Abstract
Apurinic/apyrimidinic (AP) sites are constantly formed in cellular DNA due to instability of the glycosidic bond, particularly at purines and various oxidized, alkylated, or otherwise damaged nucleobases. AP sites are also generated by DNA glycosylases that initiate DNA base excision repair. These lesions represent a significant block to DNA replication and are extremely mutagenic. Some DNA glycosylases possess AP lyase activities that nick the DNA strand at the deoxyribose moiety via a β- or β,δ-elimination reaction. Various amines can incise AP sites via a similar mechanism, but this non-enzymatic cleavage typically requires high reagent concentrations. Herein, we describe a new class of small molecules that function at low micromolar concentrations as both β- and β,δ-elimination catalysts at AP sites. Structure-activity relationships have established several characteristics that appear to be necessary for the formation of an iminium ion intermediate that self-catalyzes the elimination at the deoxyribose ring.
Apurinic/apyrimidinic (AP) sites are common DNA lesions that occur naturally through deglycosylation of DNA, loss of unstable modified nucleobases, or as an intermediate in the base excision repair (BER) pathway1,2,3,4. If left unrepaired, AP sites can inhibit DNA replication and transcription and contribute to mutagenesis5,6,7,8. In the BER pathway, AP sites are generated by DNA glycosylases that hydrolyze the bond between the damaged base and the deoxyribose1,3,9. The newly-formed AP sites can be further processed by an AP endonuclease that cleaves the phosphodiester DNA backbone on the 5′ side of AP sites2,10. This creates a 3′-hydroxyl to enable initiation of gap-filling DNA repair synthesis.
Many DNA glycosylases, including human NEIL1, NEIL2, NEIL3, NTH1 and OGG1, possess AP lyase activities that catalyze either a β- or β,δ-elimination reaction at the deoxyribose moiety9,11,12,13,14. This reaction proceeds via the formation of a transient enzyme-DNA covalent intermediate and generates DNA breaks with a phospho-α, β-unsaturated aldehyde or phosphate group at the 3′-end. In addition to these enzyme-catalyzed reactions, non-enzymatic cleavage of DNA at AP sites occurs under alkaline conditions or elevated temperatures1,15,16 and in the presence of polyamines9,15,17,18,19, histones9,20, various nucleophilic peptides21,22,23, or photoactivated metalloinserters24. For many of these compounds, the strand scission has been shown to proceed through a β-elimination of an imine intermediate and is inhibited by the presence of methoxyamine25.
Due to the biologically significant role of AP sites, previous efforts have been made to engineer effective non-enzymatic AP lyase catalysts, such as the acridine-nucleobase dimers linked by a polyamine chain26. Although these so-called “artificial nucleases” appeared to be potent in cleavage of AP site-containing DNA in vitro, no reagents or drugs have been developed based on this design for use in vivo. Thus, the availability of small molecules that possess AP lyase activity remains largely an unmet need.
In conjunction with our investigations to identify inhibitors of DNA glycosylases, screens of small molecule libraries revealed that hNEIL1 was inhibited by several purine analoques27. To potentially expand the number of lead compounds, a library of selected kinase inhibitors28 (courtesy of F. Hoffmann-La Roche Inc., Dr. Paul Gillespie) was assayed as inhibitors of hNEIL1 or hOGG1, since many of these small molecules are composed of a core purine-like structure. Although no effective inhibition of either glycosylase was observed, several of these compounds showed an unexpected ability to efficiently cleave AP site-containing DNA. Herein we describe structure-activity and mechanistic studies of this new class of small molecule catalysts for DNA strand incision.
Results and Discussion
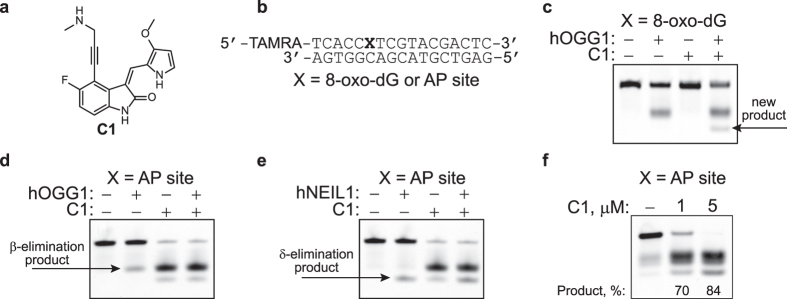
Compound C1 can cleave DNA containing AP site
The initial observation that suggested the presence of an unusual activity in a subset of the small molecules under study was made when C1 (Fig. 1a) was tested as a potential inhibitor of hOGG1. Specifically, addition of this compound to reactions of hOGG1 with a fluorescently-labeled duplex oligodeoxynucleotide containing a site-specific 8-oxo-dG adduct (Fig. 1b) resulted in an additional product that migrated faster than the β-elimination product; hOGG1 alone yielded only the β-elimination product (Fig. 1c). No nicked products were observed when 8-oxo-dG-containing DNA was incubated with C1 alone.
Figure 1. Non-enzymatic DNA cleavage at an AP site.
AP site-containing DNA was obtained by treating the corresponding dU-containing DNA with UDG. (a) Structure of a representative catalyst for DNA scission. (b) DNA substrates. (c) Scission of 8-oxo-dG-containing DNA (250 nM) in the presence of hOGG1 (100 nM) and C1 (10 μM). (d) Scission of AP site-containing DNA (250 nM) in the presence of hOGG1 (50 nM) and C1 (10 μM). (e) Scission of AP site-containing DNA (250 nM) in the presence of hNEIL1 (50 nM) and C1 (10 μM). (f) Scission of AP site-containing DNA (2 μM) by C1. The percent of products was corrected for the spontaneous cleavage. Reactions were carried out at 37 °C for 30 min (b–d) or 16 h (e).
Several mechanistic possibilities were hypothesized that could account for the formation of the new product. 1) In the presence of C1, hOGG1 had acquired the ability to catalyze a δ-elimination reaction in addition to its glycosylase and β-elimination AP lyase functions; 2) C1 could convert the β-elimination product to the δ-elimination product; and 3) AP sites formed in the glycosylase reaction, could serve as a substrate for a C1-catalyzed β,δ-elimination reaction. To address these possibilities, duplex DNA containing a single site-specific AP site was generated by treating the corresponding dU-containing DNA with uracil DNA glycosylase (UDG) (Fig. 1b) and reacted with C1 or hOGG1, either individually or in combination. As with the 8-oxo-dG-containing oligodeoxynucleotide, hOGG1 produced the expected β-elimination product, while the addition of C1 produced a mixture of two products (Fig. 1d). This product mixture was also formed when the AP-containing oligodeoxynucleotide was incubated with C1 alone. While the position of the slower migrating product band corresponded to the β-elimination product of hOGG1 (Fig. 1d), the faster migrating product band co-migrated with the known δ-elimination product of hNEIL1 (Fig. 1e). Thus, C1 promotes the β- and δ-elimination reactions at AP sites.
The cleavage reaction by C1 on AP site-containing DNA was concentration dependent (Fig. 1e and Supplementary Figure S1), with low micromolar concentrations being sufficient to observe the products. Similar to the previously designed “artificial nucleases”26, C1 showed turn-over catalysis on the DNA substrate: 1 pmol C1 generated ~1.4 pmol products in 16 h (Fig. 1f). Thus, the rate of incision by C1 on this DNA was ~1.5 × 10−3 min−1 or higher. The comparison of C1 with the currently available AP lyase reagents spermine17 and the KWKK peptide23 demonstrated that the newly-identified catalyst was at least 100-fold more efficient (Supplementary Figure S1).
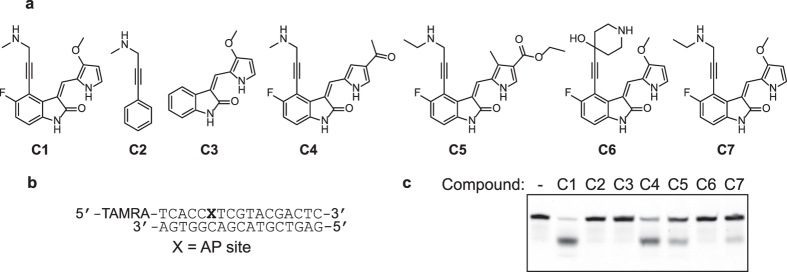
Several structural moieties in C1 are important for catalysis
Analogues of C1 were assayed to determine the contribution of specific structural features to the strand scission chemistry. We initially examined the ability of simple, commercially available amines lacking the indolinone-pyrrole moiety C2 (Fig. 2) and CS1-CS3 (Supplementary Figure S2) to incise DNA at an AP site. Formation of incision products, if any, was below the level of detection at 10 μM. Thus, the indolinone-pyrrole moiety is important for the reaction. It is possible that the indolinone-pyrrole subunit binds to the AP site29 or minor groove and positions the secondary amine to catalyze strand cleavage via a covalent intermediate with the AP site. Compounds C3 (Fig. 2) and CS4 (Supplementary Figure S2) that contained the indolinone-pyrrole moiety but lacked the secondary amine were also completely inactive. These data strongly suggested the role for the amine as the reactive functional group. The replacement of the methoxy group on the pyrrole by other substitutions, such as in C4 and C5 (Fig. 2), led to decreased activity. The reaction was also modulated by a substituent on the amino group, such that increasing the steric demands of the secondary amine decreased the amount of the product observed (compare C1, C6, and C7 (Fig. 2), and C4, CS5, and CS6 (Supplementary Figure S2)). These structure-activity analyses demonstrated that several of the individual moieties are necessary but not sufficient for cleavage, and that these moieties must act cooperatively to produce strand scission.
Figure 2. Structure-function analyses of the catalysts for DNA scission at an AP site.
(a) Structures of representative compounds. (b) DNA substrate. (c) Assay for the abilities of representative compounds (10 μM) to incise AP site-containing DNA (250 nM). Reactions were carried out at 37 °C for 30 in.
DNA structure modulates the C1-catalyzed cleavage
The importance of the indolinone-pyrrole moiety for the strand scission chemistry has suggested that the structure of the DNA substrate could affect the C1-mediated cleavage. To address these relationships, the initial rates of reaction were measured for a single-stranded DNA (the sequence as in Fig. 1a) and the corresponding double-stranded DNAs that contained either A, C, G, or T opposite the AP site. The homopolymeric oligodeoxynucleotide, 5′-TAMRA-(T)5-AP-(T)11-3′, was also examined as an unambiguous single-stranded DNA structure. These data demonstrated that although strand scission could occur in the context of single-stranded DNA, the double-stranded DNAs were the much preferred substrates for C1 (Fig. 3). It was also found that the nature of the base opposite the AP site modulated the catalysis, with the rate of AP site hydrolysis being faster opposite pyrimidines than purines. These observations are consistent with the proposal that the indolinone-pyrrole subunit interacts with DNA to occupy an empty space at the AP site. Notably, the initial rate measured for the double-stranded DNA with a C opposite the AP site, was very close to the rate observed for this substrate under conditions of limiting C1 concentration (Fig. 1f).
Figure 3. Rates of C1-catalyzed cleavage of AP site-containing DNA.
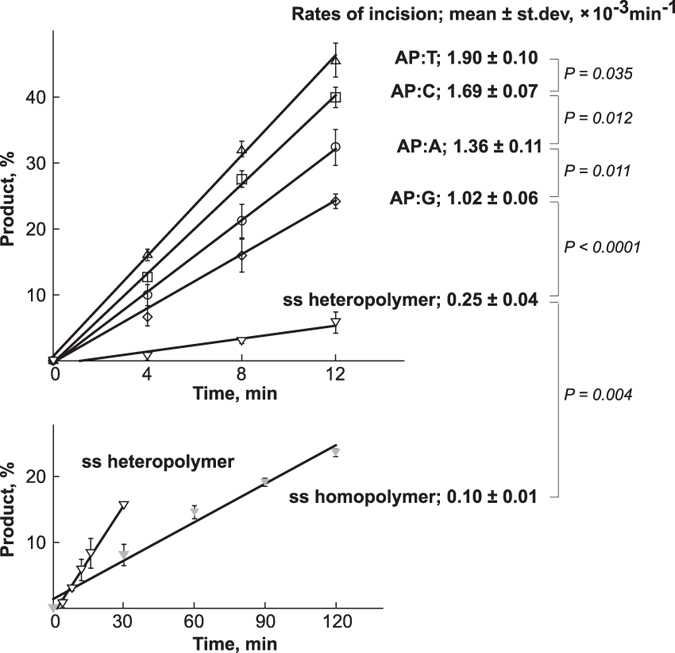
Reactions were carried out at 37 °C using 250 nM DNA and 5 μM C1. The mean initial rates with respective standard deviations were calculated from three independent experiments using KaleidaGraph 4.1 software (Synergy Software). The P values were calculated using Students’ t-test.
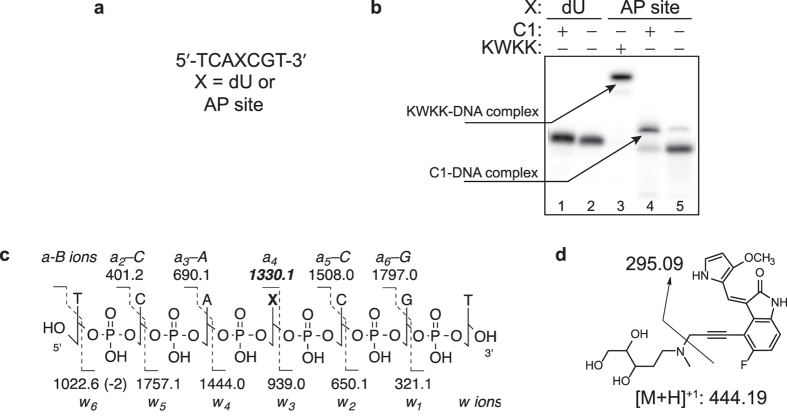
Cleavage of DNA at AP sites by C1 proceeds via an intermediate involving the secondary amine
To test for the covalent intermediate with the ring-opened, aldehydic form of the deoxyribose, an AP site-containing 32P-labeled oligodeoxynucleotide (Fig. 4a) was incubated with C1 in the presence of NaB(CN)H3. While this reductant slowly reacts with the AP lesion, it efficiently traps the imine- or iminium ion-conjugate. In a control reaction with the AP site-containing oligodeoxynucleotide and NaB(CN)H3 (Fig. 4b, lane 5), a small fraction (~3%) of the DNA manifested a decreased mobility. This low-abundance product is commonly observed in the trapping reactions23 and likely represents a complex with Tris molecules, as was previously shown in reactions with the malondialdehyde pyrimidopurinone DNA adduct30. In a positive control reaction using the lysine-tryptophan-lysine-lysine (KWKK) peptide23, the imine intermediate was trapped, as evidenced by the shift in DNA mobility (Fig. 4b, lane 3). In the presence of NaB(CN)H3 and C1, the majority of DNA (~80%) formed a complex that appeared as a species with decreased mobility (Fig. 4b, lane 4). The complex formation was not due to non-specific binding of C1 to DNA, since no shift was observed when the corresponding dU-containing oligodeoxynucleotide was tested under identical conditions (Fig. 4b, lane 1). These data were consistent with the hypothesis that the reagent was able to form a covalent iminium ion intermediate, which then positions a side group for a proton abstraction from the sugar ring.
Figure 4. Formation of an iminium ion intermediate between C1 and AP site.
(a) DNA substrates. (b) Cyanoborohydride trapping of a complex between C1 and AP site-containing DNA. (c) CID fragmentation of the reduced iminium ion intermediate. (d) Fragmentation of the reduced C1-deoxyribose conjugate after enzymatic digestion.
The reductive trapping of C1 was repeated using unlabeled AP site-containing oligodeoxynucleotide (Fig. 4a), and the product was analyzed by mass spectrometry (MS). The analysis revealed a mass consistent with the reduced DNA-C1 covalent complex (m/z 1134.64 for [M-2H]−2). Collision induced dissociation (CID) of this ion resulted in a complete set of a–B (Base) and w ions, consistent with the reduced iminium ion intermediate between C1 and the AP site (Supplementary Figure S3). In the a–B ion series, fragmentation of the C3′-O bond is normally accompanied by the neutral loss of the nucleobase. The reduced linkage between C1 and the AP site was expected to be less labile and as a result, we observed the a4 (m/z 1330.1) ion as well as the a4-B (m/z 1005.6). The oligodeoxynucleotide was also enzymatically digested and analyzed by MS (Fig. 4c and Supplementary Figure S4). A digestion product was observed with a mass consistent with the reduced C1-deoxyribose conjugate (m/z 444.19); fragmentation of this product ion gave a daughter ion with m/z 295.09, that resulted from the neutral loss of N-methyl amino-2-deoxyribitol (Fig. 4d and Supplementary Figure S4). This product was identical to that prepared from the reductive amination reaction of 2-deoxyribose and C1. These studies demonstrate that C1 cleaves AP site-containing DNA through covalent catalysis involving the secondary amine.
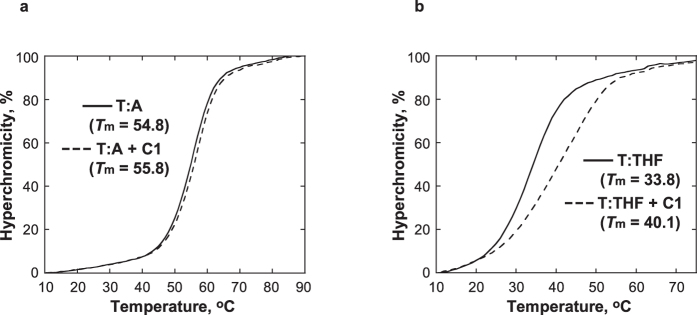
C1 increases the thermal stability of DNA containing an analogue of an AP site
The effect of C1 was studied on thermal stability of DNA containing a tetrahydrofuran (THF), structural analogue of an AP site that is incapable of undergoing the β-elimination reaction31. The Tm of the duplex DNA containing THF opposite T (T:THF) increased by 6.3 °C upon addition of 1 equivalent of C1, while the Tm of the control DNA containing a T:A pair was less affected and only increased by 1 °C (Fig. 5). While the melting curve of T:THF with C1 indicated thermal stabilization by C1, the presence of C1 also broadened the melting curve, indicating a less cooperative melting transition. Collectively, these observations suggest that C1 specifically binds to DNA at the AP site providing localized stabilization to the DNA helix. This is consistent with the proposed model of interactions between C1 and DNA in which the indolinone-pyrrole moiety occupies the space available at the AP site.
Figure 5. Modulation of DNA thermal stability by C1.
The melting temperature curves of T:A (a) and T:THF (b) were obtained using 5 μM DNA in 10 mM sodium phosphate buffer, pH 7.0, 100 mM NaCl, and 1 mM EDTA in the absence (solid line) and presence (dashed line) of 5 μM C1.
C1 has a higher affinity for DNA containing an AP site
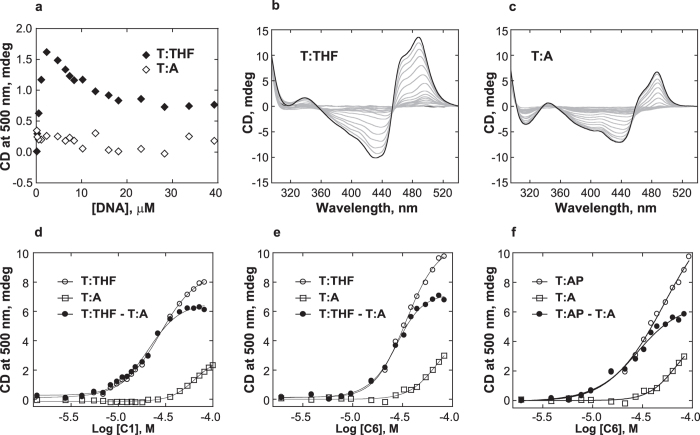
In order to examine the binding mode and affinity of C1 to AP site-containing DNA, circular dichroism (CD) analyses were conducted using both THF-containing and control T:A duplex oligodeoxynucleotides (Fig. 6). The induced CD (ICD) of C1 upon interaction with T:THF duplex was observed as a strong exciton signal, a bisignate shape with positive and negative bands relative to the absorption maximum of the free C1. This is generally indicative of the formation of dimeric or higher order complexes, either in a groove-binding or an external stacking-binding mode32. The titration of C1 with increasing T:THF concentrations (Fig. 6a and Supplementary Figure S5) revealed both non-specific and specific interactions. At the start of titration, C1 is in excess and non-specific interactions are favored. Upon addition of T:THF, the ICD at ~488 nm increased. At approximately 2 μM DNA concentration, the ICD band shifted to lower energy (~495 nm) and then decreased until the equivalence point and plateaued in excess of DNA indicating a specific interaction. Titration of C1 with the control T:A DNA (Fig. 6a and Supplementary Figure S5) did not exhibit the positive ICD around 495 nm indicating the absence of specific interactions. The forward titration of T:THF also exhibited evidence of non-specific and specific interactions (Fig. 6b). At low concentrations of C1 with DNA in excess, the ICD band was centered at ~495 nm (specific); as the concentration of C1 increased relative to DNA, the ICD intensity increased and shifted to higher energy (~488 nm), indicative of non-specific binding as expected at high ligand concentrations. The observed hypsochromic shift may indicate a change in or a more defined conformation of C1 when specifically bound to the AP site. Titration of the control DNA without an AP site also showed an ICD and evidence of non-specific binding (Fig. 6c). It is worth noting that the band at 450–520 nm only appeared at a 3-fold excess of C1 with respect to DNA, making this wavelength range the best choice to construct the binding curves. Binding isotherms were constructed and analyzed using a simple bimolecular binding model as published previously32. The nonlinear equations that resulted from the 500 nm isotherms (Fig. 6d) gave the dissociation constant (KD) of 64 μM for the control T:A DNA (non-specific binding) and 29 μM for the T:THF DNA (specific binding); the KD of C1 with THF-containing duplex was calculated to be 22 μM after subtraction to correct for the non-specific binding contribution and possible ligand-ligand interactions (Table 1). The KD values calculated at 495 nm were only slightly outside the errors of those calculated at 500 nm (Table 1).
Figure 6. CD analyses of the interactions of C1 and its structural analogue C6 with duplex DNA.
(a) Titration curves of ICD intensity at 500 nm using a constant C1 concentration (20 μM) and increasing concentrations of DNA (reverse titration). (b) CD spectra for T:THF and (c) T:A using a constant DNA concentration (10 μM) and increasing concentrations of C1 (forward titration). (d) Titration curves of ICD intensity at 500 nm versus the logarithm of the C1 concentration for T:THF and T:A (derived from the data in panels b and c). (e) Titration curves of ICD intensity at 500 nm versus the logarithm of the C6 concentration for T:THF and T:A (derived from the data in Supplementary Figure S6). (d) Titration curves of ICD intensity at 500 nm versus the logarithm of the C6 concentration for T:AP and T:A (derived from the data in Supplementary Figure S6).
Table 1. Dissociation constants (K D, μM) of C1 and C6.
| Compound | Wavelength | DNA |
||||
|---|---|---|---|---|---|---|
| T:A | T:THF | T:THF corrected for non-specific interactions | T:AP | T:AP corrected for non-specific interactions | ||
| C1 | 500 nm | 64 ± 3 | 29 ± 1 | 22 ± 1 | ND | ND |
| 495 nm | 66 ± 2 | 40 ± 1 | 26 ± 1 | ND | ND | |
| C6 | 500 nm | 69 ± 14 | 35 ± 1 | 28 ± 1 | 44 ± 5 | 28 ± 2 |
| 495 nm | 70 ± 14 | 42 ± 2 | 28 ± 1 | 62 ± 15 | 26 ± 3 | |
The numbers represent the KD values and the corresponding errors obtained from fitting the experimental data to a sigmoidal curve.
Further, the possibility has been considered that the affinity of C1 could be different for DNA containing a natural AP site instead of THF. To address this question, we tested structurally related but inactive compound C6 (Fig. 2) in CD analyses with DNA containing either THF or an AP site that was created from dU by UDG treatment. The CD spectra observed for C6 in forward titration experiments using the T:THF and T:A DNAs (Supplementary Figure S6) were generally similar to that observed for C1 (Fig. 6b,c). The binding isotherms were constructed (Fig. 6e) and analyzed as above. The KD of C6 with THF-containing duplex, corrected for the non-specific interactions was calculated to be 28 μM (Table 1), which is only ~25% higher than the corresponding KD calculated for C1. Thus, C6 seems to be an appropriate model for studying interactions of C1 with DNA. Forward titration of C6 was then performed using DNA that contained the UDG-derived AP site (Supplementary Figure S6). Analyses of binding isotherms revealed that the affinity of C6 to this DNA was essentially identical to THF-containing DNA (Fig. 6f and Table 1). Considering that deoxyribose at a natural AP site predominantly exists in the ring-closed, THF-like form31, the latter result was not surprising. It can be anticipated that prior to formation of a covalent intermediate, the affinity of C1 to AP site-containing DNA would be comparable to that measured for THF-containing DNA. Thus, the stronger binding of C1 to DNA containing an AP site compared to the control DNA was consistent with the thermal stability data and together supports the proposal that the indolinone-pyrrole subunit occupies an empty space at the AP site of DNA. It is likely that the affinity of C1 for the AP site contributes to its lyase activity and provides a clear advantage over C2, CS1, CS2, and CS3, which lack the indolinone-pyrrole moiety (Fig. 2 and Supplementary Figure S2).
Conclusions
The AP DNA strand scission catalysts described in this study represent a new class of compounds that can be utilized to cleave AP sites under physiological conditions. Based on this core structure, molecules can be designed to improve selectivity of AP site incision opposite different bases and potentially for different sequence contexts. Due to recent progress in the development of high-throughput screening techniques, these compounds could be rapidly advanced to be efficient reagents to cleave AP sites that are created through depurination or the BER pathway in cells and organisms. The optimized versions of C1 may also have therapeutic applications, particularly in combination with many common anticancer drugs that either damage DNA, such as alkylating agents33, or target DNA repair, such as PARP34,35 or AP endonuclease36 inhibitors. It is worth mentioning that the 3′ ends created via β- or β,δ-elimination cannot be utilized by DNA polymerases without prior repair37. Biological consequences of the conversion of AP sites into such DNA strand breaks are expected to be complex and vary depending on the cellular capacity to repair or tolerate different types of DNA damage. The possible outcomes may include an increased therapeutic efficacy of anti-cancer treatments (more efficient cell killing) and diminished drug-induced mutagenesis. The cleavage of DNA at AP sites may be particularly beneficial for the treatment of cancers that have defects in mechanisms for repair of DNA strand breaks, such as BRCA deficient cancers34,35.
Methods
Materials
T4 polynucleotide kinase and UDG were purchased from New England Biolabs Inc. (Ipswich, MA). His-tagged human NEIL1 was purified according to a previously developed method38. His-tagged human OGG1 was purified by the same method as described for hNEIL1 using a DNA construct that was kindly provided by Dr. Gregory Verdine (Harvard University, Cambridge, MA).
The small molecule compounds C1, C3-C7, and CS4-CS8 were provided by F. Hoffmann-La Roche Inc. The amines lacking the indolinone-pyrrole moiety C2 and CS1-CS3 were purchased from Sigma-Aldrich (St. Louis, MO). The compounds were dissolved in DMSO at concentrations ranging from 5 mM to 50 mM and stored at −20 °C.
An oligodeoxynucleotide containing an internal 8-oxo-dG and a TAMRA fluorophore on the 5′-end (5′-TAMRA-TCACC(8-oxo-dG)TCGTACGACTC-3′) was synthesized and purified in-house using previously described methodology39. All other oligodeoxynucleotides were purchased from Integrated DNA Technologies (Coralville, IA). Oligodeoxynucleotides containing an internal dU and a TAMRA fluorophore on the 5′-end (5′-TAMRA-TCACC(dU)TCGTACGACTC-3′ and 5′-TAMRA-(T)5(dU)(T)11-3′) or an internal tetrahydrofuran (THF) (5′-TCACC(THF)GTCGTA-3′) or dU (5′-TCACC(dU)GTCGTA-3′) were HPLC-purified by the manufacturer.
DNA cleavage assays
To create double-stranded DNA substrates for the incision reactions, lesion-containing TAMRA-labeled oligodeoxynucleotides were combined with the corresponding complementary oligodeoxynucleotides in a buffer composed of 20 mM Tris-HCl, pH 7.5, and 100 mM KCl, and the mixtures were heated for 2 min at 90 °C and slowly cooled to 4 °C. The AP sites were created by incubating the dU-containing double-stranded DNA substrates (2.5 μM) with UDG (0.5 units/μl) for 30 min at 37 °C. The cleavage reactions were conducted with 250 nM double-stranded DNA substrates in 20 mM Tris-HCl, pH 7.5, 100 mM KCl, 0.1% (w/v) bovine serum albumin, and 0.01% (v/v) Tween-20. Concentrations of DNA glycosylases, the KWKK peptide (Sigma-Genosys), spermine (Sigma-Aldrich), C1, and related compounds are given in the figures or figure legends. Following incubation at 37 °C for 30 min, four volumes of formamide were added to the reactions, and DNAs were resolved through a 15% polyacrylamide gel in the presence of 8 M urea. To measure the initial rates of incision, aliquots were collected at time points that were empirically determined to be in the linear range of the product formation. The gel images were captured with the FluorChem M system (Protein Simple) using a 534 nm LED light source and 593 nm emission filter. The intensities of DNA bands were measured using Image StudioTM Lite version 4.0 software (LI-COR). The data were plotted and analyzed using KaleidaGraph 4.1 software (Synergy Software).
Trapping reactions
To prepare radioactively-labeled AP site-containing DNA for the trapping reactions, the corresponding dU-containing oligodeoxynucleotide 5′-TCA(dU)CGT-3′ (10 μM) was incubated with T4 polynucleotide kinase (1 unit/μl) in the presence of [γ-32P]-ATP (6000 Ci/mmol, PerkinElmer, Inc.) for 1 h at 37 °C, and AP sites were generated as described above. To test for the formation of the imine- or iminium ion-mediated complexes, 32P-labeled dU- or AP site-containing oligodeoxynucleotides (250 nM) were incubated with C1 (10 μM) or KWKK (2 mM) in 100 mM Hepes-K, pH 7.3, and 50 mM NaB(CN)H3 at 18 °C overnight. Reactions were terminated by the addition of NaBH4 (100 mM final). DNAs were passed through P-6 Bio-Spin columns (Bio-Rad, Hercules, CA), mixed with an equal volume of a gel-loading solution [95% (v/v) formamide, 20 mM EDTA, 0.02% (w/v) bromophenol blue, and 0.02% (w/v) xylene cyanol], and resolved through a 20% polyacrylamide gel in the presence of 8 M urea. Radioactively-labeled DNAs were visualized using a PhosphorImager screen (GE Healthcare).
Preparation of the DNA-C1 complexes for analyses by MS was done by incubation of dU-containing oligodeoxynucleotide (40 μM) with UDG (0.5 units/μl) for 2 h at 37 °C and subsequent reaction of DNA (16 nmoles) with C1 (640 nmoles) in the presence of 50 mM NaB(CN)H3 in 100 mM Hepes-K, pH 7.3, at 37 °C overnight. An authentic standard of the reduced C1-deoxyribose conjugate was prepared as follows: C1 (400 μg) and deoxyribose (2.8 mg) were dissolved in 200 μl of 50 mM potassium phosphate buffer, pH 7.0, and the reaction was stirred at 37 °C overnight. NaB(CN)H3 was added to reach a concentration of 50 mM, and the reaction was stirred for 3 h at 37 °C. The reaction was analyzed utilizing electrospray ionization (ESI) - liquid chromatography (LC) - MS monitoring m/z 444.2 corresponding to the desired product.
Mass spectrometry
MS analyses were performed in the Vanderbilt University facility on a Waters Acquity UPLC system (Waters, Milford, MA) connected to a Finnigan LTQ mass spectrometer (ThermoElectron) equipped with an Ion Max API source and a standard electrospray probe using a Phenomenex Luna column (1 μm, 1.0 mm × 100 mm). LC conditions were as follows: buffer A contained 10 mM NH4CH3CO2 and buffer B contained CH3CN. The following gradient program was used with a flow rate of 70 μl/min: isocratic at 100% A for 1 min then: 1 to 4 min, linear gradient from 100% A to 95% A/5% B (v/v); 4 to 6 min, linear gradient to 80% A/20% B (v/v); 6 to 7 min, linear gradient to 70% A/30% B (v/v); 7 to 8 min, linear gradient to 60% A/40% B (v/v); 8–9 min, linear gradient to 50% A/50% B (v/v); 9 to 13 min, linear gradient to 100% B; 13 to 15 min, linear gradient to 100% A; 15 to 16 min, isocratic at 100% A, isocratic at 100% A from 16 to 18 min.
The temperature of the column was maintained at 50 °C and the samples (15 μl) were infused with an auto-sampler. The electrospray conditions were as follows: source voltage 4 kV, source current 100 μA, N2 was used as the auxiliary gas and the flow-rate setting was 20, sweep gas flow-rate setting 5, sheath gas flow setting 34, capillary voltage −49 V, capillary temperature 350 °C, and tube lens voltage −18 V. No CID offset was employed. MS-MS conditions were as follows: normalized collision energy 25%, activation Q 0.250, and activation time 30 ms. The isolation width in MS-MS was 2 Da. The automatic gain control settings in full MS and MSn were 10000. The maximum injection time in full MS and MSn were 10 ms and 40 ms, respectively. The MS data were acquired in negative mode. The number of μscan used for data acquisition in full MS and MSn modes was 2 and 1, respectively. Product ion spectra were acquired over the range m/z 200–2000. The ions were selected for CID analysis and the elucidation of the CID fragmentations of the candidate oligodeoxynucleotide sequence was done with the aid of the Mongo Oligo Mass Calculator (v. 2.06) from The RNA Institute, State University of New York at Albany (http://mods.rna.albany.edu/Masspec-Toolbox). After the oligodeoxynucleotide sequence was identified, the proposed sequence was purchased from Midland Certified Reagents (Midland, TX) and subjected to the same LC-ESI-MS-MS analysis in order to compare the CID spectra.
LC-ESI-MS of the enzymatic digestion reactions was performed on an Acquity ultra-performance liquid chromatography system interfaced to a Finnigan LTQ mass spectrometer operating in ESI positive mode and using a a Phenomenex Luna column C18 (1 mm × 250 mm). The following gradient program was used with a flow rate of 60 μl/min: A: 10 mM NH4COOH, B: Acetonitrile, temperature of the column 50 °C; from 0 to 10 min linear gradient from 100% A to 20% A/80% B (v/v); from 10 min to 14 min linear gradient to 100% B; from 14 min to 17 min isocratic at 100% B; from 17 min to 20 min linear gradient to 100% A.
DNA melting
The DNA melting curves were obtained using a Varian Cary400 spectrophotometer equipped with a programmable heating block. Double-stranded DNAs (5 μM) containing either THF opposite T or a control A:T pair were prepared in 10 mM sodium phosphate buffer (pH 7.0), 100 mM NaCl, and 1 mM EDTA by heating the solution at 95 °C for 5 min then cooling to 15 °C at a rate of 1 °C/min. The melting curves were determined in the absence or presence of 1 equivalent of C1. The UV absorption at 260 nM was monitored as a function of temperature. The temperature was increased at a rate of 1 °C/min from 10 to 90 °C. The melting temperature (Tm) was determined as the inflection point of a sigmoidal function used to fit the melting curve: y = Af + [(Ai − Af)/(1 + exp((x − x0)/dx)], where Ai is the initial percent hyperchromicity value or low temperature horizontal asymptote when DNA is in the double-stranded state, Af is the final percent hyperchromicity value or high horizontal asymptote when DNA is in the melted state, x0 is the point of inflection, and dx is the change in temperature corresponding to the most significant change in percent hyperchromicity. The data were fitted using KaleidaGraph 4.0 software (Synergy Software).
Circular dichroism
Double-stranded DNAs containing either THF or A opposite T were prepared in 10 mM sodium phosphate buffer (pH 7.0), 100 mM NaCl and 1 mM EDTA as described above. To prepare corresponding AP site-containing DNA, the dU-containing oligodeoxynucleotide was reacted with UDG (0.5 unit/μl of reaction) for 2 h at 37 °C. UDG was removed utilizing a Vivaspin centrifugal concentrator, MWCO 20 K (Sigma), and the filtrate was lyophilized. DNA was reconstituted at a final concentration of 10 μM in the buffer described above and annealed with the complement strand by heating solution at 75 °C for 2 min and slow cooling to room temperature.
The CD titrations were performed on Jasco J-810 spectropolarimeter equipped with a thermoelectrically controlled, single-cell holder. CD spectra were collected using bandwidth 1 nm, response time 0.25 s, speed 200 nm/min, sensitivity 100 mdeg, scan accumulation 5–7, and temperature of 15 °C. Binding constants were determined by performing nonlinear least squares fitting32. The λ selected to construct binding isotherms (500 nm) was where the contribution of non-specific binding was not significant and where DNA did not contribute to the observed signal. The data were therefore treated as a simple bimolecular binding reaction, where the CD signal versus the logarithm of total ligand concentration was plotted and a sigmoid fit (similar to equation used for Tm determination) was used to determine the dissociation constant. The reverse titration where C1 was titrated with DNA showed a more complex binding interaction and were treated as the formalism described by Drake40. The data were fitted using KaleidaGraph 4.0 software (Synergy Software).
Additional Information
How to cite this article: Minko, I. G. et al. Catalysts of DNA Strand Cleavage at Apurinic/Apyrimidinic Sites. Sci. Rep. 6, 28894; doi: 10.1038/srep28894 (2016).
Supplementary Material
Acknowledgments
Research reported in this publication was partially supported by the National Institutes of Health, the National Cancer Institute (award numbers T32 CA106195, P01 CA160032, and P30 CA068485) and the National Institute of Environmental Health Sciences (P30 ES000267 and T32 ES0007060). This investigation was made possible by the generous gift of a kinase inhibitor library and ongoing support and consultation from Dr. Paul Gillespie, F. Hoffmann-La Roche Inc.
Footnotes
Author Contributions I.G.M., T.M.H., C.J.R., A.K.M. and R.S.L. designed the study, I.G.M., A.C.J., A.R.D.L., F.G. and N.D. conducted experiments and analyzed the data, I.G.M., C.J.R. and R.S.L. wrote the manuscript, T.M.H. and A.K.M. provided a critical review of the manuscript.
References
- Lindahl T. Instability and decay of the primary structure of DNA. Nature 362, 709–715 (1993). [DOI] [PubMed] [Google Scholar]
- Verly W. G., Paquette Y. & Thibodeau L. Nuclease for DNA apurinic sites may be involved in the maintenance of DNA in normal cells. Nat. New Biol. 244, 67–69 (1973). [DOI] [PubMed] [Google Scholar]
- Nakamura J. et al. Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer Res. 58, 222–225 (1998). [PubMed] [Google Scholar]
- Atamna H., Cheung I. & Ames B. N. A method for detecting abasic sites in living cells: age-dependent changes in base excision repair. Proc. Natl. Acad. Sci. USA 97, 686–691 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb L. A. Apurinic sites as mutagenic intermediates. Cell 40, 483–484 (1985). [DOI] [PubMed] [Google Scholar]
- Kroeger K. M., Goodman M. F. & Greenberg M. M. A comprehensive comparison of DNA replication past 2-deoxyribose and its tetrahydrofuran analog in Escherichia coli. Nucleic Acids Res. 32, 5480–5485 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S. L., Lee S. K., Johnson R. E., Prakash L. & Prakash S. The stalling of transcription at abasic sites is highly mutagenic. Mol. Cell. Biol. 23, 382–388 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerasooriya S., Jasti V. P. & Basu A. K. Replicative bypass of abasic site in Escherichia coli and human cells: similarities and differences. PLoS One 9, e107915 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly V. & Verly W. G. Possible roles of β-elimination and δ-elimination reactions in the repair of DNA containing AP (apurinic/apyrimidinic) sites in mammalian cells. Biochem. J. 253, 553–559 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsley W. S., Penhoet E. E. & Linn S. Human endonuclease specific for apurinic/apyrimidinic sites in DNA. Partial purification and characterization of multiple forms from placenta. J. Biol. Chem. 252, 1235–1242 (1977). [PubMed] [Google Scholar]
- Schrock R. D. 3rd. & Lloyd R. S. Reductive methylation of the amino terminus of endonuclease V eradicates catalytic activities. Evidence for an essential role of the amino terminus in the chemical mechanisms of catalysis. J. Biol. Chem. 266, 17631–17639 (1991). [PubMed] [Google Scholar]
- Dodson M. L., Michaels M. L. & Lloyd R. S. Unified catalytic mechanism for DNA glycosylases. J. Biol. Chem. 269, 32709–32712 (1994). [PubMed] [Google Scholar]
- McCullough A. K. et al. The reaction mechanism of DNA glycosylase/AP lyases at abasic sites. Biochemistry 40, 561–568 (2001). [DOI] [PubMed] [Google Scholar]
- Wallace S. S. DNA glycosylases search for and remove oxidized DNA bases. Environ. Mol. Mutagen. 54, 691–704 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh P. J. & Knowland J. Novel reagents for chemical cleavage at abasic sites and UV photoproducts in DNA. Nucleic Acids Res. 23, 1664–1670 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama H. et al. Chemistry of thermal degradation of abasic sites in DNA. Mechanistic investigation on thermal DNA strand cleavage of alkylated DNA. Chem. Res. Toxicol. 7, 673–683 (1994). [DOI] [PubMed] [Google Scholar]
- Male R., Fosse V. M. & Kleppe K. Polyamine-induced hydrolysis of apurinic sites in DNA and nucleosomes. Nucleic Acids Res. 10, 6305–6318 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakilas A. G., Bennett P. V. & Sutherland B. M. High efficiency detection of bi-stranded abasic clusters in γ-irradiated DNA by putrescine. Nucleic Acids Res. 30, 2800–2808 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe Y. S. & Sasaki S. DNA cleavage at the AP site via β-elimination mediated by the AP site-binding ligands. Bioorg. Med. Chem. 24, 910–914 (2016). [DOI] [PubMed] [Google Scholar]
- Zhou C., Sczepanski J. T. & Greenberg M. M. Mechanistic studies on histone catalyzed cleavage of apyrimidinic/apurinic sites in nucleosome core particles. J. Am. Chem. Soc. 134, 16734–16741 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behmoaras T., Toulme J. J. & Helene C. A tryptophan-containing peptide recognizes and cleaves DNA at apurinic sites. Nature. 292, 858–859 (1981). [DOI] [PubMed] [Google Scholar]
- Pierre J. & Laval J. Specific nicking of DNA at apurinic sites by peptides containing aromatic residues. J. Biol. Chem. 256, 10217–10220 (1981). [PubMed] [Google Scholar]
- Kurtz A. J., Dodson M. L. & Lloyd R. S. Evidence for multiple imino intermediates and identification of reactive nucleophiles in peptide-catalyzed β-elimination at abasic sites. Biochemistry 41, 7054–7064 (2002). [DOI] [PubMed] [Google Scholar]
- Zeglis B. M., Boland J. A. & Barton J. K. Targeting abasic sites and single base bulges in DNA with metalloinsertors. J. Am. Chem. Soc. 130, 7530–7531 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liuzzi M. & Talpaert-Borle M. A new approach to the study of the base-excision repair pathway using methoxyamine. J. Biol. Chem. 260, 5252–5258 (1985). [PubMed] [Google Scholar]
- Fkyerat A., Demeunynck M., Constant J.-F., Michon P. & Lhomme J. A new class of artificial nucleases that recognize and cleave apurinic sites in DNA with great selectivity and efficiency. J. Am. Chem. Soc. 115, 9952–9959 (1993). [Google Scholar]
- Jacobs A. C. et al. Inhibition of DNA glycosylases via small molecule purine analogs. PLoS One 8, e81667 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. et al. United States Patent No 6130239 (2000).
- Khan G. S., Shah A., Ziaur R. & Barker D. Chemistry of DNA minor groove binding agents. J. Photochem. Photobiol. B. 115, 105–118 (2012). [DOI] [PubMed] [Google Scholar]
- Niedernhofer L. J. et al. Temperature-dependent formation of a conjugate between tris(hydroxymethyl)aminomethane buffer and the malondialdehyde-DNA adduct pyrimidopurinone. Chem. Res. Toxicol. 10, 556–561 (1997). [DOI] [PubMed] [Google Scholar]
- Takeshita M., Chang C. N., Johnson F., Will S. & Grollman A. P. Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J. Biol. Chem. 262, 10171–10179 (1987). [PubMed] [Google Scholar]
- Garbett N. C., Ragazzon P. A. & Chaires J. B. Circular dichroism to determine binding mode and affinity of ligand-DNA interactions. Nat. Protoc. 2, 3166–3172 (2007). [DOI] [PubMed] [Google Scholar]
- Puyo S., Montaudon D. & Pourquier P. From old alkylating agents to new minor groove binders. Crit. Rev. Oncol. Hematol. 89, 43–61 (2014). [DOI] [PubMed] [Google Scholar]
- Bryant H. E. et al. Specific killing of BRCA2-deficient tumors with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- Farmer H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]
- Wilson D. M. 3rd. & Simeonov A. Small molecule inhibitors of DNA repair nuclease activities of APE1. Cell. Mol. Life Sci. 67, 3621–3631 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres S. N., Schellenberg M. J., Wallace B. D., Tumbale P. & Williams R. S. Recognition and repair of chemically heterogeneous structures at DNA ends. Environ. Mol. Mutagen. 56, 1–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy L. M. et al. Human polymorphic variants of the NEIL1 DNA glycosylase. J. Biol. Chem. 282, 15790–15798 (2007). [DOI] [PubMed] [Google Scholar]
- Mullah B. & Andrus A. Automated synthesis of double dye-labeled oligonucleotides using tetramethylrhodamine (TAMRA) solid supports. Tetrahedron Lett. 38, 5751–5754 (1997). [Google Scholar]
- Strat D., Missailidis S. & Drake A. F. A novel methodological approach for the analysis of host-ligand interactions. Chem. Phys. Chem. 8, 270–278 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.