Abstract
Disrupting Notch signaling ameliorates experimental liver fibrosis. However, the role of individual Notch ligands in liver damage is unknown. We investigated the effects of Delta-like ligand 4 (Dll4) in liver disease. DLL4 expression was measured in 31 human liver tissues by immunohistochemistry. Dll4 function was examined in carbon tetrachloride– and bile duct ligation–challenged mouse models in vivo and evaluated in hepatic stellate cells, hepatocytes, and Kupffer cells in vitro. DLL4 was expressed in patients' Kupffer and liver sinusoidal endothelial cells. Recombinant Dll4 protein (rDll4) ameliorated hepatocyte apoptosis, inflammation, and fibrosis in mice after carbon tetrachloride challenge. In vitro, rDll4 significantly decreased lipopolysaccharide-dependent chemokine expression in both Kupffer and hepatic stellate cells. In bile duct ligation mice, rDll4 induced massive hepatic necrosis, resulting in the death of all animals within 1 week. Inflammatory cell infiltration and chemokine ligand 2 (Ccl2) expression were significantly reduced in rDll4-receiving bile duct ligation mice. Recombinant Ccl2 rescued bile duct ligation mice from rDll4-mediated death. In patients with acute-on-chronic liver failure, DLL4 expression was inversely associated with CCL2 abundance. Mechanistically, Dll4 regulated Ccl2 expression via NF-κB. Taken together, Dll4 modulates liver inflammatory response by down-regulating chemokine expression. rDll4 application results in opposing outcomes in two models of liver damage. Loss of DLL4 may be associated with CCL2-mediated cytokine storm in patients with acute-on-chronic liver failure.
In multicellular organisms, the Notch signaling pathway is critical for cell-cell communication, which determines organ development and cell fate, in particular cell proliferation, differentiation, and apoptosis.1, 2, 3 In humans, four Notch receptors (NOTCH 1, 2, 3, and 4) and five membrane-anchored canonical ligands [Jagged (JAG) 1 and 2 and Delta-like ligand (DLL) 1, 3, and 4] have been identified.1 Soluble Notch ligands have also been described, one belongs to the cold shock protein family.4 On binding to their receptors, Notch ligands initiate signaling and consequently lead to ADAM (a disintegrin- and metalloprotease)-mediated cleavage of the extracellular receptor domain. Subsequently, γ-secretase cleavage sets free the Notch intracellular domain. The Notch intracellular domain translocates into the nucleus and forms a complex with the DNA-binding protein CSL [CBF1/recombination signal binding protein J κ (RBPjκ)/Su(H)/Lag-1] to activate transcription of primary target genes, such as HES gene family members (eg, HES1 and HES5) in mammals. These, in turn, act as Notch effectors to modulate cellular functions. Because of the existence of four Notch receptors and more than five ligands in mammals, the functions of Notch signaling pathway under (patho-)physiological conditions are complex and highly dependent on disease- and cell-specific context.5 The ubiquity and versatility of Notch signaling has recently been demonstrated, when it was shown that increased expression of Jag1 was able to rescue the Duchenne muscular dystrophy phenotype in a canine model.6
Notch receptors and ligands are constitutively expressed in various human tissues, including the liver.2 Mutations in the JAG1 or NOTCH2 genes cause Alagille syndrome, a multisystemic congenital disease characterized by defective development of interlobular bile ducts and vessel architecture.7, 8 Most patients with Alagille syndrome do not develop overt fibrosis.9 Overexpression of JAG1 and NOTCH3 is found in small neovessels and bile ductules of diseased livers (eg, with primary biliary cirrhosis and primary sclerosing cholangitis).10, 11 These findings suggest that Notch signaling might be associated with periportal fibrosis by participating in aberrant neovascularization and ductular reaction. Besides its critical role in Alagille syndrome, Notch signaling synergizes with the Wnt signaling pathway to determine lineage differentiation of hepatic progenitor cells during liver regeneration.12, 13
Notch activation has also been implicated in experimental liver fibrosis. In a rat model of carbon tetrachloride (CCl4) challenge, blocking Notch signaling by γ-secretase inhibitor significantly attenuated liver fibrosis, suggesting that even nonselective interference with Notch signaling is a viable antifibrotic strategy.14 Recently, a study based on mice with RBPjκ knockdown in macrophages revealed that Notch signaling activation in macrophages is necessary for liver fibrosis induced by CCl4 and bile duct ligation (BDL).15 However, detailed knowledge on functions of Notch family members in liver inflammation and fibrosis is incomplete, especially in experimental models. Because the Notch inhibitors tested in experimental and human clinical trials are hitherto non-specific and may have adverse effects on proliferating cells of the gastrointestinal tract,16, 17, 18 we set out to more specifically test for novel targets within the Notch signaling pathway. Thus, we explored potential functions of Notch ligands, particularly delta-like ligand 4 (Dll4), in liver inflammation and fibrosis.
Materials and Methods
Reagents
Human and mouse recombinant Dll4 (1506-D4-050-CF and 1389-D4-050-CF), mouse recombinant chemokine ligand 2 (Ccl2; 479-JE-010), and transforming growth factor (TGF)-β (240-B-010) proteins, and Ccl2 enzyme-linked immunosorbent assay kit (DY479-05) were purchased from R&D (Minneapolis, MN). Dulbecco's modified Eagle's medium, penicillin/streptomycin, and l-glutamine were obtained from Cambrex (Verviers, Belgium). Fetal bovine serum was from Invitrogen (Karlsruhe, Germany). Lipopolysaccharide (LPS) and JSH-23 (NF-κB inhibitor, J4455-5MG) were from Sigma (Munich, Germany). Viromer Red was from Lipocalyx (Halle, Germany). The primer sequences and antibodies used are shown in Table 1, Table 2.
Table 1.
Primers Used for Real-Time Quantitative PCR
| Gene | Forward sequence | Reverse sequence |
|---|---|---|
| Hes1 | 5′-AGAAGAGGCGAAGGGCAAGAAT-3′ | 5′-AGGTCATGGCGTTGATCTGG-3′ |
| Hes5 | 5′-CGCATCAACAGCAGCATAGAG-3′ | 5′-TGGAAGTGGTAAAGCAGCTTC-3′ |
| Col1a1 | 5′-CATGTTCAGCTTTGTGGACCT-3′ | 5′-GCAGCTGACTTCAGGGATGT-3′ |
| Col1a2 | 5′-AAGGGTGCTACTGGACTCCC-3′ | 5′-TTGTTACCGGATTCTCCTTTGG-3′ |
| Acta1 | 5′-GGACGTACAACTGGTATTGTGC-3′ | 5′-CGGCAGTAGTCACGAAGGAAT-3′ |
| Tnfa | 5′-CTGAACTTCGGGGTGATCG-3′ | 5′-GGCTACAGGCTTGTCACTCG-3′ |
| Ifng | 5′-GCCACGGCACAGTCATTGA-3′ | 5′-TGCTGATGGCCTGATTGTCTT-3′ |
| Il1a | 5′-CGAAGACTACAGTTCTGCCATT-3′ | 5′-GACGTTTCAGAGGTTCTCAGAG-3′ |
| Il6 | 5′-CTGCAAGAGACTTCCATCCAG-3′ | 5′-AGTGGTATAGACAGGTCTGTTGG-3′ |
| Il10 | 5′-TCTTTCAAACAAAGGACCAGC-3′ | 5′-ACCCAGGGAATTCAAATGC-3′ |
| Il12 | 5′-CAATCACGCTACCTCCTCTTTT-3′ | 5′-CAGCAGTGCAGGAATAATGTTTC-3′ |
| Il2 | 5′-TCCAGAACATGCCGCAGAG-3′ | 5′-CCTGAGCAGGATGGAGAATTACA-3′ |
| Il4 | 5′-GAAGCCCTACAGACGAGCTCA-3′ | 5′-ACAGGAGAAGGGACGCCAT-3′ |
| Il13 | 5′-TGGGTCCTGTAGATGGCATTG-3′ | 5′-AGACCAGACTCCCCTGTGCA-3′ |
| Tgfb | 5′-GCAACAACGCCATCTATGAG-3′ | 5′-ACGCCAGGAATTGTTGCTAT-3′ |
| Ccl2 | 5′-CTTCTGGGCCTGCTGTTCA-3′ | 5′-CAGCCTACTCATTGGGATCA-3′ |
| Ccl5 | 5′-AGATCTCTGCAGCTGCCCTCA-3′ | 5′-GGAGCACTTGCTGCTGGTGTAG-3′ |
| Cxcl9 | 5′-TGGGCAGAAGTTCCGTCTTG-3′ | 5′-ATTACCGAAGGGAGGTGGACAACG-3′ |
| Ccl8 | 5′-TCTACGCAGTGCTTCTTTGCC-3′ | 5′-AAGGGGGATCTTCAGCTTTAGTA-3′ |
| Cxcl10 | 5′-AAATCATCCCTGCGAGCCTATC-3′ | 5′-GGAGCCCTTTTAGACCTTTTTTGG-3′ |
| Fas | 5′-TGTTTTCCCTTGCTGCAGAC-3′ | 5′-TCCTCAGCTTTAAACTCTCGGA-3′ |
| DR5 | 5′-CTAACCCAGCCCATAATCGT-3′ | 5′-AGAGAATGGTTGGAATGGCT-3′ |
| Tnfr1 | 5′-CTTCATTCACGAGCGTTGTC-3′ | 5′-CAGTGACCCCTGATGGATGT-3′ |
| Actb | 5′-GTGCTATGTTGCTCTAGACTTCG-3′ | 5′-ATGCCACAGGATTCCATACC-3′ |
Table 2.
Primary Antibodies Used in the Study
| Antibody name | Host | Product no. | Company |
|---|---|---|---|
| Jagged1 | Goat | sc-6011 | Santa Cruz Biotechnology (Dallas, TX) |
| Jagged2 | Rabbit | sc-5604 | Santa Cruz Biotechnology |
| Dll1 | Rabbit | sc-9102 | Santa Cruz Biotechnology |
| Dll3 | Rabbit | sc-67269 | Santa Cruz Biotechnology |
| Dll4 | Rabbit | sc-28915 | Santa Cruz Biotechnology |
| Notch1 | Goat | sc-6014 | Santa Cruz Biotechnology |
| Notch2 | Rabbit | ab8926 | Abcam (Cambridge, UK) |
| Notch3 | Rabbit | sc-5593 | Santa Cruz Biotechnology |
| Notch4 | Rabbit | ab23427 | Abcam |
| Ccl-2 | Rabbit | LS-C211039-100 | LS-Bio (Seattle, WA) |
| α-SMA | Mouse | M0851 | Dako (Hamburg, Germany) |
| Cleaved-caspase3 | Rabbit | 9664 | Cell Signaling (Danvers, MA) |
| F4/80 | Rat | ab6640 | Abcam |
| CD16 | Mouse | Ab74512 | Abcam |
| Stabilin2 | Mouse | MAB3645 | R & D |
| Myeloperoxidase | Rabbit | AB45977 | Abcam |
| p65 | Rabbit | Sc-372 | Santa Cruz Biotechnology |
| p50 | Mouse | Sc-8414 | Santa Cruz Biotechnology |
| Histone H1 | Mouse | Sc-8030 | Santa Cruz Biotechnology |
| GAPDH | Rabbit | Ab9485 | Abcam |
| CD45 | Rabbit | ab10558 | Abcam |
α-SMA, α-smooth muscle actin; Ccl, chemokine ligand; dll, delta-like ligand; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Patients' Liver Tissue Specimen
Twenty-six resected liver tissues with hepatitis B virus (HBV)-related cirrhosis [10 from patients with compensated cirrhosis and 16 with acute-on-chronic liver failure (ACLF)] and five with hepatolithiasis were obtained from The First Affiliated Hospital, College of Medicine, Zhejiang University (Hangzhou, China). In addition, five specimens with hepatic hemangioma were used as healthy controls. The study was approved by the Ethics Committee of The First Affiliated Hospital, College of Medicine, Zhejiang University. Written informed consent was obtained from all patients.
Cells
Isolation of primary mouse hepatic stellate cells (HSCs), Kupffer cells (KCs), and hepatocytes was performed as previously described.19, 20, 21 JS-1 cells (mouse HSC line) were used as published.22 RAW 264.7 cells (murine macrophage cell line) were grown in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum and 2 mmol/L l-glutamine.
Animal Models
Male C57BL/6 wild-type mice were used for CCl4 challenge and BDL operation. In the CCl4 model, 24 mice were divided into four groups: sham, recombinant Dll4 (rDll4), CCl4, and CCl4 + rDll4 group (n = 6 for each group). In the CCl4 group, mice were i.p. injected with CCl4 (0.5 mL/kg, diluted 1:4 in mineral oil, twice weekly) for 4 weeks. In the CCl4 + rDll4 group, mice were i.p. injected with CCl4 plus 6.25 μg/kg rDll4 twice weekly for 4 weeks. Sham-treated mice received mineral oil only. Animals were sacrificed 24 hours after the last CCl4 injection.
BDL operation was performed as described.23 Mice were divided into BDL, and BDL + rDll4 groups. To assess additional effects of Ccl2 in animals receiving rDll4 + BDL, different dosages of mouse recombinant Ccl2 (rCcl2, 1 and 5 μg/kg) were i.p. injected 1 day after rDll4 administration twice weekly (n = 6 for each dosage).
All animal protocols were performed in full compliance with the guidelines for animal care and approved by the Animal Care Committee of Zhejiang University.
Serum ALT and AST Measurement
Serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels in experimental animal studies were quantified with a Hitachi 7600–110 automatic analyzer (Tokyo, Japan).
Histology, IHC, and Immunofluorescence Staining
Liver tissues were fixed in 4% formaldehyde and embedded in paraffin. Sections (4 μm thick) were stained with hematoxylin and eosin, Sirius red, and Masson trichrome. Immunohistochemistry (IHC) and immunofluorescence staining were performed as previously described.24 Briefly, the slides were deparaffinized in xylene and rehydrated in a dilution series of graded ethanol to distilled water. Antigen retrieval was performed by microwave treatment in EDTA buffer (1 mmol/L, pH 8.0) for 10 minutes. The slides were incubated with 3% H2O2 for 30 minutes at room temperature. After washing with phosphate-buffered saline three times, slides were incubated with primary antibodies at 4°C overnight (Table 2). The next day, the slides were washed with phosphate-buffered saline three times, followed by incubating with EnVision peroxidase-labeled anti-rabbit or anti-mouse antibodies (Dako) for 1 hour at room temperature. Peroxidase activity was detected with diaminobenzidine. The slides were counterstained with hematoxylin. Immunoreactivity was visualized under light microscopy.
For immunofluorescence staining, secondary antibodies, Alexa 633-rabbit anti-mouse IgG and Alexa 488-donkey anti-rabbit IgG (Molecular Probes/Invitrogen, Karlsruhe, Germany), were applied for 30 minutes at room temperature. Samples were mounted using Dako-Cytomation Fluorescence Mounting Medium. The slides were inspected and images were taken on a confocal microscope (Leica, Heidelberg, Germany). Sections without primary antibodies were used as negative controls.
Semiquantification for staining was performed using Leica Quantimet imaging software version 4 (Leica, Heidelberg, Germany).
RNA Isolation and Real-Time Quantitative PCR (qPCR)
Total RNA was purified from cells or tissues with the High Pure RNA isolation kit (Roche Diagnostics GmbH, Mannheim, Germany). cDNA was synthesized from 1 μg RNA with the Transcriptor First Strand cDNA synthesis kit (Roche Diagnostics). Real-time quantitative PCR (qPCR) was performed on an ABIPrism 7700 sequence detection system (Applied Biosystems, Foster City, CA) with TaqMan probes and TaqMan universal PCR master mix, No AmpErase UNG. The transcript numbers in each sample were quantified in duplicate. Data are shown as the means ± SEM of triplicate experiments.
Hydroxyproline Content Assay
Hydroxyproline content was determined as previously described.25
Western Blot Analysis
Radioimmunoprecipitation assay buffer (1 × Tris-buffer saline, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) was used to lyse cultured cells in the presence of complete protease inhibitor cocktail (Roche Diagnostics) and phosphatase inhibitors (Sigma-Aldrich). Subcellular fractionation was performed according to manufacturer's instructions (Genetex).
Nuclear protein concentration was determined using the DC protein assay (Bio-Rad, Munich, Germany). Proteins (20 μg per sample) were separated by 12% SDS-PAGE and transferred to nitrocellulose membrane (Pierce, Rockford, IL). Non-specific binding was blocked with 5% milk in Tris-buffered saline with Tween 20 before incubation with primary antibodies overnight at 4°C. Horseradish peroxidase–linked anti-rabbit, anti-mouse, or anti-goat antibodies (Santa Cruz Biotechnology, Dallas, TX) were used as secondary antibodies. Membranes were developed using Chemi-Smart (Vilber Lourmat, Eberhardzell, Germany). Densitometry was calculated with ImageJ software version 1.49 (NIH, Bethesda, MD).
Luciferase Assay
Mouse Ccl2 promoter reporter construct (pcDNA3.1-CCL2-Luc) containing a 540-bp fragment of the 5′-flanking region of the CCL2 gene starting from 20-bp 5′ of the transcription start site was kindly provided by Dr. Sze-Kwan Lin (National Taiwan University Hospital, Taipei, Taiwan).26 Luciferase substrate was purchased from Promega (Mannheim, Germany). Viromer Red [Lipocalyx, Halle (Saale), Germany] was used for transfection. Firefly luciferase activity was detected and normalized to protein content using Tecan infinite M200 (Thermo Scientific, Waltham, MA). Normalized values were expressed as the means ± SEM of three independent experiments.
Enzyme-Linked Immunosorbent Assay
The amounts of Ccl2 in the culture supernatants were quantified by enzyme-linked immunosorbent assay, according to the manufacturer's instructions (R&D Systems). Briefly, 2 × 105 cells were cultured and subjected to different treatments. The culture supernatant mixed with reagent diluent was added to anti-Ccl2 antibody–coated wells. Biotinylated goat anti-mouse Ccl2 detection antibody was added and incubated at room temperature for 2 hours, followed by incubation with streptavidin-horseradish peroxidase for 20 minutes. Then, colorimetric (tetramethylbenzidine) solution (Abcam, ab171524) was added for another 20 minutes. Finally, the reaction was terminated using stop solution (Abcam, ab171529), and the relative absorbance was measured.
Statistical Analysis
Variables were summarized as means ± SEM and depicted graphically as means ± SEM. Parametric data were analyzed using two-tailed unpaired t-test. Kaplan-Meier survival analysis was used to evaluate mortality of mice after BDL operation. P < 0.05 was considered significant.
Results
Elevated DLL4 and JAG1 Expression in Liver Sinusoids of Patients with Cirrhosis
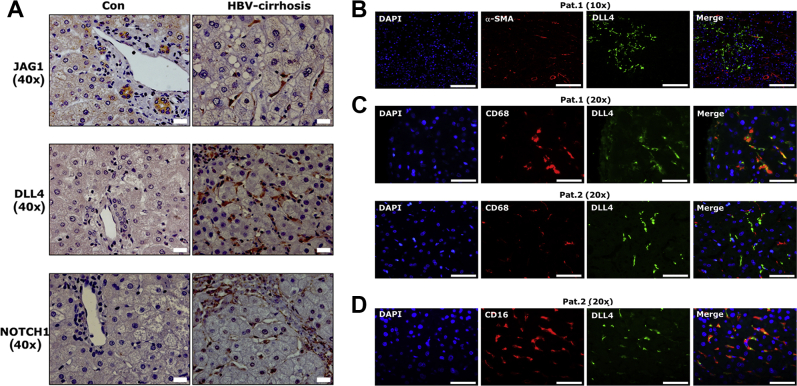
We determined protein expression of five conventional Notch ligands (JAG1, JAG2, DLL1, DLL3, and DLL4) and four receptors (NOTCH 1, 2, 3, and 4) by IHC in five healthy control livers, five liver specimens with hepatolithiasis, and ten cirrhotic liver tissues with HBV infection. In five control specimens, only JAG1, but no other Notch ligands or receptors, were detected (Supplemental Table S1). Positive JAG1 staining was localized in cholangiocytes (Figure 1A). As in control liver tissues, JAG1 was the only Notch ligand expressed in cholangiocytes of five patients with hepatolithiasis. In addition, NOTCH1 and NOTCH2 expression was observed in cholangiocytes of these patients. In both healthy controls and patients with hepatolithiasis, none of the Notch ligands and receptors was detectable in sinusoidal cells (Supplemental Table S1). In HBV-associated cirrhotic liver tissue, Notch ligands JAG1, DLL3, and DLL4 as well as receptors NOTCH 1, 2, 3, and 4 showed intense positive staining in distinct liver cells (Supplemental Table S1). Among expressed ligands/receptors, positive staining of JAG1, DLL4, and NOTCH1 was localized in sinusoidal cells (Figure 1A).
Figure 1.
Expression of Delta-like ligand 4 (DLL4) in sinusoidal cells. A: Immunohistochemical staining shows that Jagged (JAG)1, DLL4, and NOTCH1 localize in sinusoidal cells in cirrhotic patients with hepatitis B virus infection. B–D: Costaining for DLL4 and α-smooth muscle actin (α-SMA)/CD68/CD16 was performed in four cirrhotic patients with high levels of DLL4 positivity in sinusoids. Representative patients show costaining for DLL4 and α-SMA (B), DLL4 and CD68 (C), and DLL4 and CD16 (D). Scale bars: 20 μm (A); 50 μm (B–D). Con, control; HBV, hepatitis B virus; Pat., patient.
To further clarify the sinusoidal source of DLL4 expression, immunofluorescence costaining with DLL4 and α-smooth muscle actin, CD68, stabilin2, and CD16, markers for myofibroblasts, KCs, and liver sinusoidal endothelial cells (LSECs), respectively, was performed in four patients with high levels of DLL4 expression in sinusoids. Costaining demonstrated that DLL4 was not expressed in α-smooth muscle actin–positive activated HSCs in sinusoids of the four patients examined (Figure 1B). In two patients, DLL4 was localized in CD68-positive KCs, whereas in the remaining two patients, DLL4 was not expressed in CD68-positive cells (Figure 1C). In the latter two patients, DLL4-positive cells expressed CD16, a marker of LSEC (Figure 1D), although these DLL4-positive sinusoidal cells did not express stabilin2, another marker of LSECs (data not shown).
Intraperitoneal rDll4 Application Reduces Liver Fibrosis in CCl4-Exposed Mice
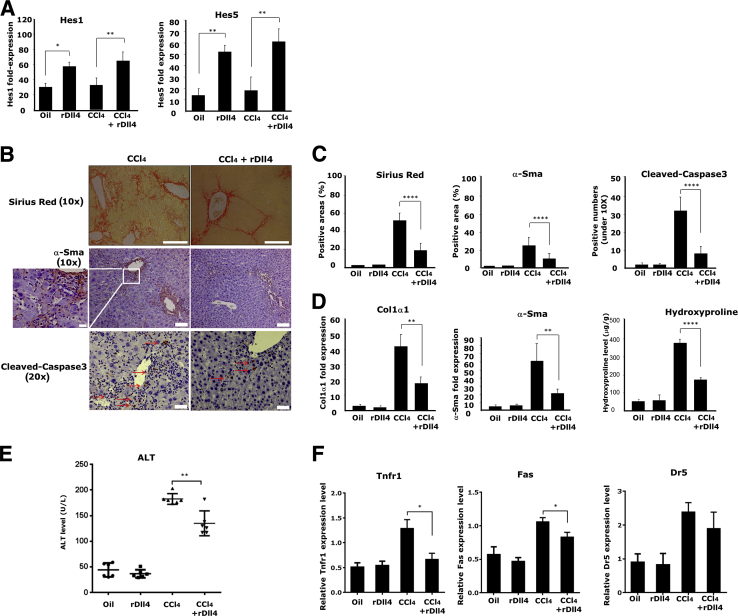
To assess the role that extracellular Dll4 plays in liver fibrogenesis, we i.p. administered murine recombinant Dll4 protein (rDll4) to mice exposed to CCl4 for 4 weeks. rDll4 application markedly induced hepatic expression of two primary Notch target genes, Hes1 and Hes5 (Figure 2A), indicating that Notch signaling pathways in the liver were activated after rDll4 application. Histologically, rDll4 application markedly reduced fibrosis and the number of α-smooth muscle actin (Sma) positive myofibroblasts (Figure 2, B and C). qPCR analyses further demonstrated that rDll4 application significantly decreased CCl4-induced hepatic transcripts of collagen 1α1 and α-Sma mRNA (Figure 2D). Consistent with these findings, CCl4-exposed livers had markedly up-regulated hydroxyproline content that was reduced in animals receiving rDll4 (Figure 2D). rDll4 injection alone had no discernible effect on the aforementioned indices when compared with control mice.
Figure 2.
Recombinant Delta-like ligand 4 (rDll4) attenuates carbon tetrachloride (CCl4)-induced liver fibrosis and hepatocyte apoptosis. A: mRNA levels of Hes1 and Hes5 in mouse liver tissues from different groups was quantified by real-time quantitative PCR (qPCR). B: Sirius red and α-smooth muscle actin (α-Sma) staining were used to assess fibrosis and myofibroblasts. Red arrows depict cleaved-caspase3 positive cells. Boxed area is highly magnified on the left to highlight α-Sma positive cells in CCl4-challenged mice. Immunohistochemistry for cleaved-caspase3 was performed to examine apoptosis of liver cells in different groups. C: Semiquantification for Masson, α-Sma, and cleaved-caspase3 staining was performed using Leica microscope imaging software. D: mRNA expression of collagen1α1 and α-Sma in mouse liver was quantified by qPCR. Hydroxyproline content assay was performed to evaluate collagen content in CCl4-challenged mice. E: Serum alanine aminotransferase (ALT) levels were measured in mice. F: mRNA expression of death receptors (Tnfr1, Fas, Trail-R/Dr5) was quantified using qPCR. Except staining and serum ALT measurement, other experiments were repeated at least three times. Data are presented as means ± SEM (A and C–E). n = 6 per group (A). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗∗P < 0.0001. Scale bars: 200 μm (B, top panels); 100 μm (B, middle panels); 50 μm (B, bottom panels); 20 μm (B, magnification).
rDll4 Application Reduces Hepatocyte Apoptosis and Expression of Death Receptors in CCl4-Exposed Mice
High rates of apoptosis is a feature of chronic liver diseases and closely associated with liver inflammation and fibrosis.27 Intraperitoneal administration of rDll4 in CCl4-exposed mice resulted in less high serum ALT levels, suggesting that rDll4 protects hepatocytes from CCl4-induced cell death (Figure 2E). IHC analysis revealed that cleaved-caspase3 was reduced in CCl4-challenged mice after rDll4 application (Figure 2, B and C). Furthermore, we quantified the transcripts of liver-specific death receptors Fas, tumor necrosis factor receptor 1 (Tnfr1), and tumor necrosis factor-related apoptosis-inducing ligand receptor (Trail-R/Dr5) in CCl4-challenged mice. Dll4 application resulted in significant down-regulation of Fas and Tnfr1 mRNA (Figure 2F). rDll4 injection alone did not show impact on the indices compared with control mice.
rDll4 Application Attenuates Liver Inflammation in CCl4-Challenged Mice
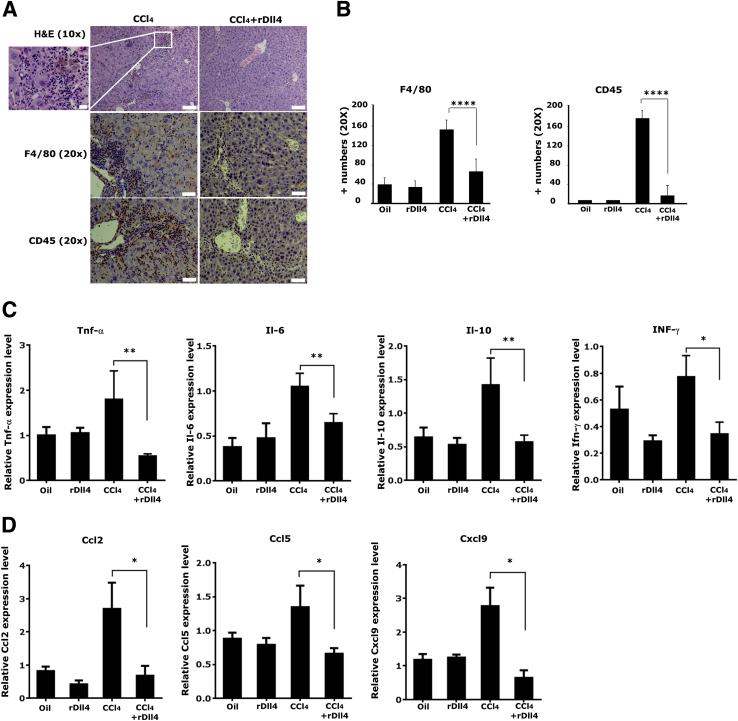
Next, the effect of rDll4 on inflammatory cell infiltration was investigated. Hematoxylin and eosin staining and IHC staining for F4/80 and CD45 (macrophage and leukocyte markers, respectively) showed that CCl4-induced inflammation was significantly reduced by rDll4 application (Figure 3, A and B).
Figure 3.
Recombinant Delta-like ligand 4 (rDll4) decreases carbon tetrachloride (CCl4) -induced inflammatory cell recruitment and cytokine/chemokine expression. A: Hematoxylin and eosin staining was performed to monitor inflammation in CCl4-induced liver damage. Boxed area is highly magnified on the left to highlight inflammatory cell infiltration in CCl4-challenged mice. F4/80 and CD45 staining were performed to visualize inflammatory cells. B: Semiquantification for F4/80 and CD45 staining was performed using Leica microscope imaging software. mRNA expression of inflammatory cytokines (C) and chemokines (D) was measured by real-time quantitative PCR (qPCR). qPCR measurements were repeated at least three times. Data are presented as means ± SEM (B and C). n = 6 in each group (A). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗∗P < 0.0001. Scale bars: 100 μm (A, top panels); 50 μm (A, middle and bottom panels); 20 μm (A, magnification).
Subsequently, we quantified the transcripts of inflammation-associated cytokines (Tnf-α, Ifn-γ, Il-1α, Il-2, Il-4, Il-6, Il-10, Il-12α, Il-13, and Tgf-β1) and chemokines (Ccl2, Ccl5, Ccl8, Cxcl9, and Cxcl10) by qPCR. rDll4 application markedly inhibited CCl4-induced mRNA expression of Tnf-α, Il-6, Il-10, and Ifn-γ (Figure 3C), whereas it did not significantly affect levels of Il-1α, Il-12α, and Tgf-β1 (data not shown). Expression levels of Il-2, Il-4, and Il-13 were low in all groups (data not shown). In addition to cytokines, rDll4 application significantly inhibited CCl4-induced transcripts of chemokines Ccl2, Ccl5, and Cxcl9 (Figure 3D). Transcript abundance of Ccl8 and Cxcl10 was not influenced by rDll4 (data not shown). rDll4 injection alone did not change the aforementioned parameters when compared with control mice.
rDll4 Administration Significantly Inhibits Chemokine Synthesis in KCs and HSCs
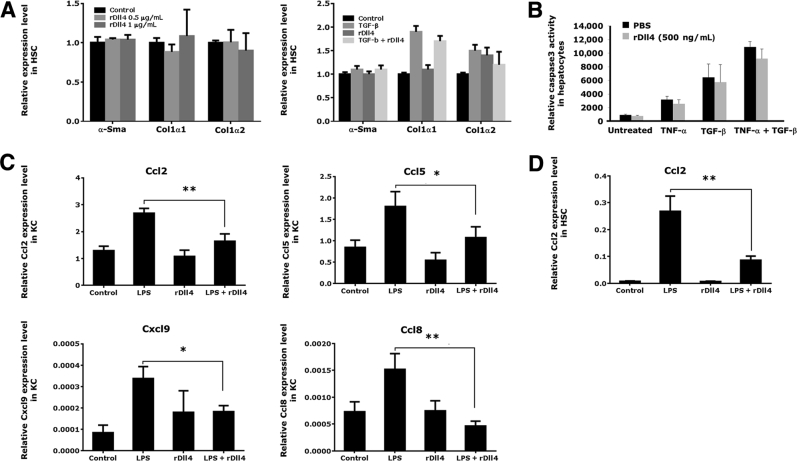
To clarify whether the antifibrotic effect of Dll4 is mediated through inhibition of HSC functions, we subjected primary mouse HSCs to rDll4. In HSCs 3 days after isolation, qPCR analyses showed that rDll4 incubation did not influence mRNA expression of α-Sma, collagen 1α1, and collagen 1α2. The Notch ligand also had no significant effect on TGF-β–induced mRNA expression of collagens 1α1 and 1α2 (Figure 4A).
Figure 4.
The effects of recombinant Delta-like ligand 4 (rDll4) in hepatic stellate cells (HSCs), hepatocytes, and Kupffer cells (KCs). A: rDll4 does not affect expression of collagen 1α1 and α-smooth muscle actin (α-Sma) in HSCs: real-time quantitative PCR (qPCR) was performed to quantify mRNA levels of collagen 1α1 and α-Sma in primary mouse HSCs (day 3 after primarily isolating from mouse liver) with or without 5 ng/mL transforming growth factor (Tgf)-β/or 500 ng/mL rDll4 incubation. B: rDll4 does not affect Tgf-β– and tumor necrosis factor (Tnf)-α–mediated hepatocyte apoptosis in vitro. Caspase3 activity was measured in primary mouse hepatocytes with or without 20 ng/mL TNF-α/TGF-β for 48 hours. C: Primary mouse KCs were treated with lipopolysaccharide (LPS) for 16 hours before they received rDll4 incubation for 6 hours. qPCR was performed to quantify mRNA levels of chemokines in KCs. D: JS-1 cells were treated with LPS for 16 hours before they received rDll4 incubation for 6 hours. qPCR was performed to quantify mRNA levels of chemokine ligand (Ccl)2 in JS-1 cells. All in vitro experiments were repeated at least three times. Data are presented as means ± SEM (A–D). ∗P < 0.05, ∗∗P < 0.01. PBS, phosphate-buffered saline.
Next, the effects of rDll4 on TNF-α or TGF-β–mediated apoptosis were investigated in primary mouse hepatocytes. Incubation with rDll4 did not prevent hepatocytes from TNF-α– or TGF-β–induced apoptosis, as quantified by caspase-3 activity (Figure 4B).
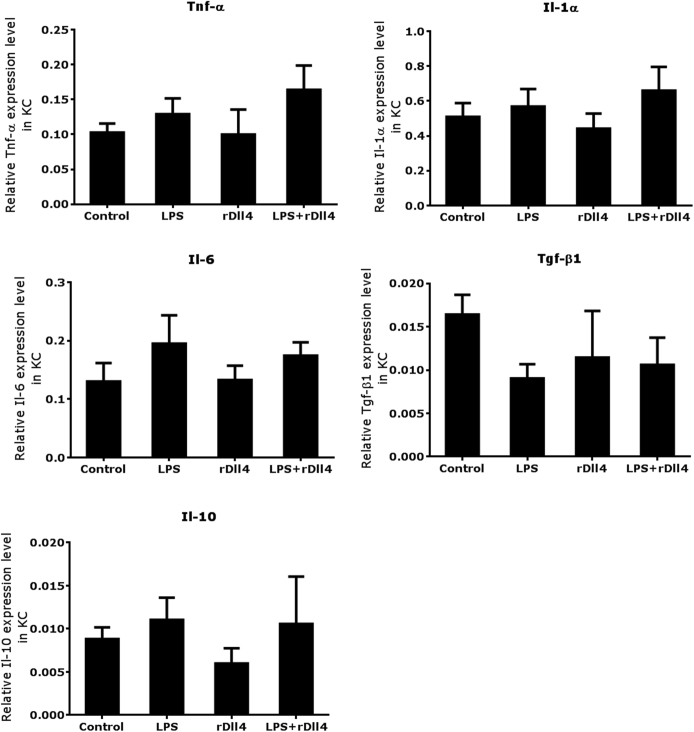
Furthermore, we examined whether rDll4 application regulates LPS-induced cytokine expression in primary mouse KCs. qPCR analyses demonstrated that LPS challenge for 16 hours slightly increased transcript numbers of Tnf-α, Il-1α, Il-6, and Il-10, but decreased transcripts of Tgf-β in KCs. Coincubation with rDll4 had no effect on expression levels of these cytokines in KCs (Supplemental Figure S1).
Subsequently, we determined the effect of rDll4 on LPS-induced chemokine expression in primary mouse KCs. KCs exposed to LPS for 16 hours increased transcript numbers of chemokines, including Ccl2, Ccl5, Ccl8, and Cxcl9. Administration of rDll4 significantly decreased mRNA expression of these chemokines in KCs (Figure 4C). Consistent with the finding in KCs, rDll4 incubation also reduced LPS-induced transcripts of Ccl2 in JS-1 cells (Figure 4D). However, in JS-1 cells, LPS-mediated expression of other chemokines (eg, Ccl5, Ccl8, and Cxcl9) was not altered by rDll4 administration (data not shown).
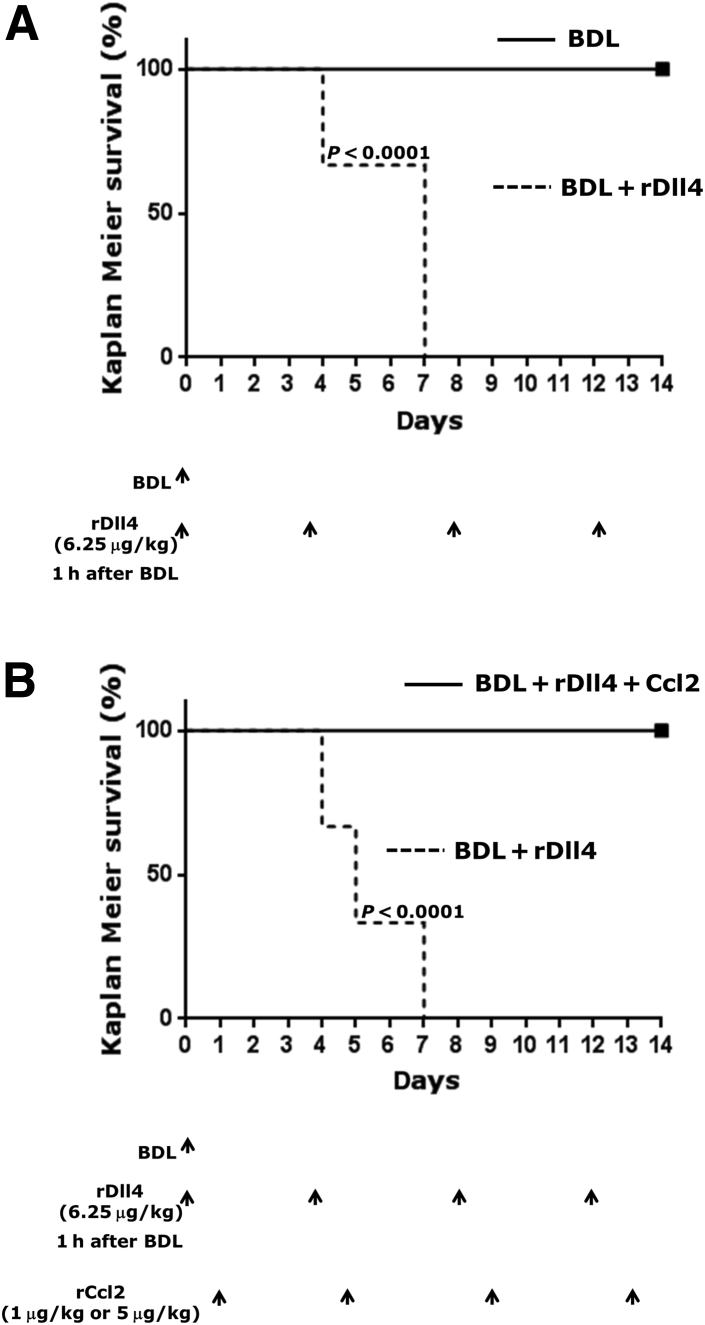
rDll4 Injection in BDL Mice Induces Massive Hepatic Necrosis and Results in Early Death
When effects of rDll4 were assessed in mice after BDL, all ligated mice died within 1 week after the second rDll4 application, whereas all BDL animals not receiving rDll4 survived. The outcome was confirmed in three separate experiments. The first rDll4 application was administered 1 hour after BDL operation and the second 4 days after surgery. A total of 18 BDL mice (six for each experiment) received rDll4, six died within 4 days after BDL and 12 on day 7 after surgery. All 18 BDL mice without rDll4 application survived (Figure 5A).
Figure 5.
Mortality of recombinant Delta-like ligand 4 (rDll4)-treated bile duct ligation (BDL) mice with or without recombinant chemokine ligand 2 (rCcl2) administration. A: All experimental mice received BDL and were divided into two groups: BDL and BDL + rDll4 (6.25 μg/kg). The first rDll4 application was administered 1 hour after BDL operation and the second 4 days after surgery. This experiment was repeated three times. Each experiment included 12 mice. B: Eighteen mice received BDL and were divided into three groups: BDL + rDll4 (6.25 μg/kg) and BDL + rDll4 + rCcl2 (rDll4: 6.25 μg/kg + rCcl2: 1 μg/kg; and rDll4: 6.25 μg/kg, + rCcl2: 5 μg/kg). Both rDll4 and rCcl2 were i.p. injected. The first rDll4 injection was performed 1 hour after mice receiving BDL. The rCcl2 injection was performed 1 day after rDll4 administration. Kaplan-Meier analyses show mortality of mice (P < 0.0001). Arrows depict the time points at which the mice received BDL or injection with rDll4 and/or rCcl2. n = 6 for each group (A and B).
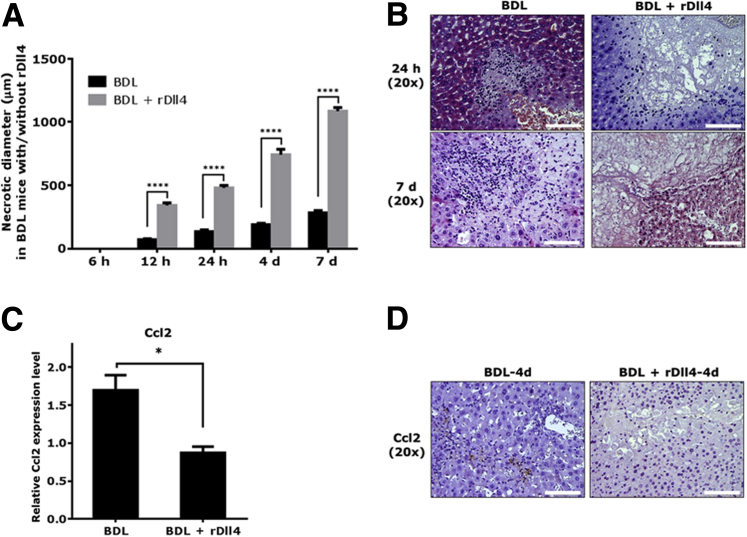
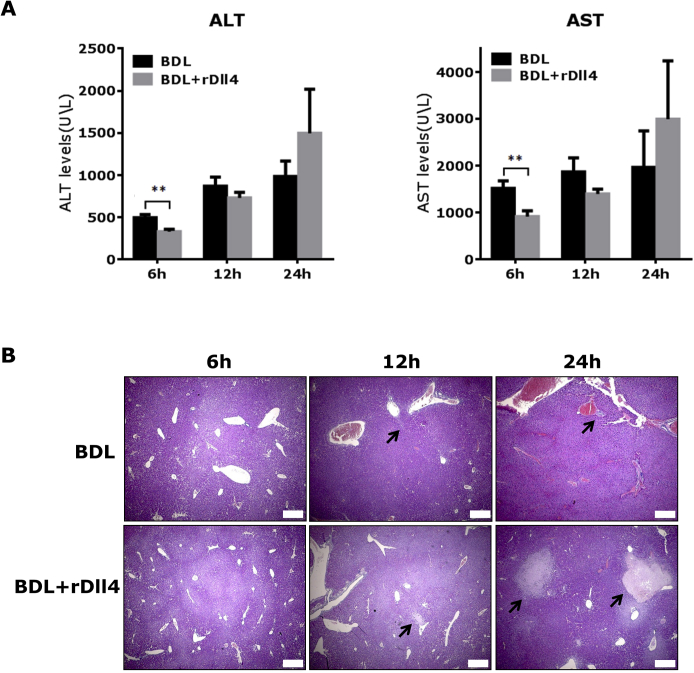
To explore the role that rDll4 plays in the early phase of liver damage after BDL, we assessed mouse liver damage at 6, 12, and 24 hours after BDL (n = 6 for each group). At 6 hours after BDL, serum ALT and AST levels were significantly lower in mice with rDll4 application compared with those without (P < 0.01) (Supplemental Figure S2A). Twenty-four hours after BDL, mice receiving rDll4 showed a trend toward higher serum ALT and AST levels when compared with control BDL animals (P > 0.05) (Supplemental Figure S2A), suggesting more serious hepatocyte damage in rDll4-receiving animals at this time point. Histologically, there were no conspicuous bile infarcts occurring in mouse livers at 6 hours after BDL, irrespective of rDll4 application (Supplemental Figure S2B). At 12 and 24 hours after BDL, both groups presented bile infarcts; however, the size of necrotic areas in mice receiving rDll4 was significantly larger when compared with those without rDll4 application (Supplemental Figure S2B and Figure 6A).
Figure 6.
Recombinant Delta-like ligand 4 (rDll4) rapidly causes massive hepatic necrosis in bile duct ligation (BDL) mice. A: The diameter of bile infarcts at different time points was calculated in BDL-administered mice with or without rDll4 injection. For each liver, diameters were measured in three areas with maximum bile infarct. B: Liver tissues from representative mice (BDL versus BDL + rDll4) showing bile infarct at 24 hours and at day 7 after BDL. Real-time quantitative PCR (C) and immunohistochemical (D) analyses reveal that rDll4 administration reduces expression of chemokine ligand (Ccl)2 in mice after 4 days of BDL. Data are presented as means ± SEM (A and C). n = 6 in each group (A). ∗P < 0.05, ∗∗∗∗P < 0.0001. Scale bars = 100 μm (B and D).
We calculated the maximal diameter of BDL-dependent bile infarcts in the two groups of mice at different time points after operation. The maximum diameters of bile infarcts were at: 12 hours, 80 μm; 24 hours, 150 μm; 4 days, 200 μm; and 7 days, 300 μm. In contrast, the maximum diameters of bile infarcts in rDll4-receiving mice after BDL were as follows: 12 hours, 360 μm; 24 hours, 500 μm; 4 days, 780 μm; and 7 days, >1000 μm (Figure 6A). At 4 and 7 days after BDL, rDll4-receiving mice presented with typical histomorphological features of massive hepatic necrosis [ie, diffuse panlobular (panacinar) and multilobular necrosis of >60% to 70%] (Figure 6B).
rDll4-Injected BDL Mice Show Reduced Inflammatory Cell Infiltrate in Areas of Bile Infarcts
In contrast to control BDL mice with cells infiltrating the bile infarcts, rDll4-receiving mice displayed less inflammatory cell infiltrate in areas of bile infarcts (Figure 6B). Subsequently, mRNA expression of chemokines (Ccl2, Ccl5, Ccl8, Cxcl9, and Cxcl10) was quantified in liver tissues from BDL mice with or without rDll4 administration by qPCR. Four days after BDL, rDll4 application significantly reduced mRNA expression of Ccl2 (Figure 6C), but not of the other tested chemokines (data not shown). IHC staining confirmed reduced expression of Ccl2 protein in BDL mice receiving rDll4 (Figure 6D).
Ccl2 Administration Rescues rDll4-Administered BDL Mice from Rapid Death
To explore the hypothesis that reduced expression of Ccl2 is relevant for the poor outcome of BDL mice after rDll4 injection, two dosages of rCcl2 (1 and 5 μg/kg) were applied i.p. to mice undergoing BDL 1 day after rDll4 application. In contrast to BDL mice receiving rDll4 only that died within 1 week after operation, all mice receiving rCcl2 survived until 2 weeks after BDL, the latest observation time point, even when these mice received rDll4 twice weekly (Figure 5B).
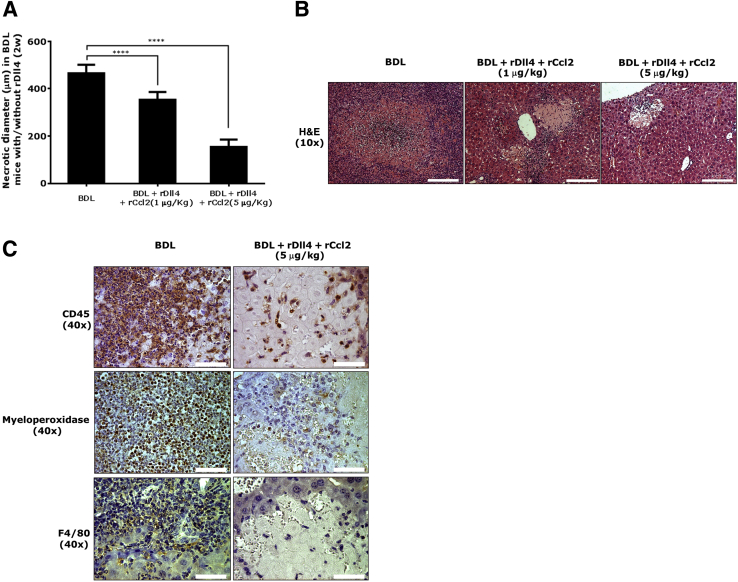
Histologically, the maximum necrotic diameter of BDL mice 2 weeks after operation reached 500 μm, whereas this parameter in the BDL mice receiving both rDll4 and rCcl2 (1 or 5 μg/kg) was 400 and 200 μm, respectively (Figure 7A). Compared with the early phase defined as less than a week after BDL, inflammatory cell infiltrates in liver tissues 2 weeks after BDL operation were more pronounced (Figure 6B and Figure 7B). IHC staining for CD45, myeloperoxidase (a neutrophil marker), and F4/80 positive cells showed that neutrophils were the predominant inflammatory cell type in the center of bile infarcts, whereas macrophages localized to the periphery of necrotic areas and portal tracts (Figure 7C). BDL mice receiving both rDll4 and rCcl2 exhibited smaller areas of bile infarcts as well as fewer inflammatory cells (Figure 7, B and C).
Figure 7.
Recombinant chemokine ligand (Ccl)2 rescue recombinant Delta-like ligand 4 (rDll4)-treated bile duct ligation (BDL) mice from death. A: The diameter of bile infarcts was calculated in mice 2 weeks after BDL with or without rDll4 and recombinant Ccl2 (rCcl2) application. For each liver, diameters were measured in three areas with maximum bile infarct. B: Hematoxylin and eosin (H&E) staining showing inflammatory cell infiltration in BDL mice with or without rDll4 and rCcl2 injection. C: Immunohistochemistry for CD45, myeloperoxidase, and F4/80 showing leukocyte, neutrophil, and macrophage infiltration in BDL mice with or without rDll4 and rCcl2 injection. Data are presented as means ± SEM. n = 6 in each group (A). ∗∗∗∗P < 0.0001. Scale bars: 200 μm (B); 50 μm (C).
Expression of DLL4 and CCL2 Is Inversely Correlated in HBV-Related Cirrhotic Livers
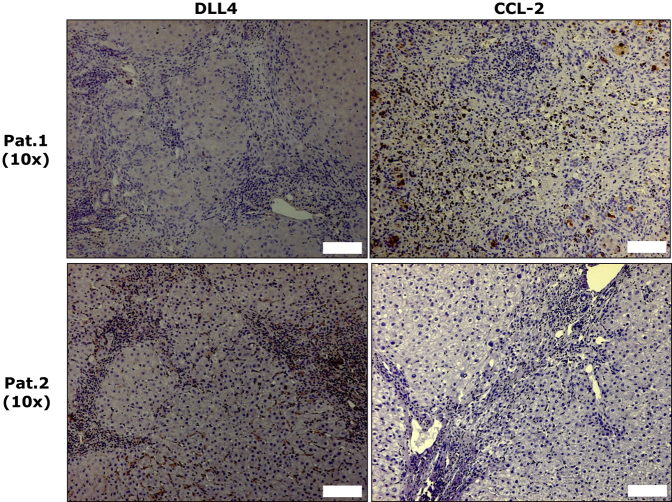
We also assessed the association between the abundance of DLL4 and CCL2 proteins in 26 liver specimens with chronic HBV infection (10 with cirrhosis and 16 with ACLF) by IHC. Among 10 cirrhotic patients, four with quiescent cirrhosis expressed neither DLL4 nor CCL2. In the remaining six patients with active cirrhosis (inflammatory grades ≥ 2), IHC analyses showed that in four expressing DLL4 protein CCL2 expression was lacking, whereas two with immunopositivity for CCL2 displayed DLL4 negative staining (Figure 8 and Supplemental Table S2). All liver tissues with ACLF displayed a strong CCL2 immunopositivity, but none of them had DLL4-positive cells (Supplemental Table S2).
Figure 8.
Delta-like ligand 4 (DLL4) had an inverse association with chemokine ligand (CCL)2 in liver tissues of hepatitis B virus (HBV)-related cirrhotic patients. Two representative patients show DLL4 and CCL2 expression in HBV-related cirrhotic patients. Scale bars = 100 μm. Pat., patient.
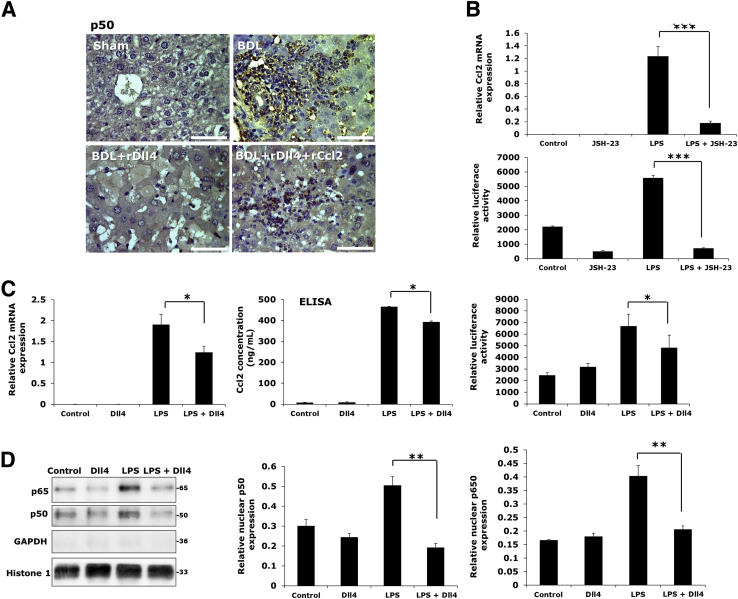
rDll4 Down-Regulates Ccl2 Expression through Inhibition of the NF-κB Pathway
Up-regulation of chemokines during an inflammatory response in tissue is mostly dependent on NF-κB activation.28 In mice after BDL, IHC confirmed up-regulation of p50 (NFκB1) in inflammatory cells, particularly in those surrounding necrotic areas (Figure 9A). The rDll4-receiving BDL mice had low p50 protein expression, whereas the BDL mice receiving both rDll4 and rCcl2 expressed high amounts of p50 in inflammatory cells (Figure 9A).
Figure 9.
Recombinant Delta-like ligand 4 (rDll4) inhibits expression of chemokine ligand (Ccl)2 by decreasing nuclear translocation of NF-κB. A: Immunohistochemistry for p50 (NF-κB1) in different groups of mice as indicated. Bile duct ligation (BDL) mice: One week after BDL. B: RAW264.7 cells were incubated with JSH-23, an NF-κB inhibitor, for 30 minutes before exposure to lipopolysaccharide (LPS) for 24 hours. Real-time quantitative PCR (qPCR) was performed to quantify mRNA levels of Ccl2 in RAW264.7 cells. RAW264.7 cells were transfected with the Ccl2-promoter-reporter construct using Viromer Red. Firefly luciferase activity was detected in RAW264.7 cells with indicated treatment. C: RAW264.7 cells were incubated with LPS for 16 hours before they received rDll4 incubation for 6 hours. qPCR was performed to quantify mRNA levels of chemokines in macrophages. Enzyme-linked immunosorbent assay (ELISA) was performed to detect levels of Ccl2 in cell supernatants. Firefly luciferase activity was detected to measure expression of Ccl2-promoter-reporter activity. D: Western blot was used to examine protein expression of p50 and p65 in RAW264.7 cells with LPS and/or rDll4 administration. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as loading control. Histone-1 was used as nuclear loading control. Quantification was performed with ImageJ software. All in vitro experiments were repeated at least three times. Data are presented as means ± SEM (B–D). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. Scale bars = 50 μm.
In vitro administration of JSH-23, an NF-κB inhibitor, in RAW264.7 macrophages 30 minutes before exposure to LPS remarkably inhibited LPS-induced mRNA expression of Ccl2 and luciferase activity of a Ccl2 promoter-reporter construct (Figure 9B). Consistent with the effects of rDll4 in KCs, rDll4 as well significantly lowered LPS-dependent up-regulation of transcript numbers, protein secretion, and promoter activity of Ccl2 in RAW264.7 macrophages (Figure 9C). The similarity of the effects seen with rDll4 or an NF-κB inhibitor on the LPS response in macrophages led to the hypothesis that rDll4 might exert its role via blockade of the NF-κB pathway. Western blot analyses showed that rDll4 incubation markedly decreased nuclear expression of p50 and p65 (Figure 9D), although rDll4 did not alter cytoplasmic levels of p65 (data not shown).
Discussion
This study initially analyzed the expression of Notch ligands and canonical receptors in patients with HBV-related cirrhosis and patients with hepatolithiasis. Given that inflammatory cells [eg, KCs, lymphocytes, or HSCs (key collagen-producing cells)] reside in liver sinusoids, we focused our investigation on the expression of Notch ligands/receptors in sinusoidal cells. Two Notch ligands, DLL4 and JAG1, and one receptor, NOTCH1, were expressed in sinusoidal cells of some patients with HBV-related cirrhosis. The results indicate that activation of the Notch signaling pathway in response to liver damage is dependent on disease etiology. Further analyses based on costaining revealed that DLL4 was expressed in KCs or LSECs, but not activated HSCs, in some cirrhotic patients with HBV infection, suggesting a close association between this Notch ligand and inflammation. Although it has been shown that disrupting Notch signaling affects experimental liver damage and fibrosis,14, 15 the role of individual Notch ligands in liver disease has not been examined to date. Furthermore, different Notch ligands (eg, JAG1 and DLL4) can elicit opposing effects in disease setting (eg, angiogenesis).29 Thus, assessing the effects of individual Notch ligands in different disease models will help hepatologists to understand the role of Notch signaling in liver disease. In this study, we focused on DLL4. We investigated the effects of Dll4 in two classic mouse models of liver damage induced by CCl4 injections and BDL in vivo and in HSCs, hepatocytes, and KCs in vitro.
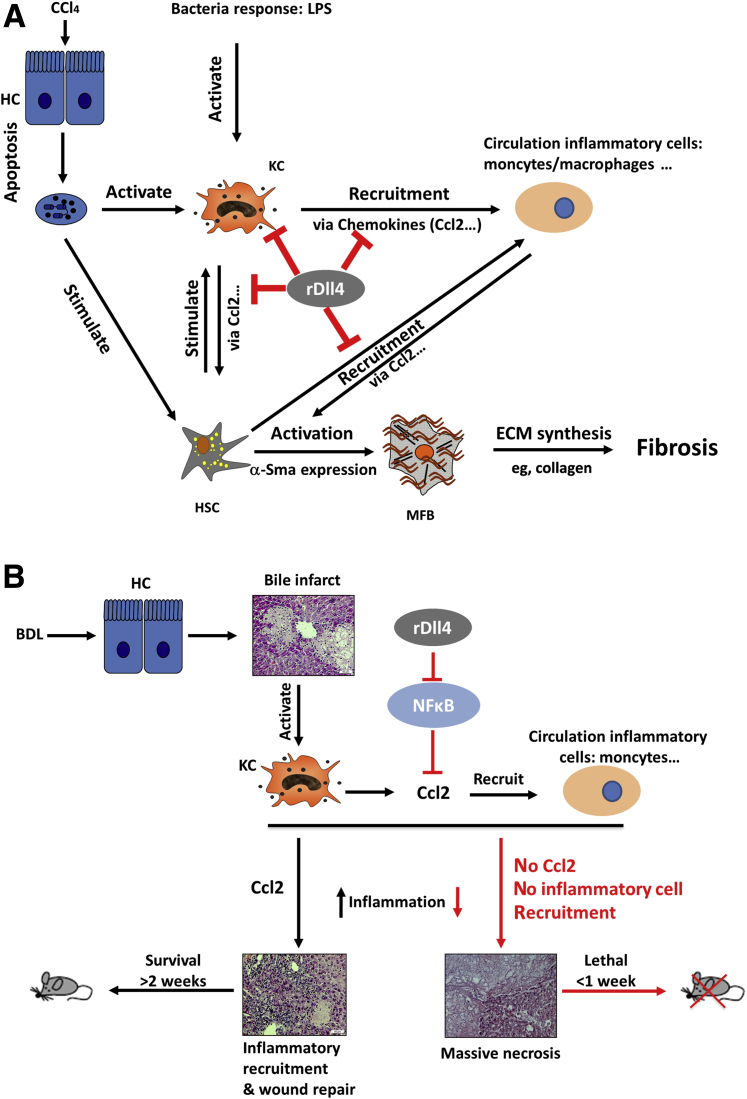
Similar to the situation in humans, Dll4 is not expressed in the normal mouse liver. CCl4 challenge for 4 weeks did not change Dll4 expression (data not shown). Thus, we adopted injection of recombinant Dll4 to assess whether ectopic expression of this Notch ligand influences liver damage. Interdependence between hepatocyte death, inflammation, and fibrosis determines the outcome of liver damage.30 In the chronic CCl4 mouse model, i.p. administration of rDll4 comprehensively ameliorated hepatocyte apoptosis, inflammation, and fibrosis. However, in vitro experiments on the basis of cultured hepatocytes, KCs, and HSCs were not completely consistent with these findings. In vitro, rDll4 neither influenced hepatocyte apoptosis, HSC activation, and collagen I synthesis, nor LPS-dependent cytokine expression in KCs. Interestingly, rDll4 significantly decreased LPS-dependent chemokine expression (eg, Ccl2) in both KCs and HSCs. The results suggest that inhibition of chemokine expression is the main mechanism by which rDll4 ameliorates liver damage in the chronic CCl4 mouse model (Figure 10A).
Figure 10.
Schemes summarizing the effects of recombinant Delta-like ligand 4 (rDll4) in carbon tetrachloride (CCl4)- (A) and bile duct ligation (BDL)- (B) dependent liver damage. Three major aspects are associated with mechanisms of liver fibrogenesis: death of hepatocytes, inflammation, hepatic stellate cell (HSC) activation, and collagen synthesis. In the chronic CCl4-induced fibrotic models, rDll4 comprehensively inhibited each of these. However, in vitro experiments clearly show that rDll4 has no direct effect on hepatocyte apoptosis, HSC activation, and collagen production. rDll4 remarkably inhibits chemokines, particularly chemokine ligand (Ccl)2, both in vivo and in vitro. In the bile duct ligation (BDL) model, rDll4 application has a fatal effect, visualized by the survival curves, through inhibition of Ccl2. The BDL mice receiving rDll4 are rescued when they are subjected to rCcl2 application. Thus, rDll4 improves liver fibrosis in chronic CCl4 models via inhibition of chemokines. HC, hepatocyte; HSC, hepatic stellate cells; KC, Kupffer cell; LPS, lipopolysaccharide; MFB, myofibroblasts.
We also assessed the effects of rDll4 administration after BDL. Without bile leakage or rupture of a biliary cyst, mice survive BDL for at least 2 weeks, although the bile infarct sizes increase over time.31 The maximum diameters of bile infarcts reach approximately 500 μm at 2 weeks after surgery. At this time point, a large number of inflammatory cells (eg, neutrophils and macrophages) infiltrate necrotic areas, engulf hepatocyte debris, and confine injury. Meanwhile, biliary liver fibrosis has been firmly established. In line with patients with hepatolithiasis, Dll4 expression is undetectable in these mice (data not shown). Thus, we examined the effects of rDll4 administrations in this model. Unexpectedly, three successive BDL experiments resulted in the same outcome with rapid death of the animals because of development of massive hepatic necrosis after two rDll4 applications. As observed in the CCl4 model, rDll4 application strongly inhibited the expression of Ccl2 after BDL. Histological analyses revealed that few inflammatory cells were recruited to areas of bile infarcts. Thus, we speculated that the absence of Ccl2 diminishes inflammatory cell recruitment to a degree that allows bile infarcts to quickly develop into unrestricted hepatic massive necrosis and liver failure. This hypothesis was confirmed when we performed a rescue experiment by injecting recombinant Ccl2 into rDll4-treated BDL mice: all rDll4-receiving mice survived for at least 2 weeks after BDL. These results suggest that the lethal effect of rDll4 in BDL mice is mediated by Ccl2 inhibition (Figure 10B).
Our data highlight the key role of etiology in determining the outcome of intervention: rDll4 inhibits Ccl2 expression in both models. But although in the CCl4 model, rDll4-mediated chemokine inhibition decreases inflammatory cell infiltration and thus attenuates hepatocyte death and fibrosis, the opposite is true in the BDL model. Herein, limiting bile infarcts requires massive recruitment of inflammatory cells to clear the debris of dead liver cells. Ccl2 inhibition by rDll4 blocks recruitment of circulating inflammatory cells into necrotic areas. Consequently, bile infarcts progress into massive hepatic necrosis. Given the discrepancies between various animal models and patients with chronic liver disease, whether and how DLL4 interacts with chemokines (eg, CCL2) in the human liver is still not clear.
ACLF is a newly recognized disease entity and characterized by acute deterioration of hepatic functions, multiple-organ failure, and deranged systemic inflammatory responses.32, 33, 34 In the progress of ACLF, recruitment of circulating monocytes and macrophages by chemokines, particularly CCL2, is a key event underlying systemic cytokine storm.35 In this study, liver tissue specimens from 16 patients with HBV-related ACLF were included. All patients demonstrated high CCL2 protein expression in the liver, whereas no DLL4 expression was found. Compared with ACLF patients, HBV patients with compensated cirrhosis usually had DLL4 expression in the liver. The expression levels of DLL4 were associated with the degree of inflammation (Supplemental Table S2). It would be interesting to know whether an excessive level of CCL2 induced by loss of DLL4 is a critical event in the formation of the cytokine storm in ACLF patients. The underlying molecular mechanisms are being investigated.
The role of Dll4 in mediating chemokine expression has previously been reported in other organs.36, 37, 38, 39 Results are conflicting because of different experimental conditions.36, 37, 38, 39 So far, it is not clear how Dll4 regulates chemokine expression in liver disease. The present study shows that rDll4 has an inhibitory effect on chemokine expression in KCs in vitro, inhibiting Ccl2, Ccl5, Ccl8, and Cxcl9. However, rDll4-mediated inhibition of chemokine expression in vivo is etiology dependent: rDll4 application significantly inhibits mRNA expression of Ccl2, Ccl5, Ccl8, and Cxcl9 in CCl4 challenged mice, whereas rDll4 merely decreased Ccl2 expression after BDL. In addition, the effect of rDll4 on cultured HSCs was selective: only Ccl2 expression was blocked by rDll4 after LPS exposure. Considering that NF-κB is a key inflammatory mediator,28 we analyzed NF-κB expression in the BDL model and its link with Dll4 and Ccl2 in vitro. It has been well recognized that NF-κB activation results in the up-regulation of chemokines.28 In the BDL model, Ccl2 expression is associated with activation of NF-κB and inflammatory cell infiltration. Application of rDll4 in BDL mice inhibited expression of Ccl2, activation of NF-κB, and inflammatory cell infiltration. Application of rCcl2 restored activation of NF-κB and inflammatory cell infiltration in BDL mice receiving rDll4 injection. In vitro, both rDll4 and JSH-23, an NF-κB inhibitor, remarkably inhibited LPS-induced expression of Ccl2 in macrophages. These results indicate a close link between expression of Ccl2 and activation of NF-κB. Thus, we speculated that rDll4 might inhibit expression of Ccl2 through interference with NF-κB activation. The hypothesis was supported by Western blot analyses: rDll4 decreased nuclear translocation of NF-κB proteins p50 and p65 in macrophages.
A similar finding has been reported for inflammatory diseases in the kidney, where genetic ablation of Notch3 resulted in diminished damage and lack of NF-κB activation.40, 41 Physical contact between cells induces the Notch signaling pathway. In liver sinusoids, KCs, LSECs, HSCs, lymphocytes, and hepatocytes contact each other and have a capacity to induce DLL4 expression in some disease setting. In this study, we found that DLL4 was expressed in either KCs or LSECs in some patients with HBV-related cirrhosis. We also found that only NOTCH1 was expressed in sinusoids of these patients. It is unclear whether NOTCH1 mediates Dll4 signaling in these damaged livers.
Taken together, Dll4 modulates liver damage via inhibition of chemokine expression. Administration of rDll4 results in different outcomes depending on the etiology of liver damage. Attenuating chemokine expression by rDll4 application results in compelling anti-inflammatory/antifibrotic effects in CCl4-dependent chronic liver damage, whereas rDll4-mediated Ccl2 inhibition incites massive hepatic necrosis in BDL mice via disrupting recruitment of inflammatory cells. Loss of DLL4 may be associated with CCL2-mediated cytokine storm in ACLF patients.
Our study shows how, with due knowledge of the intricate roles receptors and ligands play in the respective tissue, the Notch pathway offers intriguing points of organ- and disease-specific interference. It is conceivable that therapeutically triggered light up-regulation of DLL4 (eg, by an interacting, ligand-type small molecule) might stop the progression of liver fibrosis toward cirrhosis if removal of the damaging agent is not an option. Alternatively, suppressing the cytokine storm by administration of rDLL4 during ACLF might reduce the concomitant massive hepatic necrosis and thereby rescue the liver in some patients. Up-regulation of Jag1 has recently been shown to rescue the Duchenne muscular dystrophy phenotype in a canine model, offering a new path of treating this devastating condition.6 We anticipate that these results will encourage activities to design small molecules that affect Notch signaling with the ultimate aim of etiology-guided molecular medicine.
Acknowledgments
We thank Drs. Scott L. Friedman and Astrid Schmieder for the JS-1 and RAW 264.7 cell lines, respectively.
H.-L.W. conceived and designed the study; B.D., Q.L., S.M., H.L., J.L., and H.-L.W. collected patient data, performed experiments, and analyzed and interpreted data; Z.S., C.X., C.Y., Y.L., B.D., Q.L., S.M., H.L., P.R.M., and H.-L.W. collected animal model data, performed experiments, and analyzed and interpreted data; Y.L., B.D., J.H., O.P., T.F., C.M., I.I., P.R.M., A.M., C.S.-G., B.G., and H.-L.W. collected in vitro data, performed experiments, and analyzed and interpreted data; H.-L.W. wrote the manuscript; Y.L., R.L., I.I., V.Z., F.L., P.R.M., H.L., P.t.D., H.G.A., J.L., B.G., M.P.E., S.D., Y.L., and H.-L.W. edited the manuscript.
Footnotes
Supported by the Returned Overseas Chinese Scholars, State Education Ministry (Starting Research Foundation for the Returned Overseas Chinese Scholars, J20050337491010-G50523), People's Republic of China (H.-L.W.), Federal Ministry of Education and Research Virtual Liver and FP7 Marie Curie International Initial Training Networks in the Liver (P.t.D. and S.D.), SFB854/TP01, Me1365/7-2 and 9-1 (P.R.M.), Germany/China (German Academic Exchange Service — China Scholarship Council) Joint Research Program 2010/2011 (Z.S., C.X., Y.Li, C.M., S.D.), the intramural program of National Institute on Alcohol Abuse and Alcoholism, NIH (B.G.), the National Key Basic Research and Development Program of China-973 Program (2012CB524905; Y.Li), National Science and technology support program of China (2012BAI06B04; Y.Li), Zhejiang provincial laboratory animal science and technology program of China (2011C37088; Z.S.), German Egyptian Research Long-Term Scholarship of DAAD/Egypt government (B.D.), and Federal Ministry of Education and Research – Virtual Liver Network (S.D.).
Z.S, Y.Liu, and B.D. contributed equally to this work.
Y.Li and H.-L.W. contributed equally to this work as senior authors.
Disclosures: None declared.
Current address of Y.Liu, Boehringer Ingelheim Pharma GmbH & Co. KG, Biberach, Germany.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2016.03.010.
Contributor Information
Youming Li, Email: zlym@zju.edu.cn.
Hong-Lei Weng, Email: honglei.weng@medma.uni-heidelberg.de.
Supplemental Data
Supplemental Figure S1.
rDll4 does not alter lipopolysaccharide (LPS)-dependent cytokine expression in Kupffer cells (KCs). Primary mouse KCs were treated with LPS for 16 hours before they were incubated with rDll4 for 6 hours. Real-time quantitative PCR was performed to quantify mRNA levels of cytokines (Tnf-α, Il-1α, Il-6, Tgf-β1, and Il-10) in KCs. These results were confirmed in three independent experiments. Data are presented as means ± SEM.
Supplemental Figure S2.
The role of rDll4 in the early phase of liver damage after bile duct ligation (BDL). A: Serum alanine aminotransferase (ALT) and asparate aminotransferase (AST) levels were quantified in two groups of BDL mice with or without rDll4 application at different time points after BDL surgery. B: Representative images of hematoxylin and eosin staining show bile infarcts in the two groups of mice at 6, 12, and 24 hours after surgery. Arrows highlight bile infarcts. Data are presented as means ± SEM (A). n = 6 for each group (A). ∗∗P < 0.01. Scale bars = 250 μm.
References
- 1.Miele L. Notch signaling. Clin Cancer Res. 2006;12:1074–1079. doi: 10.1158/1078-0432.CCR-05-2570. [DOI] [PubMed] [Google Scholar]
- 2.Geisler F., Strazzabosco M. Emerging roles of Notch signaling in liver disease. Hepatology. 2015;61:382–392. doi: 10.1002/hep.27268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morell C.M., Fiorotto R., Fabris L., Strazzabosco M. Notch signalling beyond liver development: emerging concepts in liver repair and oncogenesis. Clin Res Hepatol Gastroenterol. 2013;37:447–454. doi: 10.1016/j.clinre.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Rauen T., Raffetseder U., Frye B.C., Djudjaj S., Muhlenberg P.J., Eitner F., Lendahl U., Bernhagen J., Dooley S., Mertens P.R. YB-1 acts as a ligand for Notch-3 receptors and modulates receptor activation. J Biol Chem. 2009;284:26928–26940. doi: 10.1074/jbc.M109.046599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kopan R., Ilagan M.X. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vieira N.M., Elvers I., Alexander M.S., Moreira Y.B., Eran A., Gomes J.P., Marshall J.L., Karlsson E.K., Verjovski-Almeida S., Lindblad-Toh K., Kunkel L.M., Zatz M. Jagged 1 rescues the Duchenne muscular dystrophy phenotype. Cell. 2015;163:1204–1213. doi: 10.1016/j.cell.2015.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L., Krantz I.D., Deng Y., Genin A., Banta A.B., Collins C.C., Qi M., Trask B.J., Kuo W.L., Cochran J., Costa T., Pierpont M.E., Rand E.B., Piccoli D.A., Hood L., Spinner N.B. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 8.Oda T., Elkahloun A.G., Pike B.L., Okajima K., Krantz I.D., Genin A., Piccoli D.A., Meltzer P.S., Spinner N.B., Collins F.S., Chandrasekharappa S.C. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- 9.Fabris L., Cadamuro M., Guido M., Spirli C., Fiorotto R., Colledan M., Torre G., Alberti D., Sonzogni A., Okolicsanyi L., Strazzabosco M. Analysis of liver repair mechanisms in Alagille syndrome and biliary atresia reveals a role for notch signaling. Am J Pathol. 2007;171:641–653. doi: 10.2353/ajpath.2007.070073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nijjar S.S., Crosby H.A., Wallace L., Hubscher S.G., Strain A.J. Notch receptor expression in adult human liver: a possible role in bile duct formation and hepatic neovascularization. Hepatology. 2001;34:1184–1192. doi: 10.1053/jhep.2001.29399. [DOI] [PubMed] [Google Scholar]
- 11.Nijjar S.S., Wallace L., Crosby H.A., Hubscher S.G., Strain A.J. Altered Notch ligand expression in human liver disease: further evidence for a role of the Notch signaling pathway in hepatic neovascularization and biliary ductular defects. Am J Pathol. 2002;160:1695–1703. doi: 10.1016/S0002-9440(10)61116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boulter L., Govaere O., Bird T.G., Radulescu S., Ramachandran P., Pellicoro A., Ridgway R.A., Seo S.S., Spee B., Van Rooijen N., Sansom O.J., Iredale J.P., Lowell S., Roskams T., Forbes S.J. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med. 2012;18:572–579. doi: 10.1038/nm.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spee B., Carpino G., Schotanus B.A., Katoonizadeh A., Vander Borght S., Gaudio E., Roskams T. Characterisation of the liver progenitor cell niche in liver diseases: potential involvement of Wnt and Notch signalling. Gut. 2010;59:247–257. doi: 10.1136/gut.2009.188367. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y., Zheng S., Qi D., Guo J., Zhang S., Weng Z. Inhibition of Notch signaling by a gamma-secretase inhibitor attenuates hepatic fibrosis in rats. PLoS One. 2012;7:e46512. doi: 10.1371/journal.pone.0046512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He F., Guo F.C., Li Z., Yu H.C., Ma P.F., Zhao J.L., Feng L., Li W.N., Liu X.W., Qin H.Y., Dou K.F., Han H. Myeloid-specific disruption of RBP-J ameliorates hepatic fibrosis by attenuating inflammation through cylindromatosis in mice. Hepatology. 2015;61:303–314. doi: 10.1002/hep.27394. [DOI] [PubMed] [Google Scholar]
- 16.Dill M.T., Duong F.H., Vogt J.E., Bibert S., Bochud P.Y., Terracciano L., Papassotiropoulos A., Roth V., Heim M.H. Interferon-induced gene expression is a stronger predictor of treatment response than IL28B genotype in patients with hepatitis C. Gastroenterology. 2011;140:1021–1031. doi: 10.1053/j.gastro.2010.11.039. [DOI] [PubMed] [Google Scholar]
- 17.van Es J.H., van Gijn M.E., Riccio O., van den Born M., Vooijs M., Begthel H., Cozijnsen M., Robine S., Winton D.J., Radtke F., Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 18.Yan M., Callahan C.A., Beyer J.C., Allamneni K.P., Zhang G., Ridgway J.B., Niessen K., Plowman G.D. Chronic DLL4 blockade induces vascular neoplasms. Nature. 2010;463:E6–E7. doi: 10.1038/nature08751. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y., Meyer C., Muller A., Herweck F., Li Q., Mullenbach R., Mertens P.R., Dooley S., Weng H.L. IL-13 induces connective tissue growth factor in rat hepatic stellate cells via TGF-beta-independent Smad signaling. J Immunol. 2011;187:2814–2823. doi: 10.4049/jimmunol.1003260. [DOI] [PubMed] [Google Scholar]
- 20.Schäfer S., Zerbe O., Gressner A.M. The synthesis of proteoglycans in fat-storing cells of rat liver. Hepatology. 1987;7:680–687. doi: 10.1002/hep.1840070411. [DOI] [PubMed] [Google Scholar]
- 21.Hu J., Srivastava K., Wieland M., Runge A., Mogler C., Besemfelder E., Terhardt D., Vogel M.J., Cao L., Korn C., Bartels S., Thomas M., Augustin H.G. Endothelial cell-derived angiopoietin-2 controls liver regeneration as a spatiotemporal rheostat. Science. 2014;343:416–419. doi: 10.1126/science.1244880. [DOI] [PubMed] [Google Scholar]
- 22.Guo J., Loke J., Zheng F., Hong F., Yea S., Fukata M., Tarocchi M., Abar O.T., Huang H., Sninsky J.J., Friedman S.L. Functional linkage of cirrhosis-predictive single nucleotide polymorphisms of Toll-like receptor 4 to hepatic stellate cell responses. Hepatology. 2009;49:960–968. doi: 10.1002/hep.22697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canbay A., Higuchi H., Bronk S.F., Taniai M., Sebo T.J., Gores G.J. Fas enhances fibrogenesis in the bile duct ligated mouse: a link between apoptosis and fibrosis. Gastroenterology. 2002;123:1323–1330. doi: 10.1053/gast.2002.35953. [DOI] [PubMed] [Google Scholar]
- 24.Weng H.L., Liu Y., Chen J.L., Huang T., Xu L.J., Godoy P., Hu J.H., Zhou C., Stickel F., Marx A., Bohle R.M., Zimmer V., Lammert F., Mueller S., Gigou M., Samuel D., Mertens P.R., Singer M.V., Seitz H.K., Dooley S. The etiology of liver damage imparts cytokines transforming growth factor beta1 or interleukin-13 as driving forces in fibrogenesis. Hepatology. 2009;50:230–243. doi: 10.1002/hep.22934. [DOI] [PubMed] [Google Scholar]
- 25.Jamall I.S., Finelli V.N., Que Hee S.S. A simple method to determine nanogram levels of 4-hydroxyproline in biological tissues. Anal Biochem. 1981;112:70–75. doi: 10.1016/0003-2697(81)90261-x. [DOI] [PubMed] [Google Scholar]
- 26.Kok S.H., Hong C.Y., Kuo M.Y., Wang C.C., Hou K.L., Lin Y.T., Galson D.L., Lin S.K. Oncostatin M-induced CCL2 transcription in osteoblastic cells is mediated by multiple levels of STAT-1 and STAT-3 signaling: an implication for the pathogenesis of arthritis. Arthritis Rheum. 2009;60:1451–1462. doi: 10.1002/art.24452. [DOI] [PubMed] [Google Scholar]
- 27.Malhi H., Gores G.J. Cellular and molecular mechanisms of liver injury. Gastroenterology. 2008;134:1641–1654. doi: 10.1053/j.gastro.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosh S., Hayden M.S. New regulators of NF-kappaB in inflammation. Nat Rev Immunol. 2008;8:837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- 29.Benedito R., Roca C., Sorensen I., Adams S., Gossler A., Fruttiger M., Adams R.H. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137:1124–1135. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 30.Wynn T.A. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y., Meyer C., Xu C., Weng H., Hellerbrand C., ten Dijke P., Dooley S. Animal models of chronic liver diseases. Am J Physiol Gastrointest Liver Physiol. 2013;304:G449–G468. doi: 10.1152/ajpgi.00199.2012. [DOI] [PubMed] [Google Scholar]
- 32.Arroyo V., Moreau R., Jalan R., Gines P. Acute-on-chronic liver failure: a new syndrome that will re-classify cirrhosis. J Hepatol. 2015;62:S131–S143. doi: 10.1016/j.jhep.2014.11.045. [DOI] [PubMed] [Google Scholar]
- 33.Moreau R., Jalan R., Gines P., Pavesi M., Angeli P., Cordoba J., Durand F., Gustot T., Saliba F., Domenicali M., Gerbes A., Wendon J., Alessandria C., Laleman W., Zeuzem S., Trebicka J., Bernardi M., Arroyo V. Acute-on-chronic liver failure is a distinct syndrome that develops in patients with acute decompensation of cirrhosis. Gastroenterology. 2013;144:1426–1437. doi: 10.1053/j.gastro.2013.02.042. e1429. [DOI] [PubMed] [Google Scholar]
- 34.Li H., Xia Q., Zeng B., Li S.T., Liu H., Li Q., Li J., Yang S.Y., Dong X.J., Gao T., Munker S., Liu Y., Liebe R., Xue F., Li Q.G., Chen X.S., Liu Q., Zeng H., Wang J.Y., Xie Q., Meng Q.H., Wang J.F., Mertens P.R., Lammert F., Singer M.V., Dooley S., Ebert M.P., Qiu D.K., Wang T.L., Weng H.L. Submassive hepatic necrosis distinguishes HBV-associated acute on chronic liver failure from cirrhotic patients with acute decompensation. J Hepatol. 2015;63:50–59. doi: 10.1016/j.jhep.2015.01.029. [DOI] [PubMed] [Google Scholar]
- 35.Jalan R., Gines P., Olson J.C., Mookerjee R.P., Moreau R., Garcia-Tsao G., Arroyo V., Kamath P.S. Acute-on chronic liver failure. J Hepatol. 2012;57:1336–1348. doi: 10.1016/j.jhep.2012.06.026. [DOI] [PubMed] [Google Scholar]
- 36.Williams C.K., Segarra M., Sierra Mde L., Sainson R.C., Tosato G., Harris A.L. Regulation of CXCR4 by the Notch ligand delta-like 4 in endothelial cells. Cancer Res. 2008;68:1889–1895. doi: 10.1158/0008-5472.CAN-07-2181. [DOI] [PubMed] [Google Scholar]
- 37.Al Haj Zen A., Oikawa A., Bazan-Peregrino M., Meloni M., Emanueli C., Madeddu P. Inhibition of delta-like-4-mediated signaling impairs reparative angiogenesis after ischemia. Circ Res. 2010;107:283–293. doi: 10.1161/CIRCRESAHA.110.221663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reynolds N.D., Lukacs N.W., Long N., Karpus W.J. Delta-like ligand 4 regulates central nervous system T cell accumulation during experimental autoimmune encephalomyelitis. J Immunol. 2011;187:2803–2813. doi: 10.4049/jimmunol.1100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukuda D., Aikawa E., Swirski F.K., Novobrantseva T.I., Kotelianski V., Gorgun C.Z., Chudnovskiy A., Yamazaki H., Croce K., Weissleder R., Aster J.C., Hotamisligil G.S., Yagita H., Aikawa M. Notch ligand delta-like 4 blockade attenuates atherosclerosis and metabolic disorders. Proc Natl Acad Sci U S A. 2012;109:E1868–E1877. doi: 10.1073/pnas.1116889109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Djudjaj S., Chatziantoniou C., Raffetseder U., Guerrot D., Dussaule J.C., Boor P., Kerroch M., Hanssen L., Brandt S., Dittrich A., Ostendorf T., Floege J., Zhu C., Lindenmeyer M., Cohen C.D., Mertens P.R. Notch-3 receptor activation drives inflammation and fibrosis following tubulointerstitial kidney injury. J Pathol. 2012;228:286–299. doi: 10.1002/path.4076. [DOI] [PubMed] [Google Scholar]
- 41.El Machhour F., Keuylian Z., Kavvadas P., Dussaule J.C., Chatziantoniou C. Activation of Notch3 in glomeruli promotes the development of rapidly progressive renal disease. J Am Soc Nephrol. 2015;26:1561–1575. doi: 10.1681/ASN.2013090968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.