary antibody (red). The specimen was treated with GA after immunostain and only briefly digested in order to retain GFP and DsRed fluorescence. (l) Zoom-in of boxed region in k showing close apposition of an ER tubule with two mitochondria. (m) Cross-sectional profile of boxed region in l. All distances and scale bars are in pre-expansion units. Scale bars, (a, i, j, k) 2 µm, (b, c, g, h, l) 500 nm, (e, f) 5 µm.
As with the original ExM report, we observed fine details in the images of expanded specimens which were hidden in images of the unexpanded specimens (Fig. 2 a–d). The cross-sectional profile of expanded microtubules yields an average Gaussian-fitted full width at half maximum (FWHM) of 79 ± 9 nm (mean ± SD (standard deviation), Supplementary Fig. 2). This 79 nm width is consistent with a convolution of the double-peaked cross-sectional profile of indirectly immunolabeled microtubules5,6 measured by localization microscopy (i.e., STORM, PALM, etc.)2,3 and an estimated ~65 nm expansion-corrected lateral spatial resolution. The uniformity of expansion is remarkably good across the sample, and an analysis of distortions between corresponding pre-expansion and post-expansion images recorded by confocal microscopy showed that distortions were generally below 100 nm (root mean square distance) over length scales of up to 30 µm (Supplementary Fig. 3). A comparison of expansion fidelity using DNA-labeled secondary antibodies also yielded similar results (Supplementary Fig. 4). Note that all distances and scale bars for expanded specimens in this report have been divided by their respective, measured expansion factors of 4–4.2 and that all distances and scale bars therefore refer to pre-expansion dimensions.
In a second approach, we found that treatment of conventionally immunostained cultured cells with glutaraldehyde (GA) also yielded excellent fluorescence retention after digestion (Supplementary Fig. 1). Although GA post-fixation is a commonly used procedure in immunofluorescence assays, GA crosslinking is also well-known for use in linking proteins or enzymes to polyacrylamide gels.7 Correlated pre-expansion localization microscopy and post-expansion confocal microscopy measurements using GA treatment of immunostained cells revealed that distortions were generally below 25 nm (root mean square distance) over length scales of up to 20 µm (Supplementary Fig. 5). Microtubule cross sectional profiles had an average Gaussian-fitted FWHM of 80 ± 7 nm (mean ± SD, Supplementary Fig. 2), indicating a spatial resolution of ~65 nm as before. A three-color stain of an early anaphase PtK1 cell produced clear images of the mitotic spindle and distinctly resolved attachments between kinetochore-fiber microtubule bundles and chromosomes with good expansion fidelity (Fig. 2 e–j, Supplementary Figs. 6–8). Although the DNA stain TO-PRO-3 is quenched by the polymerization reaction, we were able to stain DNA after expansion through a brief incubation step with the dye (see methods). A panel of GA-treated immunostained cells for a variety of cytoskeletal structures and sub-cellular organelles are shown in Supplementary Fig. 9.
Conventionally immunostained cells treated with either MA-NHS or GA showed 3–4× brighter signal after expansion compared to untreated cells using DNA-labeled antibodies (Supplementary Fig. 10). Although fluorescence retention post-expansion was somewhat better using DNA-labeled antibodies than with MA-NHS or GA treatment of conventional antibodies (~90% compared to ~70%, Supplementary Fig. 11), we found that pre-expansion specimens were ~4× brighter with conventional antibodies than with DNA-antibodies (data not shown). We believe the higher brightness likely results from the ability to conjugate more of the small fluorophore molecules (~600 g/mol) to an antibody than the comparably large single-stranded oligonucleotides (~6000 g/mol) before compromising the antibody’s binding ability.
We observed in cultured cells that GA-treated specimens tolerated digestion times as short as 30 minutes with low distortion, while MA-treated specimens required longer digestion times to avoid distortion (~12–18 hours, Supplementary Figs. 13, 14). This observation led us to ask whether fluorescent protein signals could themselves be retained for imaging after digestion and expansion. Gratifyingly, we found that cells treated with GA retained intrinsic fluorescence signal from fluorescent proteins (GFP, DsRed) targeted to various structures when using a short (~30 min) digestion step (Fig. 2 k–m, Supplementary Fig. 11). The use of long digestion times (>12 hours), or the omission of GA treatment, resulted in little retained fluorescent protein (FP) signal (Supplementary Figs. 15, 16). Hybrid experiments using a mixture of FP and antibody stains are straightforward (Fig. 2 k–m).
The above methods extended well to brain tissue. The treatment of conventionally immunostained 100 µm-thick THY1-YFP-H mouse brain slices with MA-NHS (Fig. 3) or GA retained antibody fluorescence, although we prefer the MA-NHS treatment because treatment with GA leads to high levels of background fluorescence (Supplementary Fig. 17).8 Complete, high-fidelity expansion in tissue required a lower MA-NHS concentration than in cultured cells (1 mM for 60 min), presumably due to physical differences in the specimens. We therefore advise validation (through correlative imaging pre- and post-expansion) and possible optimization of these procedures or their variations before applying them to uncharacterized specimens which may have different properties.
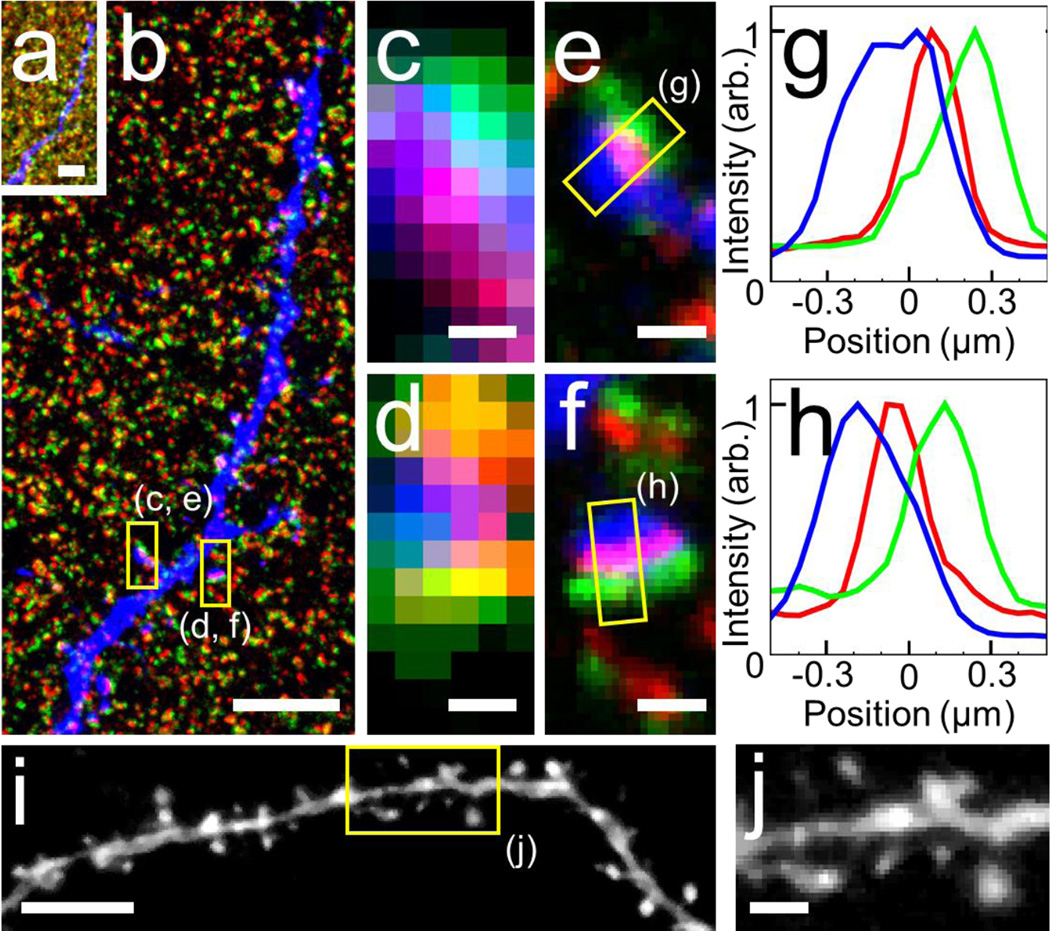
Figure 3.
Confocal (a–f) and epifluorescence (i, j) images of expanded mouse brain tissue using the MA-NHS treatment method. (a) Single pre-expansion focal plane of a THY1-YFP-H mouse brain slice indirectly immunostained for YFP (blue), the presynaptic marker Bassoon (green), and the postsynaptic marker Homer (red) using conventional secondary antibodies. (b) The same area in a after expansion, displayed with the relative size compared to a, in order to show the relative amount of physical expansion. Zoom-in of the boxed regions in b before expansion (c, d) and after expansion (e, f), revealing that the presynaptic and postsynaptic markers are well-resolved and aligned with dendritic spines. (g, h) Cross-sectional profiles of the boxed regions in e, f. (i) Epifluorescence image of a neuron in an expanded THY1-YFP-H mouse brain slice using YFP itself as the fluorescence reporter; image was recorded using a 20× 0.45 NA objective lens. The specimen was treated with MA-NHS and only briefly (1 h) digested in order to retain FP fluorescence. (j) Zoom in of the boxed region in i showing clearly resolved dendritic spines. All distances and scale bars correspond to pre-expansion dimensions. Scale bars, (a, b) 5 μm, (c, d, f, g) 500 nm, (i) 4 μm, (f) 1 μm.
We immunostained THY1-YFP-H brain slices for YFP-expressing neurons and the pre- and postsynaptic markers Bassoon and Homer using conventional secondary antibodies (Fig. 3 a–f) and treated with MA-NHS before gelation, digestion, and expansion. Presynaptic and postsynaptic densities were well-resolved and junctions between synapses and dendritic spines were clearly observable (Fig. 3 a–f, Supplementary Fig. 18, Supplementary Video 1). Over length scales of up to 30 µm we observed distortions generally below 0.2 µm (Supplementary Fig. 19). By decreasing the digestion time for MA-treated mouse brain tissue to 1 h (rather than 12–18 hours), we were able to preserve intrinsic YFP fluorescence in expanded brain tissue and we were easily able to observe dendritic spines on a neurite even using a rudimentary epifluorescence microscope equipped with a 20× 0.45 NA air objective lens (Fig. 3 i–j). Omission of MA-NHS treatment results in very weak intrinsic YFP fluorescence levels (Supplementary Fig. 20) as also pointed out by Boyden and coworker.1
The MA-NHS and GA polymer-linking steps resemble the formaldehyde-acrylamide linking strategy used in the CLARITY tissue clearing procedure to link protein amines to a polyacrylamide gel, although labeling of amines on the specimen with MA-NHS and GA is over an order of magnitude faster, requiring an incubation of 60 minutes (or less) as opposed to one or more days in CLARITY.9,10 While MA is almost certainly being incorporated covalently into the polymer, the linking mechanism of GA-treated specimens is less obvious. GA exists in aqueous solution as a complex equilibrium distribution of monomeric and polymeric forms which contain aldehyde and alkene groups.11 Both aldehydes and alkene groups on GA could in principle become covalently linked to the acrylamide polymer. Additionally, it is possible that the GA polymer could become linked to the gel by topological (mechanical) entanglement with the acrylamide polymer,12 or a combination of covalent and topological mechanisms. We favor MA-NHS treatment for brain tissue due to its lower background signal and GA treatment for cultured cells due to its generality with both immunolabeled specimens and fluorescent proteins. Supplementary Table 1 summarizes all stain procedures and imaging conditions used in this report.
Boyden and coworkers pointed out that not all organic fluorophores survive the polymerization step (e.g., several cyanine fluorophores do not survive) and they recommended use of Alexa 488, TAMRA or Atto 565, and Atto 647N.1 To these we add that Alexa 405, Atto 488, Alexa 532, Alexa 546, Alexa 568, GFP, YFP, DsRed, Hoechst 33342, and SYBR Gold also survive polymerization (Fig. 2, Supplementary Fig. 21). We found that Alexa 546 provided some advantages over TAMRA or Atto 565 owing to the larger net charge on Alexa 546 and the ability to label antibodies with a larger number of Alexa 546 molecules without severe fluorescence quenching. Additionally, fluorophores may be introduced post-digestion to avoid quenching or bleaching by the polymerization reaction, such as by incubating the gel with labeled streptavidin for a specimen that has been labeled with a biotin-labeled secondary antibody, or through incubation with DNA-binding fluorophores such as TO-PRO-3, etc. (Fig. 2 e–j, Supplementary Fig. 21).
We have introduced and characterized new polymer-linking methods for expansion microscopy which now enable the use of conventional fluorophore-labeled antibodies and FPs and should help to rapidly disseminate the ExM to a large and growing community of researchers applying super-resolution techniques to a wide range of biological questions. The methods improve the brightness of immunostained specimens compared to DNA-conjugated antibodies while making use of conventional secondary antibodies that are in many cases already available in research laboratories. Given the choice, we generally prefer immunostaining of FPs due to its enhancement of signal brightness. However, the use of intrinsic FP signals with ExM creates flexibility in multi-channel situations when compatible antibody species may not be available or when FPs are separable spectrally, but not antigenically (e.g., CFP-YFP). The use of intrinsic FP signals may also provide advantages when antibody penetration into thick samples is limited.
Expansion microscopy is a highly attractive imaging modality owing to its compatibility with conventional microscopes and conventional probes, its robust multicolor and 3D capabilities, and its optical clearing properties for thick tissues.1 While the method is limited to fixed specimens whose mechanical properties do not prevent expansion, the currently achieved ~65 nm resolution is sufficient to answer a wide range of biological questions and is likely to improve with further development.
METHODS
Reagents and reagent preparation
Unconjugated secondary antibodies were purchased from Jackson Immunoresearch (West Grove, PA, USA) including donkey anti-rat (712-005-151), donkey anti-rabbit (711-005-152), donkey anti-mouse (715-005-151), and donkey anti-chicken (703-005-155). An Alexa Fluor 488 conjugated donkey anti-rat antibody (712-545-150) was purchased from Jackson Immunoresearch. Primary antibodies are listed as follows: Rat anti-alpha tubulin (MA1-80017, Thermo Fisher Scientific, Waltham, MA, USA), Rabbit anti-detyrosinated tubulin (ab48389, Abcam, Cambridge, MA, USA), Mouse anti-HEC1 (ab3613, Abcam), Rabbit anti-TOM20 (sc-11415, Santa Cruz Biotechnology, Santa Cruz, CA, USA), Rabbit anti-GFP (A31857, Life Technologies, Carlsbad, CA, USA), Chicken anti-GFP (A10262, Thermo Fisher Scientific), Rabbit anti-Homer1 (160003, Synaptic Systems, Goettingen, Germany), Mouse anti-Bassoon (ab82958, Abcam). Bovine serum albumin (BSA) was purchased from Santa Cruz Biotechnology. NHS-functionalized (amine-reactive) dyes and biotin were obtained from Sigma-Aldrich, (Atto 488, Atto 565, Atto 647N, St. Louis, MO, USA) or Thermo Fisher Scientific (Alexa Fluor 405, Alexa 488, Alexa 532, Alexa Fluor 546, Alexa Fluor 647, EZ-link NHS-PEG-4-Biotin). Dyes were obtained in 1 mg aliquots from the suppliers, dissolved at a concentration of ~100 mg/mL in anhydrous DMSO, sub-aliquoted into anhydrous DMSO at 1 and 10 mg/mL, and stored at −20 °C. NAP-5 size-exclusion chromatography columns were obtained from GE Healthcare (Little Chalfont, Buckinghamshire, United Kingdom) and were reused ten or more times by washing with 5 mL aqueous 1 M sodium hydroxide between uses and storage at 4 °C in phosphate-buffered saline (PBS) containing 2 mM sodium azide for up to several months. Methacrylic acid N-hydroxy succinimidyl ester (MA-NHS), anhydrous dimethyl sulfoxide (DMSO), sodium bicarbonate, PIPES salt (for buffer), ethylene diamine tetraacetic acid (EDTA), magnesium chloride, Triton X-100, and sodium borohydride were obtained from Sigma-Aldrich. MA-NHS was dissolved in anhydrous DMSO at a concentration of 1 M and stored at −20 °C until used. Paraformaldehyde (32%) and glutaraldehyde (50%) were obtained from Electron Microscopy Sciences (Hatfield, PA, USA). All DNA was purchased from Integrated DNA Technologies (Coralville, IA, USA). DNA stains including Hoescht 33342 (NucBlue Live), SYBR Gold, and TO-PRO-3 were purchased from Life Technologies. Tetramethylethylenediamine (TEMED, 17919) and ammonium persulfate (APS, 17874) were purchased from Thermo Fisher Scientific. 4-hydroxy-TEMPO (97%, 176141), and sodium acrylate (97%, 408220) were purchased from Sigma-Aldrich. 40% acrylamide (1610140) and 2% bis bis-acrylamide (1410142) solutions were purchased from Bio-Rad Laboratories (Hercules, CA, USA).
Preparation of fluorophore-labeled antibodies and streptavidin
Fluorophore-conjugated antibodies or streptavidin were prepared as follows. To 40 µL of unconjugated protein (~1.3 mg/mL IgG, or 1 mg/mL streptavidin) was added 5 µL of aqueous 1 M sodium bicarbonate (pH ~8.3) and 1 µL of NHS-dye stock in DMSO. These reagents were allowed to react at room temperature (22 °C) for ~30 minutes. During the reaction, a NAP-5 size-exclusion chromatography column, for purification of labeled antibody from free dye, was equilibrated by flowing ~10 mL of PBS through each column. The ~50 µL reaction was loaded onto the column followed by flowing through and discarding 650 µL of PBS and flowing through and keeping 300 µL eluate. The eluate was characterized by absorption spectroscopy by measuring the average concentration of dye and average concentration of antibody according to the instructions provided by the dye manufacturers. Care was taken to avoid adding more than ~5 % DMSO to the antibody solution to avoid disturbing the antibody in all antibody-labeling reactions. The obtained dye to protein ratios are listed in Supplementary Table 1. The DNA-antibody conjugate was prepared using 5′ amine modified DNA (TAC GCC CTA AGA ATC CGA ACT TTA CGC CCT AAG AAT CCG AAC) according to the protocol described previously1 (updated protocols available at expansionmicroscopy.org). The tri-functional linker was prepared from 5′ acrydite and 3′ amine modified DNA (GTT CGG ATT CTT AGG GCG TA), reacted with a tenfold molar excess of Atto 488 NHS for 1 h at pH 8.3, and purified by cold ethanol precipitation.
Fluorescence microscopes
Confocal microscopy was performed on a Leica SP5 inverted confocal scanning microscope at the UW Biology Imaging Core (Fig. 2, Supplementary Figs. 2–8, 21) using a 63× 1.2 NA water lens (Leica, Nussloch, Germany), or an Olympus upright FV1000 (Fig. 3, Supplementary Figs. 18, 19) with a 25× 1.0 NA SCALE objective. Conventional widefield epifluorescence imaging was performed on an inverted Nikon Ti-S microscope configured with a 10× 0.25 NA air objective lens (Nikon, Melville, NY, USA), 20× 0.45 NA air objective lens (Nikon), or a 60× 1.2 NA water-immersion objective lens (Nikon). The widefield microscope was illuminated using a four-channel light emitting diode source (LED4D120, Thorlabs, Newton, NJ, USA) using a multiband filter set (LF405/488/532/635-A-000, Semrock, Rochester, NY, USA) and images were captured with a Zyla 5.5 sCMOS camera (Andor, Windsor, CT, USA) (Supplementary Figs. 1, 9–17, 20). Localization microscopy (Supplementary Fig. 5) was performed on a homebuilt Nikon Ti-U system configured for total internal reflection fluorescence using a Nikon CFI Plan Apo Lambda 100× 1.45 NA objective and a 647-nm diode-pumped solid-state laser source (MPB Communications, Pointe-Claire, QC, Canada). A 405-nm solid state laser (Obis, Coherent) was used for activation to increase the rate of fluorophore blinking. Localization images were acquired on an EMCCD (iXon Ultra 897, Andor) operating at 200 frames per second. A custom-built focus lock using an objective nanopositioner (Nano F-100S, Mad City Labs, Madison, WI, USA) and a 940-nm diode laser (LP-940, Thorlabs) was used to control axial drift.
Cell culture
BS-C-1 and Ptk1 cells were obtained from ATCC and both tested negative for mycoplasma using 4′,6-diamidino-2-phenylindole dihydrochloride. Cell lines obtained from ATCC were used without additional authentication. BS-C-1 cells were cultured in EMEM (ATCC, 30-2003, Manassas, VA, USA) containing penicillin and streptomycin (PS, 15140-122, Life Tech.), 10 % FBS (FB22-500, Serum Source International, Charlotte, NC, USA), and non-essential amino acids (NEAA, 11140-050, Life Tech.). PtK1 cells were cultured in RPMI (11875-093, Life Tech.) containing PS, 10 % FBS and NEAA. Cells were maintained at 37 °C environment with 5 % CO2.
Immunostaining of cultured cells
See also Supplementary Table 1 for a summary and detailed list of concentrations and reagents for the preparation of all imaged specimens.
Immunostaining of BS-C-1 cells was conducted as follows. Cells were seeded at a density of ~50,000 cells per well of a 24-well plate containing a 12 mm #1.5 coverglass and incubated overnight. Cells were optionally extracted for 30s with PEM (0.1 M PIPES pH 7, 1 mM EDTA, 1 mM MgCl2) containing 0.5 % Triton-X-100 immediately prior to fixation. The extraction step is important for high-quality stains of cytoskeletal structures, but was not used on stains of organelle structures where treatment with detergent would likely destroy the structure (see Supplementary Table 1). Specimens were fixed for 10 minutes in a solution containing 3.2 % paraformaldehyde and 0.1 % glutaraldehyde in PEM (for microtubules) or PBS (for organelles), followed by brief washing in PBS and reduction in an aqueous solution of 10 mM sodium borohydride for 5 minutes. After reduction, samples were washed three times with PBS and then incubated with blocking/permeabilization buffer (PBS with 3 % BSA and 0.5 % Triton X-100) for 30 minutes. Specimens were then incubated with primary antibodies in blocking/permeabilization buffer for 45 minutes, washed three times with PBS, and incubated for 45 minutes with secondary antibodies in blocking/permeabilization buffer. After three more washes with PBS, cells were treated with either GA or MA-NHS. GA-treatment consisted of a 10 minute, room-temperature incubation with 0.25% GA in PBS followed by washing three times with PBS. MA-NHS-treatment consisted of a 60 minute, room-temperature incubation with 25 mM MA-NHS in PBS followed by washing three times with PBS. For correlative pre-expansion localization microscopy and post-expansion widefield imaging of fixed BS-C-1 cells in Supplementary Fig. 5, a tertiary antibody immunostain was performed including steps for: primary rat anti-tubulin, secondary Alexa 647 mouse anti-rat, tertiary Atto 488 donkey anti-mouse antibody and finally GA treatment.
Immunostaining of PtK1 cells was conducted using a variation of the above protocol for BS-C-1 cells, but with the following differences. Cells were incubated with rat anti-tubulin and mouse anti-HEC1 primary antibodies overnight at 4 °C. After washing, cells were incubated at room temperature for 45 minutes with secondary antibodies consisting of donkey anti-rat secondary antibody labeled with Atto 488 and a donkey anti-mouse secondary antibody that was dually labeled Alexa 546 and biotin. After secondary labeling, samples were treated with GA as described above for BS-C-1 cells. Prior to post-ExM imaging, the expanded samples were incubated with 2 μg/mL Alexa 546 labeled streptavidin in PBS containing 3% BSA for one hour. After contracting during this incubation, the gel was allowed to re-expand to full size in DI water. Additionally, immediately prior to pre- and post-ExM imaging, cells were incubated with 1 μM TO-PRO-3 in water for 15 minutes.
Transfection of cultured cells
BS-C-1 cells were dissociated and concentrated to ~106 cells/ml by centrifugation at 90-G for 10 min and resuspended in Solution SF (Lonza, Basel, Switzerland). A 100 µL volume of cells was mixed with 5 µg of plasmid: pAcGFP1-Mito (Clontech, Mountain View, CA, USA) in Supplementary Figs. 9 e and 16, or pAc-GFPC1-Sec61β (a gift from Tom Rapoport, Addgene plasmid# 15108) in Supplementary Figs. 9 f and 15, or Sec61Bβ and pDsRed2-Mito (BD Biosciences, Franklin Lakes, NJ, USA) in Fig. 2 k, I. The cells were then electroporated in an electrode cuvette with pulse code X-001 in a Lonza Amaxa nucleofector, immediately resuspended in warm media, and plated in a 24-well plate as described above. After 24–48 hours, the cells were fixed with paraformaldehyde and glutaraldehyde (Supplementary Figs. 15, 16 a, or paraformaldehyde only in 16 b–c), or fixed and immunostained for outer mitochondrial membrane (Fig. 2 k, l) or with anti-GFP (Supplementary Fig. 9 e–f) as described above.
Mouse brain tissue dissection and preparation
All animal experiments were carried out in accordance with the Institutional Animal Care and Use Committee at the University of Washington. Mice (strain C57BL/6) were anesthetized with isoflurane and perfused transcardially with PBS, followed by paraformaldehyde (PFA, 4% w/v in PBS). Brains were dissected out, postfixed in 4% PFA in PBS at 4 °C for one hour and washed in PBS. Then, the brains were sliced to 100 µm thickness using a vibratome. All mice used in this work were between the ages of 1 and 4 months at the time of dissection. Both male and female mice were used.
Immunostaining of tissue slices
100 µm thick mouse brain slices were first incubated in blocking/permeabilization buffer (3% BSA and 0.1% Triton X-100 in PBS) for 6–12 h at 4 °C. The tissue was then incubated in primary antibody diluted into blocking/permeabilization buffer for at least 24 h at 4 °C and was then washed three times in blocking/permeabilization buffer (20 min each). Tissues were then incubated with secondary antibody diluted into blocking/permeabilization buffer for 24 h at 4 °C and afterwards were washed three times with PBS (20 min each). The brain slices were then either treated with 0.1% GA in PBS or 1 mM MA-NHS in PBS for 1 h at room temperature followed by three washes with PBS. Tissue slices that were not immunostained (samples with fluorescent protein signal preserved) were simply treated with GA or MA-NHS. See also Supplementary Table 1 for a summary and detailed list of concentrations and reagents for the preparation of all imaged specimens.
Gelation, digestion, and expansion of cultured cell specimens
Fixed cell samples on 12 mm round coverglass were incubated in monomer solution (1× PBS, 2 M NaCl, 2.5% (w/w) acrylamide, 0.15% (w/w) N,N’-methylenebisacrylamide, 8.625% (w/w) sodium acrylate) for ~1 minute at room temperature prior to gelation. Concentrated stocks of ammonium persulfate (APS) and tetramethylethylenediamine (TEMED) at 10% (w/w) in water were diluted in monomer solution to concentrations of 0.2% (w/w) for gelation, with the initiator (APS) added last. The gelation solution (~70 µl) was placed in a 1 mm deep, 1 cm diameter Teflon well and the coverglass was placed on top of the solution with cells face down. Gelation was allowed to proceed at room temperature for 30 min. The coverglass and gel were removed with tweezers and placed in digestion buffer (1× TAE buffer, 0.5% Triton X-100, 0.8 M guanidine HCl) containing 8 units/mL Proteinase K (EO0491, Thermo or P8107S, New England BioLabs, Ipswich, MA, USA) added freshly. Unless otherwise indicated, gels were digested at 37 °C for various amounts of time as follows: MA-treated cells were digested overnight, GA-treated cells were digested for 30 min to 1 h, and fluorescent protein samples were digested for 30 min maximum. The gels (sometimes still attached to the coverglass) were removed from digestion buffer and placed in ~50 mL DI water to expand. Water was exchanged every 30 min until expansion was complete (typically 3–4 exchanges).
Post expansion labeling of expanded cultured cell specimens with streptavidin
Expanded cultured cell specimens initially immunostained with biotin-modified antibodies were submerged in a streptavidin solution (2 µg/mL) in PBS containing 3% BSA for 45 min. The contracted gels were then washed and re-expanded in DI water.
Gelation, digestion, and expansion of mouse tissue specimens
Tissue samples were incubated in monomer solution at 4 °C for 45 min prior to gelation. Tissue was gelled with the same solution as cells but with the addition of 4-hydroxy-2,2,6,6-tetramethylpiperidin-1-oxyl (4-hydroxy-TEMPO) at a concentration of 0.01% (w/w) from a 1% (w/w) stock as an inhibitor to allow complete diffusion of the monomers throughout the tissue. The glass slide with the sample and a #1.5 coverglass on top separated by spacers (one #1 coverglass) on either side of the tissue was used as a gelation chamber. The samples were allowed to gel for 2–2.5 h at 37 °C. Excess gel around the samples was removed, the glass around the samples was cut to leave the tissue on a small glass square, and the samples were placed in digestion buffer with 8 units/mL and were allowed to digest at 37 °C for various amounts of time: stained samples were digested overnight and fluorescent protein samples were digested for 1 h. The gels were removed from the digestion solution (using the glass square to support the gel) and placed in DI water to expand. Gradually increasing the amount of water helped prevent the gels from folding.
Expanded specimen handling
Expanded gels were cut to fit on coverglass (2–4 cm edge-length rectangles) excess water was removed and then gently placed on coverglass substrates for imaging. When possible, gels were immobilized using a small amount of cyanoacrylate glue on the periphery after wicking away excess water from the edges.
Correlative Localization Microscopy and ExM
Pre-expansion localization microscopy images of Alexa 647 labeled microtubules were acquired at 200 Hz for ~80,000 frames at ~2 kW/cm2 in an oxygen scavenging switching buffer (100 mM Tris pH 8, 10 % glucose (w/w), 0.5 mg/mL glucose oxidase, 40 μg/mL catalase, and 143 mM 2-mercaptoethanol). After localization microscopy, samples were washed to remove the switching buffer, gelled, digested, and expanded as described above. During gelation, the Alexa 647 signal was destroyed, however the Atto 488 from the tertiary antibody remained fluorescent for widefield epifluorescence imaging.
Image Processing
Expanded cell culture confocal z-stacks were aligned frame by frame using an automated rigid registration routine in Mathematica in order to correct for minor lateral drift during acquisition. Mitotic spindle confocal z-stacks of PtK1 cells were processed to remove peripheral non-specific adsorption of the HEC1 antibody as follows: A binary 3D mask of the kinetochore attachments was generated by binarizing the kinetochore channel and retaining connected-component features larger than 100 voxels and within 1 μm of the outer surface of the chromosomes. The kinetochore binary mask was then dilated by three pixels and multiplied by the original channel data. The processing was performed to clarify the maximum intensity projections in Fig. 2 f, i, j, but had little effect on the individual z-sections as shown in detail in Supplementary Fig. 7. Localization microscopy images were analyzed as described previously.5 Registration of pre- and post-expansion correlative images were carried out in the open-source software Elastix, using rigid (similarity) and non-rigid (B-spline) transformations to determine the expansion factor and quantify distortions. Details, including example data and processing scripts, are included in the Supplemental Protocol.
Reproducibility
All experiments were carried out ≥3 times including all sample preparation and analysis, except as noted below. Representative data for each experiment are shown. Experiments for Supplementary Figs. 5, 10 c, 11, 12, and 16 b were performed only once.
Supplementary Material
Acknowledgments
This work is supported by the University of Washington (J.C.V.), a Burroughs-Wellcome Career Award at the Scientific Interface (J.C.V.), an NSF Graduate Research Fellowship DGE-1256082 (T.J.C.), and by NIH grants EY10699 and EY17101 (R.O.L.W.). The authors would like to thank Prof. Linda Wordeman for providing the PtK1 cell line and anti-HEC1 antibody, for access to an electroporator, and for helpful discussions. The authors would also like to thank Kaori Oda for performing the cardiac perfusion of mice.
Footnotes
AUTHOR CONTRIBUTIONS
T.J.C., A.R.H., H.O., R.O.L.W., and J.C.V. designed the experiments. T.J.C., A.R.H., H.O., H.-J.K., and G.J.T. performed the experiments and analysis. T.J.C., A.R.H., and J.C.V. wrote the paper and all authors commented on the manuscript. J.C.V. supervised the project.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Chen F, Tillberg PW, Boyden ES. Expansion microscopy. Science. 2015;347:543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 4.Hern DL, Hubbell JA. Incorporation of adhesion peptides into nonadhesive hydrogels useful for tissue resurfacing. J. Biomed. Mater. Res. 1998;39:266–276. doi: 10.1002/(sici)1097-4636(199802)39:2<266::aid-jbm14>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 5.Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olivier N, Keller D, Gönczy P, Manley S. Resolution Doubling in 3D-STORM Imaging through Improved Buffers. PLoS ONE. 2013;8:e69004. doi: 10.1371/journal.pone.0069004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weston PD, Avrameas S. Proteins coupled to polyacrylamide beads using glutaraldehyde. Biochem. Biophys. Res. Commun. 1971;45:1574–1580. doi: 10.1016/0006-291x(71)90200-2. [DOI] [PubMed] [Google Scholar]
- 8.Lee K, Choi S, Yang C, Wu H-C, Yu J. Autofluorescence generation and elimination: a lesson from glutaraldehyde. Chem. Commun. 2013;49:3028. doi: 10.1039/c3cc40799c. [DOI] [PubMed] [Google Scholar]
- 9.Chung K, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang B, et al. Single-Cell Phenotyping within Transparent Intact Tissue through Whole-Body Clearing. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Migneault I, Dartiguenave C, Bertrand MJ, Waldron KC. Glutaraldehyde: behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. Biotechniques. 2004;37:790–806. doi: 10.2144/04375RV01. [DOI] [PubMed] [Google Scholar]
- 12.Kim D, Park K. Swelling and mechanical properties of superporous hydrogels of poly(acrylamide-co-acrylic acid)/polyethylenimine interpenetrating polymer networks. Polymer. 2004;45:189–196. doi: 10.1163/156856204322793575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.