Abstract
The V-ATPase is a proton pump consisting of a membrane-integral V0 sector and a peripheral V1 sector, which carries the ATPase activity. In vitro studies of yeast vacuole fusion and evidence from worms, flies, zebrafish and mice suggested that V0 interacts with the SNARE machinery for membrane fusion, that it promotes the induction of hemifusion and that this activity requires physical presence of V0 rather than its proton pump activity. A recent in vivo study in yeast has challenged these interpretations, concluding that fusion required solely lumenal acidification but not the V0 sector itself. Here, we identify the reasons for this discrepancy and reconcile it. We find that acute pharmacological or physiological inhibition of V-ATPase pump activity de-acidifies the vacuole lumen in living yeast cells within minutes. Time-lapse microscopy revealed that de-acidification induces vacuole fusion rather than inhibiting it. Cells expressing mutated V0 subunits that maintain vacuolar acidity were blocked in this fusion. Thus, proton pump activity of the V-ATPase negatively regulates vacuole fusion in vivo. Vacuole fusion in vivo does, however, require physical presence of a fusion-competent V0 sector.
Vacuoles of yeast cells have served as a model to study important aspects of membrane fusion in eukaryotic cells. Key elements of the vacuolar fusion apparatus have been identified by genetic screens1,2,3,4,5,6 and an in vitro system reconstituting fusion of purified vacuoles served to elucidate important mechanistic aspects7,8,9. Reconstitution experiments with pure proteins provided reduced fusion systems allowing to study the contributions of vacuolar SNAREs, the tether complex HOPS and the Rab-GTPase Ypt7 separately10,11,12,13,14. Studies on the fusion of isolated yeast vacuoles indicated a physical role of the membrane-integral sector of the V-ATPase, V0, in vacuole fusion that could be separated from its function in proton pumping. They also showed that V0 interacts with vacuolar SNARE proteins7,9,15,16.
Data from the endo-lysosomal system and at the plasma membrane in several other systems confirmed this finding and suggested that stimulation of fusion processes by V0 is a widespread phenomenon. Unbiased genetic screens identified V0 alleles causing defects in the fusion of synaptic vesicles, multivesicular bodies and phagosomes15,17,18,19,20. Targeted approaches in vertebrates implicated V0 in secretion of insulin, neurotransmitters and catecholamines21,22,23,24. All these studies provide evidence that the observed fusion defects depend on the physical presence of V0 and on its interaction with SNAREs. In none of the cases could they be explained by a loss of V-ATPase proton pump function. A recent study addressing the role of V0 for yeast vacuole fusion in vivo challenged the conclusions from the studies mentioned above and suggested that fusion required only acidity of the organelle lumen but not the V0 sector25.
Care must be taken to distinguish acidification-dependent and purely physical roles of the V-ATPase in every trafficking reaction under study. In yeast, a genetic separation of the functions of V0 in proton translocation and vacuole fusion was achieved by a random mutagenesis screen which produced point mutants in the V0 proteolipid subunits c (Vma3), c’ (Vma11) and c” (Vma16) and by protein fusions between c and c” subunits, which largely maintain proton pump activity but show strong reductions in fusion activity in vitro15,26. The substitutions that inactivate vacuole fusion are all in the transmembrane domains of the different V0 proteolipids, i.e. at sites where the affected amino acids could not directly interact with any V1 subunit. Therefore, their effect was explained by an altered conformation of V0 that allows proton pump activity but does not support the activity of V0 in vacuole fusion.
In living yeast cells, vacuoles fuse during organelle transmission in mitosis. Vacuoles produce tubulo-vesicular inheritance structures that are transmitted to daughter cells and fuse there27. A direct, real-time assay of this fusion event in vivo poses challenges because the process is slow and poorly synchronized. Therefore, vacuole morphology at steady state is frequently used as a substitute. This is based on the assumption that, upon a strong block of vacuole fusion, vacuolar fragments should accumulate over time, leading to an overall fragmented appearance of the vacuolar compartment. In line with this, vacuolar fusion problems often correlate with a fragmented vacuolar phenotype2,6. Vacuole inheritance and fusion occur also during mating. Upon mating, the zygote divides and its first diploid daughter cell receives vacuolar membranes from both mating partners, which fuse in the daughter27. Vacuolar material also flows from the bud to the mothers but this process has not been investigated in detail. Yeast mating has recently been used to analyze the in vivo requirements of vacuole fusion in daughters. The results led the authors to conclude that vacuole fusion in vivo required only vacuolar acidity and not the physical properties of V0. These results challenged in vitro studies from several laboratories that had suggested that the fusion of isolated vacuoles in vitro requires the physical presence of V0 and not V-ATPase pump function7,9,15,16,28,29,30.
In order to address this contradiction between these in vivo and in vitro observations we investigated the behaviour of vacuoles upon loss of the vacuolar proton pump function in vivo. We relied on video microscopy to analyze the morphological changes that follow the acute inactivation of V-ATPase proton pump activity and the resulting loss of vacuolar acidity. Our observations support, in good agreement with all in vitro results, the physical and pump-independent role of the V0 sector to promote vacuole fusion and suggest that the proton gradient counteracts vacuole fusion in vivo. We resolve the discrepancy between our observations and those of a previous in vivo study25.
Results
V0 reversibly associates with the peripheral V1 sector in order to form the V-ATPase holo-enzyme, which is active as a proton pump (Fig. 1). V1 carries the ATPase activity. In absence of V1, V0 cannot conduct protons but it assembles normally and is targeted to vacuoles31,32 (Suppl. Fig. 1). Thus, if a fusion defect is due to a loss of proton pump activity it should become equally visible in a V1 mutant, which eliminates pump activity as effectively as a V0 mutant but leaves V0 intact. If V0 deletion shows additional defects compared to a V1 deletion, those cannot be accounted for by defects in proton pump activity, but they are consistent with a physical role of V0.
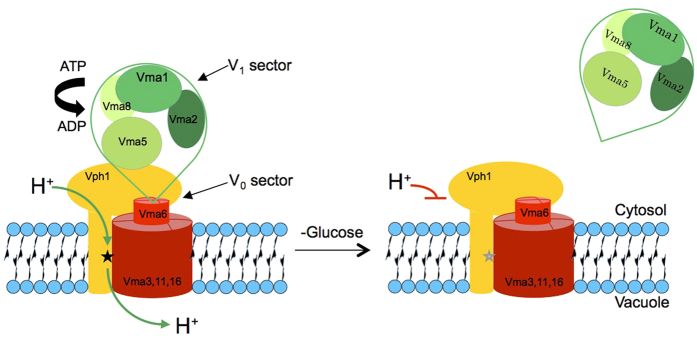
Figure 1. Organisation of V-ATPase and its glucose-dependent dissociation.
V-ATPase is composed of a peripheral sector V1 (green), which carries the ATPase activity, and the proton-conducting, membrane-integral V0 sector (red and yellow). Only the V1 and V0 sector subunits used in this work are indicated. The black star in the V0 a-subunit Vph1 represents the residue R735. Glucose withdrawal releases V1 from V0 and renders it soluble in the cytosol. Proton conductance by V0 is blocked in this state.
Vacuoles in pump-deficient V-ATPase V1 mutants fuse into a single organelle
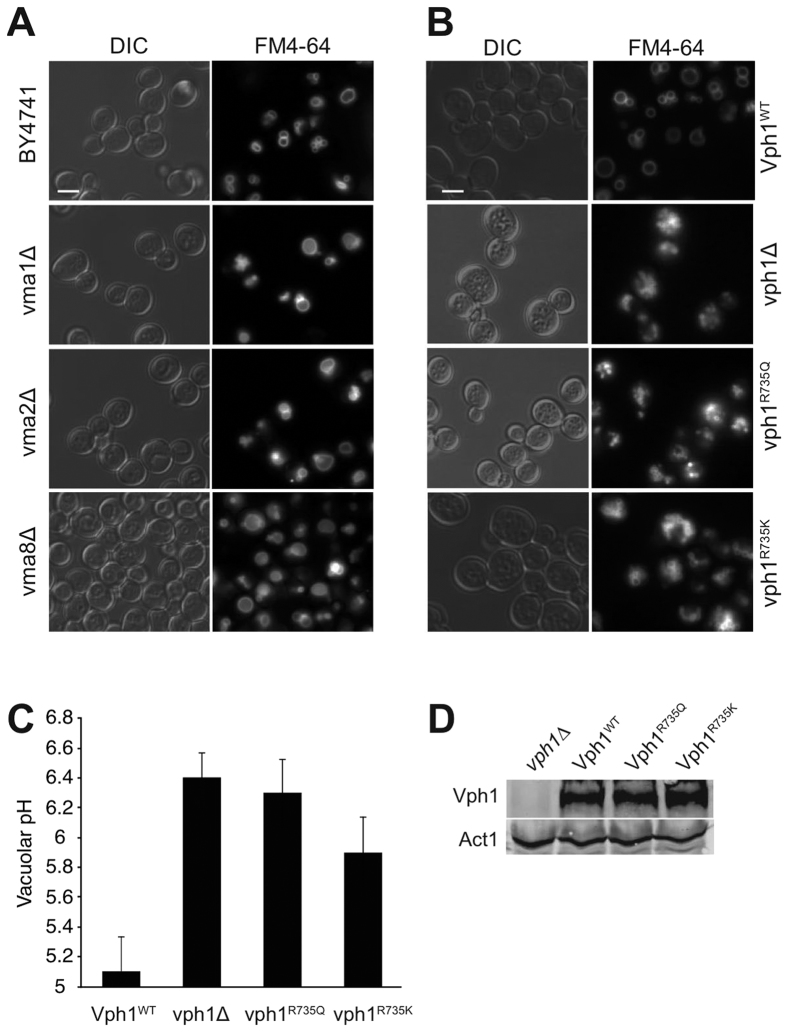
Vacuole fusion problems are often correlated with vacuolar fragmentation in vivo6,33,34. Using this simple in vivo criterion for vacuolar fusion activity we assessed vacuolar structure in a series of V1 and V0 mutants. We used several mutants in which one of the eight V1 subunits had been deleted, which abolishes proton pump function but leaves the V0 sector assembled and un-modified35. Under logarithmic growth, V1 deletion mutants showed a large vacuole whereas the isogenic wildtype (BY4741) showed several smaller vacuoles per cell (Fig. 2A). By contrast, mutants lacking the vacuolar isoform of the V0 a-subunit Vph1 showed strong vacuolar fragmentation, as did cells expressing a vph1R735Q or a vph1R735K allele (Fig. 2B,D). Since the vph1R735Q allele abolishes V-ATPase pump activity but maintains an assembled V0 sector, the presence of fragmented vacuoles had been taken as an indication that vacuole fusion requires V-ATPase pump activity25 rather than SNARE-dependent conformational changes of V015. A strong caveat for this interpretation is that vph1R735Q interferes with the physiological dissociation of V1 from V0, which is triggered by glucose depletion36. Since R735 is in one of the transmembrane regions of the a-subunit, it could not directly interact with any V1 subunit. Its effect on V0/V1 association is therefore best explained by an influence on the conformation of V0, which is quite flexible and dependent on Vph137,38. This is consistent with the phenotype of the vph1R735K mutant, which also shows fragmented vacuoles. However, in contrast to vph1R735Q, vph1R735K does support growth in alkaline media39, suggesting that V-ATPase remains at least partially functional as a pump. We found the vacuolar pH in vph1R735K cells to be 5.9 whereas it was 6.3 in vph1R735Q mutants, 6.4 in vph1Δ cells and 5.1 in wildtype cells (Fig. 2C). Coonrod et al. had concluded that the fusion defect of vph1Δ vacuoles in vivo could be rescued by expressing an artificial proton pump that lowered vacuolar pH by less than 0.2 pH units, from 6.6 to 6.4. If this were true, vph1R735K vacuoles should be sufficiently acidified to fuse into a large, intact organelle. That this is not observed is consistent with an influence of R735 substitutions on vacuole fusion via the conformation or interactions of V0.
Figure 2. The morphology produced by vph1 mutations is atypical among mutants lacking V-ATPase pump activity.
BY4741 cells were grown in buffered media to logarithmic phase, stained with FM4-64, and visualized by fluorescence microscopy and DIC. Scale bar: 5 μm. (A) Cells with deletions in various V1 subunits. (B) vph1Δ cells reconstituted with plasmids expressing the indicated alleles. (C) Cells from panel B were loaded with BCECF-AM and vacuolar pH was determined by fluorescence spectroscopy. Error bars represent Standard Deviation. (n = 9–22) (D) Total cell extracts of the cells from panel B were analyzed by SDS-PAGE and Western blotting against Vph1 and actin (Act1).
Acute inactivation of V-ATPase induces vacuole fusion in vivo
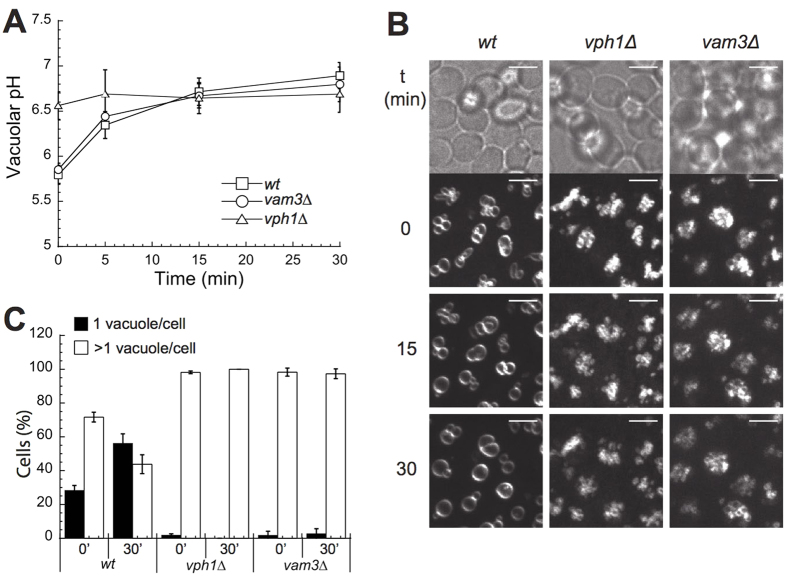
The fusion of vacuoles into a single large vacuole in V1 mutants, which lack V-ATPase pump activity entirely, suggested that the lack of pump activity might actually favor vacuole fusion in vivo. We addressed this hypothesis by time-lapse fluorescence microscopy following acute inactivation of the V-ATPase. Concanamycin A is a highly potent and specific cell-permeable inhibitor of V-ATPases that allows to rapidly inactivate this proton pump in living cells. We added Concanamycin A to logarithmically growing cells in which vacuoles had been stained by the vital dye FM4-64. Cells were immobilized in chamber slides so that we could follow the changes in vacuolar structure by video-microscopy over the next 30 min. In parallel, we measured changes in vacuolar pH over time. Under these conditions, wildtype cells showed a vacuolar pH of around 5.6 and several smaller vacuoles (Fig. 3). Addition of concanamycin A led to a progressive increase in vacuolar pH to 6.5 after 15 min and to 6.7 after 30 min (Fig. 3A). In good correlation to the loss of vacuolar acidity, the vacuoles in many cells began to fuse after 10 min and this process was completed for essentially all cells in the population within 30 min (Fig. 3B,C). We also tested cells lacking the vacuolar SNARE Vam3, a core protein of the vacuolar fusion machinery. vam3Δ cells showed a vacuolar pH of 5.8. Addition of concanamycin A neutralized their pH to 6.7 but the vacuoles of vam3Δ cells did not fuse (Fig. 3B,C).
Figure 3. Vacuole fusion after addition of the pumping inhibitor concanamycin A.
(A) Vacuoles of the indicated BY4741 cells were loaded with BCECF-AM and vacuolar pH was measured before and at different time points after addition of concanamycin (A). (B) Logarithmically grown cells stained with the vacuolar dye FM4-64 were immobilized in chamber slides as described and overlaid with 200 μl SC media. 200 μl media containing 4 μM concanamycin A (from a 200 μM stock in DMSO) were added to each well (final concentration 2 μM). At the indicated time points, image stacks were acquired using a confocal spinning disc setup. Maximum projections of the stacks are shown. Scale bar: 5 μm. (C) Quantification of the experiments in panel B. A number of 60–170 cells were classified according to the number of vacuoles per cell (n = 4).
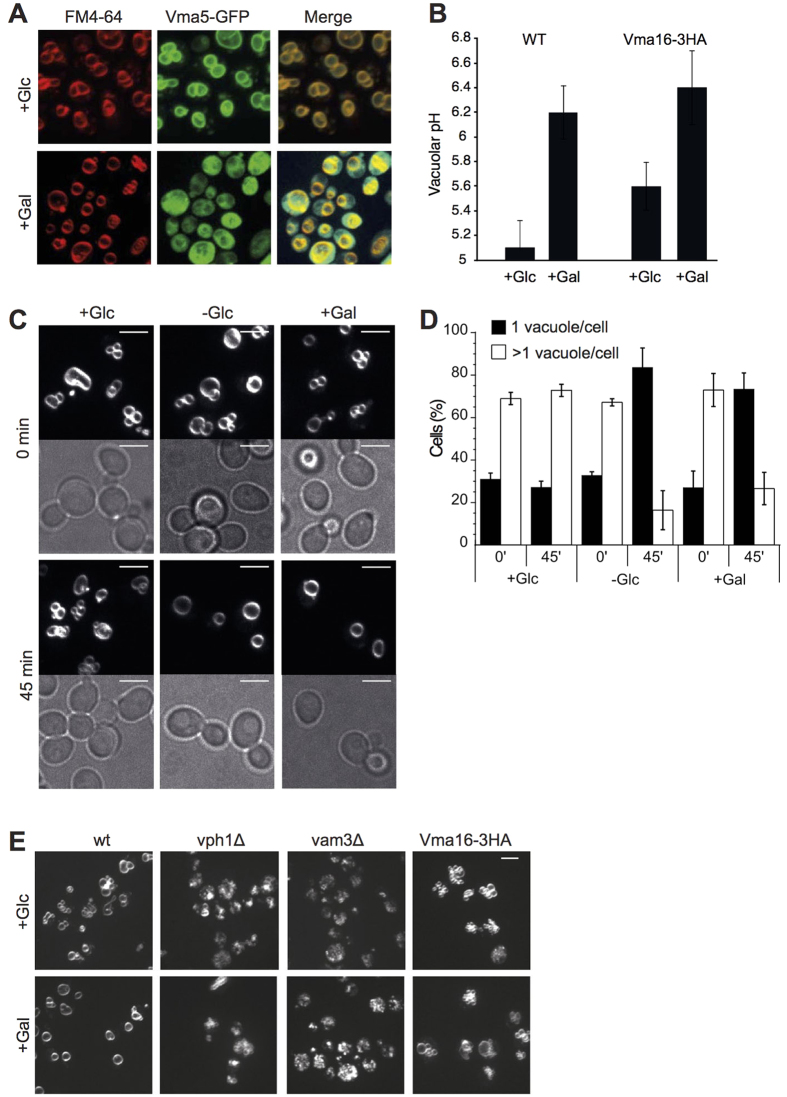
As an alternative, non-pharmacological approach to inactivate vacuolar proton pump activity we used acute glucose withdrawal. In case of glucose shortage, yeast cells rapidly dissociate their V1 sectors from V0 in order to inactivate their V-ATPases, leading to de-acidification of the vacuole40,41,42. Switching from the preferred substrate glucose to other carbon sources, such as galactose or amino acids, requires an adaptation phase, which entails a transient bottleneck in carbon metabolism and downregulation of V-ATPase activity. We assessed the changes of vacuolar structure and vacuolar pH under these conditions. Cells growing on buffered medium with glucose (SC+Glc) were shifted to either SC-Glc (without glucose), SC+Gal (with galactose) or SC+Glc, and analyzed by fluorescence microscopy (Fig. 4). Withdrawal of glucose, or its replacement by galactose, led to a displacement of the V1 subunit Vma5-GFP from vacuoles into the cytosol (Fig. 4A). Vacuoles of wildtype cells were de-acidified from pH 5.1 to pH 6.2 (Fig. 4B) and fused into a single large organelle (Fig. 4C,D). The transition from several clustered vacuoles to a single fused vacuole was usually direct, suggesting that over this short time period re-fission of vacuoles hardly occurs (Fig. 4C, Suppl. Movies 1 and 2). Glucose withdrawal did not induce vacuoles fusion in cells lacking Vph1 or the SNARE Vam3 (Fig. 4E). We also tested V0 mutants that have a functional proton pump but are deficient for vacuole fusion in vitro, such as a strain in which the V0 proteolipids Vma16 and Vma3 are expressed as a single fusion protein (vma16-3HA)26. Upon shift from glucose to galactose, vma16-3HA cells showed vacuolar de-acidification from pH 5.6 to pH 6.4 (Fig. 4B). However, their vacuoles fused only poorly (Fig. 4E), consistently with the low vacuolar fusion activity that this mutant had shown in vitro (Strasser et al.15). Thus, both pharmacological and metabolic reduction of V-ATPase proton pump function induce vacuole fusion. This fusion depends on vacuolar SANREs and requires physical properties of V0 that are perturbed in the pump-active mutant vma16-3HA.
Figure 4. Vacuoles coalesce upon shift to media without glucose.
(A) Effect of glucose withdrawal on V-ATPase assembly. Logarithmically growing BY4741 cells expressing Vma5-GFP were labeled with FM4-64, shifted from glucose media (+Glc) to media containing galactose (+Gal) and analyzed by fluorescence microscopy immediately or after 45 min incubation at 30 °C. (B) Vacuolar de-acidification in response to withdrawal of glucose. The indicated cells were grown as in (A) and vacuolar pH was measured before and after shift to galactose. Error bars represent Standard Deviation (n = 6). (C) Logarithmically growing cells, labeled with FM4-64, were shifted from glucose media to media containing no glucose (-Glc), galactose (+Gal) or glucose (+Glc, control). Cells were analyzed by fluorescence microscopy immediately or after 45 min of incubation at 30 °C. Scale bars: 5 μm. (D) Quantification of experiments as shown in (C). 50–220 cells were classified according to the number of vacuoles per cell (n = 3). (E) Response of mutants to glucose withdrawal. The indicated strains were subjected to a shift from glucose to galactose-containing media as in C and analyzed by fluorescence microscopy. Only FM4-64 fluorescence is shown.
Vacuole fusion in zygotes is independent of the vacuolar R-SNARE Nyv1
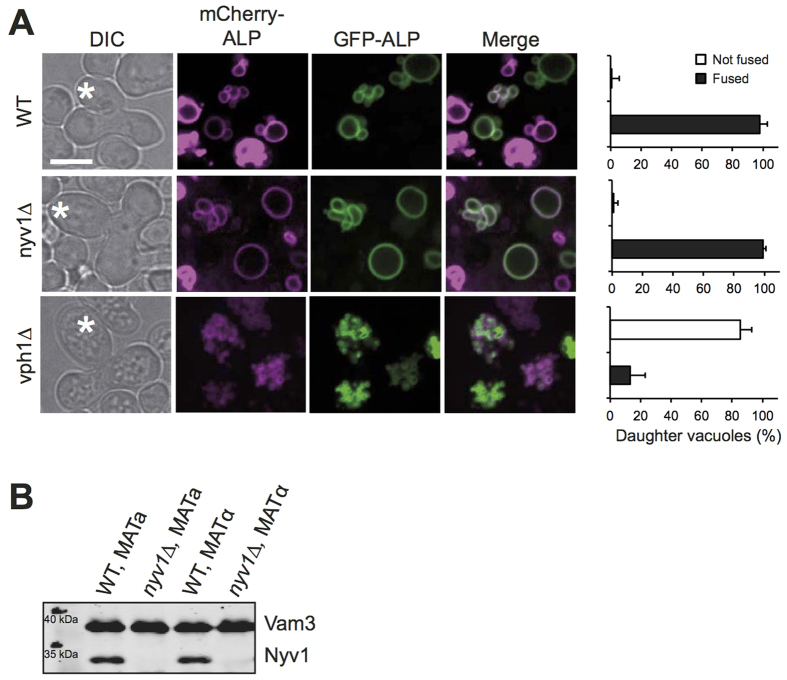
A recent in vivo study had concluded that vacuolar acidity rather than a physical contribution of V0 is necessary and sufficient to induce vacuole fusion25. In order to understand the qualitative differences between the in vivo observations in this study and ours, we revisited the in vivo approaches taken by Coonrod et al. These authors relied on yeast mating to study vacuole fusion. Upon mating, a yeast zygote divides and its first diploid daughter cell receives vacuolar vesicles from both mating partners, which fuse in the daughter27. Enzymatic and microscopic assays had been set up as readouts for this fusion event. The mating partners were genetically modified such that one partner carried a plasmid expressing a pro-alkaline phosphatase (pro-ALP) in its vacuoles and the other partner expressed the vacuolar maturase Pep4. Alternatively, the vacuoles of the two mating partners were labeled by differentially colored fluorescent proteins. Vacuole fusion in the daughter results in co-localization of the two fluorescent proteins and it gives Pep4 access to the pro-ALP, which is thereby converted into its mature form. We repeated the mating experiments described by Coonrod et al., using their fluorescently labeled mating partners that carried the vacuolar alkaline phosphatase ALP labeled with mCherry or GFP, respectively. We observed efficient co-localization of both fluorophores in daughters of wildtype zygotes but not in those of vph1Δ zygotes (Fig. 5A), confirming their observations25. However, both fluorophores efficiently co-localized also in daughters lacking the vacuolar R-SNARE Nyv1, suggesting that this protein is not required for vacuole fusion during mating. The lack of Nyv1 in these cells had been verified by Western blotting (Fig. 5B). This result is consistent with a previous observation that mixing of vacuole content during mating is Nyv1-independent1. It contradicts Coonrod et al., who reported that vacuole fusion during mating was Nyv1-dependent25. However, these authors had assessed the effect of Nyv1 only via the maturation of pro-ALP by Pep4.
Figure 5. Vacuole fusion upon mating.
(A) Plasmids expressing the PHO8 gene (ALP) fused to mCherry or GFP were introduced into isogenic a- and α-strains of BY4741 cells or the indicated nyv1Δ or vph1Δ mutants. a- and α-cells were grown over night in HC-Leu medium. In the morning, the cultures were diluted to OD600nm = 0.2, the respective a and α cells were mixed and incubated for 4–6 h at 30 °C. Upon emergence of the first diploid daughters (labeled by an asterisk), the cells were analyzed by spinning disc fluorescence microscopy. The frequency of daughter cells showing complete co-localization of mCherry and GFP in their vacuoles was determined from 90 cells (n = 3). (B) Vacuoles were isolated from the wildtype and nyv1Δ cells used in A. Their proteins were analyzed by SDS-PAGE and Western blotting against Vam3 and Nyv1.
Vacuole fusion during mating cannot be assayed by maturation of vacuolar pro-enzymes
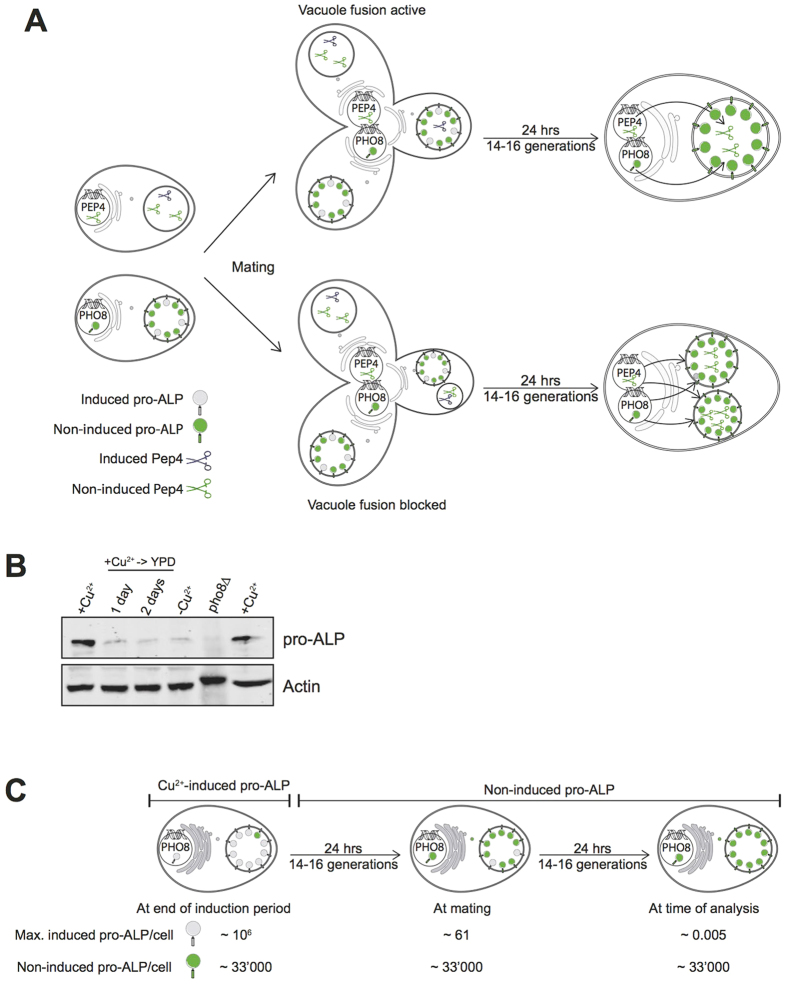
We tried to understand the discrepancies between our microscopic observations and the results from the enzymatic Pep4/ALP assay published by Coonrod et al. by analyzing their assay procedure. The enzymatic assay relies on the assumption that the pro-ALP activated by vacuole fusion in the diploid offspring stems from the haploid mating partner and was not produced during the diploid phase, between mating and the final analysis of ALP activity (Fig. 6A). This is critical because mating takes many hours and the cytosols and genomes of the mating partners are already mixed in the diploids long before the daughter cell emerges. As a consequence, the diploids can express both pro-ALP and PEP4 and deliver them to the nascent daughter that serves to analyze vacuole fusion. Coonrod et al. tried to circumvent this problem by expressing pro-ALP and PEP4 from the copper-inducible CUP1 promoter, which allows to accumulate these proteins prior to mating and repress further expression during and after mating. However, since mating is slow and not quantitative, the mating products had to be selected before ALP maturation was measured by Western blotting. This selection via complementary auxotrophic markers requires a 24 h culture period that allows only zygotes to grow. The CUP1 promoters are repressed during this period. However, the diploids divide, leading to tremendous dilution of the pre-accumulated pro-ALP and Pep4.
Figure 6. Limitations of the in vivo fusion assay employed by Coonrod et al.
(A) The figure illustrates the mating procedure and the potential fate of proteins expressed from plasmids carrying the PHO8 gene (producing pro-ALP) or the PEP4 gene (producing its maturase proteinase (A) under control of the inducible CUP1 promotor. Induced pro-ALP is indicated by grey circles, pro-ALP resulting from constitutive, non-induced background expression is indicated by green circles. Induced Pep4 is indicated by grey scissors, Pep4 resulting from non-induced background expression by black scissors. For discussion, see main text. (B) Dilution of induced pro-ALP over time. KEBY136 cells25 expressing PHO8 from the CUP1 promotor were grown on SC-URA plus 50 μM CuCl2 for 24 h and then shifted to YPD without copper. Aliquots of cells were withdrawn before and 24 and 48 h after the shift and analyzed by SDS-PAGE and Western blotting against ALP and actin. Samples for background controls were taken from cells that had never been induced by copper and from pho8Δ cells that lacked the PHO8-expressing plasmid (KEBY136). (C) Conservative estimation of the abundance of induced pro-ALP relative to pro-ALP resulting from non-induced expression of PHO8 from the CUP1 promotor, resulting from the published mating assay for vacuole fusion25. The first 24h period represents a period of cultivation of the cells used to chase pro-ALP from the biosynthetic pathway. The second 24 h culture period that follows mating is used to select for diploid offspring.
A further important constraint is that, like every inducible promoter, the CUP1 promoter does not provide an absolutely tight on/off switch. Non-induced expression from the CUP1 promotor occurs at 1/10–1/30 of the rate achieved upon copper induction25,43,44. This creates a low but constitutive background expression of pro-ALP and Pep4. Both proteins resulting from this background expression are delivered to all vacuoles in the diploids. But the assay can only work if the pro-ALP induced and accumulated before mating gives rise to the major fraction of ALP observed after mating, i.e. if it remains significantly above the level of constitutive background expression. We verified this by Western blotting of ALP (Fig. 6B). Induction by copper (Cu2+) generated a strong signal, as published25. Within 24 h after transfer to medium without copper, this signal declined to a background level of approximately 8% that remained constant during futher growth of the cells for up to 92 h. It was equal to the signal observed before induction (-Cu2+). This signal was absent in extracts from cells lacking pro-ALP (pho8Δ). Thus, cell division dilutes the pre-accumulated pro-ALP to background levels in less than 24 h. Since the mating assay includes two growth periods for the diploids of 24 h each, both of which occur after the CUP1- pro-ALP construct has been shut down, the ALP signal measured after mating is dominated by the constitutively expressed ALP that was produced in the diploid.
This conclusion is strengthened by a conservative estimation of the fate of the induced pro-ALP during the maturation-based mating assay (Fig. 6C). The generation time of yeast in rich media is around 90–100 minutes, i.e. a cultivation period of 24 h allows 14 to 16 divisions. After the cessation of copper induction, the cells grow in rich media for 24 h, they are mated for 3–5 h and then cultivated for yet another 24 h in order to select for the diploids25. During both 24 h incubations, the cells divide 14–16 times. This dilutes the pro-ALP that has been produced during the preceding induction period by a factor of 228 to 232, i.e. more than 200 million-fold. The most abundantly expressed proteins in yeast are present at <106 copies per cell21. Even if we assume that induced expression of the CUP1-pro-ALP construct were exceptionally efficient and reached this level, the dilution of the initially induced 106 pro-ALP molecules by division would leave 106/214 < 61 molecules/cell after the first 24 h growth period and 106/228 < 0.005 molecules/cell after the selection for diploids. Furthermore, the half-life of ALP is <9 h in yeast45. Protein degradation over 48 h will hence diminish ALP further, bringing its maximal abundance to 61/22 < 16 molecules per cell after 24 h and to 0.005/25 < 1.5*10−4 at the time of analysis at 48 h. At the same time, non-induced, constitutive background expression from the CUP1 promoter occurs in every newly produced cell, creating a constant background level of around 1/30 of the inducible level, equivalent to 1/30 * 106 = 33'000 molecules per cell. It exceeds the level of pro-ALP accumulated prior to mating by a factor of more than 6*106. Thus, the pool of pro-ALP that is finally assayed stems entirely from constitutive background expression. Its major fraction, 1-(16/33'000) ≫ 99.9%, will have been produced after mating and will have been delivered to the vacuoles by biosynthetic transport, independently of vacuole-vacuole fusion (Fig. 6A,B). The same applies to the maturation enzyme Pep4. Here, the effect of background level expression is even more severe because few Pep4 molecules biosynthetically delivered to a vacuole should suffice to activate vacuolar hydrolases and the pro-ALP contained in it.
Thus, both our experimental results and theoretical considerations indicate that the mating assay as conceived by Coonrod et al. cannot report vacuole fusion in the zygotes because its signal is dominated by biosynthetic trafficking of constitutively expressed pro-ALP to the vacuoles. This might explain how these authors could observe defective ALP maturation with various mutants, such as vam3Δ, which lacks the vacuolar Qa-SNARE that is necessary for biosynthetic delivery to vacuoles46. However, we cannot rationalize how Coonrod et al. could have observed Nyv1-dependence for pro-ALP maturation because biosynthetic delivery routes towards vacuoles are Nyv1-independent47 and also vacuole fusion in zygotes is Nyv1-independent, as observed by us and by others1.
The pyrophosphate-driven proton pump GFP-AVP1 alters vacuole structure in vivo
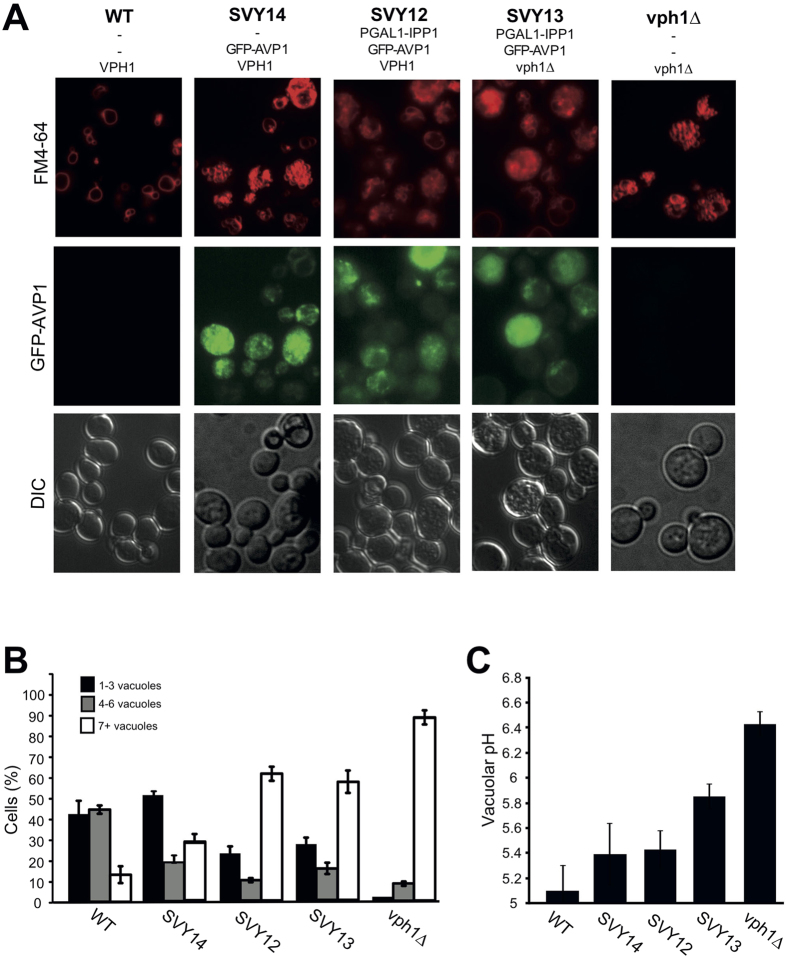
An ideal way of proving that the only contribution of V-ATPase to vacuole fusion is its proton pump function would be to replace V-ATPase by an unrelated proton pump, such as a proton-pumping pyrophosphatase from Arabidopsis thaliana (AVP1). In an attempt to re-establish vacuolar acidity independently of the V-ATPase, Coonrod et al. expressed GFP-AVP1 in vph1Δ cells. AVP1 pump activity depends on hydrolysis of pyrophosphate, a metabolite that is toxic to yeast cells. Pyrophosphate accumulates to levels supporting AVP1 activity only upon downregulation of the endogenous essential pyrophosphatase Ipp148. Coonrod et al. reported that they had created ipp1Δ mutants expressing GFP-AVP1 from a high-copy plasmid48. Coonrod et al. were unable to send us these ipp1Δ cells expressing GFP-AVP1 (LGY 253, LGY254) that provide the central argument for their conclusions. We had independently tried to create such a strain and could not obtain viable cells, presumably because Ipp1 is essential and the detoxification of pyrophosphate by GFP-AVP1 alone is insufficient for cell survival. This matches the experience of Perez-Castineira et al., who did not analyze the effects of GFP-AVP1-expression in ipp1Δ cells but used mutants in which IPP1 was expressed from a regulatable GAL promotor48,49,50. Even in the repressed state, residual expression from the Gal promotor appears to produce a minimal amount of Ipp1 that remains necessary for survival in presence of GFP-AVP1. Therefore, we used these strains with Gal-regulated IPP1 to revisit the effects of GFP-AVP1 on vacuole morphology. The GFP-AVP1 plasmid was, however, identical to the one used by Coonrod et al. While vph1Δ cells showed fragmented vacuoles, expression of GFP-AVP1 in these cells yielded a heterogeneous population, with 30% of the cells showing 1–3 large vacuoles and >50% maintaining fragmented vacuolar morphology (strain SVY13; Fig. 7A,B). This matches previous observations, which had been taken as evidence that the re-acidification of vph1Δ vacuoles by GFP-AVP1 had rescued vacuole fusion25. As a control, we expressed GFP-AVP1 in cells proficient for VPH1 and for IPP1, which maintained vacuolar acidification via V-ATPase. Expression of GFP-AVP1 changed vacuolar morphology in these VPH1-proficient and IPP1-proficient cells, increasing the frequency of cells with very numerous, highly fragmented vacuoles from <10% to 30% (strain SVY14; Fig. 7A,B). Additional downregulation of IPP1 (strain SVY12) exacerbated this effect, yielding a vacuolar morphology quite similar to the distribution observed in vph1Δ expressing AVP1. Thus, expression of GFP-AVP1 and downregulation of IPP1 strongly perturbs vacuolar morphology, even in V-ATPase-proficient cells. This renders it problematic to ascribe the segregation of the vph1Δ population into cells with fragmented and non-fragmented vacuoles to a rescue of vacuolar acidity and fusion by GFP-AVP1.
Figure 7. Effect of GFP-AVP1 expression on vacuolar structure and pH.
(A) Vacuolar morphology. V-ATPase proficient cells (BY4741) or vph1Δ cells, and cells expressing GPF-AVP1 in these backgrounds (SVY12, SVY13, SVY14), were grown at pH 5.5 to logarithmic phase, stained with FM4-64, and analyzed by spinning disc fluorescence microscopy. (B) Classification of the cells shown in panel A according to the number of vacuoles per cell. 200–400 cells were analyzed per strain (n = 3–5). (C) Vacuolar pH was measured for the cells used in (A). (n = 7–9).
Next, we tested the effects of GFP-AVP1 on vacuolar pH. As reported25, GFP-AVP1 expression in vph1Δ rescued vacuolar acidity partially, from pH 6.4 to pH 5.8, whereas the vacuolar pH in the wildtype was 5.1 under the same conditions (Fig. 7C). Seeking to explain the re-appearance of larger vacuoles in the vph1Δ/GFP-AVP1 strain as a consequence of the re-activation of vacuolar fusion by AVP1-dependent vacuole acidification, two interpretations are possible: First, an acidification to pH of 5.8 might already support vacuole fusion. If this were true, the effects of numerous V0 mutants that are deficient for vacuole fusion in vivo and in vitro15 could not be explained by a lack of vacuolar acidification. Our measurements of such mutants yielded for example pH = 5.6 for vma16-3HA (Fig. 4B). Nevertheless, this mutant maintains fragmented vacuoles in vivo (see above) and shows drastically lower fusion activity in vitro15. Thus, if pH 5.8 were necessary and sufficient for vacuole fusion, the fusion defect of this mutant supports a physical role of V0 in vacuole fusion in vivo that is independent of its role in proton pumping. The second possibility is that the expression of AVP1 is heterogeneous, with half of the cells expressing GFP-AVP1 insufficiently, showing higher-than-average pH and keeping fragmented vacuoles, and half of them expressing it strongly, allowing lower-than-average vacuolar pH and fusion. The latter hypothesis had been put forward25 but it had not been experimentally verified. We analyzed the intensity of GFP-AVP1 expression by fluorescence microscopy. Cells presenting a large vacuole usually showed barely detectable levels of GFP-AVP1 whereas cells showing high GFP-AVP1 signals typically had fragmented vacuoles (Suppl. Figs 2 and 3).This observation contradicts the assumption that the subpopulation of vph1Δ/GFP-AVP1 cells that shows a large vacuole does so because it expresses AVP1 more highly than the average and has more acidic vacuoles. Also in the wildtype background, the presence of a large vacuole was inversely correlated with the GFP-AVP1 signal (Suppl. Fig. 4).
Expression of GFP-AVP1 interferes with vacuole fragmentation
Due to this contradiction we explored the possibility that expression of GFP-AVP1 as such could impact vacuolar morphology in ways independent of vacuolar acidity, for example via vacuole fusion and fragmentation2,3,6,30,34,51,52,53,54. We challenged cells with 0.5 M NaCl, which induces rapid and synchronous fragmentation of wildtype vacuoles, following an ordered series of steps with different protein requirements55. In V-ATPase-proficient cells expressing GFP-AVP1 (SVY12), we could only analyze the part of the population that contained large vacuoles. Expression of GFP-AVP1 prevented the vacuoles in these cells from fragmenting upon salt treatment, and even from forming membrane invaginations that precede the actual fission event (Fig. 8A,B). This suggests that GFP-AVP1 interferes with vacuolar fragmentation. These observations indicate that the introduction of GFP-AVP1 itself has side effects on vacuolar structure and functionality that manifest themselves even in presence of an intact V-ATPase. Therefore, GFP-AVP1 is not a suitable tool to substitute for V-ATPase proton pump function in studies aiming at an analysis of vacuole fusion via the structure of the organelle in vivo.
Figure 8. Effect of GFP-AVP1 on vacuolar fission activity.
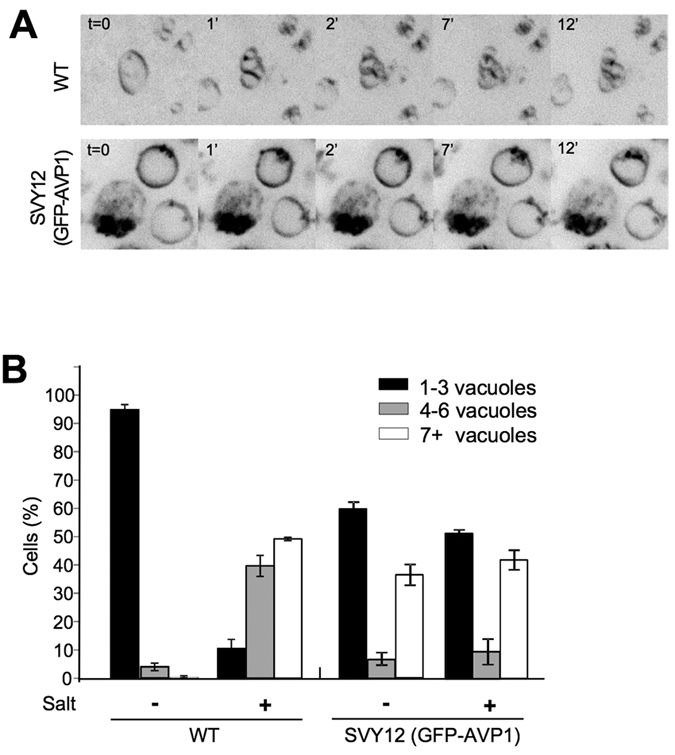
(A) AVP1 expression impairs vacuole fragmentation. Wildtype cells (BY4741) and cells expressing GFP-AVP1 (SVY12) were grown logarithmically in SC medium and labeled with FM4-64. NaCl was added to a final concentration of 0.5 M NaCl (t = 0 min) to induce vacuole fragmentation. During the following 12 min of incubation at 30 °C, the cells were analyzed by taking stacks of confocal images. Maximum-intensity projections of FM4-64 fluorescence in the stacks are shown, covering 6 μm in the z-direction. (B) Classification of the cells shown in panel A according to the number of vacuoles per cell. 200–400 cells were analyzed per strain (n = 3–5).
Discussion
Vacuole fusion can be assessed in different ways in vivo. Steady state vacuole structure has served as a readout because reductions in vacuolar fusion activity often coincide with a fragmentation of the vacuole in vivo2,3,6. We analyzed vacuole structure in vivo using time-lapse microscopy and acute inactivation of V-ATPase pump function by glucose withdrawal or by concanamycin A, a drug that blocks V-ATPase-dependent proton pumping but not vacuole fusion in vitro when used at appropriate low concentrations9,56. This allowed us to directly observe that loss of the vacuolar proton gradient induces vacuole fusion on the time scale of minutes. Since the reported experiments of Coonrod et al. had led these authors to the opposite conclusion, we revisited their experiments and performed additional controls on the assays used in their study.
Coonrod et al. judged the influence of the proton gradient on vacuole fusion via steady state vacuole morphology, relying on the vph1R735Q substitution in V0 to abolish V-ATPase pump function. Unlike vph1R735Q, which shows strongly fragmented vacuoles, all V1 deletion mutants eliminate V-ATPase pump function but they maintain an intact V0 sector31,35 and do not show fragmented vacuoles. Instead, they tend to show a fusion of vacuoles into a single, large organelle. The same is true for wildtype cells in which V-ATPase pump activity has been acutely reduced by pharmacological inhibition or by physiological V0/V1 dissociation following glucose depletion. Thus, the typical result of suppression of V-ATPase-dependent proton pumping is vacuole fusion. Coonrod et al. assumed that the vph1R735Q mutant left V0 structurally unchanged. This assumption may not be justified given that vph1R735Q interferes with the physiological dissociation of the V0/V1 interaction that occurs upon glucose withdrawal from yeast cells32,36. Since R735 is located in the transmembrane region of Vph1 it cannot directly interact with any V1 subunit57,58,59. vph1R735Q must hence influence V1–V0 dissociation indirectly, probably via an effect on V0 conformation. This interpretation is consistent with numerous observations indicating that V0 and its subunits can assume alternative conformations7,37,38,56,58,60,61. Therefore, the vph1R735Q allele cannot be used to dissociate physical and proton pump-related roles of V0 in vacuole fusion in yeast.
Coonrod et al. also assayed vacuole fusion by mating cells. The vacuoles of the mating partners were labeled with different fluorophores and co-localization of these fluorophores in the vacuole of the first diploid daughter forming from the zygotes was taken as a readout for fusion. Co-localization was not observed in experiments with the vph1R735Q mutant. Due to the constraints of this mutant that have been outlined above, these data cannot argue in favour of or against a physical or pump-related function of V0 in vacuole fusion. We were surprised to observe that, in contrast to cells lacking the vacuolar Q-SNARE (vam3Δ), cells lacking the cognate R-SNARE (nyv1Δ) did fuse their vacuoles in the daughter. This suggests that vacuoles in daughters emerging from zygotes fuse independently of Nyv1, although this protein promotes in vivo vacuole-vacuole fusion in other stages of the yeast life cycle, for example when haploid cells grow in the vegetative life cycle1,51,62. Nyv1 might be substituted for by another R-SNARE. A likely candidate is Ykt6, which binds to Vam3 and can replace Nyv1 under certain conditions in vitro28,63. We could not test the requirement for Ykt6 in the mating assay because Ykt6 is essential for cell growth and survival64.
Whereas microscopic analysis shows that vacuole fusion in the daughter of nyv1Δ zygotes proceeds as in wildtypes, assay of the same reaction via the maturation of pro-ALP by proteinase A had suggested that vacuole fusion was quantitatively blocked in nyv1Δ daughters25. We cannot provide a rational explanation how these authors could have observed Nyv1-dependent maturation of pro-ALP. These data are rendered even less plausible by the fact that, as we have shown above experimentally and theoretically, the ALP signals in this assay stem entirely from pro-ALP and proteinase A that have been produced in the diploid offspring and not before mating. Therefore, the pro-ALP maturation assay cannot report vacuole-vacuole fusion in daughter cells. It might at best measure effects on biosynthetic transport to the vacuole, but even then the reported effect of nyv1Δ cells remains unexplained because these biosynthetic transport routes are Nyv1-independent47.
An argument for the notion that vacuolar acidification was necessary and sufficient for vacuole fusion was also derived from the partial suppression of vacuolar fragmentation in vph1Δ mutants by expression of the heterologous pyrophosphate-driven proton pump AVP125. This experiment cannot provide a compelling argument because, as our control experiments demonstrate, GFP-AVP1 itself severely distorts vacuolar morphology and also interferes with vacuole fission. This is critical because vacuole structure in vivo is determined by an equilibrium of fusion and fission activity, i.e. a diminution of fission activity can rescue vacuole fragmentation51,52,53,65. GFP-AVP1 produces these effects even in V-ATPase-proficient cells with acidified vacuoles, suggesting that GFP-AVP1 perturbs vacuole structure independently of a rescue of vacuolar acidification. We assume that GFP-AVP1 might perturb the metabolism of pyrophosphate, which is a key metabolic intermediate for many nucleotide-dependent reactions. This is underlined by the fact that deletion or downregulation of the yeast pyrophosphatase gene IPP1 is lethal49,66. Downregulation or deletion of IPP1 is, however, necessary in order to let pyrophosphate accumulate to levels high enough to fuel GFP-AVP1 and allow it to work as a proton pump48. These levels may be detrimental to the cells and affect vacuolar functionality and structure. Although we could not obtain the ipp1Δ strain and hence the strain background is not identical to the one used by Coonrod et al., our results nevertheless indicate that the effects of GFP-AVP1 on vacuole structure cannot be attributed to a rescue of vacuolar pH.
In sum, our experimental controls and theoretic considerations leave no valid argument supporting a positive role for vacuolar acidification in vacuole-vacuole fusion in vivo. By contrast, our in vivo results are in line with a series of in vitro studies on vacuole fusion which had demonstrated a physical requirement of V0 for fusion but which had failed to detect significant effects of pharmacological or genetic ablation of V-ATPase pump activity1,7,9,15,51,52,56. The results from our in vivo analysis of vacuole fusion are also supported by a series of studies in living cells from numerous other systems. In Drosophila melanogaster, the effect of R735 substitutions in the a subunit v100 had been analyzed20, an experiment that Coonrod et al. had proposed as a means to re-evaluate the role of acidification V0-dependent fusion reactions in non-yeast systems. The v100R755A allele affects the same residue as the vph1R735Q allele in yeast and it does not support acidification. However, v100R755A does support fusion at the presynaptic membrane and on endosomes20. In Caenorhabditis elegans, secretion of multivesicular bodies is inhibited by point mutations of the V0 a-subunit that preserve proton pump function. Knockout of the catalytic V1 sector, which eliminates V-ATPase proton pumping, did not have this effect18, separating the secretion defect from V-ATPase pump function. In glia of zebrafish, phagosome-lysosome fusion is inhibited by ablation of the V0 a-subunit Atp6v0a1 although these organelles maintain their acidification19. In yeast, numerous point mutants in the V0 proteolipids support proton pump function while leading to significant vacuole fragmentation15,41,42 in vivo. Also covalent fusion of two proteolipid subunits of V0 by a peptide linker located at the lumenal face of the membrane (in Vma16-3HA), i.e. at the opposite side of where fusion is initiated, induced vacuole fragmentation in vivo. Also this construct supports proton pump function in vivo and in vitro15,27,67 but interferes with vacuole fusion in vivo that is triggered by an acute loss of proton pump activity. In chromaffin cells, rapid photo-inactivation of V0 subunits reduces the frequency of regulated exocytic events and the shape of the exocytic spikes, suggesting that V0 is in contact with the fusion pore. Acute ablation of V1 subunits by photo-inactivation or pharmacological inhibition of the V-ATPase did not have these effects24,25, indicating that they cannot be attributed to a loss of proton pump function. Finally, V0 proteolipids interact with SNAREs and this interaction is required for regulated exocytosis but not for proton pumping22.
In sum, a substantial body of work, stemming from in vivo studies in a variety of organisms and cell types and from in vitro studies on the fusion of vacuoles7,9,15,16, supports the notion that V0 contributes physically to membrane fusion and separates this activity from its role in proton pumping. Our direct microscopic observations of vacuoles in vivo are consistent with these findings and indicate that the proton pumping activity even counteracts vacuole fusion. However, while published studies collectively make a clear case for a physical and pump-independent role of V0 in membrane fusion1,7,9,15,51,52,56 we should not generally exclude effects of acidification. These can be observed for various trafficking reactions68 and they can even synergize with physical effects. The latter has been elegantly demonstrated by a recent study in chromaffin cells which proposed that V-ATPase disassembles when the lumen of secretory vesicles is well acidified24. This triggered V0/V1 dissociation should liberate V0 sectors to physically support the fusion process only in vesicles which are acidified and therefore can be expected to be loaded with transmitters. Thus, many interesting facets of V-ATPase function in membrane trafficking remain to be discovered and progress on this question will depend on the development of further approaches to manipulate the physical and pump-related functions of the V-ATPase at different stages of a trafficking reaction.
Materials and Methods
Strains and culture conditions
Culture media were either synthetic complete (SC) or yeast extract/peptone/dextrose (YPD; 2% glucose), buffered to pH 5.5 with 50 mM MES buffer. Cultures were shaken at 30 °C and 150 rpm. Strains carrying expression plasmids were grown on the corresponding SC dropout medium selecting for the auxotrophic marker. Strains and plasmids used in this study are listed in Tables 1 and 2, respectively. The SVY9 strain was generated as described48,50. The deletion mutant SVY13 was generated by replacing VPH1 with a nourseothricin (clonNat) cassette from plasmid pFA6a-natNT269 using SVY9 as parental strain.
Table 1. List of strains.
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Euroscarf |
| BY4242 vma1Δ | vma1Δ::kanMX4 | Euroscarf |
| BY4741 vph1Δ | vph1Δ::kanMX4 | Euroscarf |
| BY4742 vph1∆ | vph1Δ::kanMX4 | Euroscarf |
| BY4741 vam3Δ | vam3Δ::kanMX4 | Euroscarf |
| BY4742 vam3Δ | vam3Δ::kanMX4 | Euroscarf |
| BY4741 vma1Δ | vma1Δ::kanMX4 | Euroscarf |
| BY4741 vma2Δ | vma2Δ::kanMX4 | Euroscarf |
| BY4741 vma8Δ | vma8Δ::kanMX4 | Euroscarf |
| BY4741 VPH1-R735Q | BY4741 vph1Δ pRS416-Vph1R735Q (URA3) | This study |
| BY4741 VPH1-R735K | BY4741 vph1Δ pRS416-Vph1R735K (URA3) | This study |
| BY4741 16-3HA | vma16Δ::natNT2 pep4Δ::kanMX4 pRS316-Vma16-Vma3 (URA3) | Strasser et al.15 |
| BY4741 Vma5-GFP | BY4741 Vma5-GFP (HIS3) | This study |
| SVY9 | BY4741 ipp1::PGAL1-IPP1-TRP1 | This study |
| SVY14 | BY4741, pTcGFP-AVP1 (URA3) | This study |
| SVY12 (p:AVP1) | SVY9, pTcGFP-AVP1 (URA3) | This study |
| SVY13(vph1Δ+ p:AVP1) | SVY12, vph1Δ::natNT2 | This study |
| BY4741 nyv1∆ | nyv1Δ::kanMX4 | Euroscarf |
| BY4742 nyv1∆ | nyv1Δ::kanMX4 | Euroscarf |
| KEBY136 | MATα ura3-52 leu2-3 112 his4-519 ade6 gal2 pep4-3 pho8∆-X | Coonrod et al.25 |
| KEBY136 CUP1-PHO8 | KEBY136 pTC2 (CUP1-PHO8) | Coonrod et al.25 |
Table 2. List of plasmids.
| Plasmid | Description | Source |
|---|---|---|
| pLG230 | Yeast expression plasmid (CEN, LEU2) encoding GFP-ALP from the CPY promoter | Coonrod et al.25 |
| pMP2 | Yeast expression plasmid (CEN, LEU2) encoding mCherry-ALP from the VPH1 promoter with ALP terminator | Coonrod et al.25 |
| Vph1wt | pRS416 Vph1 | This study |
| Vph1R735Q | pRS416 Vph1R735Q | This study |
| Vph1R735K | pRS416 Vph1R735K | This study |
| Vph1-GFP | pRS416 Vph1-GFP | This study |
| Vma16-3HA (pLG146) | pRS316 PromVMA3 VMA16::3xHA::VMA3 | Flannery et al.26 |
| pTcGFP-AVP1 | GFP-AVP1 | Perez-Castineira et al.48 |
| pTC2 | Yeast expression plasmid (CEN, URA3) encoding PHO8 (alkaline phosphatase; ALP) from the CUP1 promoter | Coonrod et al.25 |
Plasmid construction
pRS416-PVph1-Vph1 carries a VPH1 gene under the control of the endogenous promoter (the region 500 bp upstream of the ORF). The VPH1 gene in the vector contains two silent mutations introducing a unique PacI site. vph1R735Q and vph1R735K alleles were produced by the Quick Change mutagenesis protocol using the primers: R735K, TCCTATTTAAAGTTATGGGCCTTATCATTGGC and GGCCCATAACTTTAAATAGGATGCAGTGTGCG; R735Q, TCCTATTTACAATTATGGGCCTTATCATTGGC and GGCCCATAATTGTAAATAGGATGCAGTGTGCG. The mutations were transferred into the non-mutagenized vector using PacI and ClaI restriction nucleases and the presence of the mutations was verified by sequencing. The Vph1-GFP fusion protein vector was generated by placing an EGFP cassette at the 3′ end of the Vph1 ORF.
Mating Assay
Cells from the two mating types (BY4741 and BY4742) were transformed with a plasmid encoding either mCherry-ALP or GFP-ALP as described in Coonrod et al. Transformants were grown in SC-LEU pH 5.5. Crosses were set up by diluting each mating type to an OD of 0.2 and incubating the samples at 30 °C with gentle mixing. Microscopy pictures were recorded on a confocal spinning disk microscope as described below after 4–6 hours of incubation.
Vacuolar pH measurements
Vacuolar pH was measured as described25,40. Cells were grown in SC or YPD medium buffered to pH 5.5 with 50 mM MES to an OD600nm of 0.8–1.2. Cells were sedimented by centrifugation (3'000 × g, 2 min, 20 °C) and resuspended in 100 μl of the same medium containing 50 μM BCECF-AM (2′,7′.bis-(2-carboxyethyl)-5-(and-6)-carboxyfluoresceine acetoxymethyl ester, Molecular Probes). The suspension was incubated at 30 °C for 30 min. Cells were washed twice in the same medium and finally resuspended at an OD600nm of 1.0. For end-point measurements, 50 μl of the cells were added to 150 μl of 50 mM MES buffer pH 5.5 in a black 96-well plate. At the same time, a calibration curve was recorded for each strain. To this purpose, 50 μl of the cell suspension was mixed with 50 μl of 50 mM MES buffer pH 5.5 and 100 μl of 2x calibration buffer (50 mM MES, 50 mM HEPES, 50 mM KCl, 50 mM NaCl, 0.2 M ammonium acetate, 10 mM NaN3, 10 mM 2-deoxyglucose, 50 μM FCCP) adjusted to pH values between 5.0 and 8.0. Fluorescence measurements were recorded in a Spectramax Gemini XS fluorescence plate reader (Molecular Devices) with excitation wavelengths of 450 and 490 nm and emission wavelength of 535 nm. The ratio of fluorescence intensities (490 nm/450 nm) was used to estimate the vacuolar pH by comparison with the calibration curves. For kinetic experiments, the labeled cell suspension was incubated with a V-ATPase inhibitor or DMSO at 30 °C. A calibration curve was obtained at the beginning and at the end of the experiment to ensure that the changes observed did not result from the ageing of the labeled suspension.
Vacuole staining and microscopy
Yeast cultures were grown over night at 30 °C in YPD or SC media (buffered to pH 5.5 with 50 mM MES) to an OD600nm of 0.2 to 1.2. Cells were harvested at 3000 × g, resuspended in 2–3 ml of fresh media at an OD600nm of 0.1 to 0.2. FM4-64 was added from a 10 mM stock in DMSO to a final concentration of 10 μM and cells were incubated at 30 °C for 1 h. After washing two times with media, cells were shaken for 1.5 to 2.5 h at 30 °C in the same media as before, but without FM4-64.
For confocal imaging, we used a Perkin-Elmer UltraView Vox Confocal Spinning Disk Setup on an inverted Zeiss Microscope with a 100x oil immersion objective (NA 1.41) and two Hamamatsu ORCA-Flash 4.0 cameras. FM4-64 was visualized in fast acquisition mode with excitation by the 488 nm laser. For stack imaging, 9 focal planes with a distance of 0.5 μm were acquired at each time point. Non-confocal microscopy was performed on an inverted Leica DMI6000B fluorescence microscope equipped with a 100x/1.4 NA lens and a Hamamatsu C10600-10B camera (ORCA-R2). Images were processed using ImageJ. Stack projections were obtained using the maximum intensity function. Brightness and contrast were linearly adjusted. Exposure of the cells to excitation light was kept at a minimum in order to avoid bleaching and light-induced fusion of vacuoles, which can occur upon prolonged illumination.
Cell immobilization for live-microscopy of yeast cells
Lab-Tek 8-well chamber slides (Nunc, 155411) were coated with 35 μl of a 1 mg/ml aqueous solution of concanavalin (freshly prepared from a 10x frozen stock). After air drying the slides, FM4-64 stained cultures were harvested for 3 min at 3000 × g, most of the supernatant was removed, and cells were resuspended in 50–100 μl of the remaining medium. 25 μl of cells were pipetted into each well of the coated chamber slide, and allowed to settle for 5 to 10 min at 25 °C. After 3 washes with 400 μl medium, 200 μl medium was added and cells were transferred to the microscope.
Alternatively, cells were immobilized on Sticky-Slide VI slides (from Ibidi) after coating of the coverslip with concanavalin A as described above. Cells were grown in YPD pH 5.5 and perfused at a flow rate of 10 μl/min with intermittent breaks to allow binding of the cells. Then, cells were kept under constant flow of medium at 20 μl/min. Then, the medium was changed to either YPD pH 5.5 (Glc-Glc) or YPGal pH 5.5 (Glc-Gal). After 10 min under the same flow, image acquisition was started using the confocal microscope set up described above. The 488 nm laser was used at minimal power (7%) and acquisition time was kept low (30 ms) to minimize light-induced damage of the cells. A picture was recorded every 12 seconds.
Glucose deprivation
Cells were grown at 30 °C over night to an OD600nm = 0.2–1.2 in buffered SC media with 2% glucose. Cultures were stained with FM4-64 and chased as described above. Cultures were divided into three 500 μl aliquots, cells were harvested at 5000 rpm for 10 s in a micro-centrifuge, washed twice, and resuspended in SC medium containing 2% glucose, no glucose, or 2% galactose instead of glucose. Cells were harvested as before, washed once in the respective media, resuspended in 500 μl media, and immediately observed under the microscope. Samples were further incubated at 30 °C and analyzed by microscopy.
Quantification of vacuole morphology
Vacuole morphology was quantified by counting cells in each indicated category from images taken with Perkin-Elmer UltraView Vox Confocal Spinning Disk or Zeiss Axioplan fluorescence microscope. Numbers of cells in each category are shown as a percentage of the total number of cells counted, and the SEM is indicated.
Additional Information
How to cite this article: Desfougères, Y. et al. Organelle acidification negatively regulates vacuole membrane fusion in vivo. Sci. Rep. 6, 29045; doi: 10.1038/srep29045 (2016).
Supplementary Material
Acknowledgments
We thank Aurelio Serrano for the AVP1 plasmid (pTcGFPAVP1), Josè Perez-Castineira for the kind support in generating the SVY12 and SVY13 yeast strains and Tom Stevens, Nia Bryant and Katerine Bowers for having sent numerous published strains and plasmids25. We are grateful for assistance by Véronique Comte. This work was supported by ERC grant n. 233458 and SNF grant 144258 to AM.
Footnotes
Author Contributions Y.D., S.V., R.G. and M.R. performed the experiments. A.M., Y.D., S.V. and M.R. analyzed data and wrote the manuscript.
References
- Takita Y., Engstrom L., Ungermann C. & Cunningham K. W. Inhibition of the Ca(2+)-ATPase Pmc1p by the v-SNARE protein Nyv1p. J Biol Chem 276, 6200–6206 (2001). [DOI] [PubMed] [Google Scholar]
- Wada Y., Ohsumi Y. & Anraku Y. Genes for directing vacuolar morphogenesis in Saccharomyces cerevisiae. I. Isolation and characterization of two classes of vam mutants. J Biol Chem 267, 18665–18670 (1992). [PubMed] [Google Scholar]
- Wada Y. & Anraku Y. Genes for directing vacuolar morphogenesis in Saccharomyces cerevisiae. II. VAM7, a gene for regulating morphogenic assembly of the vacuoles. J Biol Chem 267, 18671–18675 (1992). [PubMed] [Google Scholar]
- Rothman J. H., Yamashiro C. T., Kane P. M. & Stevens T. H. Protein targeting to the yeast vacuole. Trends Biochem Sci 14, 347–350 (1989). [DOI] [PubMed] [Google Scholar]
- Sato T. K., Rehling P., Peterson M. R. & Emr S. D. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol Cell 6, 661–671 (2000). [DOI] [PubMed] [Google Scholar]
- Seeley E. S., Kato M., Margolis N., Wickner W. & Eitzen G. Genomic analysis of homotypic vacuole fusion. 13, 782–794 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C. et al. Trans-complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature 409, 581–588 (2001). [DOI] [PubMed] [Google Scholar]
- Wickner W. Yeast vacuoles and membrane fusion pathways. EMBO J 21, 1241–1247 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer M. J., Reese C., Buhler S., Peters C. & Mayer A. Vacuole membrane fusion: V0 functions after trans-SNARE pairing and is coupled to the Ca2+-releasing channel. J Cell Biol 162, 211–222 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima J., Hickey C. M., Xu H., Jun Y. & Wickner W. Reconstituted membrane fusion requires regulatory lipids, SNAREs and synergistic SNARE chaperones. EMBO J 27, 2031–2042 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroupe C., Hickey C. M., Mima J., Burfeind A. S. & Wickner W. Minimal membrane docking requirements revealed by reconstitution of Rab GTPase-dependent membrane fusion from purified components. Proc Natl Acad Sci USA 106, 17626–17633 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchi P. C. & Zick M. Membrane Fusion Catalyzed by a Rab, SNAREs, and SNARE Chaperones Is Accompanied by Enhanced Permeability to Small Molecules and by Lysis doi: 10.1091/mbc.E11-08-0680 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Jun Y., Thompson J., Yates J. & Wickner W. HOPS prevents the disassembly of trans-SNARE complexes by Sec17p/Sec18p during membrane fusion. EMBO J 29, 1948–1960 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick M. & Wickner W. The tethering complex HOPS catalyzes assembly of the soluble SNARE Vam7 into fusogenic trans-SNARE complexes. doi: 10.1091/mbc.E13-07-0419 (2013). [DOI] [PMC free article] [PubMed]
- Strasser B., Iwaszkiewicz J., Michielin O. & Mayer A. The V-ATPase proteolipid cylinder promotes the lipid-mixing stage of SNARE-dependent fusion of yeast vacuoles. EMBO J 30, 4126–4141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreelatha A. et al. Vibrio effector protein VopQ inhibits fusion of V-ATPase-containing membranes. Proc Natl Acad Sci USA doi: 10.1073/pnas.1413764111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiesinger P. R. et al. The v-ATPase V0 subunit a1 is required for a late step in synaptic vesicle exocytosis in Drosophila. Cell 121, 607–620 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liégeois S., Benedetto A., Garnier J.-M., Schwab Y. & Labouesse M. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J Cell Biol 173, 949–961 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri F. & Nüsslein-Volhard C. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell 133, 916–927 (2008). [DOI] [PubMed] [Google Scholar]
- Williamson W. R., Wang D., Haberman A. S. & Hiesinger P. R. A dual function of V0-ATPase a1 provides an endolysosomal degradation mechanism in Drosophila melanogaster photoreceptors. J Cell Biol 189, 885–899 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S. et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003). [DOI] [PubMed] [Google Scholar]
- Di Giovanni J. et al. V-ATPase membrane sector associates with synaptobrevin to modulate neurotransmitter release. Neuron 67, 268–279 (2010). [DOI] [PubMed] [Google Scholar]
- Sun-Wada G.-H. et al. The a3 isoform of V-ATPase regulates insulin secretion from pancreatic beta-cells. J Cell Sci 119, 4531–4540 (2006). [DOI] [PubMed] [Google Scholar]
- Poëa-Guyon S. et al. The V-ATPase membrane domain is a sensor of granular pH that controls the exocytotic machinery. J Cell Biol 203, 283–298 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coonrod E. M. et al. Homotypic Vacuole Fusion in Yeast Requires Organelle Acidification and Not the V-ATPase Membrane Domain. Dev Cell 27, 462–468 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannery A. R., Graham L. A. & Stevens T. H. Topological characterization of the c, c’, and c” subunits of the vacuolar ATPase from the yeast Saccharomyces cerevisiae. J Biol Chem 279, 39856–39862 (2004). [DOI] [PubMed] [Google Scholar]
- Weisman L. S. Organelles on the move: insights from yeast vacuole inheritance. Nat Rev Mol Cell Biol 7, 243–252 (2006). [DOI] [PubMed] [Google Scholar]
- Thorngren N., Collins K. M., Fratti R. A., Wickner W. & Merz A. J. A soluble SNARE drives rapid docking, bypassing ATP and Sec17/18p for vacuole fusion. EMBO J 23, 2765–2776 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese C., Heise F. & Mayer A. Trans-SNARE pairing can precede a hemifusion intermediate in intracellular membrane fusion. Nature 436, 410–414 (2005). [DOI] [PubMed] [Google Scholar]
- Takeda K. et al. The vacuolar V1/V0-ATPase is involved in the release of the HOPS subunit Vps41 from vacuoles, vacuole fragmentation and fusion. FEBS Lett 582, 1558–1563 (2008). [DOI] [PubMed] [Google Scholar]
- Kane P. M., Kuehn M. C., Howald-Stevenson I. & Stevens T. H. Assembly and targeting of peripheral and integral membrane subunits of the yeast vacuolar H(+)-ATPase. J Biol Chem 267, 447–454 (1992). [PubMed] [Google Scholar]
- Kane P. M. & Smardon A. M. Assembly and regulation of the yeast vacuolar H+-ATPase. J Bioenerg Biomembr 35, 313–321 (2003). [DOI] [PubMed] [Google Scholar]
- LaGrassa T. J. & Ungermann C. The vacuolar kinase Yck3 maintains organelle fragmentation by regulating the HOPS tethering complex. J Cell Biol 168, 401–414 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt H., Perz A. & Ungermann C. An overexpression screen in Saccharomyces cerevisiae identifies novel genes that affect endocytic protein trafficking. Traffic 12, 1592–1603 (2011). [DOI] [PubMed] [Google Scholar]
- Graham L. A., Hill K. J. & Stevens T. H. VMA8 encodes a 32-kDa V1 subunit of the Saccharomyces cerevisiae vacuolar H(+)-ATPase required for function and assembly of the enzyme complex. J Biol Chem 270, 15037–15044 (1995). [DOI] [PubMed] [Google Scholar]
- Qi J. & Forgac M. Cellular environment is important in controlling V-ATPase dissociation and its dependence on activity. J Biol Chem 282, 24743–24751 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couoh-Cardel S., Hsueh Y.-C., Wilkens S. & Movileanu L. Yeast V-ATPase Proteolipid Ring Acts as a Large-conductance Transmembrane Protein Pore. Sci Rep 6, 24774 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare D. K. et al. An expanded and flexible form of the vacuolar ATPase membrane sector. Structure 14, 1149–1156 (2006). [DOI] [PubMed] [Google Scholar]
- Kawasaki-Nishi S., Nishi T. & Forgac M. Arg-735 of the 100-kDa subunit a of the yeast V-ATPase is essential for proton translocation. Proc Natl Acad Sci USA 98, 12397–12402 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diakov T. T. & Kane P. M. Regulation of vacuolar proton-translocating ATPase activity and assembly by extracellular pH. J Biol Chem 285, 23771–23778 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra K. J. & Kane P. M. Reversible association between the V1 and V0 domains of yeast vacuolar H+-ATPase is an unconventional glucose-induced effect. Mol Cell Biol 18, 7064–7074 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane P. M. & Parra K. J. Assembly and regulation of the yeast vacuolar H(+)-ATPase. J Exp Biol 203, 81–87 (2000). [DOI] [PubMed] [Google Scholar]
- Etcheverry T. Induced expression using yeast copper metallothionein promoter. Meth Enzymol 185, 319–329 (1990). [DOI] [PubMed] [Google Scholar]
- Peña M. M., Koch K. A. & Thiele D. J. Dynamic regulation of copper uptake and detoxification genes in Saccharomyces cerevisiae. Mol Cell Biol 18, 2514–2523 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiano R., Nagaraj N., Fröhlich F. & Walther T. C. Global proteome turnover analyses of the Yeasts S. cerevisiae and S. pombe. Cell Rep 9, 1959–1965 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T., Rieder S. E. & Emr S. D. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol 138, 517–529 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G. & Stevens T. H. The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. 10, 1719–1732 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Castiñeira J. R., Hernández A., Drake R. & Serrano A. A plant proton-pumping inorganic pyrophosphatase functionally complements the vacuolar ATPase transport activity and confers bafilomycin resistance in yeast. Biochem J doi: 10.1042/BJ20110447 (2011). [DOI] [PubMed] [Google Scholar]
- Serrano-Bueno G. et al. Inorganic pyrophosphatase defects lead to cell cycle arrest and autophagic cell death through NAD+ depletion in fermenting yeast. J Biol Chem 288, 13082–13092 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake R., Serrano A. & Perez-Castiñeira J. R. N-terminal chimaeras with signal sequences enhance the functional expression and alter the subcellular localization of heterologous membrane-bound inorganic pyrophosphatases in yeast. Biochem J 426, 147–157 (2010). [DOI] [PubMed] [Google Scholar]
- Baars T. L., Petri S., Peters C. & Mayer A. Role of the V-ATPase in regulation of the vacuolar fission-fusion equilibrium. 18, 3873–3882 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaillat L., Baars T. L. & Mayer A. Cell-free Reconstitution of Vacuole Membrane Fragmentation Reveals Regulation of Vacuole Size and Number by TORC1. 23, 881–895 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C., Baars T. L., Buhler S. & Mayer A. Mutual control of membrane fission and fusion proteins. Cell 119, 667–678 (2004). [DOI] [PubMed] [Google Scholar]
- Weisman L. S. Yeast Vacuole Inheritance and Dynamics. http://dx.doi.org/10.1146/annurev.genet.37.050203.103207 37, 435–460 (2003). [DOI] [PubMed]
- Zieger M. & Mayer A. Yeast vacuoles fragment in an asymmetrical two-phase process with distinct protein requirements. 23, 3438–3449 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller O., Neumann H., Bayer M. J. & Mayer A. Role of the Vtc proteins in V-ATPase stability and membrane trafficking. J Cell Sci 116, 1107–1115 (2003). [DOI] [PubMed] [Google Scholar]
- Toei M., Toei S. & Forgac M. Definition of membrane topology and identification of residues important for transport in subunit a of the vacuolar ATPase. Journal of Biological Chemistry 286, 35176–35186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki-Nishi S., Nishi T. & Forgac M. Interacting helical surfaces of the transmembrane segments of subunits a and c’ of the yeast V-ATPase defined by disulfide-mediated cross-linking. J Biol Chem 278, 41908–41913 (2003). [DOI] [PubMed] [Google Scholar]
- Oot R. A. & Wilkens S. Subunit interactions at the V1-Vo interface in the yeast vacuolar ATPase. J Biol Chem doi: 10.1074/jbc.M112.343962 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel N. Neurotransmitter release: the dark side of the vacuolar-H+ATPase. Biol Cell 95, 453–457 (2003). [DOI] [PubMed] [Google Scholar]
- Kartner N., Yao Y., Bhargava A. & Manolson M. F. Topology, glycosylation and conformational changes in the membrane domain of the vacuolar H(+) -ATPase a Subunit. J. Cell. Biochem. doi: 10.1002/jcb.24489 (2013). [DOI] [PubMed] [Google Scholar]
- Rohde J., Dietrich L., Langosch D. & Ungermann C. The transmembrane domain of Vam3 affects the composition of cis- and trans-SNARE complexes to promote homotypic vacuole fusion. J Biol Chem 278, 1656–1662 (2003). [DOI] [PubMed] [Google Scholar]
- Ungermann C. et al. Three v-SNAREs and two t-SNAREs, present in a pentameric cis-SNARE complex on isolated vacuoles, are essential for homotypic fusion. J Cell Biol 145, 1435–1442 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew J. A. et al. Ykt6p, a prenylated SNARE essential for endoplasmic reticulum-Golgi transport. J Biol Chem 272, 17776–17783 (1997). [DOI] [PubMed] [Google Scholar]
- Alpadi K. et al. Dynamin-SNARE interactions control trans-SNARE formation in intracellular membrane fusion. Nature communications 4, 1704 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano A., Perez-Castiñeira J. R., Baltscheffsky M. & Baltscheffsky H. H+-PPases: yesterday, today and tomorrow. IUBMB Life 59, 76–83 (2007). [DOI] [PubMed] [Google Scholar]
- Wang Y., Cipriano D. J. & Forgac M. Arrangement of subunits in the proteolipid ring of the V-ATPase. J Biol Chem 282, 34058–34065 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y., Sun-Wada G.-H., Tabata H. & Kawamura N. Vacuolar-type proton ATPase as regulator of membrane dynamics in multicellular organisms. J Bioenerg Biomembr 40, 53–57 (2008). [DOI] [PubMed] [Google Scholar]
- Janke C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.