Summary
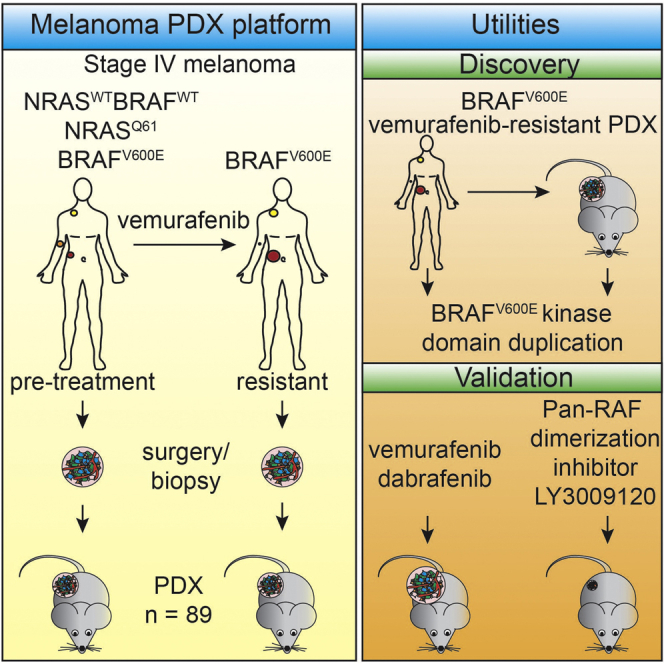
The therapeutic landscape of melanoma is improving rapidly. Targeted inhibitors show promising results, but drug resistance often limits durable clinical responses. There is a need for in vivo systems that allow for mechanistic drug resistance studies and (combinatorial) treatment optimization. Therefore, we established a large collection of patient-derived xenografts (PDXs), derived from BRAFV600E, NRASQ61, or BRAFWT/NRASWT melanoma metastases prior to treatment with BRAF inhibitor and after resistance had occurred. Taking advantage of PDXs as a limitless source, we screened tumor lysates for resistance mechanisms. We identified a BRAFV600E protein harboring a kinase domain duplication (BRAFV600E/DK) in ∼10% of the cases, both in PDXs and in an independent patient cohort. While BRAFV600E/DK depletion restored sensitivity to BRAF inhibition, a pan-RAF dimerization inhibitor effectively eliminated BRAFV600E/DK-expressing cells. These results illustrate the utility of this PDX platform and warrant clinical validation of BRAF dimerization inhibitors for this group of melanoma patients.
Graphical Abstract
Highlights
-
•
Patient-derived xenograft (PDX) platform comprises 89 metastatic melanoma tumors
-
•
Platform includes several pre-vemurafenib and vemurafenib-resistant PDXs
-
•
Duplication of the BRAFV600E kinase domain is identified as a resistance mechanism
-
•
Pan-RAF dimerization inhibitor LY3009120 eliminates melanoma cells with this duplication
Kemper et al. have built a platform composed of 89 metastatic melanoma xenografts. Using this collection as a resource, they identified a BRAFV600E protein harboring a duplicated kinase domain. A pan-RAF dimerization inhibitor suppresses expansion of PDXs expressing this BRAF mutant.
Introduction
Until 5 years ago, treatment options for metastatic melanoma were limited to chemotherapy, which did not significantly improve patient survival. However, the genetic characterization of melanoma (Davies et al., 2002) has prompted the development of therapies targeting specifically the oncogenic drivers of the disease. Approximately half of the patients diagnosed with metastatic melanoma harbor an activating mutation in BRAF, most commonly T1799A. This encodes the BRAFV600E protein, which renders these patients eligible for treatment with selective BRAF inhibitors. In clinical trials, the first of these inhibitors, vemurafenib, elicited partial or complete tumor regression in the majority of patients (Chapman et al., 2011, Flaherty et al., 2010).
Despite these promising results, while some patients show remarkable durable responses to BRAF inhibition (BRAFi), the majority show an initial response to vemurafenib but eventually develop resistance (Solit and Rosen, 2014). Accounting for this are a plethora of resistance mechanisms, including reactivation of the MAPK pathway, the PI3K/AKT pathway, or both (Nazarian et al., 2010, Paraiso et al., 2011, Poulikakos et al., 2011, Shi et al., 2012, Shi et al., 2014a, Das Thakur et al., 2013, Wagenaar et al., 2014, Wagle et al., 2011). As the majority of resistance mechanisms cause reactivation of the MAPK pathway (Van Allen et al., 2014), a logical next step was to determine the clinical benefit of combinatorial treatment of a MEK inhibitor (e.g., trametinib or cobimetinib) with a BRAF inhibitor (vemurafenib or dabrafenib). The response to such combined treatment was significantly more durable than what was seen for single BRAFi (Larkin et al., 2014, Robert et al., 2015). Still also in the combination treatment setting, resistance again limited overall survival benefit (Hugo et al., 2015, Wagle et al., 2014). Because of these major challenges, there is a dire need to develop more effective (combinatorial) treatment regimens.
Resistance to targeted drugs is mostly studied in in vitro cell models (Basile et al., 2012, Nazarian et al., 2010, Poulikakos et al., 2011, Vergani et al., 2011), but the use of long-term cultured cancer cell lines has several limitations. First, they do not reliably predict the clinical effect of therapeutics (Burchill, 2006, Voskoglou-Nomikos et al., 2003). Second, the monolayer character of cell culture does not recapitulate the 3D interactions between stromal and cancer cells, which influence not only the dynamics of tumor progression but also therapy response (Straussman et al., 2012). Moreover, the establishment of 2D cell lines from human cancers can induce irreversible changes, including genetic aberrations (De Witt Hamer et al., 2008), alterations in gene expression (Daniel et al., 2009), or dependencies on certain signaling pathways (Clement et al., 2007). Importantly, these properties are not restored upon xenografting of cell lines (Daniel et al., 2009).
A clinically useful model would therefore be to implant tumor fragments derived from patients immediately into mice, thereby cancelling the opportunity for tumors to acquire alterations resulting from in vitro culturing. Such patient-derived xenografts (PDXs) already have been established for several tumor types, including colon cancer (Bertotti et al., 2011), pancreatic cancer (Rubio-Viqueira et al., 2006), breast cancer (DeRose et al., 2011), and melanoma (Einarsdottir et al., 2014, Girotti et al., 2016, Monsma et al., 2015, Das Thakur et al., 2013). Some of these PDX models proved successful in large-scale assessment of the effect of several (combinatorial) therapies and to identify stratification markers, discriminating subgroups of tumors divergently responding to targeted therapy (Bertotti et al., 2011, Gao et al., 2015).
Therefore, we have established a model for metastatic melanoma in which the complex interactions between tumors and at least some components of the tumor microenvironment are maintained. Here we present a collection comprising 89 PDXs established from human metastatic melanomas, which were acquired either before the start of therapy or after the emergence of resistance. This comprehensive PDX platform was analyzed for biomarkers, chromosomal aberrations, RNA expression profiles, mutational spectrum, genetic heterogeneity, and targeted drug resistance patterns. In addition, we tested the utility of this platform as a limitless source of patients’ tumor material by screening for resistance mechanisms. Collectively, our results demonstrate that this platform is highly suitable for studying resistance, discovering additional drug targets for companion treatment, and for preclinical studies of human melanoma in general.
Results
Establishment of a PDX Platform for Metastatic Melanoma
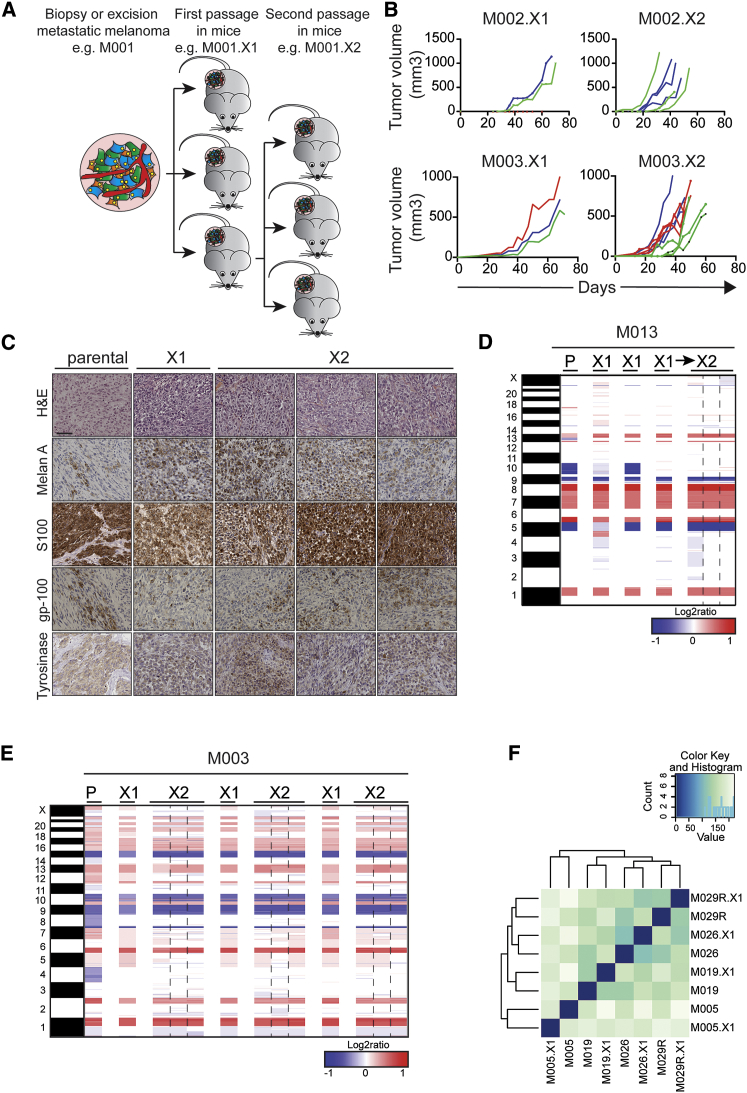
Tumor specimens were obtained from patients diagnosed with BRAFV600E, NRASQ61, or BRAFWTNRASWT metastatic melanoma during surgery or by fine-needle biopsy of mainly subcutaneous lesions. Tumor fragments were immediately transplanted subcutaneously into immunodeficient NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice with a success rate of 86%. This yielded a total of 89 PDXs, comprising 73 BRAFV600E, 10 NRASQ61, and 6 BRAFWTNRASWT xenografts (Table S1). Our attempt to generate cell lines from these PDX samples was successful in 30% of the cases (Table S1). After a first passage in mice (.X1), tumor fragments or digests could be propagated in vivo (.X2, Figure 1A) or cryopreserved for later use. Passaging in mice led to a gradual increase in the tumor outgrowth kinetics (Figure 1B), probably due to loss of human stromal components, consistent with PDXs derived from other tumor types (DeRose et al., 2011, Monsma et al., 2012).
Figure 1.
Stable Melanoma Marker Expression, Chromosomal Aberrations, and Gene Expression upon In Vivo Passaging of PDXs
(A) Tumor fragments derived from biopsies or surgical excisions were transplanted subcutaneously into NSG mice. After first passage (.X1), PDX fragments were passaged into a next set of mice (.X2).
(B) Speed of tumor outgrowth during passaging, shown for two different PDXs. Colors represent single PDX (.X1) passaged into a next set of three mice (.X2).
(C) H&E stainings and IHC stainings for MelanA, S100, gp-100, and tyrosinase were performed on the parental tumor and two subsequent passages of PDXs (X1 and X2). Scale bars indicate 100 μm.
(D) Copy-number profiles, based on array CGH, show the parental tumor (P) M013 and two PDX passages (the X2 passage was established from the third X1 PDX).
(E) Copy-number profiles, based on array CGH, show the parental tumor (P) M003 and two PDX passages (the X2 passages are next to their own X1-passaged PDX). Blue, deletion; red, amplification.
(F) Hierarchical clustering of RNA-sequencing data performed on patient samples and PDXs derived of these samples, after filtering as described in the Supplemental Experimental Procedures, is shown.
Stable Melanoma Marker Expression, Chromosomal Aberrations, and Gene Expression upon In Vivo Passaging of PDXs
We next determined the effect of passaging PDXs in vivo on melanoma marker expression, chromosomal aberrations, and RNA profiles. First, immunohistochemistry (IHC) for melanoma markers Melan-A, S100, gp-100, and tyrosinase, commonly used for the clinical diagnosis of melanoma, showed that melanoma marker expression remained stable when the patient’s tumor was compared to two consecutive passages in vivo as PDXs (Figure 1C; Figure S1).
Second, chromosomal aberrations, analyzed by array comparative genomic hybridization (array CGH), revealed that the genetic heterogeneity was captured in the PDXs (Figures 1D and 1E): passaging of M013 in mice revealed that at least some of the genetic aberrations found in the parental tumor were heterogeneous. For instance, the loss of chromosome 10, which was observed in the patient’s tumor, could be detected in one of three first-passage PDXs only, suggesting that chromosome 10 had not been lost in all parental tumor cells (Figure 1D). This is consistent with recent studies by us and others showing that melanomas are highly heterogeneous (Kemper et al., 2015, Shi et al., 2014b, Van Allen et al., 2014). Also passaging of M003 in mice revealed a variable pattern, which can be explained either by heterogeneity in the original tumor or by loss of genomic variation upon in vivo passaging. Importantly, after the first passage, the chromosomal content was stable and highly similar to the parental chromosomal content (Figure 1E).
Third, we analyzed gene expression patterns, comparing patient samples to their corresponding PDXs. We obtained samples for PDX establishment from four different patients, with tumor samples taken either before the start of vemurafenib treatment or after a patient had acquired therapy resistance (indicated by R). RNA was isolated from both patients’ tumors and PDXs and gene expression profiling was done by RNA sequencing. Hierarchical clustering of all samples was performed, resulting in a clustering that revealed a high concordance in gene expression between the patient samples and their corresponding PDXs (Figure 1F).
Together, these data show that, during the passaging of human melanomas in mice, the phenotypic and genetic characteristics are well preserved, yielding a collection of PDXs closely resembling the original patients’ tumors.
Clinical History of Patients from Matched Pre- and Post-vemurafenib PDX Pairs
We acquired a set of six matched PDX pairs, representing tumor material from patients both before the start of treatment with vemurafenib and after resistance had occurred (indicated by R). We illustrated the treatment schedule, the location of the lesion from where material was obtained for xenografting, and the specific response of these lesions to vemurafenib in Figure S2. In four patients, the resistant lesions, from which post-treatment PDXs were derived, either initially responded to vemurafenib before acquiring resistance (M009R, M026R, and M048R1; Figure S2) or emerged as new lesions during treatment (M029R and M048R2; Figure S2). Therefore, this group of PDXs was labeled acquired resistant. One matched pair (M005) was derived from a patient who was still responding to vemurafenib when the post-treatment sample was obtained, and was therefore categorized as on treatment. The sixth PDX pair was acquired from a patient (M019) who progressed immediately on vemurafenib treatment, qualifying these melanomas as intrinsic resistant (Figure S2).
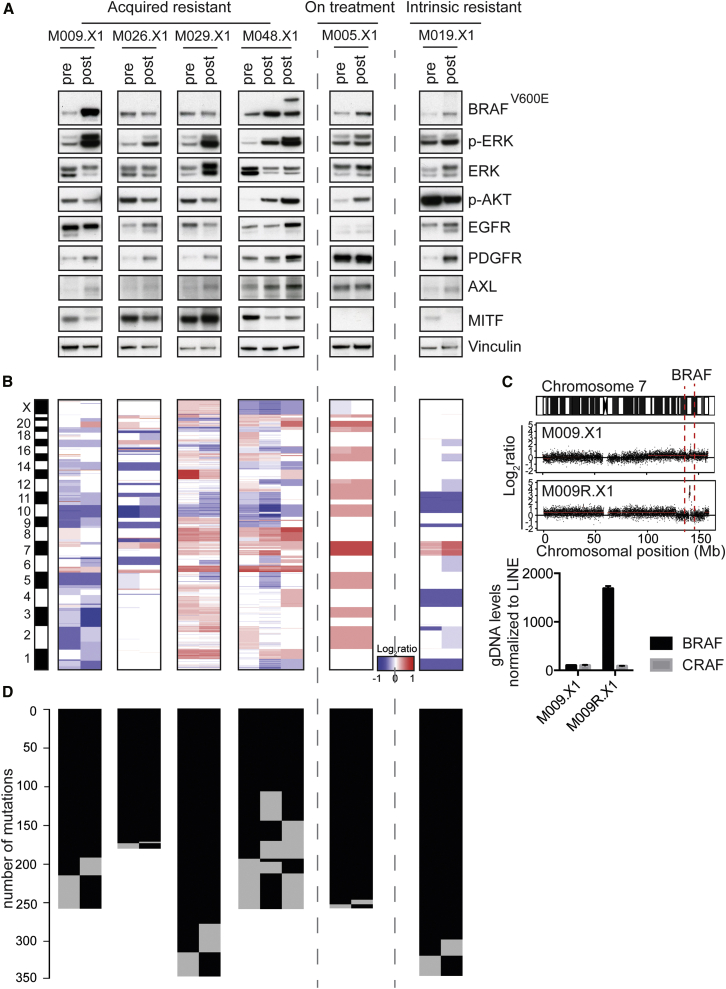
Vemurafenib-Resistant PDXs Commonly Display MAPK Pathway Reactivation
Several groups, including ours, have demonstrated that the MAPK pathway and, to a lesser extent, the PI3K pathway are commonly reactivated in vemurafenib-resistant melanomas (Kemper et al., 2015, Shi et al., 2014b, Van Allen et al., 2014). To determine whether this was recapitulated in our matched PDX series, we performed immunoblotting for phosphorylated ERK (p-ERK) and AKT (p-AKT). Indeed, all four acquired resistant PDXs from the matched pair set had reactivated p-ERK (Figure 2A), which was confirmed by IHC (Figure S3A). Only one PDX pair, namely M048, also showed reactivated p-AKT (Figure 2A). As expected, the PDX of the intrinsically resistant patient M019 already showed elevated p-ERK and p-AKT levels in the pre-treatment setting, while p-ERK was relatively low in the PDX from the on-treatment patient (Figure 2A; Figure S3A).
Figure 2.
Vemurafenib-Resistant PDXs Commonly Display MAPK Pathway Reactivation and Have Distinct Resistance Mechanisms
(A) Immunoblotting of all six matched PDX pairs for factors in the MAPK pathway in pre-vemurafenib and post-relapse PDXs. Asterisks indicate PDXs that were xenografted in mice that received PLX4720 chow.
(B) Copy-number profiles for matched PDX pairs, determined by CopywriteR, are shown.
(C) Amplification of the genomic region containing BRAFT1799A was identified in M009R.X1 (top panel). Validation of this amplification was performed by qPCR on gDNA (bottom panel). CRAF was included as a negative control. CT values were normalized to LINE.
(D) Mutation matrix for matched PDX pairs, comparing pre-vemurafenib and post-relapse tumors. Number of mutations is indicated; each black line represents one mutation.
Previously, we and others have shown that acquired resistance to BRAFi is effectively brought about by the loss of expression of microphthalmia-associated transcription factor (MITF) (Konieczkowski et al., 2014, Müller et al., 2014). Consistent with this, we observed decreased expression of MITF in three acquired resistant post-vemurafenib PDXs (M009R.X1, M026R.X1, and M048R.X1; Figure 2A). Reduced expression of MITF coincides with upregulation of one or multiple receptor tyrosine kinases (RTKs), such as EGFR, PDGFR, and, most commonly, AXL (Konieczkowski et al., 2014, Müller et al., 2014), which was well recapitulated in these matched acquired resistant PDXs (Figure 2A). Only M029R.X1 displayed increased expression of MITF expression upon acquired resistance (Figure 2A), which also previously has been reported as a resistance mechanism to MAPK pathway inhibition (Garraway et al., 2005, Müller et al., 2014).
In conclusion, our series of matched PDXs captures the resistance biomarkers that are commonly seen in drug-resistant melanomas in patients.
Identification of Resistance Mechanisms in Matched PDX Pairs
To uncover the cause of resistance in the matched PDX pairs, we performed whole-exome sequencing (WES) on genomic DNA (gDNA) derived from all matched PDXs. The level of tumor infiltration by mouse stroma was determined by a mouse pathologist via analysis of formalin-fixed paraffin-embedded (FFPE) H&E stainings of these PDXs, revealing that most PDXs contained >80% of tumor cells (Table S2). XenofilteR (R.J.C.K. and O.K., unpublished data) was used to remove all sequence reads that originated from mouse DNA. Copy-number profiles were generated from the filtered WES data by CopywriteR (Kuilman et al., 2015) (Figure 2B). This analysis revealed that pre- and post-vemurafenib PDXs had highly similar DNA copy-number profiles, although some variation was observed. This could result from inter-tumor heterogeneity, as most pre- and post-vemurafenib PDXs were not derived from the same patient lesion (Figure S2B). When analyzing copy-number aberrations (CNAs) in more detail, we detected a BRAFT1799A amplification, an established resistance mechanism (Shi et al., 2014b, Das Thakur et al., 2013, Van Allen et al., 2014), in the resistant M009R.X1, but not in the pre-treatment PDX (Figure 2C, top panel). This amplification was validated by qPCR on gDNA (Figure 2C, bottom panel). None of the other resistant PDXs displayed an amplification of BRAFT1799A (Figures S3B and S3C).
Next we analyzed the presence of mutations in the matched PDX pairs (Figure 2D). The BRAFi resistance-inducing mutation NRASQ61K (Nazarian et al., 2010) was detected in two of the post-vemurafenib PDXs (M026R.X1 and M029R.X1) and was confirmed by Sanger sequencing (Figures S4A and S4B). In M048R2.X1, a mutation in AKT3 (L51R) (Catalogue of Somatic Mutations in Cancer [COSMIC]: COSM309035) was detected and confirmed by Sanger sequencing (Figure S4C). This mutation has not been validated yet as a cause of resistance, but two other previously described resistance-conferring mutations, namely AKT3E17K and AKT1Q79K (Shi et al., 2014a, Shi et al., 2014b), are located within the same pleckstrin homology (PH) domain as AKT3L51R. These mutations induce (re)localization of AKT to the membrane, causing constitutive activation of the PI3K/AKT pathway (Parikh et al., 2012). The M048R2.X1 PDX, harboring this AKT3L51R mutation, indeed displays highly activated AKT (Figure 2A), suggesting that this mutation activates the kinase activity.
In summary, we have identified BRAFT1799A amplification and NRASQ61K and AKT3L51R mutations as the likely causes for vemurafenib resistance in our matched acquired resistant PDX pairs, capturing the mutational spectrum seen in resistant human melanomas.
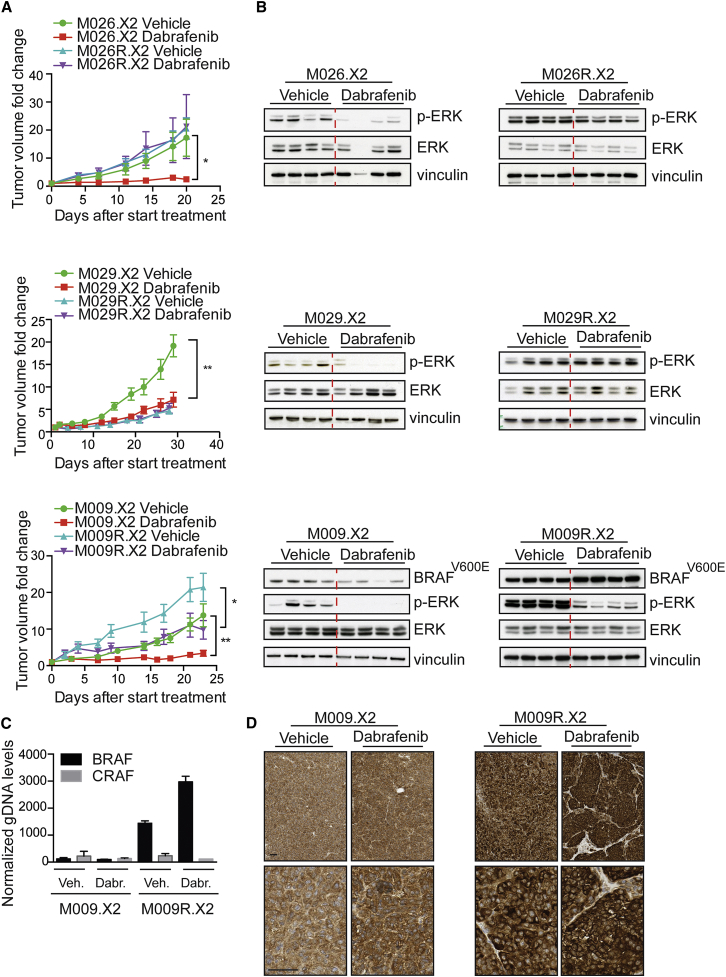
Validation of Resistance to BRAFi In Vivo in Matched PDX Pairs
The next step was to confirm that PDXs derived from vemurafenib-naive BRAFV600E lesions were responsive to BRAFi in vivo, in contrast to PDXs from vemurafenib-resistant melanomas. We first analyzed the response to the BRAFi dabrafenib of the matched M026 PDX pair. The treatment-naive M026.X2 melanoma was highly sensitive to BRAFi, resulting in reduced growth and a decrease in p-ERK abundance upon BRAFi (Figures 3A and 3B; Figure S4D). In contrast, M026R.X2, a PDX derived from a vemurafenib-resistant lesion from the same patient in which we identified a NRASQ61K mutation as the resistance mechanism (Figures S4A and S4B), was completely resistant to BRAFi (Figure 3A). Consistently, p-ERK levels of M026R.X2 were unaffected by BRAFi (Figure 3B). A similar pattern was observed for the matched M029 PDX pair: treatment-naive melanoma M029.X2 responded well to BRAFi along with p-ERK inhibition, whereas tumor outgrowth of NRASQ61K mutant M029R.X2 and its p-ERK levels were unaffected by BRAFi (Figures 3A and 3B; Figure S4D). Of note, the growth rate of the (untreated) M029R.X2 was much slower than that of its treatment-naive counterpart M029.X2.
Figure 3.
Validation of Resistance to BRAFi In Vivo in Matched PDX Pairs
(A) Tumor dynamics of matched PDX pairs upon treatment with 30 mg/kg dabrafenib (n = 8 tumors/group). Graphs represent fold change in tumor volume relative to the tumor volume at treatment initiation. Unpaired t test was performed at the last time point (∗p < 0.05 and ∗∗p < 0.01). Error bars indicate SD.
(B) Immunoblotting for p-ERK on matched PDX pairs, treated with and without dabrafenib (each lane represents a tumor derived from an individual mouse), is shown.
(C) The BRAFT1799A amplification was validated by qPCR on gDNA. CRAF was included as a negative control. CT values were normalized to LINE. Error bars indicate SD.
(D) Stainings for BRAFV600E on M009.X2 and M009R.X2, treated with and without BRAF inhibitor. Scale bars indicate 100 μm.
This behavior was different for matched pair M009.X2/R.X2, in which the presence of the BRAFT1799A amplification correlated with therapy resistance. For this PDX set, expansion of the treatment-naive M009.X2 melanoma was reduced upon BRAFi and p-ERK levels decreased, as expected (Figures 3A and 3B; Figure S4D). However, BRAFi also slowed down the growth and decreased p-ERK levels of M009R.X2, which was derived from a vemurafenib-resistant lesion (Figures 3A and 3B; Figure S4D). This coincided with two interesting observations. First, M009R.X2 grew much faster than M009.X2. Therefore, in spite of the notable effect of BRAFi, M009R.X2 continued to grow exponentially. Second, expression of BRAFV600E, although already highly expressed in M009R.X2 when compared to M009.X2, was even further increased upon BRAFi (Figures 3C and 3D). The cause for this is unknown but could reflect either a dosage change in the BRAFT1799A gene or selection for a subpopulation with super-amplification of BRAFT1799A. Additionally, this would suggest that the BRAF amplification and the resulting resistance can be dynamic, as has been suggested by others (Das Thakur et al., 2013).
These results demonstrate concordance between drug responses in patients and their corresponding PDXs, and they illustrate that the therapy response can be either stable or dynamic.
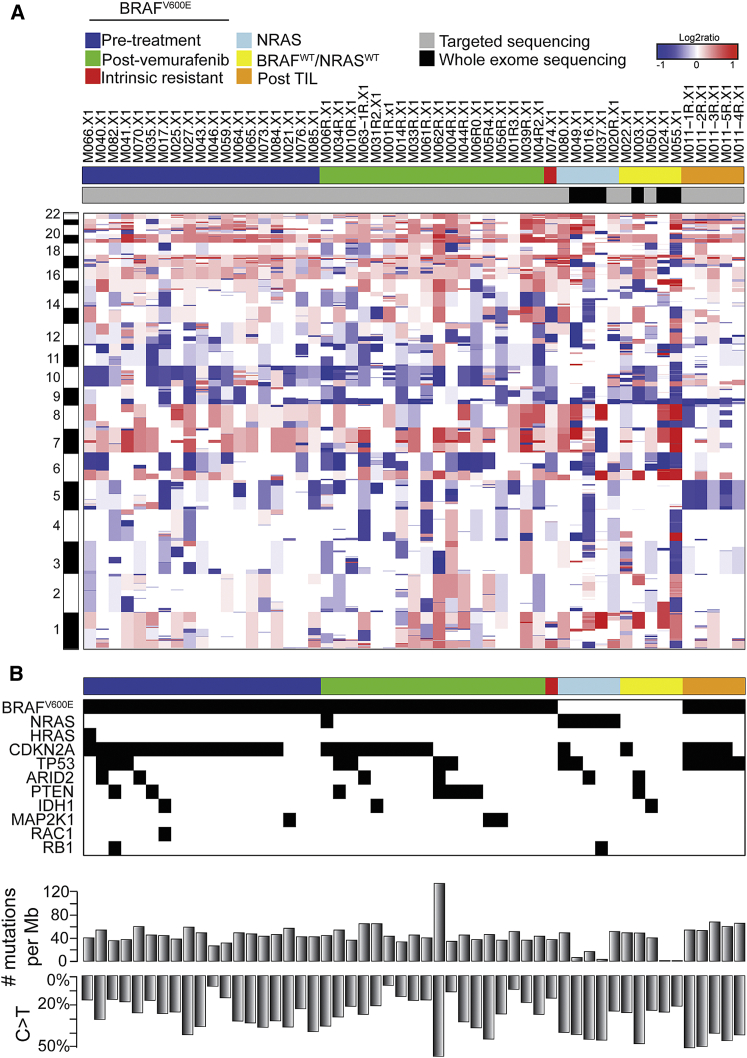
Mutational Characterization of the PDX Panel
In addition to six matched PDX pairs, the collection comprises an additional 76 melanoma PDXs. Targeted sequencing using a 360-cancer gene panel was performed for more than half of these. The 47 PDXs comprise the following: (1) BRAFV600E PDXs derived from vemurafenib-naive or -resistant melanomas, (2) NRASQ61 PDXs, (3) BRAFWT/NRASWT PDXs, and (4) a set of five PDXs derived from different lesions of one patient who received tumor-infiltrating lymphocyte (TIL) therapy. Additionally, for six PDXs, comprising three NRASQ61 and three BRAFWTNRASWT PDXs, WES was performed.
Using CopywriteR, we generated copy-number profiles from the sequencing data (Figure 4A). These profiles revealed a CNA pattern typical of melanoma, including gain of chromosome 7, where BRAF is located, and loss of chromosome 10, which harbors the tumor suppressor gene PTEN, both established drivers of melanomagenesis. Next we analyzed which mutations are present in our PDX panel, focusing on mutations in the 15 driver genes on which the molecular classification of melanomas has been described previously (Cancer Genome Atlas Network, 2015) (Figure 4B). For 11 of 15 driver genes, mutations were observed in the 53 PDX samples. The complete list of mutations identified by targeted sequencing is provided in Table S3.
Figure 4.
Mutational Characterization of the PDX Panel
(A) Targeted sequencing for 360 cancer genes was performed on 47 PDXs, and WES was performed on an additional six PDXs. Copy number profiles were derived from these data. PDXs were grouped in pre-BRAF inhibitor treatment (pre-treatment), BRAF inhibitor-resistant (post-vemurafenib), intrinsic resistant, NRASQ61 mutant (NRAS), and BRAFWTNRASWT PDXs and PDXs derived from five different lesions for a single patient after tumor-infiltrating lymphocytes therapy (post-TIL).
(B) Mutation matrix for 11 known melanoma driver genes. Total number of mutations per 1 Mb and the percentage of mutations with a C > T are indicated below the mutation matrix.
PDXs Derived from Vemurafenib-Resistant Melanomas Harbor a Plethora of Established Clinical Resistance Mechanisms
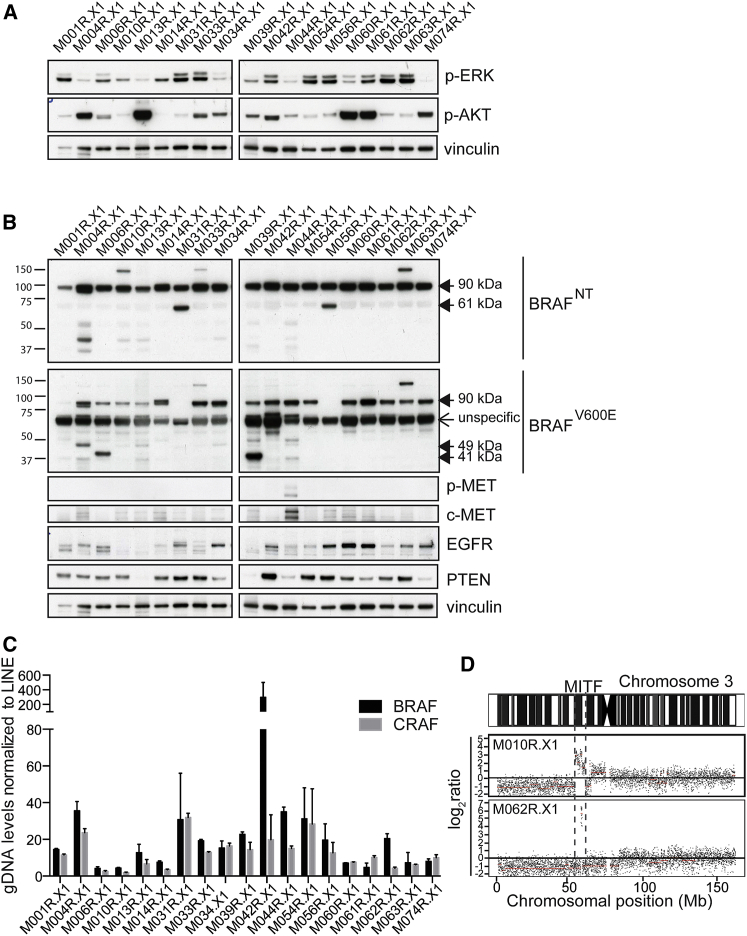
In the panel of 53 sequenced PDXs (Table S4), 19 were derived from BRAFV600E metastatic melanomas that had acquired resistance to vemurafenib. We analyzed this PDX set for the presence of known resistance mechanisms. As discussed above, resistance to vemurafenib is commonly associated with reactivation of the MAPK pathway and/or the PI3K/AKT pathway. Concordantly, when we determined the corresponding biomarker levels in these PDXs, we found that, in most, either p-ERK or p-AKT was highly induced (Figure 5A; Figure S5A).
Figure 5.
PDXs Derived from Vemurafenib-Resistant Melanomas Harbor a Plethora of Established Clinical Resistance Mechanisms
(A) Immunoblotting for p-ERK and p-AKT to detect reactivation of the MAPK pathway and/or the PI3K/AKT pathway in the post-vemurafenib PDXs. Vinculin was used as a loading control.
(B) Immunoblotting to detect previously described resistance mechanisms in post-vemurafenib PDXs. Vinculin was used as a loading control.
(C) Analysis of BRAFT1799A amplification by qPCR on gDNA of all post-vemurafenib PDXs is shown.
(D) DNA copy-number profiles revealed amplification of the region containing MITF in two independent PDXs.
Next we analyzed BRAF expression by immunoblotting to detect alternative splice variants, which also are known to render melanomas refractory to BRAFi. These variants splice out the RAS-binding domain (RBD) of BRAF (encoded by exons 3–5), inducing dimerization of BRAF and downstream signaling without the need to be activated by RAS (Poulikakos et al., 2011). Several splice variants have been described to induce resistance, i.e., 61-kDa (exons 4–10), 48-kDa (exons 2–8), and 41-kDa (exons 2–10) variants (Poulikakos et al., 2011). Using two different antibodies, recognizing either the N-terminal (BRAFNT, epitope encoded by exons 2–3) or the V600E region (BRAFV600E, epitope in exon 15), we detected these different splice variants in several PDXs (Figure 5B).
We also identified other previously detected and validated resistance mechanisms in the post-vemurafenib PDX panel, including hyperactivation of c-MET (Vergani et al., 2011), EGFR overexpression (Sun et al., 2014), loss of PTEN (Paraiso et al., 2011) (Figure 5B), amplification of BRAFT1799A (Shi et al., 2014b) (Figure 5C), and amplification of MITF (Garraway et al., 2005, Van Allen et al., 2014) (Figure 5D). Additionally, using the targeted sequencing data, we found known BRAF inhibitor resistance-conferring mutations, including MAP2K1E203K (Nikolaev et al., 2011), BRAFL505H (Wagenaar et al., 2014), NRASQ61K (Nazarian et al., 2010), and PIK3CAE545K (Shi et al., 2014b) (Table S5).
Through a combination of mutational data with biochemical analyses, the cause of resistance was resolved for 13 of the 19 PDXs derived from patients with acquired resistance to vemurafenib. This also revealed that some PDXs harbor multiple resistance mechanisms (Table S5). For example, M056R.X1 harbored a MAP2K1E203K mutation, alternative BRAF splicing, as well as EGFR overexpression, indicating that resistance mechanisms can be heterogeneous (Kemper et al., 2015, Shi et al., 2014b). Our data show that this heterogeneity is captured and maintained in PDXs.
To summarize, this set of post-vemurafenib PDXs harbors a wide range of resistance mechanisms, closely recapitulating clinical samples. Some PDXs harbored multiple resistance mechanisms, a phenomenon commonly seen in melanoma, indicating that the heterogeneity of resistance can be maintained in these PDX models.
PDXs Derived from Vemurafenib-Resistant Patients Express a BRAFV600E Mutant Harboring a Kinase Domain Duplication
Despite the advances made in targeted therapies for BRAFV600E melanoma, drug resistance continues to be a major obstacle for achieving durable clinical responses. To illustrate the utility of this PDX platform, we set out to screen for resistance mechanisms. We focused on those that can be discovered at the protein level, in other words, using analyses requiring substantial amounts of melanoma tumor material. Thus, we took advantage of our PDX collection serving as an unlimited source for this purpose.
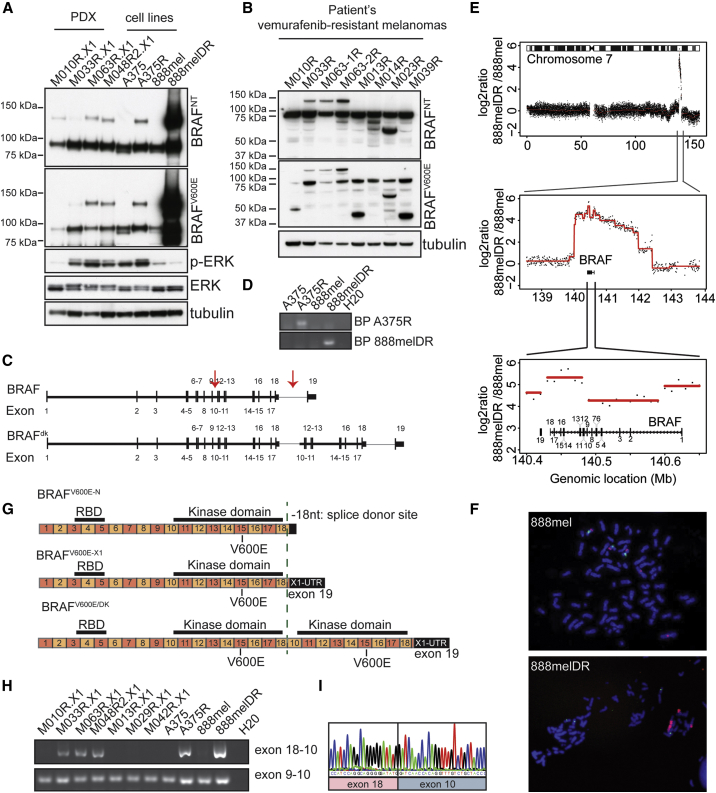
In addition to the previously identified resistance mechanisms described above, we noted that several PDXs derived from vemurafenib-resistant melanomas expressed an unusual BRAFV600E protein, which migrated in an SDS-PAGE gel at an apparent molecular weight of ∼140 kDa. It was recognized by both the BRAFV600E- and the BRAFNT-specific antibodies (Figure 2A, M048R2.X1; Figure 5B, M010R.X1, M033R.X1, and M063R.X1). Strikingly, none of the pre-treatment PDXs showed expression of this abnormal BRAFV600E protein (Figure S5B). Furthermore, we discovered similar BRAF proteins in two cell lines that had acquired resistance to MAPK pathway inhibition in vitro (PLX4720-resistant A375R and dabrafenib/trametinib double-resistant [DR] 888mel) (Figure 6A). Importantly, this abnormal BRAFV600E protein also was detected in corresponding patients samples of the PDXs expressing this ∼140-kDa BRAFV600E protein (Figure 6B), excluding that this was due to an in vitro or PDX artifact.
Figure 6.
Resistance Mechanism to BRAFi Involving Duplication of the Kinase Domain of BRAFV600E Discovered in PDX Panel
(A) Immunoblotting for BRAF using a set of four PDXs and two in vitro generated melanoma cell lines resistant for PLX4720 (A375R) or dabrafenib/trametinib (888melDR). Tubulin is used as a loading control. BRAFNT, antibody recognizing an epitope encoded by exons 2–3 of BRAF; BRAFV600E, antibody recognizing specifically the BRAFV600E epitope.
(B) Immunoblotting for BRAF on patient samples of vemurafenib-resistant lesions is shown.
(C) Representation of BRAFV600E and BRAFV600E/DK at the genomic level. Arrows indicate the introns where the breakpoints are localized. Black vertical bars, exons; horizontal bars, introns.
(D) Validation of specific genomic breakpoints is shown.
(E) DNA copy-number alterations in the 888melDR relative to 888mel cell line for chromosome 7, with magnification of the amplified region and further magnification of the BRAF locus, are shown.
(F) Fluorescence in situ hybridization (FISH) of 888mel and 888melDR cell line, using either a BRAF probe (red) or a chromosome 7 centromere probe (green), is shown.
(G) Illustration of BRAFV600E- and BRAFV600E/DK-encoding mRNA. Upper row indicates BRAFV600E with normal 3′ UTR (BRAFV600E−N), middle row indicates BRAFV600E with alternative X1 3′ UTR (BRAFV600E−X1), and bottom row indicates BRAFV600E/DK. Green dashed line indicates splice donor site localized within exon 18, which can be used for alternative 3′ UTR splicing.
(H) PCR product using a forward primer in exon 18 and a reverse primer in exon 10 validates the presence of BRAFV600E/DK. As a control, primers in exon 9 (forward) and exon 10 (reverse) were used.
(I) Sanger sequencing of PCR product obtained in (H) is shown.
To unmask the identity of this apparently common ∼140-kDa BRAFV600E protein, we performed whole-genome sequencing (WGS). We observed that resistant melanoma cells harbored genomic rearrangements and partial duplication of the BRAF genomic locus: the first breakpoint was located in the intron between exons 9 and 10 of BRAFT1799A, while the second breakpoint had occurred in the intron between exons 18 and 19. The resulting duplication contained exons 10–18 of the BRAFT1799A gene, which harbors the kinase domain (Figure 6C; Figures S6A–S6F). This was confirmed by PCR using primers specifically for each breakpoint (Figure 6D). We also observed a larger amplification on chromosome 7 of 888melDR, raising the possibility that the locus containing BRAFT1799A with the kinase domain duplication was amplified specifically during resistance acquisition (Figures 6E and 6F; Figures S6C and S6D).
To confirm that the duplication of the BRAFT1799A kinase domain-encoding region results in the production of a BRAFV600E protein harboring a duplication of its kinase domain (hereafter BRAFV600E/DK), we performed RNA sequencing on the four PDXs that expressed the ∼140-kDa protein. This revealed that, in three of the four cases, the fusion between exon 18 and exon 10 had occurred at 18 bp 5′ to the stop codon in exon 18 and at the start at exon 10 (Figure 6G). This location in exon 18 recently was identified as a splice donor site (Figure 6G), which is used by melanoma cells to splice to an alternative X1 3′ UTR localized in exon 19 (Marranci et al., 2015). Melanoma cells can thus express BRAF with a normal 3′ UTR (BRAF-N) or with an alternative X1 3′ UTR (BRAF-X1, Figure 6G).
This raised the possibility that, resulting from the genomic rearrangement, the BRAFV600E/DK-expressing tumors use this alternative splice donor site in exon 18 to splice to the next exon 10. We validated this predicted fusion on mRNA by RT-PCR, using a forward primer in exon 18 and a reverse primer in exon 10 (Figure 6H). This confirmed the presence of BRAFV600E/DK in three PDXs (M033R.X1, M063R.X1, and M048R2.X1) that showed the ∼140-kDa band on immunoblotting (Figure 6A), as well as in two vemurafenib-resistant cell lines (A375R and 888melDR). Also, we confirmed the location of the fusion by Sanger sequencing (Figure 6I). Furthermore, using RNA-sequencing data of an independent set of MAPK pathway inhibition- resistant melanomas (Hugo et al., 2015), we identified the BRAFV600E/DK in four of 44 resistant tumors (Table S6), in which no other genomic mechanism was found to explain the acquired resistance.
To validate the identity and configuration of BRAFV600E/DK independently at the protein level, we performed immunoprecipitation (IP) for BRAF on the 888melDR cell line, which showed very high expression of the endogenous BRAFV600E/DK protein. The samples were run on an SDS-PAGE gel from which the 140-kDa band was excised and subjected to mass spectrometry. The result confirmed the findings from the genetic analyses (Figure S7). Taken together, our data indicate that the genomic breakpoint can occur anywhere between exons 18 and 19 and that the tumors use an alternative splice donor site within exon 18 to splice to the next exon (which is exon 10 in the case of the BRAFV600E/DK). This configuration allows for the natural stop codon in exon 18 to remain intact, as it is removed by splicing, resulting in an in-frame kinase domain duplication.
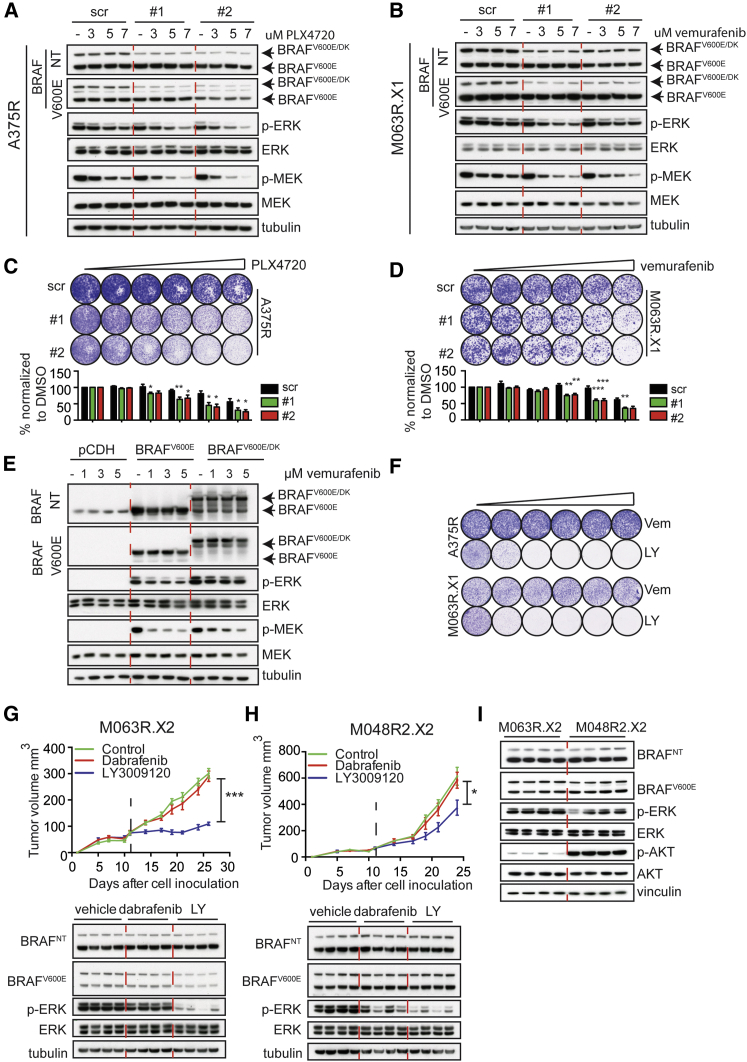
BRAFV600E/DK Is Responsible for Resistance to BRAFi
This is a hitherto unidentified mechanism that may cause resistance to BRAFi. Therefore, we examined whether specific depletion of BRAFV600E/DK, but not BRAFV600E, would result in restoration of BRAF inhibitor sensitivity. We designed small hairpin RNAs (shRNAs) specifically targeting the region between exons 18 and 10 in the BRAFV600E/DK-encoding mRNA. As expected, these shRNAs depleted the BRAFV600E/DK protein from the A375R or M063R.X1 cells, but not the BRAFV600E protein (Figures 7A and 7B). Only when BRAFV600E/DK was silenced was BRAFi capable of decreasing p-ERK levels. Functional validation of this observation in a colony formation assay revealed that BRAFV600E/DK depletion increased sensitivity to BRAFi, in a dose-dependent manner. These results indicate that specific knockdown of BRAFV600E/DK sensitizes tumor cells to BRAFi (Figures 7C and 7D).
Figure 7.
BRAFV600E/DK Is Responsible for Resistance to BRAFi
(A and B) Immunoblotting of A375R (A) or M063R.X1 (B) cells infected with either scrambled shRNAs (scr) or two different shRNAs specifically targeting the BRAFV600E/DK. Cells were treated with indicated concentrations of inhibitor.
(C and D) Colony formation assays with A375R (C) or M063R.X1 (D) cells infected with either scrambled shRNAs (scr) or two different shRNAs specifically targeting the BRAFV600E/DK-encoding RNA (1 and 2). Cells were treated for 7 days with control vehicle or 1, 3, 5, 7, or 10 μM PLX4720 or vemurafenib. Graphs depict the normalization of six independent experiments. Unpaired t test was performed for each concentration of drug (∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001). Error bars indicate SEM.
(E) Immunoblotting of HEK293T cells transfected with empty vector or a vector with BRAFV600E or BRAFV600E/DK, treated with vehicle or 1, 3, or 5 μM vemurafenib, is shown.
(F) Treatment of A375R and PDX-derived cell line M063R.X1 with increasing concentrations of vemurafenib (0.25−5 μM) or pan-RAF inhibitor LY3009120 (10 nM−1 μM) is shown.
(G and H) Treatment of two PDXs (M048R2.X2 and M063R.X2) that express the BRAFV600E/DK with 30 mg/kg dabrafenib or 15 mg/kg LY3009120 (n = 8 tumors/group). Graphs represent tumor volume and dashed lines indicate start of treatment. Unpaired t test was performed at the last time point (∗p < 0.05 and ∗∗∗p < 0.005). Error bars indicate SD. Lower part depicts the immunoblotting for p-ERK and BRAF on M048R2.X2 and M063R.X2, treated with either dabrafenib or LY3009120 (each lane represents a tumor derived from an individual mouse).
(I) Immunoblotting for basic levels (vehicle treated) of p-ERK and p-AKT in M063R.X2 and M048R2.X2 (each lane represents a tumor derived from an individual mouse) is shown.
To determine whether BRAFV600E/DK hyperactivates ERK, we overexpressed its cDNA in HEK293T cells. As expected, this resulted in the expression of a 140-kDa protein, which co-migrated with the BRAFV600E/DK protein seen in resistant PDXs and cell lines (Figures 6A and 7E). Indeed, BRAFV600E/DK expression resulted in hyperactivation of ERK (Figure 7E). In agreement with the idea that BRAFV600E/DK constitutively fuels the MAPK pathway even in the presence of BRAF inhibitor, we observed that, upon exposure to vemurafenib, BRAFV600E/DK-expressing cells maintained higher levels of active ERK than cells producing BRAFV600E (Figure 7E).
BRAFV600E/DK Melanomas Are Sensitive to Pan-RAF Dimerization Inhibition
Finally, having established that BRAFV600E/DK accounts for BRAFi resistance in melanoma, we wished to identify a treatment capable of targeting cells harboring this mutant. Recently, Peng et al. (2015) have shown that a new pan-RAF dimerization inhibitor (LY3009120) inhibits various forms of BRAF dimers, including the previously described p61 BRAF isoform (Peng et al., 2015, Poulikakos et al., 2011). We treated two cell lines (A375R and PDX-derived cell line M063R.X1), both of which harbored BRAFV600E/DK, with LY3009120. Compared to treatment with vemurafenib, these cell lines were highly sensitive to treatment with LY3009120 (Figure 7F).
Next we tested the effect of the LY3009120 compound in vivo, using two PDXs (M063R.X2 and M048R2.X2) that had acquired the BRAFV600E/DK as resistance mechanism (Figure 6A). Both PDXs were treated either with the BRAF inhibitor dabrafenib, to which they were completely resistant (Figures 7G and 7H), or the pan-RAF dimerization inhibitor LY3009120. LY3009120 treatment of M063R.X2 resulted in an effective inhibition of p-ERK and thereby stable disease (Figure 7G). In contrast, upon LY3009120 treatment, M048R2.X2 displayed delayed tumor outgrowth only (Figure 7H). As shown previously, resistance mechanisms in PDX samples can be heterogeneous (Kemper et al., 2015, Van Allen et al., 2014). Moreover, above we described that M048R2.X2 harbors an AKT3L51R mutation in addition to the BRAFV600E/DK mutation (Figure S4C), which conceivably explains the reduced sensitivity to the pan-RAF dimerization inhibitor: the AKT3L51R mutation fuels the PI3K/AKT pathway, which may reduce the effect of BRAFi. Indeed, AKT signaling was much more active in the M048R2.X2 PDX (Figure 7I), explaining its only partial sensitivity to LY3009120.
These results warrant clinical validation of LY3009120 for treating melanoma patients harboring a BRAFV600E/DK mutation. More generally, these findings highlight one key feature of our PDX platform.
Discussion
Here we present a comprehensive and well-characterized PDX collection comprising 89 metastatic human melanomas, including a set of matched pre-treatment/post-relapse pairs. Tumor tissue for this platform was derived from BRAFV600E, NRASQ61, and BRAFWT/NRASWT metastatic melanomas. Samples were acquired before the start of targeted therapy and/or after resistance had occurred. By analyzing biomarker expression, chromosomal aberrations, and RNA expression, we demonstrate that these PDXs recapitulate the key characteristics of the corresponding patients’ tumors. In addition, we show that PDXs derived from vemurafenib-resistant melanomas harbor a plethora of established clinical resistance mechanisms: (1) we have identified previously established resistance-causing mutations, amplifications, and protein expression changes in a cohort of treatment-refractory PDXs; (2) we show that resistance to targeted therapy is maintained in PDXs; (3) we present evidence for reversal and adaption of drug response similar to what is seen in the clinic; and (4) we observed genetic heterogeneity in resistance mechanisms in PDXs, similar to what is observed in the clinic. Although not explored here, adding to the versatility of the PDX platform, several fundamental aspects of melanoma progression, like phenotype switching (Hoek et al., 2006, Verfaillie et al., 2015), are better studied in an in vivo setting. Finally, to illustrate the power and utility of this platform, we have taken advantage of its limitless tumor resource, in contrast to patients’ biopsies. Screening in PDX cell lysates for BRAF proteins with aberrant molecular weights, we identified a kinase domain duplication mutant of BRAFV600E, which drives vemurafenib resistance.
While several resistance mechanisms have been identified previously thanks to the efforts of many laboratories (Nazarian et al., 2010, Paraiso et al., 2011, Poulikakos et al., 2011, Shi et al., 2012, Shi et al., 2014a, Das Thakur et al., 2013, Wagenaar et al., 2014, Wagle et al., 2011), we show here that a specific genetic duplication encoding the kinase domain of BRAFV600E causes resistance to MAPK pathway inhibition. Remarkably, the genomic rearrangement, which results from intronic breaks between exons 9 and 10 and between exons 18 and 19, leaves the original stop codon present in exon 18 intact. As melanoma cells seem to preferentially use the alternative splice donor site in exon 18 to splice to the alternative X1 3′ UTR in exon 19 (and thus use the X1 stop codon) (Marranci et al., 2015), this conceivably explains why cells carrying the genomic duplication of the kinase domain use this splice donor site to produce the BRAFV600E/DK protein. This may imply that the splicing event can be induced by drug exposure; this would provide a selective advantage, whereas expression of BRAFV600E/DK in the absence of BRAFi would not.
Previously, others have shown that duplication of the kinase domain of RTKs, like EGFR (Gallant et al., 2015, Ozer et al., 2010) or FGFR1 (Zhang et al., 2015), drives oncogenicity in glioblastoma, lung cancer, and gliomas, respectively. Recently, wild-type BRAF with a duplicated kinase domain was detected in a neuroblastoma tumor after chemotherapy (Eleveld et al., 2015), but such an event has not yet been implicated in acquired resistance to targeted therapy. Further research will be required to unravel the mechanism of how BRAFV600E/DK functions at a molecular level. Of note, we have attempted to overexpress the BRAFV600E/DK in BRAFV600E and NRASWTBRAFWT melanoma cell lines, but we observed that cells quickly shut down the expression of this mutant, thereby precluding the study of any functional consequences. Melanomas harboring BRAFV600E/DK to drive resistance have adapted over the course of acquiring resistance to express optimal levels of BRAFV600E and BRAFV600E/DK, and perhaps additional rewiring of signaling networks. Apparently, for this particular mutation, this is difficult to recapitulate in the lab by introducing this mutant freshly into melanoma cells that previously were not dependent on it. This is not a general pattern; for example, we have shown previously that ectopic expression of MEK1T55insdelRT is easily achieved and drives drug resistance (Kemper et al., 2015).
Using an inhibitor that targets the BRAFV600E homodimers (Peng et al., 2015), we were able to eliminate BRAFV600E/DK melanoma cells and effectively inhibited tumor growth of PDXs in vivo. As we identified the BRAFV600E/DK as a resistance mechanism in ∼10% of PDXs and patient samples, this pan-RAF dimerization inhibitor may offer a clinical opportunity for this particular subgroup of patients who have required resistance to (combined) inhibitors of the BRAF pathway, arguing that screening for this subset of patients could be beneficial.
We conclude that the PDX platform presented here reflects the original melanomas in patients very well with respect to several key characteristics, including resistance mechanisms. This platform therefore provides a relevant and comprehensive suite of well-characterized treatment-naive and -resistant melanomas, representing an invaluable toolbox for studying fundamental aspects of melanoma biology and for the development, validation, and optimization of melanoma (combinatorial) treatments, to further improve the perspective on melanoma patients.
Experimental Procedures
Patient Samples, Animals, and PDXs
The collection and use of human tissue was approved by the Medical Ethical Review Board of the Antoni van Leeuwenhoek. Animal experiments were approved by the animal experimental committee of the institute and performed according to Dutch law. Human tumor tissue was obtained either by excision during surgery or using a 14-gauge biopsy needle. Tumor fragments of ∼5 mm3 were used for subcutaneous transplantation into NSG mice, which was performed under anesthesia. Before reaching the maximum allowed tumor size, mice were sacrificed, tumors were removed, and tumor pieces were (1) fixed in formalin and embedded in paraffin; (2) snap-frozen and stored at −80°C for further analyses; (3) cryopreserved in 10% fetal calf serum (FCS) in DMSO and stored at −80°C for additional passages; and (4) re-transplanted into a new set of NSG mice. Treatment was performed with dabrafenib (Abmole, 30 mg/kg daily) or LY3009120 (Selleck, 15 mg/kg twice daily [b.i.d.]) by oral gavage. Inhibitors were dissolved in DMSO and further diluted in the vehicle 0.5% hydroxypropylmethylcellulose (Sigma-Aldrich) and 0.2% Tween 80 in (pH 8.0) distilled H20.
IHC
PDX pieces were fixed in formalin and embedded in paraffin. Slides were stained for H&E, S100 (Z031129, DakoCytomation), gp-100 (MS-264-S0, Thermo Scientific), melanA (M719629, DakoCytomation), tyrosinase (T311, 9319, Cell Signaling Technology), and p-ERK1/2 (E10, 4370, Cell Signaling Technology) by our in-house Animal Pathology facility. The NKI-AVL Core Facility Molecular Pathology & Biobanking (CFMPB) provided the NKI-AVL Biobank patient material and performed the BRAFV600E (VE1, Spring Bioscience) staining according to the manufacturer’s protocol.
Immunoblotting and Antibodies
Immunoblotting was performed as described previously (Possik et al., 2014). The following antibodies were used: p-ERK1/2 (E10, 9106), ERK1/2 (9102), p-MEK (41G9, 9154), MEK (L38C12, 4694), p-AKT (D9E, 4060), and p-MET (Tyr1234, 3077) from Cell Signaling Technology; BRAFV600E (VE1) from Spring Bioscience; B-RAF (F7), EGFR (1005), MET (C-28), PDGFR (C20), AXL (C-20), and PTEN (A2B1) from Santa Cruz Biotechnology; MITF (ab12039) from Abcam; and vinculin (V9131) from Sigma.
Cell Culture, Transfection, and Virus Production
Melanoma cell lines and HEK293T cells were cultured in DMEM containing fetal bovine serum (FBS) (Sigma), 2 mM glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin (all Gibco) under standard conditions. Resistant cell lines were generated by treatment with increasing concentrations of PLX4720 (Selleck Chemicals, up to 3 μM) or the combination of dabrafenib (GSK2118436, Abmole, up to 0.5 μM)/trametinib (GSK1120212, S2673, Selleck Chemicals, up to 50 nM). Transfections and production of lentivirus were performed as described previously (Vredeveld et al., 2012). A375R cells were infected and selected with puromycin (1 μg/ml). For the colony formation assays, 20,000 cells were seeded in six-well plates and indicated concentrations of PLX4720, vemurafenib, or LY3009120 were added the next day. Cells were stained by 0.1% crystal violet in 50% methanol and 50% H20. After staining, de-staining by 10% acetic acid was used to quantify the number of stained cells. Color intensity was measured at 590 nm and values were normalized to DMSO control. For immunoblotting, cells were treated for 24 hr with the indicated concentrations of BRAFi.
qPCR, Sanger Sequencing, and Validation of the Presence/Cloning of BRAFV600E/DK and shRNA Generation
All primers and hairpin sequences are described in Table S7. BRAFV600E/DK was cloned from the 888melDR cell line into a TOPO TA-cloning vector (450071, Invitrogen); we used the restriction site XbaI and SwaI to clone BRAFV600E/D into the pCDH vector. BRAFV600E/DK shRNAs were designed at the junction region between exons 18 and 10.
Statistical Testing
The data of in vivo experiments were analyzed at the last time point by an unpaired t test using the Prism software. For the colony formations assays, six independent experiments were performed. Data were normalized to the scr control for each concentration of the drug. Unpaired t tests were used (in Prism software) to compare the effect between the scr and the two hairpins for each concentration of the drug.
Author Contributions
K.K., O.K., X.K., and D.S.P. designed and managed all experiments. K.K., X.K., P.C.-S., and A.S. performed all in vitro and in vivo experiments. O.K., T.K., R.J.C.K., and Y.S.J. performed the bio-informatics analysis. O.B.B. and A.F.M.A. performed the mass spectrometry analysis. C.S. was responsible for the RNA sequencing of the patient samples. T.N.M.S., C.U.B., and J.B.H. were responsible for the patient data and the acquisition of the patient samples. F.W. and D.L.v.d.V. performed the volumetric measurements on the computed tomography (CT) scans, which was supervised by E.E.V. D.J.A. and U.M. were responsible for DNA sequencing. K.K., O.K., and D.S.P. wrote the manuscript. All authors revised and approved the manuscript. The project was supervised by D.S.P.
Acknowledgments
We would like to express our gratitude to the patients and their relatives for their cooperation in this study. We thank Xiaohang Qiao for her assistance with the BRAF IP-mass spectrometry experiment and we thank Ji-Ying Song for assessing the PDX H&E slides. We also acknowledge Iris de Rink, Arno Velds (Central Genomics Facility of the NKI), the NKI High Performance Computing, and the NKI-AVL CFMPB. D.S.P. and K.K. are members of the EuroPDX consortium. This work was financially supported by a grant from the Dutch Cancer Society (NKI-2013-5799; K.K., P.C.-S., J.B.H., and D.S.P.), European Research Council (ERC Synergy Grant “Combat Cancer”; X.K., O.K., and D.S.P.), Netherlands Organization for Scientific Research (NWO) through a VIDI grant (723.012.102, A.F.M.A.) and as part of the National Roadmap Large-scale Research Facilities of the Netherlands (project 184.032.201; O.B.B. and A.F.M.A.), Cancer Research UK and The Wellcome Trust (WT098051, D.J.A.), The Josephine Nefkins Foundation (E.E.V.) and Barcode for Life (E.E.V.), and a Queen Wilhelmina Award by the Dutch Cancer Society (D.S.P.).
Published: June 16, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.05.064.
Supplemental Information
References
- Basile K.J., Abel E.V., Aplin A.E. Adaptive upregulation of FOXD3 and resistance to PLX4032/4720-induced cell death in mutant B-RAF melanoma cells. Oncogene. 2012;31:2471–2479. doi: 10.1038/onc.2011.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertotti A., Migliardi G., Galimi F., Sassi F., Torti D., Isella C., Corà D., Di Nicolantonio F., Buscarino M., Petti C. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1:508–523. doi: 10.1158/2159-8290.CD-11-0109. [DOI] [PubMed] [Google Scholar]
- Burchill S.A. What do, can and should we learn from models to evaluate potential anticancer agents? Future Oncol. 2006;2:201–211. doi: 10.2217/14796694.2.2.201. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman P.B., Hauschild A., Robert C., Haanen J.B., Ascierto P., Larkin J., Dummer R., Garbe C., Testori A., Maio M., BRIM-3 Study Group Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement V., Sanchez P., de Tribolet N., Radovanovic I., Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr. Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel V.C., Marchionni L., Hierman J.S., Rhodes J.T., Devereux W.L., Rudin C.M., Yung R., Parmigiani G., Dorsch M., Peacock C.D., Watkins D.N. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 2009;69:3364–3373. doi: 10.1158/0008-5472.CAN-08-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Thakur M., Salangsang F., Landman A.S., Sellers W.R., Pryer N.K., Levesque M.P., Dummer R., McMahon M., Stuart D.D. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–255. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M.J., Bottomley W. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- De Witt Hamer P.C., Van Tilborg A.A.G., Eijk P.P., Sminia P., Troost D., Van Noorden C.J.F., Ylstra B., Leenstra S. The genomic profile of human malignant glioma is altered early in primary cell culture and preserved in spheroids. Oncogene. 2008;27:2091–2096. doi: 10.1038/sj.onc.1210850. [DOI] [PubMed] [Google Scholar]
- DeRose Y.S., Wang G., Lin Y.-C., Bernard P.S., Buys S.S., Ebbert M.T.W., Factor R., Matsen C., Milash B.A., Nelson E. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med. 2011;17:1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einarsdottir B.O., Bagge R.O., Bhadury J., Jespersen H., Mattsson J., Nilsson L.M., Truvé K., López M.D., Naredi P., Nilsson O. Melanoma patient-derived xenografts accurately model the disease and develop fast enough to guide treatment decisions. Oncotarget. 2014;5:9609–9618. doi: 10.18632/oncotarget.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleveld T.F., Oldridge D.A., Bernard V., Koster J., Daage L.C., Diskin S.J., Schild L., Bentahar N.B., Bellini A., Chicard M. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat. Genet. 2015;47:864–871. doi: 10.1038/ng.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty K.T., Puzanov I., Kim K.B., Ribas A., McArthur G.A., Sosman J.A., O’Dwyer P.J., Lee R.J., Grippo J.F., Nolop K., Chapman P.B. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant J.-N., Sheehan J.H., Shaver T.M., Bailey M., Lipson D., Chandramohan R., Red Brewer M., York S.J., Kris M.G., Pietenpol J.A. EGFR kinase domain duplication (EGFR-KDD) is a novel oncogenic driver in lung cancer that is clinically responsive to afatinib. Cancer Discov. 2015;5:1155–1163. doi: 10.1158/2159-8290.CD-15-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Korn J.M., Ferretti S., Monahan J.E., Wang Y., Singh M., Zhang C., Schnell C., Yang G., Zhang Y. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015;21:1318–1325. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]
- Garraway L.A., Widlund H.R., Rubin M.A., Getz G., Berger A.J., Ramaswamy S., Beroukhim R., Milner D.A., Granter S.R., Du J. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Girotti M.R., Gremel G., Lee R., Galvani E., Rothwell D., Viros A., Mandal A.K., Lim K.H.J., Saturno G., Furney S.J. Application of sequencing, liquid biopsies, and patient-derived xenografts for personalized medicine in melanoma. Cancer Discov. 2016;6:286–299. doi: 10.1158/2159-8290.CD-15-1336. [DOI] [PubMed] [Google Scholar]
- Hoek K.S., Schlegel N.C., Brafford P., Sucker A., Ugurel S., Kumar R., Weber B.L., Nathanson K.L., Phillips D.J., Herlyn M. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- Hugo W., Shi H., Sun L., Piva M., Song C., Kong X., Moriceau G., Hong A., Dahlman K.B., Johnson D.B. Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell. 2015;162:1271–1285. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper K., Krijgsman O., Cornelissen-Steijger P., Shahrabi A., Weeber F., Song J.-Y., Kuilman T., Vis D.J., Wessels L.F., Voest E.E. Intra- and inter-tumor heterogeneity in a vemurafenib-resistant melanoma patient and derived xenografts. EMBO Mol. Med. 2015;7:1104–1118. doi: 10.15252/emmm.201404914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczkowski D.J., Johannessen C.M., Abudayyeh O., Kim J.W., Cooper Z.A., Piris A., Frederick D.T., Barzily-Rokni M., Straussman R., Haq R. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 2014;4:816–827. doi: 10.1158/2159-8290.CD-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T., Velds A., Kemper K., Ranzani M., Bombardelli L., Hoogstraat M., Nevedomskaya E., Xu G., de Ruiter J., Lolkema M.P. CopywriteR: DNA copy number detection from off-target sequence data. Genome Biol. 2015;16:49. doi: 10.1186/s13059-015-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J., Ascierto P.A., Dréno B., Atkinson V., Liszkay G., Maio M., Mandalà M., Demidov L., Stroyakovskiy D., Thomas L. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N. Engl. J. Med. 2014;371:1867–1876. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- Marranci A., Tuccoli A., Vitiello M., Mercoledi E., Sarti S., Lubrano S., Evangelista M., Fogli A., Valdes C., Russo F. Identification of BRAF 3’UTR Isoforms in Melanoma. J. Invest. Dermatol. 2015;135:1694–1697. doi: 10.1038/jid.2015.47. [DOI] [PubMed] [Google Scholar]
- Monsma D.J., Monks N.R., Cherba D.M., Dylewski D., Eugster E., Jahn H., Srikanth S., Scott S.B., Richardson P.J., Everts R.E. Genomic characterization of explant tumorgraft models derived from fresh patient tumor tissue. J. Transl. Med. 2012;10:125. doi: 10.1186/1479-5876-10-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsma D.J., Cherba D.M., Eugster E.E., Dylewski D.L., Davidson P.T., Peterson C.A., Borgman A.S., Winn M.E., Dykema K.J., Webb C.P. Melanoma patient derived xenografts acquire distinct Vemurafenib resistance mechanisms. Am. J. Cancer Res. 2015;5:1507–1518. [PMC free article] [PubMed] [Google Scholar]
- Müller J., Krijgsman O., Tsoi J., Robert L., Hugo W., Song C., Kong X., Possik P.A., Cornelissen-Steijger P.D.M., Foppen M.H.G. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat. Commun. 2014;5:5712. doi: 10.1038/ncomms6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R., Shi H., Wang Q., Kong X., Koya R.C., Lee H., Chen Z., Lee M.-K., Attar N., Sazegar H. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev S.I., Rimoldi D., Iseli C., Valsesia A., Robyr D., Gehrig C., Harshman K., Guipponi M., Bukach O., Zoete V. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat. Genet. 2011;44:133–139. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- Ozer B.H., Wiepz G.J., Bertics P.J. Activity and cellular localization of an oncogenic glioblastoma multiforme-associated EGF receptor mutant possessing a duplicated kinase domain. Oncogene. 2010;29:855–864. doi: 10.1038/onc.2009.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraiso K.H.T., Xiang Y., Rebecca V.W., Abel E.V., Chen Y.A., Munko A.C., Wood E., Fedorenko I.V., Sondak V.K., Anderson A.R.A. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 2011;71:2750–2760. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh C., Janakiraman V., Wu W.-I., Foo C.K., Kljavin N.M., Chaudhuri S., Stawiski E., Lee B., Lin J., Li H. Disruption of PH-kinase domain interactions leads to oncogenic activation of AKT in human cancers. Proc. Natl. Acad. Sci. USA. 2012;109:19368–19373. doi: 10.1073/pnas.1204384109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S.-B., Henry J.R., Kaufman M.D., Lu W.-P., Smith B.D., Vogeti S., Rutkoski T.J., Wise S., Chun L., Zhang Y. Inhibition of RAF Isoforms and Active Dimers by LY3009120 Leads to Anti-tumor Activities in RAS or BRAF Mutant Cancers. Cancer Cell. 2015;28:384–398. doi: 10.1016/j.ccell.2015.08.002. [DOI] [PubMed] [Google Scholar]
- Possik P.A., Müller J., Gerlach C., Kenski J.C.N., Huang X., Shahrabi A., Krijgsman O., Song J.-Y., Smit M.A., Gerritsen B. Parallel in vivo and in vitro melanoma RNAi dropout screens reveal synthetic lethality between hypoxia and DNA damage response inhibition. Cell Rep. 2014;9:1375–1386. doi: 10.1016/j.celrep.2014.10.024. [DOI] [PubMed] [Google Scholar]
- Poulikakos P.I., Persaud Y., Janakiraman M., Kong X., Ng C., Moriceau G., Shi H., Atefi M., Titz B., Gabay M.T. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C., Karaszewska B., Schachter J., Rutkowski P., Mackiewicz A., Stroiakovski D., Lichinitser M., Dummer R., Grange F., Mortier L. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 2015;372:30–39. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- Rubio-Viqueira B., Jimeno A., Cusatis G., Zhang X., Iacobuzio-Donahue C., Karikari C., Shi C., Danenberg K., Danenberg P.V., Kuramochi H. An in vivo platform for translational drug development in pancreatic cancer. Clin. Cancer Res. 2006;12:4652–4661. doi: 10.1158/1078-0432.CCR-06-0113. [DOI] [PubMed] [Google Scholar]
- Shi H., Moriceau G., Kong X., Lee M.-K., Lee H., Koya R.C., Ng C., Chodon T., Scolyer R.A., Dahlman K.B. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat. Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Hong A., Kong X., Koya R.C., Song C., Moriceau G., Hugo W., Yu C.C., Ng C., Chodon T. A novel AKT1 mutant amplifies an adaptive melanoma response to BRAF inhibition. Cancer Discov. 2014;4:69–79. doi: 10.1158/2159-8290.CD-13-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Hugo W., Kong X., Hong A., Koya R.C., Moriceau G., Chodon T., Guo R., Johnson D.B., Dahlman K.B. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solit D.B., Rosen N. Towards a unified model of RAF inhibitor resistance. Cancer Discov. 2014;4:27–30. doi: 10.1158/2159-8290.CD-13-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straussman R., Morikawa T., Shee K., Barzily-Rokni M., Qian Z.R., Du J., Davis A., Mongare M.M., Gould J., Frederick D.T. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Wang L., Huang S., Heynen G.J.J.E., Prahallad A., Robert C., Haanen J., Blank C., Wesseling J., Willems S.M. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–122. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- Van Allen E.M., Wagle N., Sucker A., Treacy D.J., Johannessen C.M., Goetz E.M., Place C.S., Taylor-Weiner A., Whittaker S., Kryukov G.V., Dermatologic Cooperative Oncology Group of Germany (DeCOG) The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verfaillie A., Imrichova H., Atak Z.K., Dewaele M., Rambow F., Hulselmans G., Christiaens V., Svetlichnyy D., Luciani F., Van den Mooter L. Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat. Commun. 2015;6:6683. doi: 10.1038/ncomms7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergani E., Vallacchi V., Frigerio S., Deho P., Mondellini P., Perego P., Cassinelli G., Lanzi C., Testi M.A., Rivoltini L. Identification of MET and SRC activation in melanoma cell lines showing primary resistance to PLX4032. Neoplasia. 2011;13:1132–1142. doi: 10.1593/neo.111102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskoglou-Nomikos T., Pater J.L., Seymour L. Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clin. Cancer Res. 2003;9:4227–4239. [PubMed] [Google Scholar]
- Vredeveld L.C.W., Possik P.A., Smit M.A., Meissl K., Michaloglou C., Horlings H.M., Ajouaou A., Kortman P.C., Dankort D., McMahon M. Abrogation of BRAFV600E-induced senescence by PI3K pathway activation contributes to melanomagenesis. Genes Dev. 2012;26:1055–1069. doi: 10.1101/gad.187252.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenaar T.R., Ma L., Roscoe B., Park S.M., Bolon D.N., Green M.R. Resistance to vemurafenib resulting from a novel mutation in the BRAFV600E kinase domain. Pigment Cell Melanoma Res. 2014;27:124–133. doi: 10.1111/pcmr.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N., Emery C., Berger M.F., Davis M.J., Sawyer A., Pochanard P., Kehoe S.M., Johannessen C.M., Macconaill L.E., Hahn W.C. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J. Clin. Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N., Van Allen E.M., Treacy D.J., Frederick D.T., Cooper Z.A., Taylor-Weiner A., Rosenberg M., Goetz E.M., Sullivan R.J., Farlow D.N. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014;4:61–68. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhou Y., Cheng C., Cui H., Cheng L., Kong P., Wang J., Li Y., Chen W., Song B. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am. J. Hum. Genet. 2015;96:597–611. doi: 10.1016/j.ajhg.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.