Abstract
Despite abundant data supporting c-Src as a metastasis-promoting oncogene, activating mutations of c-Src are rare. This suggests that trans-interacting proteins may have a critical role in regulating c-Src activation. Here, we first report the discovery of Src homology 3 (SH3) domain-binding glutamic acid-rich-like protein (SH3BGRL), a novel c-Src activator in mice. Ectopic expression of murine SH3BGRL (mSH3BGRL) strongly promoted both tumor cell invasion and lung metastasis. Molecularly, mSH3BGRL specifically bound the inactive form of c-Src phosphorylated at Tyr527, promoting Tyr416 phosphorylation of c-Src and subsequent FAK-mediated activation of ERK and AKT signaling pathways. Targeting endogenous c-Src alone was sufficient to abolish mSH3BGRL-induced cancer metastasis in vivo. Unexpectedly, human SH3BGRL (hSH3BGRL) in turn suppressed tumorigenesis and metastasis in nature. We attempted site-specific reversion of hSH3BGRL amino-acid sequence to mSH3BGRL and found V108A substitution sufficient to restore SH3BGRL function as a c-Src activator and metastasis promoter. Notably, the somatic mutation R76C of hSH3BGRL can similarly act as hSH3BGRL-V108A and mSH3BGRL in tumorigenesis and metastasis. Our results uncover an evolutionarily controversial role of SH3BGRL in driving tumor metastasis through c-Src activation, and suggests that hSH3BGRL mutation status could be relevant to cancer diagnosis and therapy.
Introduction
Cancer metastasis accounts for up to 90% of deaths from solid tumors.1 c-Src, a prometastasis oncogene, is well characterized to promote cellular invasion, migration and has been justly proposed as a therapeutic target in cancers.2, 3, 4, 5 However, the biochemical regulation of c-Src activation is not fully understood. The best-characterized Src regulator is phosphorylation, with the two major sites of phosphorylation on murine and chicken c-Src being Tyr416 (Tyr419 in humans) and Tyr527 (Tyr530 in humans). Normally, c-Src is kept inactive by autoinhibitory intramolecular interactions between (1) the SH2 domain and phosphorylated Tyr527 at the C terminus, and (2) the SH3 domain and the proline-rich SH2 kinase linker region.6 Several Src-binding proteins have been shown to compete for the SH2 and/or SH3 domains on Src, thereby relieving such intramolecular inhibition and permitting c-Src kinase activity via Tyr416 autophosphorylation.7 This in turn displaces the ‘activation loop' from the substrate-binding pocket and makes it accessible to c-Src substrates.7 Interestingly, activating mutations on the inhibitory C terminus of c-Src are rarely observed in human cancers,8, 9, 10 suggesting that binding proteins might have a dominant role in the regulation of c-Src activity. SH3BGRL is a member of the SH3 domain-binding glutamic acid-rich protein (SH3BGR) family.11, 12 With the exception of SH3BGRL3, a common feature of this family is the proline-rich sequence (PLPPQIF), which contains both the SH3- and Homer EVH1-binding motifs.13 The function of the SH3BGR family is largely unknown. SH3BGRL3 has been shown to be upregulated in response to TNF-α simulation and was able to block TNF-induced apoptosis when added exogenously to human fibroblast cell lines.14, 15, 16 Similarly, SH3BGRL was upregulated in BRCA1 mutation-positive breast tumors versus BRCA2 mutation-positive sporadic breast tumors, as well as lymphocytic infiltrate-positive versus -negative ones.17 In ER-positive breast tumors, SH3BGRL was demonstrated to be overexpressed by SAGE analysis,18 hinting its possible tumor-enhancing role. In contrast to this expected prosurvival character, SH3BGRL was reported as an inhibitor of v-Rel-induced transformation in chicken cells.13 Nonetheless, a thorough investigation of the biological function and molecular mechanism of SH3BGRL signaling is lacking. In this study, when screening the affected genes by ectopic expression of a metastatic phosphatase PRL-3,19, 20, 21 we found that SH3BGRL was consistently downregulated by PRL-3, indicating that human SH3BGRL (hSH3BGRL) could be a tumor suppressor.
To clarify the obscure functions of SH3BGRL in tumor cell metastasis, we studied the possible consequences of SH3BGRL on tumorigenesis and metastasis by, respectively, forcing murine Src homology 3 BGRL (mSH3BGRL) and human Src homology 3 BGRL (hSH3BGRL) overexpression in murine and human tumor cells. Ectopic expression of mSH3BGRL promotes tumor cell metastasis. However, the hSH3BGRL gene unexpectedly had opposite functions, and single point mutations of this gene had restored the tumorigenic and metastatic promoter in its murine ortholog. Our findings implicate SH3BGRL as a novel evolutionarily controversial player for c-Src activation in cancer metastasis, and suggest that hSH3BGRL mutation could be relevant to cancer diagnosis and therapy.
Results
mSH3BGRL enhances cellular invasiveness in vitro and metastatic potential in vivo
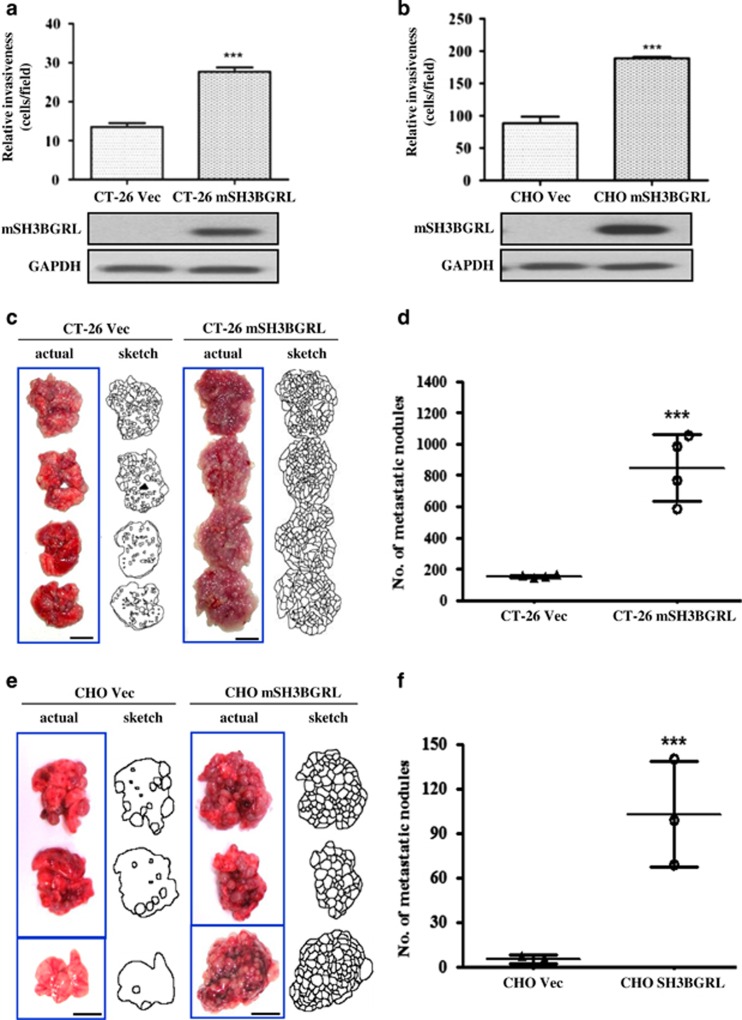
Given metastatic phosphatase PRL-3 can downregulate SH3BGRL in HCT-116 cells in our previous preliminary analysis (Supplementary Figures 1A and B), we first generated pooled colonies of murine CT-26 metastatic colorectal cancer or Chinese hamster ovary (CHO) cells stably overexpressing mSH3BGRL constructs or appropriate vector controls. Using Matrigel-coated transwell invasion assays, we observed that CT-26 cells overexpressing mSH3BGRL had a significantly higher relative invasive potential compared with control cells (Figure 1a). Similarly, non-tumorigenic CHO cells overexpressing mSH3BGRL also exhibited increased invasion potential relative to control cells (Figure 1b). These observations indicated a candidate role for SH3BGRL in promoting cellular invasion. As invasive potential promotes metastasis progression, we next directly tested these cell pairs for metastatic capacity in the well-established tail vein mouse model.22 Although CT-26 cells are naturally metastatic,23 CT-26 cells overexpressing mSH3BGRL formed significantly more lung tumor macrometastasis compared with control cells after 4 weeks of cell injection (11.4-fold increase, P<0.001; Figures 1c and d). To investigate if mSH3BGRL was alone sufficient for metastasis, we repeated this experiment using non-metastatic CHO cells. Remarkably, CHO cells overexpressing mSH3BGRL robustly formed lung macrometastatic tumors compared with CHO control cells, which largely failed to do so (18.4-fold increase, P<0.001; Figures 1e and f). Collectively, our results suggest that overexpression of mSH3BGRL robustly promotes cells invasiveness and strongly drives metastasis in vivo.
Figure 1.
mSH3BGRL enhances cell invasiveness in vitro and drives metastasis in vivo. (a) Matrigel invasion assay using CT-26 cells stably overexpressing vector (CT-26 Vec) or GFP-SH3BGRL (CT-26 mSH3BGRL). After 36 h incubation, the number of invaded cells were counted (mean±s.d., n=3, ***P<0.001). The accompanying immunoblot shows the expression of exogenous SH3BGRL, with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a loading control. (b) Similar experiment as in (a), but using CHO cells stably expressing vector (CHO Vec) or RFP-SH3BGRL (CHO mSH3BGRL) (mean±s.d., n=3, ***P<0.001). (c) In total, 1 × 106 CT-26 Vec or CT-26 SH3BGRL cells were injected intravenously into the tail vein of nude mice. After 28 days, mice were killed and their lungs were photographed. (d) Scoring for metastatic tumor nodules in (c) (mean±s.d., n=4, ***P<0.001). (e) In total, 1 × 106 CHO Vec or CHO mSH3BGRL cells were injected and analyzed after 28 days injection. (f) Scoring for metastatic tumor nodules as in (e) (mean±s.d., n=3, ***P<0.001).
mSH3BGRL specifically binds to p-c-Src 527 to promote c-Src activation
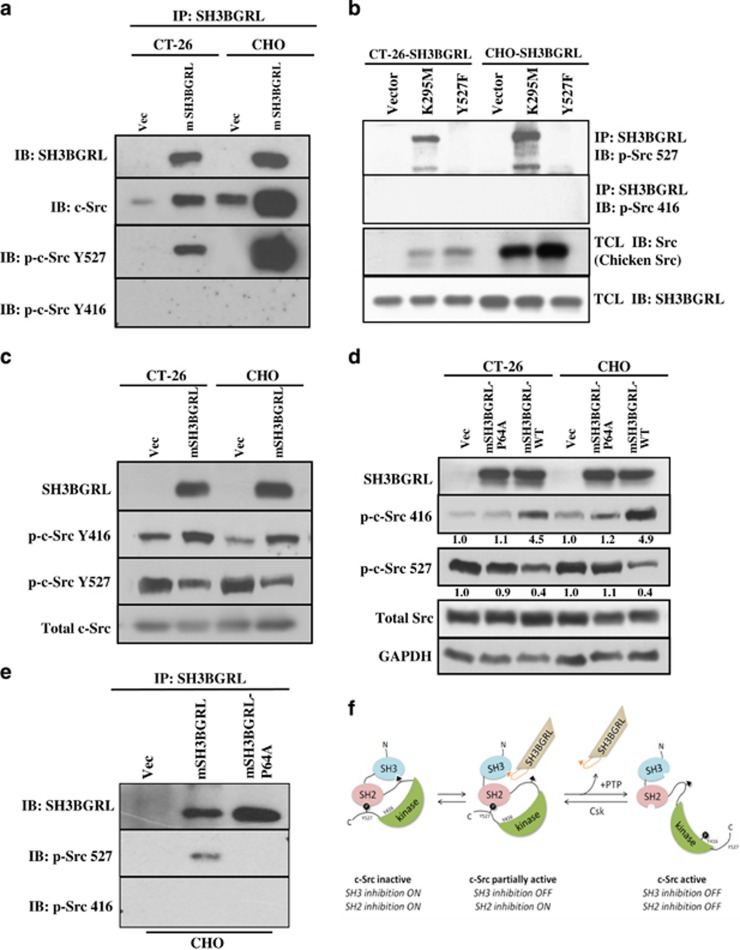
SH3BGRL contains an SH3-binding motif.13 To identify potential mSH3BGRL-binding partners involved in its prometastatic signaling, we used a protein array comprising the SH3 domains from 38 different proteins with recombinant GST-mSH3BGRL as a probe. We specifically identified the SH3 domain from c-Src as a potential mSH3BGRL-binding partner (Supplementary Figure 2). As c-Src has well-documented roles as an oncogene driving tumorigenesis and metastasis,23, 24 we validated whether mSH3BGRL could bind c-Src intracellularly. c-Src was detected in mSH3BGRL immunoprecipitates in both CT-26 and CHO cells (Figure 2a), supporting our SH3 domain array results. c-Src activity is primarily regulated by phosphorylation on two sites—Tyr527 (inactivating phosphorylation) and Tyr416 (activating phosphorylation).25 Intriguingly, mSH3BGRL specifically bound c-Src phosphorylated on Tyr527 but not c-Src phosphorylated on Tyr416 (Figure 2a). To further exclude the possibility that mSH3BGRL prefers to bind inactive c-Src (pY527) is because of the abundant phosphorylated c-Src at Y527 in cells, we respectively forced the expressions of the constitutively inactivated chicken c-Src mutant K295M and the activated Y527F, and manifested that mSH3BGRL was only precipitated with the inactive chicken c-Src, but not its abundant active mutant Y527F (Figure 2b). Moreover, immunofluorescent staining manifested the colocalization of p-Src 527 with some portions of mSH3BGRL (Supplementary Figure 3). In parallel with this, we observed an increase in the activated c-Src (pY416) with a corresponding decrease of inactive c-Src in total lysates from both CT-26 and CHO cells overexpressing mSH3BGRL (Figure 2c). These results together showed that mSH3BGRL specifically bind inactive c-Src to promote c-Src activation in cells.
Figure 2.
mSH3BGRL enhances bind to inactive c-Src and promote c-Src activation. (a) SH3BGRL immunoprecipitates from the various stable cell lines were immunoblotted with antibodies against SH3BGRL, c-Src, p-c-Src Y527 and p-c-Src Y416. (b) Immunoprecipitation of p-c-Src Y527 and p-c-Src Y416 by overexpression of chicken c-Src mutants K295M and Y527F and detected with the indicated antibodies as in (a). (c) Western blots of p-c-Src Y527 and p-c-Src Y416 in cells overexpressing mSH3BGRL with the indicated antibodies in (a). (d) Cells stably expressing vector (Vec), SH3BGRL-P64A (P64A) or SH3BGRL-WT (SH3BGRL) were immunoblotted with the indicated antibodies. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as a loading control, and the averaged relative protein expression level is quantified and shown under the particular panels. (e) Immunoprecipitation of p-c-Src Y527 and p-c-Src Y416 by overexpression of wild-type mSH3BGRL and its P64A mutants in CHO cells and detected with the indicated antibodies. (f) Schematic model for SH3BGRL-mediated c-Src activation. The SH3-binding domain from SH3BGRL competes for and displaces the inhibitory SH3-binding SH2 kinase linker region in c-Src, triggering conformational changes that promote c-Src kinase activity.
Several Src-binding proteins have shown to compete for the Src-SH3 domain to relieve autoinhibition and promote c-Src activation.6 To investigate the involvement for the proline-rich sequence of mSH3BGRL in binding and subsequent activation of c-Src, a point mutation was introduced (P64A) into this motif. This mutation has been previously demonstrated to abolish the SH3-binding domain capacity of mSH3BGRL.13 We found that only wild-type mSH3BGRL, but not mSH3BGRL-P64A mutant, obviously promoted c-Src activation (Figure 2d). Moreover, breakdown of SH3 domain of mSH3BGRL (P64A mutant) lost its binding activity to inactive c-Src (Figure 2e), strongly supporting a requirement for the SH3-binding domain of mSH3BGRL in promoting c-Src activation in vitro. Thus, we propose a model wherein mSH3BGRL mediates an early step in c-Src activation by competitively disrupting the intramolecular inhibitory interaction between SH3 domain and the proline-rich SH2 kinase linker region within c-Src, likely inducing structural change, which then promotes dephosphorylation of Tyr527 to result in full kinase activation (Figure 2f).
mSH3BGRL overexpression triggers the activation of FAK, MAPK and AKT signaling pathways downstream of c-Src
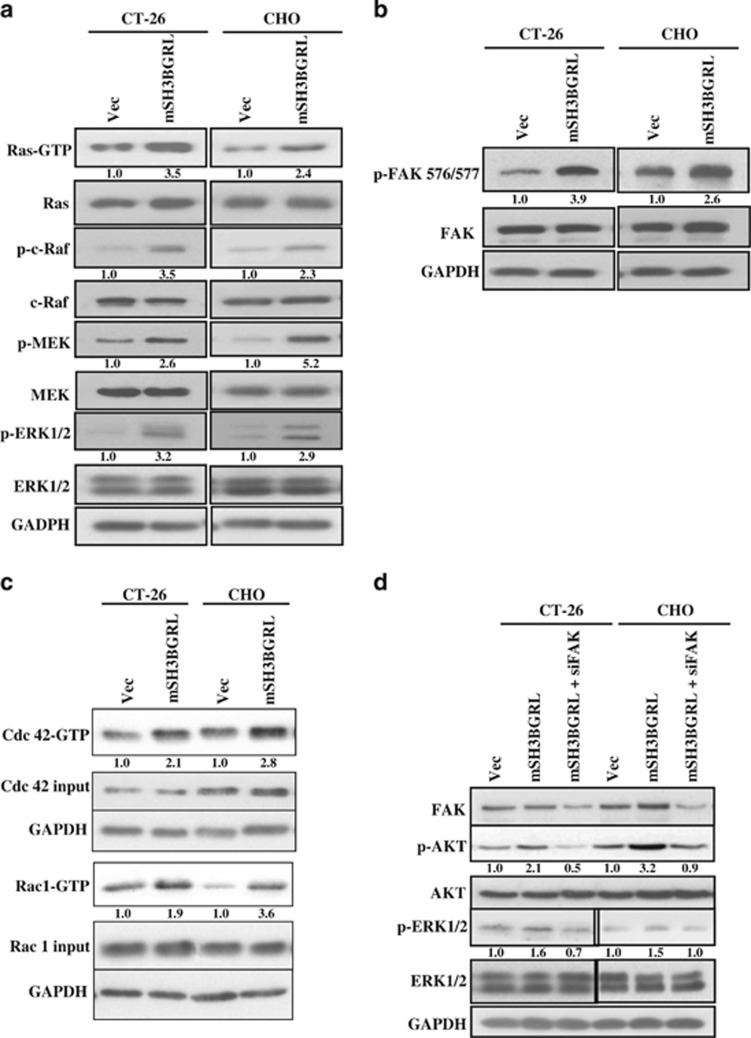
As c-Src is a pleiotropic kinase, which can activate various downstream signaling pathways, including Ras/MAPK and AKT,26, 27 we next investigated if mSH3BGRL might also regulate these downstream pathways. Analysis of activated Ras (GTP-Ras) revealed a higher activation in both CT-26 and CHO cells overexpressing mSH3BGRL (Figure 3a). Correspondingly, increased phosphorylation of key Ras effectors, including c-Raf, MEK and ERK1/2,28 were notably observed in both CT-26 and CHO cells overexpressing mSH3BGRL (Figure 3a). FAK is a key c-Src substrate with important roles in transducing signals from focal adhesions, and is frequently overexpressed in a number of different types of cancer, especially in primary invasive cancers and metastatic lesions.29 c-Src phosphorylates FAK on Tyr576/577 to promote FAK kinase activity, downstream signaling and invasion.30 In both CT-26 and CHO cells overexpressing mSH3BGRL, we noted a pronounced increase in FAK phosphorylation on Tyr576/577 (Figure 3b). As the activated c-Src/FAK signaling complex has been well characterized as an activator of Cdc42 and Rac GTPases, and the intimate regulators of cell motility,31 we further analyzed the activity of these GTPases upon forced mSH3BGRL expression. In both CT-26 and CHO cells overexpressing mSH3BGRL, an increase in active GTP-bound forms of Cdc42 and Rac1 was observed (Figure 3c).
Figure 3.
mSH3BGRL activates MAPK and AKT pathways in an FAK-dependent manner. (a) Lysates from stable cell lines were incubated with GST-Raf to enrich for GTP-bound active Ras. Eluates and total lysates were subsequently immunoblotted with various Ras/MAPK pathway-specific antibodies. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as a loading control. (b) Lysates from (a) were immunoblotted with anti-p-FAK Y576/577 or FAK antibodies. (c) Lysates prepared as in (a) were incubated with GST-PBD (to enrich for GTP-bound active Cdc42 and Rac1). Elutes and total lysates were subsequently immunoblotted with antibodies against Cdc42 and Rac1. (d) CT-26 Vec, CT-26 SH3BGRL, CHO Vec and CHO SH3BGRL cells were transfected with FAK-specific siRNA, where indicated, for 48 h and lysed. Lysates were immunoblotted with the various antibodies listed. GAPDH was served as a loading control. The averaged relative protein expression level is quantified and shown under the immediate panels.
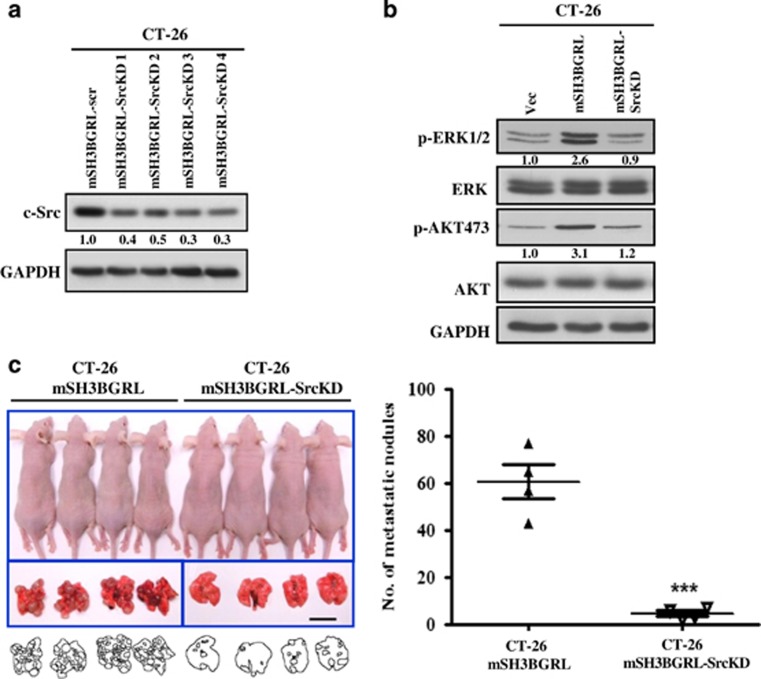
To verify the role of FAK in mSH3BGRL-associated signaling via the c-Src/FAK complex, RNA interference was used to deplete endogenous FAK or c-Src. In both CT-26 and CHO cells overexpressing mSH3BGRL, downregulation of FAK effectively restored levels of activated ERK1/2, as well as AKT to the control level, and AKT is a key PI3K effector and oncogenic kinase32 strongly activated upon mSH3BGRL overexpression (Figure 3d). Next, we stably depleted c-Src in CT-26 cells using RNA interference (Figure 4a). Using pooled knockdown cells for subsequent experiments (mSH3BGRL-SrcKD), we analyzed the status of MAPK and AKT pathway activity. c-Src depletion in CT-26 mSH3BGRL cells resulted in a evident reduction in active ERK1/2 and AKT similar to that observed in control cells (Figure 4b). Again, two Src inhibitors treatments similarly blocked ERK and AKT activation (Supplementary Figure 4). Importantly, CT-26 mSH3BGRL-SrcKD cells also formed significantly less lung tumor macrometastasis compared with CT-26 mSH3BGRL cells after 14 days of cell injection (12.4-fold decrease, P<0.001; Figure 4c). Collectively, our results suggest that mSH3BGRL promotes metastasis through activation of the c-Src activation of FAK and downstream oncogenic Ras/MAPK and AKT pathways.
Figure 4.
Knockdown of c-Src abrogates mSH3BGRL-induced metastasis in nude mice. (a) Four independent pools of CT-26 SH3BGRL cells stably expressing c-Src-specific shRNA (SH3BGRL-SrcKD 1–4) were analyzed for the expression of c-Src compared with a scrambled control (SH3BGRL-scr). glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as a loading control. (b) Lysates from exponentially growing stable cells (SH3BGRL-SrcKD is a combined pool from all four clones in (a)) were immunoblotted with various antibodies. GAPDH served as a loading control. The averaged relative protein expression level in (a) and (b) is quantified and shown under the immediate panels. (c) In total, 1 × 106 CT-26 SH3BGRL or CT-26 SH3BGRL-SrcKD cells were injected intravenously into the tail vein of nude mice. After 14 days, mice were killed and their lungs were photographed and scored for metastatic tumor nodules (mean±s.d., n=4, ***P<0.001).
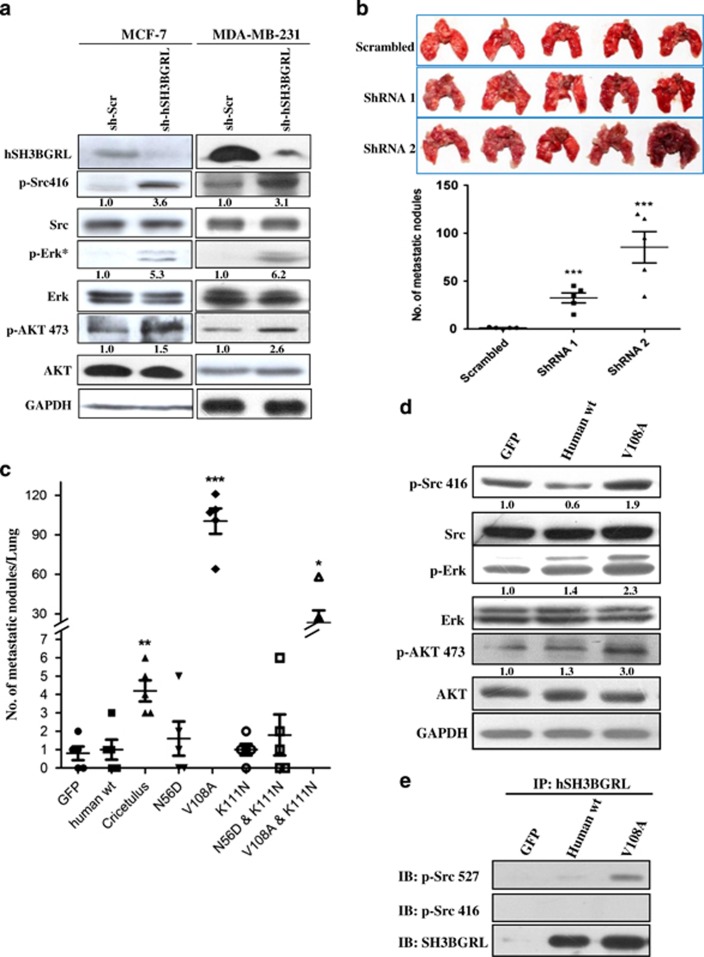
Wild-type hSH3BGRL is a metastatic suppressor in vivo, but mSH3BGRL-alike V108A mutation reverses it back to a metastatic driver
To explore the physiological relevance of murine orthologous gene, hSH3BGRL, in cancer metastasis, we silenced endogenous hSH3BGRL in the human breast cancer cell lines MCF-7 and MDA-MB-231 with specific shRNAs, respectively. Unexpectedly, hSH3BGRL knockdown resulted in c-Src, ERK1/2 and AKT activation in these cells (Figure 5a)—completely opposite to our earlier observations with mSH3BGRL (Figure 3). To further confirm these unexpected results, hSH3BGRL, in MDA-MB-231 cells was constitutively silenced by two hSH3BGRL-specific shRNAs (shRNA1 and 2) (Supplementary Figure 5A). Independent depletion of hSH3BGRL caused a significant increase in lung metastasis compared with control cells within only 13 days after tail vein injection (Figure 5b; P<0.001). In agreement with increased metastasis, subcutaneous-inoculated hSH3BGRL-depleted MDA-MB-231 cell pools also induced significant tumor burden in nude mice as early as on the 14th day postinoculation, whereas control cells had no observable tumor burden yet (Supplementary Figure 5B). Collectively, our results clearly indicated an unexpected tumor-suppresive role for hSH3BGRL, a finding completely opposite to that seen from its murine ortholog (mSH3BGRL).
Figure 5.
hSH3BGRL is a tumor suppressor, but its mutation can reverse it back to metastatic diver. (a) Lysates from MCF-7 and MDA-MB-231 cells were stably transfected with two individual hSH3BGRL-specific shRNAs as cell pools and immunoblotted with the indicated antibodies, respectively. *p-ERK was detected after serum-free starvation of the cells for 24 h. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was served as a loading control. The indicated relative protein expression level is quantified and shown under the immediate panel. (b) In total, 1x106 MDA-MB-231 Vec or MDA-MB-231 SH3BGRL knockdown cell pools were injected into the tail vein of nude mice. After 13 days, mice were killed and their lungs were photographed and scored for metastatic tumor nodules. (c) Effect of amino-acid alteration of hSH3BGRL to lung metastasis in CHO cells. In total, 1x106 stably transfected cells were injected into nude mice through the tail vein. After 19 days injection, the assayed mice were killed and the dissected lungs were analyzed. The metastatic nodules in each lung in (b) and (c) were counted. Mean±s.d., n=4 or 5, *P<0.05; **P<0.001; ***P<0.0001, unpaired Student's t-test. (d) Lysates from CHO cells stably transfected with hSH3BGRL and its mutant V108A and immunoblotted with the indicated antibodies. The averaged protein relative expression level is quantified and shown under the immediate panel. (e) Co-immunoprecipitation of hSH3BGRL V108 mutant with p-c-Src 527, rather than wild-type hSH3BGRL were analyzed by immunoblotted by the indicated antibodies.
To gain insight into the possible impact of mutations on hSH3BGRL's influence on c-Src activation and tumor metastasis, we did a sequence alignment of the amino-acid sequences of SH3BGRL orthologs from Homo sapiens, Mus musculus and Cricetulus griseus (Chinese hamster). Two conserved amino-acid substitutions were identified between the amino-acid sequences of human and either rodent species—N56D and V108A (Supplementary Figure 6A). To test the possibility that these mutation(s) might account for the antagonistic function of hSH3BGRL and mSH3BGRK, hSH3BGRL-N56D and hSH3BGRL-V108A mutants were generated to understand if these 'reverting' mutations could restore the oncogenic character seen for mSH3BGRL. Using stably transfected CHO cells injected into tail veins of nude mice for metastasis analysis, we first observed that hSH3BGRL-V108A-expressing CHO cells induced extensive lung metastasis compared with control, wild-type hSH3BGRL- or hSH3BGRL-N56D-expressing cells (Figure 5c and Supplementary Figure 6B). Xenograft model of DLD-1 colorectal cancer cells also demonstrated that ectopic expression of wild-type hSH3BGRL repressed tumor formation (Supplementary Figure 6C), whereas hSH3BGRL knockdown or overexpression hSH3BGRL-V108A in turn refueled tumorigenesis, respectively (Supplementary Figures 6D and E). Mechanistically, we found that hSH3BGRL-V108A, but not wild-type hSH3BGRL, could efficiently activate c-Src and downstream AKT and ERK (Figure 5d). Additionally, hSH3BGRL-V108A was found to interact with the inactive p-c-Src Y527 to a greater extent, compared with the wild-type hSH3BGRL (Figure 5e). Taken together, our data suggest that by the introduction of a single point mutation in hSH3BGRL (V108A) was sufficient to completely revert the metastasis-suppressive character of hSH3BGRL back to the prometastatic character of its murine ortholog.
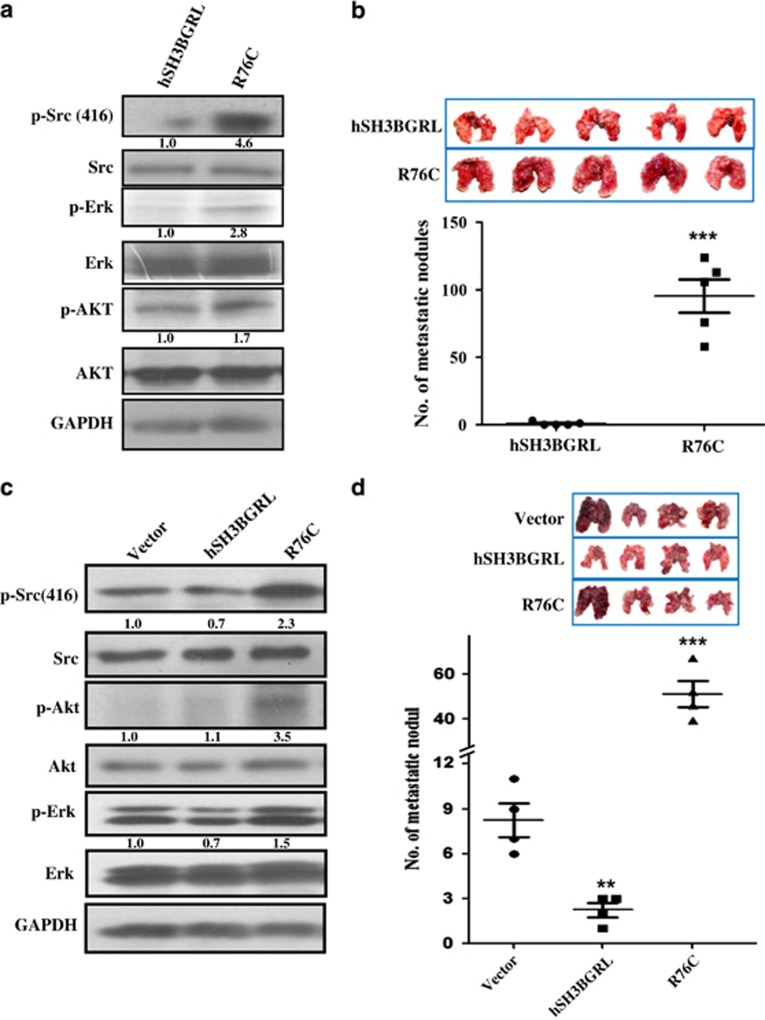
Somatic mutation of hSH3BGRL can promote metastasis
It is well documented that mutation of the classic tumor suppressor, p53, usually leads to more aggressive phenotypes.33 To determine whether hSH3BGRL has somatic mutations in tumors, we searched the publically available COSMIC database (http://www.sanger.ac.uk; http://www.cbioportal.org/public-portal/cross_cancer) and noted that hSH3BGRL has natural mutations in various tumors (Supplementary Tables 1 and 2), indicating that mutation of tumor suppressor hSH3BGRL reversely endorses it a metastatic driver. To validate this hypothesis, we overexpressed the relatively most frequent mutation (Supplementary Tables 3), R76C of hSH3BGRL, in CHO cells. In contrast to wild-type hSH3BGRL, R76C mutation can activate Src and the subsequent ERK and AKT activation, which is in line with the function of mSH3BGRL (Figure 6a). In vivo tumor formation with CHO stable cell lines containing R76C mutant overexpression also showed increased tumorigenic ability (Supplementary Figure 7). Furthermore, hSH3BGRL-R76C mutant markedly promotes CHO cell metastasis via tail injection of the cells at only 17 days postinjection, compared with the wild-type hSH3BGRL (Figure 6b).
Figure 6.
Somatic hSH3BGRL mutant R76C resembled to mSH3BGRL promotes lung metastasis. (a) Lysates from CHO cells stably transfected with hSH3BGRL or its somatic mutant R76C and immunoblotted with the indicated antibodies, respectively. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as a loading control. The indicated protein relative expression level is quantified under the immediate panel. (b) In total, 1x106 CHO hSH3BGRL- or its hSH3BGRL-R76C-overexpressing cells (R76C) were injected intravenously into the tail vein of nude mice. After 17 days, mice were killed and their lungs were photographed and scored for metastatic tumor nodules; mean±s.d., n=5, ***P<0.0001, unpaired Student's t-test. (c) Lysates from MDA-MB-231 cells stably transfected with empty vector (Vector), wild-type hSH3BGRL and its somatic mutant R76C and immunoblotted with the indicated antibodies. The averaged relative protein expression level is quantified and shown under the immediate panels. (d) Effect of hSH3BGRL and its somatic mutation R76C to lung metastasis in MDA-MB-231 cells. In total, 1x106 stably transfected cells in (c) were injected into nude mice through tail vein. After 15 days injection, the assayed mice were killed and the dissected lungs were photographed and the metastatic nodules were counted. mean±s.d., n=4, **P<0.001; ***P<0.0001, unpaired Student's t-test.
To exclude the possibility that murine cell context difference would facilitate the R76C mutant to promote metastasis, we further overexpressed hSH3BGRL-R76C mutant in MDA-MB-231 cells again to establish the stable cell pools. Western blotting results showed that like mSH3BGRL, R76C mutant can also activate Src and its downstream AKT and ERK1/2 effectors (Figure 6c), and enhanced tumor cell spreading in nude mice within 15 days after cell injection via tail veins, whereas wild-type hSH3BGRL overexpression evidently suppressed metastasis in vivo (Figure 6d). Taken together, our results obviously manifested that mutation of hSH3BGRL can reverts it as a tumor promoter or metastatic driver from a tumor suppressor.
hSH3BGRL is indeed upregulated in human tumors
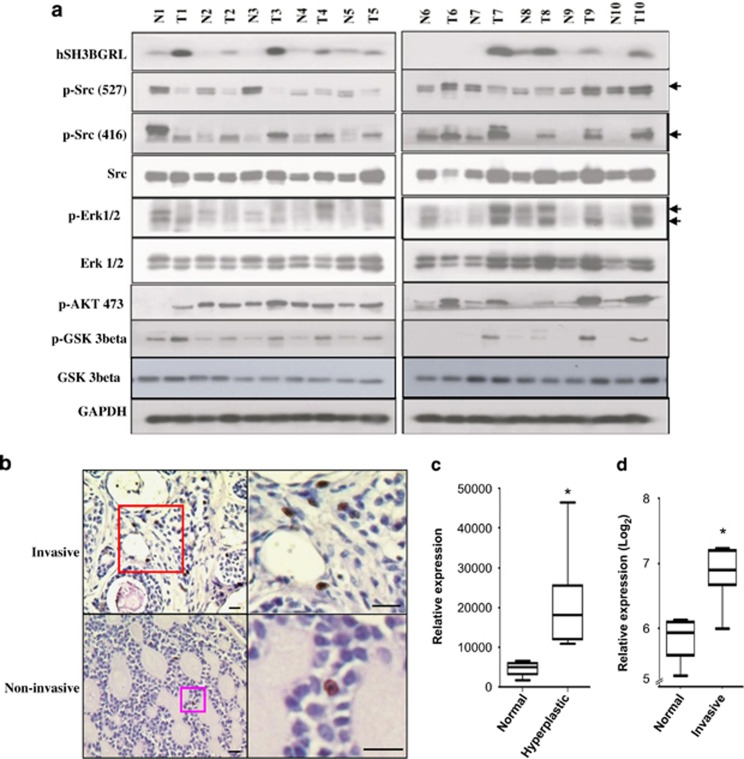
Given that mutation of hSH3BGRL can promote tumorigenesis and metastasis, we used collected breast tumor samples to investigate whether hSH3BGRL is upregulated in tumors. We produced the specific monoclonal antibody against hSH3BGRL (Supplementary Methods and Supplementary Figure 8) and checked hSH3BGRL expression in 10 pairs of fresh breast tumor samples by immunoblotting. Notably, we found that hSH3BGRL was expressed higher compared with that in the patient-matched surrounding normal tissues (Figure 7a). Additionally, hSH3BGRL upregulation in tumors accompanied with activated c-Src, AKT and ERK, and an immediate downstream effector, Gsk3β, activation further confirmed the overall activation of AKT signaling (Figure 7a). To demonstrate if hSH3BGRL might be related to other types of tumors, we analyzed another 30 oral squamous carcinoma samples and found hSH3GRL highly expressed in 7/30 invasive oral squamous carcinomas, with lower expression in noninvasive samples (Figure 7b). Next, we searched the GEO database and found that hSH3BGRL transcript expression was elevated in both epithelium of hyperplastic lobular (Figure 7c) and breast invasive tumors (Figure 7d) compared with normal breast tissues, demonstrating that increased hSH3BGRL expression might be related to malignancy progression.
Figure 7.
Upregulation of hSH3BGRL is involved in patient tumors. (a) Immunoblotting of lysates from breast tumor samples (T) and their conrresponding normal tissues (N) with the indicated antibodies, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is used as loading control. The predicted protein bands are indicated by an arrow. (b) Immunohistochemical staining of squamous oral carcinoma with monoclonal anti-hSH3BGRL antibody (Clone 246). The upper panel shows the higher grade of malignancy compared with the lower panel. Bar=25 μm. (c) hSH3BGRL mRNA expression in epithelial cells of hyperplastic enlarged lobular units of breast compared with the normal tissues. Student's t-test analysis was used, *P<0.05, n=8. Analysis was based on the information from GEO profile database (ID: GDS2739). (d) hSH3BGRL mRNA expression in breast invasive tumors compared with the normal tissues. *P<0.05, n=6. Analysis was carried out as in (c) (data from GEO ID: GDS4114).
Discussion
In this study, we report that mSH3BGRL promotes cancer metastasis as a novel activator of c-Src to simultaneously trigger downstream FAK, AKT and MAPK signaling pathways. Knockdown of c-Src completely abrogated mSH3BGRL's effect, characterizing mSH3BGRL as a novel c-Src transactivator. Unexpectedly, hSH3BGRL, its human ortholog, strongly suppressed tumor formation and metastasis—a completely reversible phenotype by a single amino-acid substitution, V108A to mimic the mSH3BGRL sequence. Importantly, somatic mutation of hSH3BGRL indeed can work in the same way.
Our results suggest that unlike the mSH3BGRL mimetic hSH3BGRL-V108A and R76C mutant, wild-type hSH3BGRL has reduced binding to the inactive form of c-Src. This might account for its reduced ability to promote c-Src activation and subsequent downstream metastasis-associated MAPK and AKT activation, as compared with mSH3BGRL. Although we did not directly observe V108A substitution in hSH3BGRL in COSMIC database, here we clearly demonstrated that somatic mutations, including R76C in hSH3BGRL can similarly function as mSH3BGRL to promote tumorigenesis and metastasis, rather than its tumor-suppressive properties of wild-type hSH3BGRL (Figure 6). The upregulation of hSH3BGRL in tumors could also be attributed to such mutation(s), although we did observe such mutations in our assayed 10 tumor samples and cell lines. Subsequent large scale of tumor sample sequencing experiments will be served to address these events.
On the other hand, we also clarified that the opposite SH3BGRL function in murine cells to human cells is not due to the difference of murine and human cellular contexts, but the mutations between them, especially like V108A substitution, as forced expression of hSH3BGRL in both murine CHO and human tumor cells hardly induce lung metastasis, compared with the control cells (Figures 5c and 6d). Additionally, murine-like hSH3BGRL-V108A and somatic mutation R76C can induce tumor metastasis in both CHO and MDA-MB-231 cells (Figures 6b and d). It is worth noting that our observations are reminiscent of the frequent upregulation of mutant p53 seen in aggressive human cancers.34, 35 For instance, regardless of the specific mutation, ectopic expression of mutant p53 variants deregulated the expression of a common pool of ~600 genes,33 lending weight to the possibility that different mutations of hSH3BGRL might also similarly result in a common acquisition of tumor-promoting property in human cancer samples, wherein there likely exists a mutation or some kinds of post-translational modification of hSH3BGRL that functions similarly as mutations, which would be further investigated.
Furthermore, our results solidly disclosed the fact that native functional difference between wild-type human and mSH3BGRL in tumorigenesis, independent of their own specific cellular contexts, indicates existence of controversial role of given evolutionarily related proteins or factors. This observation thus partly interprets the inconsistence of therapeutical outputs in clinical trials from model animals.
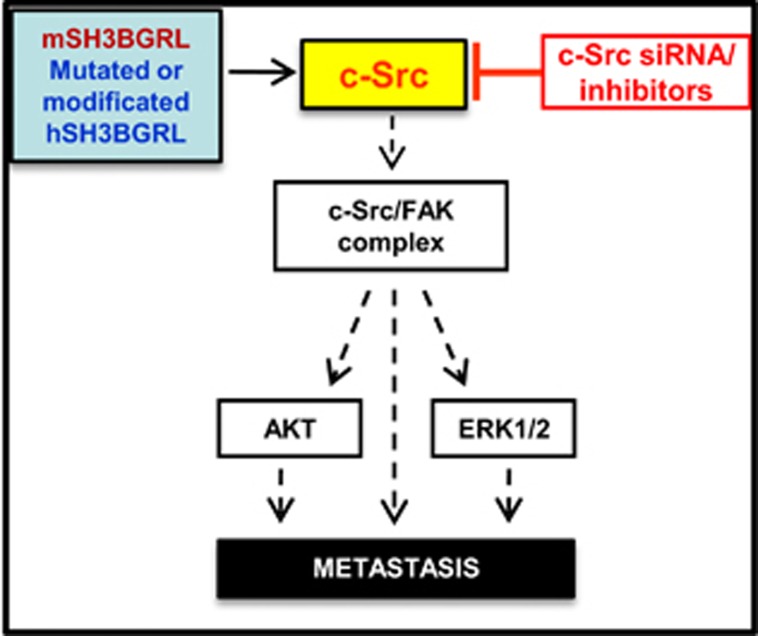
SH3BGRL is devoid of any known functional enzymatic domains.12 In light of our results, we hypothesize that it likely functions primarily as a scaffold/binding partner to regulate c-Src activation. As activating mutations on the inhibitory C terminus of c-Src are rarely observed in human cancers,8, 9, 10 binding proteins, such as SH3BGRL, might have important roles in the regulation of c-Src activity. Considering previous reports and our results herein, we propose SH3BGRL as a novel c-Src-activating scaffold with potential diagnostic and therapeutic reference against cancer metastasis by interrupting upregulated hSH3BGRL in tumors (Figure 8), rather than general blocking c-Src that can induce secondary side effects to normal cells. Although the detailed mechanism for mSH3BGRL or hSH3BGRL mutant-mediated activation of c-Src is unknown, future attempts at resolving the SH3BGRL–SH3-c-Src interaction should shed light into the structural elements involved and spur development of inhibitors of this interaction as potential drug candidates. Blocking c-Src hyperactivation by targeting SH3BGRL would be an efficient strategy to inhibit both MAPK and PI3K/AKT signaling pathways, which are frequently activated in many types of tumors.
Figure 8.
Schematic signaling pathway for mSH3BGRL or hSH3BGRL mutant-mediated metastasis. mSH3BGRL-alike hSH3BGRL mutants or with modification activate the downstream FAK/Src complex and the further AKT and Erk signals via c-Src kinase, and selectively blocking hyperactive c-Src by specific antagonists, including siRNAs could effectively delay tumor cell metastasis.
Materials and Methods
For plasmids, proteins, antibodies, oligos and detailed protocols, see Supplementary Information online.
Stable cell pools
Cells were transfected with vectors encoding DsRed-mSH3BGRL (CT-26), EGFP-SH3BGRL (CHO), hSH3BGRL or its mutations, or appropriate controls using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Transfected cells were selected 48 h later with 0.75 mg/ml hygromycin (for pDsRed-Monomer-Hyg-C1) or neomycin (for pEGFP-C1) for 3–4 weeks. Fluorescent cells were subsequently sorted from non-fluorescent cells using FACS to obtain stable cell pools. For knockdown of c-Src in CT-26 cells overexpressing SH3BGRL, four shRNA constructs against mouse Src (OriGene, Rockville, MD, USA; cat. no. TG514248) were transfected to generate stable cell pools using 1 μg/ml puromycin selection for 3–4 weeks. Similarly, two shRNA constructs against hSH3BGRL (OriGene; cat. no. TG309466) were transfected to establish MDA-MB-231 hSH3BGRL stable knockdown cell pools.
Invasion assays
These were carried out as described previously.27 Briefly, cells (1.84 × 105) were added to the upper chamber of coated transwells inserts (BD Biocoat Matrigel, BD Bioscience, San Jose, CA, USA) in a serum-free medium containing 0.1% bovine serum albumin. The total number of invaded cells in the lower chamber was counted after 48 h of incubation at 37 °C with 5% CO2 according to the manufacturer's protocol.
Identification of SH3BGRL-binding proteins
For identification of SH3BGRL-binding proteins, 25 μg of recombinant GST or GST-SH3BGRL was incubated with a commercially printed SH3 domain array derived from 38 unique proteins (SH3 Array II). Arrays were processed and analyzed according to the manufacturer's recommendations (Panomics Inc., Redwood City, CA, USA).
Immunoprecipitation and western blotting
For immunoprecipitation, clarified cell lysates were incubated with protein G-agarose beads crosslinked with anti-SH3BGRL antibody overnight at 4 °C on a rotator. After washing with lysis buffer, bound proteins were eluted and analyzed. All experiments were repeated three times and the relative expression level of key proteins is statistically analyzed by protein band intensitometry and shown as the average values. For detailed procedures, see Supplementary Information online.
siRNA experiments
For FAK siRNA (small interfering RNA) treatment, 80 pmol of siRNAs were transfected into cells using Lipofectamine 2000 (Invitrogen) in a six-well plate, according to the manufacturer's instructions. At 48 h post-transfection, cells were harvested and analyzed.
GTPase activity assays
For details of GST-PBD and GST-Raf production, see Supplementary Information. After measuring cell lysate concentration, 1 mg of total cell lysates were incubated with the PBD and Raf-conjugated Sepharose beads in the lysis buffer, followed by shaking at 4 °C for 1 h. Bound proteins were then washed five times with lysis buffer and boiled in 1 × sodium dodecyl sulfate–polyacrylamide gel electrophoresis loading buffer for 10 min before analysis.
Experimental metastasis assays
Five 10-week-old nude mice (Jackson Lab, Bar Harbor, ME, USA) in each group were injected with 1x106 cells intravenously via the tail vein and killed 4–5 weeks later. All of the tissues were examined for metastasis. Lungs were photographed for gross morphology and scored for the number of metastatic nodules using the formula N/3, where N is the diameter of each visible nodule in millimeter.
Materials and methods used are available in detail in the Supplementary section.
Acknowledgments
This work was supported by research grants from the National Natural Science Foundation of China (WH; No. 81171947), Project of International Collaboration in Science and Technology, Guangdong Province (WH; No. 2014A050503030) and The Agency of Science, Technology and Research (A* STAR), Singapore. We are also thankful to Professor Wanjin Hong, Professor Shulan Yang and Mr Mingming Zhang in our group for critical reading of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell 2006; 127: 679–695. [DOI] [PubMed] [Google Scholar]
- Johnson FM, Gallick GE. SRC family nonreceptor tyrosine kinases as molecular targets for cancer therapy. Anticancer Agents Med Chem 2007; 7: 651–659. [DOI] [PubMed] [Google Scholar]
- Liu W, Yue F, Zheng M, Merlot A, Bae DH, Huang M et al. The proto-oncogene c-Src and its downstream signaling pathways are inhibited by the metastasis suppressor, NDRG1. Oncotarget 2015; 6: 8851–8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Larroque-Lombard AL, Peyrard L, Thauvin C, Rachid Z, Williams C et al. Target modulation by a kinase inhibitor engineered to induce a tandem blockade of the epidermal growth factor receptor (EGFR) and c-Src: the concept of type III combi-targeting. PLoS One 2015; 10: e0117215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen B, Johnson FM. Regulation of SRC family kinases in human cancers. J Signal Transduct 2011; 2011: 865819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorge JD, Jakymiw A, Fujita DJ. Selected glimpses into the activation and function of Src kinase. Oncogene 2000; 19: 5620–5635. [DOI] [PubMed] [Google Scholar]
- Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell 1999; 3: 629–638. [DOI] [PubMed] [Google Scholar]
- Irby RB, Mao W, Coppola D, Kang J, Loubeau JM, Trudeau W et al. Activating SRC mutation in a subset of advanced human colon cancers. Nat Genet 1999; 21: 187–190. [DOI] [PubMed] [Google Scholar]
- Nilbert M, Fernebro E. Lack of activating c-SRC mutations at codon 531 in rectal cancer. Cancer Genet Cytogenet 2000; 121: 94–95. [DOI] [PubMed] [Google Scholar]
- Tan YX, Wang HT, Zhang P, Yan ZH, Dai GL, Wu MC et al. C-src activating mutation analysis in Chinese patients with colorectal cancer. World J Gastroenterol 2005; 11: 2351–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeo A, Mazzocco M, Arrigo P, Vidal-Taboada JM, Oliva R, Pirola B et al. Identification and characterization of a new human gene encoding a small protein with high homology to the proline-rich region of the SH3BGR gene. Biochem Biophys Res Commun 1998; 247: 302–306. [DOI] [PubMed] [Google Scholar]
- Mazzocco M, Maffei M, Egeo A, Vergano A, Arrigo P, Di Lisi R et al. The identification of a novel human homologue of the SH3 binding glutamic acid-rich (SH3BGR) gene establishes a new family of highly conserved small proteins related to Thioredoxin superfamily. Gene 2002; 291: 233–239. [DOI] [PubMed] [Google Scholar]
- Majid SM, Liss AS, You M, Bose HR. The suppression of SH3BGRL is important for v-Rel-mediated transformation. Oncogene 2006; 25: 756–768. [DOI] [PubMed] [Google Scholar]
- Berleth ES, Nadadur S, Henn AD, Eppolito C, Shiojiri S, Gurtoo HL et al. Identification, characterization, and cloning of TIP-B1, a novel protein inhibitor of tumor necrosis factor-induced lysis. Cancer Res 1999; 59: 5497–5506. [PubMed] [Google Scholar]
- Berleth ES, Henn AD, Gurtoo HL, Wollman R, Alderfer JL, Mihich E et al. A novel tumor necrosis factor-alpha inhibitory protein, TIP-B1. Int J Immunopharmacol 2000; 22: 1137–1142. [DOI] [PubMed] [Google Scholar]
- Henn AD, Berleth ES, Mihich E, Ehrke MJ. Changes in cytosolic and membrane TNF inhibitory protein-B1 (TIP-B1) levels associated with protection from TNF-induced cytotoxicity. FASEB J 2001; 15: 1315–1317. [DOI] [PubMed] [Google Scholar]
- van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002; 415: 530–536. [DOI] [PubMed] [Google Scholar]
- Abba MC, Hu Y, Sun H, Drake JA, Gaddis S, Baggerly K et al. Gene expression signature of estrogen receptor alpha status in breast cancer. BMC Genomics 2005; 6: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Bardelli A, Buckhaults P, Velculescu VE, Rago C St, Croix B et al. A phosphatase associated with metastasis of colorectal cancer. Science 2001; 294: 1343–1346. [DOI] [PubMed] [Google Scholar]
- Wang H, Quah SY, Dong JM, Manser E, Tang JP, Zeng Q. PRL-3 down-regulates PTEN expression and signals through PI3K to promote epithelial-mesenchymal transition. Cancer Res 2007; 67: 2922–2926. [DOI] [PubMed] [Google Scholar]
- Zeng Q, Dong JM, Guo K, Li J, Tan HX, Koh V et al. PRL-3 and PRL-1 promote cell migration, invasion, and metastasis. Cancer Res 2003; 63: 2716–2722. [PubMed] [Google Scholar]
- Elkin M, Vlodavsky I. Tail vein assay of cancer metastasis. Curr Protoc Cell Biol 2001; Chapter 19: Unit 19.2. [DOI] [PubMed] [Google Scholar]
- Brattain MG, Strobel-Stevens J, Fine D, Webb M, Sarrif AM. Establishment of mouse colonic carcinoma cell lines with different metastatic properties. Cancer Res 1980; 40: 2142–2146. [PubMed] [Google Scholar]
- Warmuth M, Damoiseaux R, Liu Y, Fabbro D, Gray N. SRC family kinases: potential targets for the treatment of human cancer and leukemia. Curr Pharm Des 2003; 9: 2043–2059. [DOI] [PubMed] [Google Scholar]
- Roskoski R Jr. Signaling by Kit protein-tyrosine kinase—the stem cell factor receptor. Biochem Biophys Res Commun 2005; 337: 1–13. [DOI] [PubMed] [Google Scholar]
- Arcaro A, Aubert M, Espinosa del Hierro ME, Khanzada UK, Angelidou S, Tetley TD et al. Critical role for lipid raft-associated Src kinases in activation of PI3K-Akt signalling. Cell Signal 2007; 19: 1081–1092. [DOI] [PubMed] [Google Scholar]
- Schlaepfer DD, Hunter T. Evidence for in vivo phosphorylation of the Grb2 SH2-domain binding site on focal adhesion kinase by Src-family protein-tyrosine kinases. Mol Cell Biol 1996; 16: 5623–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger R, Krebs EG. The MAPK signaling cascade. FASEB J 1995; 9: 726–735. [PubMed] [Google Scholar]
- Piguet AC, Dufour JF. PI(3)K/PTEN/AKT pathway. J Hepatol 2011; 54: 1317–1319. [DOI] [PubMed] [Google Scholar]
- Schlaepfer DD, Mitra SK. Multiple connections link FAK to cell motility and invasion. Curr Opin Genet Dev 2004; 14: 92–101. [DOI] [PubMed] [Google Scholar]
- Mitra SK, Schlaepfer DD. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr Opin Cell Biol 2006; 18: 516–523. [DOI] [PubMed] [Google Scholar]
- Schaller MD. Cellular functions of FAK kinases: insight into molecular mechanisms and novel functions. J Cell Sci 2010; 123: 1007–1013. [DOI] [PubMed] [Google Scholar]
- Garritano S, Inga A, Gemignani F, Landi S. More targets, more pathways and more clues for mutant p53. Oncogenesis 2013; 2: e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev 2012; 26: 1268–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal A, Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Res 2000; 60: 6788–6793. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.