Abstract
Receptor tyrosine kinases (RTKs) play critical roles in physiological and pathological processes, and are important anticancer drug targets. In vitro mechanistic and drug discovery studies of full-length RTKs require protein that is both fully functional and free from contaminating proteins. Here we describe a rapid cell-free and detergent-free co-translation method for producing full-length and functional ERBB2 and EGFR receptor tyrosine kinases supported by water-soluble apolipoprotein A-I based nanolipoprotein particles.
The four members of the ERBB family of mammalian receptor tyrosine kinases (EGFR/ERBB1, ERBB2/HER2, ERBB3, ERBB4) play key roles in the development and maintenance of a variety of tissues1. Overexpression and mutational activation of EGFR and ERBB2 actively contribute to the progression of both epithelial and brain cancers2,3,4. While a number of monoclonal antibodies and small molecule tyrosine kinase inhibitors have been developed as anticancer therapeutics that function by inhibiting EGFR and ERBB2 signaling5, resistance of tumor cells to ERBB-targeted therapeutic agents remains a thorny clinical problem that may be addressed with a deeper understanding of receptor activation mechanisms.
High-resolution structural studies using isolated ERBB receptor intracellular and extracellular regions have led to some insight into the mechanisms of receptor activation and signaling6,7, highlighting roles for dimerization of extracellular and intracellular domains in stimulating receptor kinase and auto-phosphorylation activities. Still, questions remain concerning the mechanisms by which extracellular ligand binding events induce activating conformational changes within the intracellular domain, highlighting the need for structural analysis of full-length receptors. Moreover, recent studies suggest that the EGFR kinase domain interacts with the plasma membrane to maintain receptor functionality8,9, raising the possibility that the membrane bilayer itself may contribute to the mechanism for propagating activating conformational changes from the extracellular portion of the receptor to the intracellular domain. Such a hypothesis underscores the need to examine the structures of full-length ERBB receptors in the context of their native membrane environment.
Isolation of full-length receptors from membranes of intact cells or tissues is complicated by the use of detergents, which are known to compromise the conformation or stability of membrane-bound proteins10,11. Moreover, the isolated receptors are often heterogeneous in post-translational modifications that may lead to multiple receptor functional states. The presence of co-purified membrane proteins and receptor-binding proteins can also affect their properties in vitro. For these reasons, novel approaches for the production and characterization of full-length receptor tyrosine kinases are needed that surmount the barriers posed by receptor truncation, detergent solubilization and sample heterogeneity.
Cell-free methods have been widely used in expressing proteins that are either toxic or unstable for in vitro study. Cell-free systems employ the protein translation machinery of model organisms to express solely the protein of interest from an input cDNA12, providing a convenient platform for the production of membrane-bound proteins13,14,15, mutant proteins and labeled proteins. Moreover, the co-expression of other proteins, or the addition of cofactors or auxiliary proteins to cell-free expression reactions, allow the properties of the expressed protein to be assessed under a variety of conditions14,15,16. However, cell-free production of large membrane proteins such as ERBB receptors is usually associated with low translation efficiency, poor protein stability and solubility.
To circumvent these issues, we have developed a nanolipoprotein particle (NLP)-based cell-free co-expression method to express soluble and functional ERBB receptors amenable for structural analysis. NLPs (10–25 nm in diameter) resemble reconstituted high-density lipoprotein (HDL) particles and are formed through spontaneous assembly of apolipoproteins and phospholipids, resulting in lipid bilayers corralled by an apolipoprotein “belt”17. The amphiphilic property of the NLPs provides a membrane mimetic that can support membrane proteins in a water-soluble environment18. When apolipoproteins and the desired membrane proteins are co-expressed in the presence of phospholipids, NLP complexes are formed containing spontaneously inserted membrane protein(s) without the need for detergents13,14,15. Mi et al. have demonstrated that intact EGFR from mammalian cellular extracts can be stably reconstituted into NLPs, and that receptors maintain function in this environment11. However, this method involved the reconstitution of heterogeneous cellular protein extracts, and required detergents to solubilize the receptors. Here we have adapted the cell-free NLP co-translation approach to produce pure solubilized ERBB2 and EGFR tyrosine kinases free from exposure to detergents using an E. coli based expression system (see illustration in Fig. 1A).
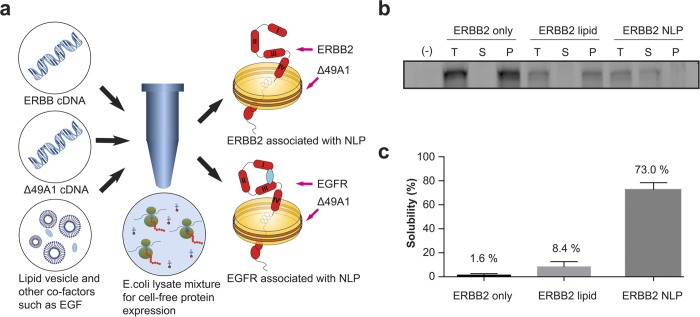
Figure 1.
(a) Schematic of cell-free co-translation of ERBB2-NLP and EGFR-NLP. (b) ERBB2 was cell-free produced in the presence and absence of DMPC or with DMPC and co-expressed Δ49A1. FluoroTect™ GreenLys (Promega) was added for visualizing newly synthesized ERBB2 protein. After 4 hours of expression, cell-free reactions were centrifuged at 14,000 rpm for 10 minutes. Small aliquots of sample before centrifuging (total, T), and the supernatant (soluble, S) and pellet (P) after centrifuging, were collected. All samples were loaded along with a cell-free reaction mixture only (-). Gel images were taken using Molecular Dynamics Typhoon 9410 Molecular Imager from GE Healthcare. Full-length versions of gel images are presented in Supplementary Figure 5. (c) The ERBB2-NLP complex shows greatly enhanced solubility as compared to protein only or protein DMPC vesicles.
Results
To facilitate protein expression using the Expressway™ Maxi Cell-Free E. coli Expression System (Life technologies), cDNAs encoding human EGFR and ERBB2 proteins were codon optimized for E. coli expression (Supplementary Figure 7) and were subcloned into expression vector pJexpress-414 (DNA2.0). Cell-free reactions using the native mammalian ERBB2 and EGFR coding sequences did not express detectable levels of protein (unpublished observations). Plasmid encoding a 6xHis-tagged version of human apolipoprotein A-I lacking the amino-terminal 49 amino acids (Δ49A1) was employed in co-translation reactions to drive the spontaneous assembly of ~300 kDa NLPs in the presence of 1,2-ditetradecanoyl-sn-glycero-3-phosphocholine (DMPC, Avanti), as previously described13.
As shown in Supplementary Figure 1, this cell-free expression system efficiently produced full-length ERBB2 and Δ49A1 proteins in the presence of DMPC. The ERBB2 and Δ49A1 proteins in this experiment were detected by including FluoroTectTM GreenLys (fluorescent lysine, Promega) in the cell-free reaction mix to visualize newly synthesized proteins, and were produced at sizes consistent with forms lacking post-translational modification (~140 kDa and ~20 kDa, respectively). Importantly, the NLP co-translation system markedly promoted the solubilization of the full-length ERBB2 protein, but only when intact nanolipoprotein particles were present. Cell-free reactions where ERBB2 alone was expressed in the absence or presence of DMPC, or co-expressed with Δ49A1 in the presence of DMPC, were centrifuged to separate soluble and insoluble receptor forms, and the total (T), soluble (S), and pellet (P) fractions were analyzed by gel fluorescence imaging (Fig. 1B). ERBB2 expressed on its own either in the absence or presence of DMPC fractionated predominantly to the pellet (Fig. 1B,C), indicative of an aggregated and likely misfolded and inactive form. However, co-translation of Δ49A1 and ERBB2 in the presence of DMPC efficiently shifted ERBB2 into the soluble fraction, strongly suggesting that the spontaneous insertion of ERBB2 into Δ49A1-induced NLPs markedly facilitates receptor solubility. To conclusively demonstrate ERBB2 association with NLPs, we purified particles by nickel affinity purification via the Δ49A1 6xHis tag and observed that essentially all of the detectable ERBB2 protein co-purified with NLPs, indicating that expressed ERBB2 successfully associated with the nanoparticle to form ERBB2-NLP complexes (Supplementary Figure 2). Yields were typically ~2 μg ERBB2 protein per milliliter reaction as determined by ELISA.
To assay the proper integration of ERBB2 into the NLPs, we conducted a carbonate extraction test. The carbonate extraction test has been widely used for testing how membrane proteins associate with a lipid bilayer19. Peripheral membrane proteins will be released from the bilayer upon treatment with 100 mM sodium carbonate (pH 11.5), whereas integral membrane proteins remain stably associated. Following carbonate extraction, approximately 3% of the ERBB2 was removed by carbonate with the remaining 97% of ERBB2 stably associated with the NLPs (Supplementary Figure 3). As a control, we cell-free co-expressed a known peripheral protein, Yersinia outer protein D (YopD) with NLP and found that about 50% of NLP associated protein was removed by carbonate. These results indicate that cell-free expressed ERBB2 is integrated into the bilayers of the NLP and therefore is carbonate resistant.
To begin to assess the functional integrity of NLP-bound ERBB2, we examined receptor auto-phosphorylation by Western blotting using an antibody that specifically recognizes ERBB2 phosphorylated at tyrosine residue 1248. The in vitro translation reactions contain both the ATP and metal ion cofactors necessary for ERBB2 kinase activity, so that blotting of a time course of ERBB2-NLP synthesis reactions revealed a close relationship between ERBB2 accumulation and auto-phosphorylation (Fig. 2A). To further test the receptor kinase activity, we treated the cell-free expressed ERBB2-NLP with calf-intestinal alkaline phosphatase (CIP) to remove the phosphate. After Ni purification that removes CIP and cell-free expression lysate components, the purified ERBB2-NLPs are then incubated with ATP, Mn2+, Mg2+ and kinase buffer to allow for re-phosphorylation. Western-blotting results indicated that the purified ERBB2-NLP is fully capable of auto-phosphorylation (Fig. 2B). These observations confirm that the cell-free expressed ERBB2 forms a correctly folded and functional intracellular kinase domain when inserted into the NLP particles.
Figure 2.
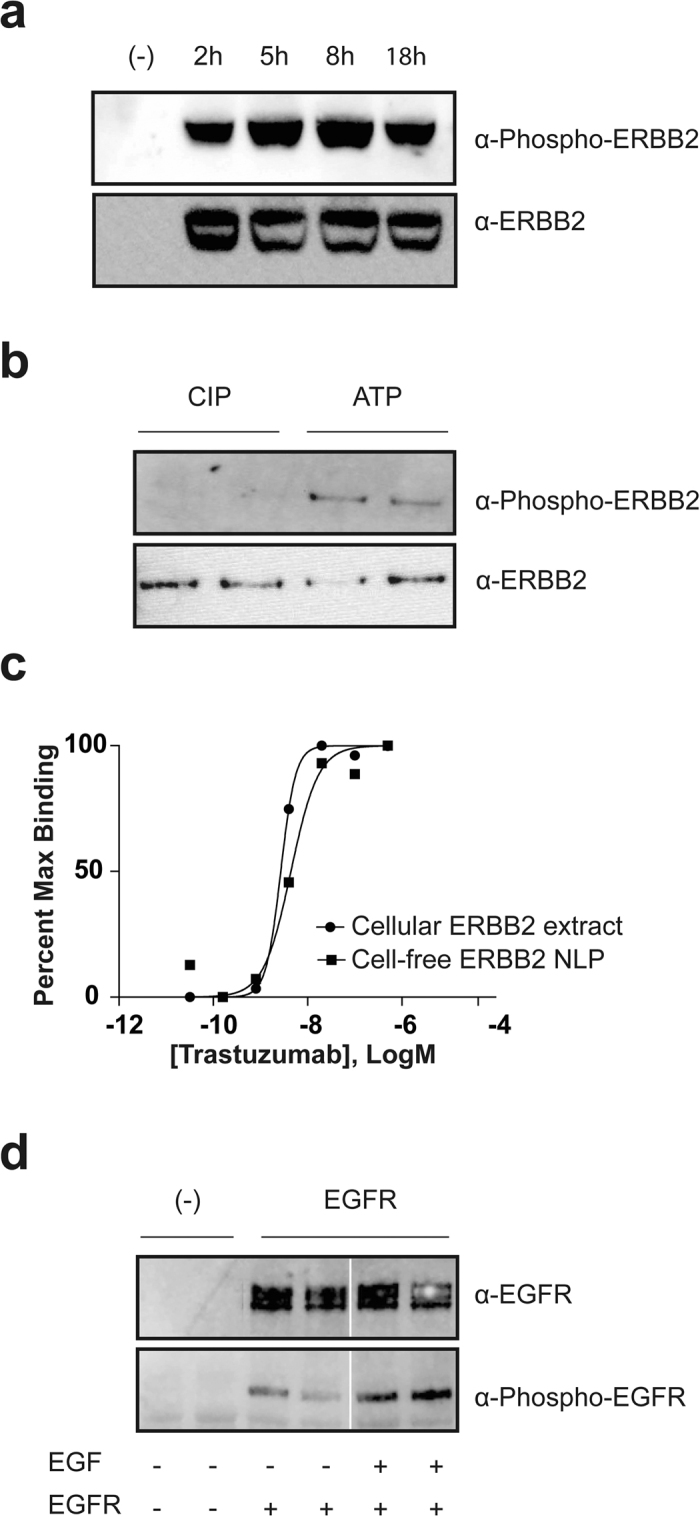
(a) NLP associated ERBB2 is tyrosine phosphorylated. Cell-free expressions were set up with and without (-) ERBB2 plasmid. Samples were collected at 2 hr, 5 hr, 8 hr and overnight (18 hr), resolved by SDS-PAGE and western blotted with anti-phospho-tyrosine ERBB2 antibody pY1248 and anti-ERBB2 antibody Ab-3 after stripping. (b) The NLP associated ERBB2 is phosphorylated independent of protein expression. Cell-free expressed ERBB2 (in replicate) was treated with calf-intestinal alkaline phosphatase (CIP) to remove the phosphate group and then Ni purified. The purified ERBB2-NLPs were incubated with ATP, Mn2+, Mg2+ and buffer to allow for re-phosphorylation. Samples were resolved by SDS-PAGE and western blotted with anti-phospho-tyrosine antibody 4G10 and anti-ERBB2 antibody Ab-3. (c) Binding of trastuzumab to cell-free ERBB2-NLPs and cellular ERBB2 extract were measured by ELISA as described. Cell-free ERBB2-NLPs or ERBB2 extracted from HEK293 cells over-expressing ERBB2 receptor were exposed to varying concentrations of trastuzumab (0–500 nM). The Kds for the trastuzumab binding to cell-free ERBB2-NLP and cellular ERBB2 extract were 4.4 nM and 2.7 nM, respectively. Each data point represents the average value of triplicate measurements. (d) NLP associated EGFR is phosphorylated, and the presence of EGF in the cell-free reaction increases the level of phosphorylation. EGFR-NLPs showed low level of phosphorylation during cell-free expression. Adding EGF, the natural ligand of EGFR, increases the phosphorylation. Cell-free expressions were set up with and without (-) EGFR plasmid, with and without EGF. After an 8 hr reaction, cell-free mixtures were resolved by SDS-PAGE and western blotted with anti-phospho-tyrosine EGFR antibody pY1110 and anti-EGFR. The white vertical line represents a splicing event of lanes from the same Western blot. Full-length versions of all western blots are presented in Supplementary Figure 6.
While the correct folding of the intracellular kinase domain of in vitro-synthesized ERBB2 is highly probable, the presence of 24 disulfide bonds in the extracellular domain of an ERBB2 monomer raises questions as to whether this domain would also fold correctly. In contrast to the other three ERBB family members, no diffusible ligand has been identified that binds to ERBB2; thus the functional integrity of the extracellular domain cannot be assessed by ligand binding or stimulation. To circumvent this issue, we compared the binding of the conformation-specific ERBB2 extracellular domain antibody trastuzumab to cell-expressed ERBB2 and in vitro-expressed ERBB2-NLPs. ELISA assays revealed that trastuzumab binds with similar affinity to ERBB2 derived from both sources (Fig. 2C), strongly suggesting that the in vitro-produced material is properly folded. It should be noted that this experiment also underscores the utility of the in vitro expression/NLP approach for rapidly testing efficacies of drugs toward mutated or altered receptors commonly found in various cancers.
We also examined the ability of the co-translational system to produce functional full-length EGFR, as indicated by the ability of the natural ligand EGF to stimulate receptor auto-phosphorylation at tyrosine residue 1110. Figure 2D demonstrates that cell-free produced EGFR-NLP phosphorylation is enhanced when synthesis reactions are carried out in the presence of EGF, indicating that the kinase and the ligand binding domains are properly folded independently. More importantly, these observations indicate that the full-length receptor signals properly and should be amenable to future mechanistic and structural studies.
To begin to examine the structural characteristics of the ERBB2-NLP complex, we carried out several small angle X-ray scattering (SAXS) experiments20, a robust technique for evaluating the conformation, dimensions, and assemblies of macromolecular complexes and particles, including membrane proteins21. The SAXS data for purified empty NLPs and purified ERBB2-NLP complexes were markedly different (Supplementary Figure 4). The primary electron density distribution shifted from being dominated by the 50–60 Å spacing set by phosphate head groups in the empty NLPs to a more globular distribution of ~125 Å, consistent with the incorporation of protein (ERBB2) into the NLPs, which is supported by the carbonate extraction data.
Discussion
We have demonstrated a method for in vitro synthesis of two full-length ERBB family receptors, ERBB2 and EGFR in a cell-free environment without the use of detergents. The co-expression and association with NLPs improves the solubility of the synthesized receptors. The ERBB2-NLP complex can be Ni affinity purified using the 6x His tag located on the scaffolding protein of the NLP, Δ49A1. Importantly, the ERBB2 protein is stably integrated into the membrane bilayer of the NLP. The cell-free produced ERBB2 is capable of auto-phosphorylation, a key feature of the ERBB family receptors. Furthermore, ERBB2 binds with the monoclonal antibody trastuzumab in an ELISA-based study, indicating the extracellular domain is correctly folded.
To achieve expression in an E. coli based system, we used codon optimized DNA sequences, which involve changes of approximately 30% of the bases (Supplementary Figure 7). We were able to obtain 2 μg ERBB2 per mL lysate, an amount which is suitable for protein functional characterization using biochemical and biophysical techniques including SDS-PAGE, Western blotting, ELISA and SAXS analysis. This method can be further optimized for obtaining high-resolution structures, such as scaling up or adding additional amino acids as well as energy to the cell-free reactions to achieve a higher protein yield. The current study focused on feasibility and function over optimizing total yield. In addition, further optimization and modification of cell-free reactions are needed to fully understand post translation modification effects, heterodimerization, and trafficking, which contribute to important receptor functionalities that are physiologically relevant. In order for E. coli based cell-free reactions to accommodate additional post translational modification, the reactions will have to be supplemented with active mammalian cell-free extracts/components that impart the desired modifications perhaps providing a viable approach to study receptor activation and phosphorylation.
Given the complexity of in vivo biogenesis pathways, it is surprising that membrane proteins synthesized in a cell-free system can insert into a membrane bilayer and stay functional. Yet this may be explained by several characteristics of the cell-free reactions. First, the presence of soluble chaperones in cell-free systems may help maintain synthesized proteins in an integration-competent conformation. In addition, the native biological membrane contains a complex mix of lipids and a large amount of proteins, some of which may inhibit/limit the unassisted access/integration of membrane protein into the lipid bilayer, while NLPs provide a more accessible surface. Last but not least, in the cells, membrane protein needs to be released from the ribosome with the help of specialized chaperoning proteins to prevent aggregation before entering the membrane. In cell-free conditions where the lipid bilayer is more readily accessible, nascent proteins may be directly integrated into the membrane more efficiently.
In conclusion, this study represents the first use of cell-free synthesis for the de novo production of functional RTKs as intact and functional full-length receptors supported by NLPs. This technology will facilitate in vitro studies of receptor signaling mechanisms with material that is free from contaminating cellular modulators that might influence receptor activity. ERBB-NLPs may provide an ideal platform for studying the interaction of receptors with the phospholipid environment, possibly a critical missing component of the ERBB signaling mechanism. ERBB-NLP technology may also be extended to the characterization of transient receptor interactions and mutant ERBB receptor forms, and could support high throughput screening of drugs that act toward the holoreceptor. In addition, cell-free produced receptors may be specifically labeled in vitro with fluorophores for Fluorescent Correlation Spectroscopy (FCS) studies of receptor homo-as well as hetero-dimerization. Finally, future developments may lead to NLP-mediated membrane protein crystallization and platforms for studying receptor interactions with specific lipids.
Methods
Cell-free expression using Expressway™ Maxi Cell-Free E. coli Expression System, Life technologies
Briefly, E. coli slyD- Extract, IVPS E. coli Reaction Buffer (-A.A.), Amino Acids (-Met), Methionine, T7 Enzyme Mix and DNA templates were added to a 1.5 mL microcentrifuge tube. Reactions were incubated in a standard shaking incubator (800 rpm) at 30 °C for 30 minutes. After that, a feed buffer made from IVPS Feed Buffer, Amino Acids (-Met) and Methionine was added to the reaction. The reactions were then incubated for 4–18 hours at 30 °C. A typical volume of a cell-free reaction was 25 to 500 μ L. After the reaction was done, the reaction tubes were placed on ice for immediate processing or analysis.
Cell-free Co-expression of membrane protein with NLP
The plasmid for Δ49A1 was previously described by the authors Cappucio et al., 2008. The genes for ERBB2 and EGFR were codon optimized for E. coli expression by DNA 2.0. The base changes that were made are shown in Supplemental Figure 7. Cell-free reactions were carried out using the Invitrogen Expressway Maxi kit following the protocols listed above. Small unilamellar vesicles of DMPC (liposomes) were prepared by probe-sonicating a 25 mg/ml water solution of DMPC until optical clarity was achieved, typically 15 min on ice, followed by two minutes of centrifugation at 14,000 rcf to remove metal contamination from the sonication probe tip. DMPC small unilamellar vesicles were added to the cell-free reaction at a final concentration of 2 mg/ml. For membrane protein and NLP co-expression, a maximum of 20 μg of membrane protein plasmid DNA and a lower concentration of Δ49A1 plasmid DNA were added to the lysate mixture. The molar ratio of DNA varied depending on the expression efficiency of the membrane protein. A typical membrane protein DNA to Δ49A1 DNA ratio was 20:1 or up to 100:1. Other additives and cofactors such as FluoroTect™ GreenLys (Promega), EGF, and protease inhibitors are added at the beginning of the reactions (Fig. 1A). The reactions were incubated at 30 °C according to the manufacturer’s instruction. The entire incubation time can be 4–18 hours. Unless otherwise stated, 8 hour expression time was used for ERBB2 and EGFR expression experiments to achieve maximum protein yield. For plasmids used in cell-free expression see Supplementary Table 1. For sequence details of codon optimized human ERBB2 and EGFR see Supplementary Figure 7.
Native purification of cell-free produced NLP complex
One ml of Superflow Ni-NTA resin (Qiagen) slurry was added to a 3 ml plastic column. Columns were equilibrated with 3 ml native lysis buffer (50 mM NaH2PO4, 300 mM NaCl, pH 8.0). Leave ~500 μl of lysis buffer on the top of the column and cap the outlet. 10 μl aliquot from the cell-free reaction is saved as Total (T). The rest of the reaction mixture was added to the prepared Ni-NTA resin and mixed for at least 1 hour at 4C on a nutator. Flow-through (FT) was collected. Column was then washed with 2 × 1 ml of native lysis buffer with 10 mM imidazole, 2 × 1 ml aliquots of native lysis buffer with 25 mM imidazole, 2 × 1 ml aliquots of native lysis buffer with 50 mM imidazole (W). Sample was eluted with 6 × 1 ml aliquots of native elution buffer, lysis buffer with 400 mM imidazole (E). Eluents were combined and dialyzed against 1xPBS using Slide-A-Lyzer Dialysis Cassettes, 10 K MWCO (Pierce), at 4 °C for 4 hours or overnight. Fresh buffer was added every hour in the first 3 hours. Samples were concentrate using Vivaspin 6 spin column (MWCO = 100 kD, Sartorius Stedim AG). Samples were centrifuged at 4000 rcf for 10 min at 4 °C to collect the concentrated sample.
ELISA based antibody binding assay
ELISA was carried out using DuoSet IC ERBB2 ELISA kit from R&D Systems. Nunc Maxisorp plates (Nunc) were coated with 100 μL anti-ERBB2 capture antibody (R&D systems) in PBS overnight at room temperature, and blocked for two hours with 5% bovine serum albumin (Sigma) in PBS. Normalized ERBB2-NLP samples were added to antibody-coated Nunc plates to bind for 2 hours at room temperature. Plates were rinsed with wash buffer (R&D systems) three times. A serial dilution of antibody (trastuzumab) were added to the plates in PBS and allowed to bind for 2 hours. The wash steps were repeated again and anti-human secondary antibody-HRP conjugate was added (Santa Cruz) at 1/5000 dilution in PBS-Tween with 5% BSA for 1 hour. Plates were then rinsed three times with PBS-Tween. One hundred μL of substrate (R&D Systems) was added to the wells and incubated in the dark for 10 minutes. Then 50 μL of 2N sulfuric acid was added to stop the reaction. Absorption at 450 nm was measured using the Spectramax M2e plate reader from Molecular Devices. Each data point represents the average value of triplicate measurements. The binding affinity for each antibody was calculated from the binding curves using GraphPad Prism, dose response analysis, nonlinear regression fit function (Fig. 2B).
SDS-PAGE
SDS-PAGE was run using NuPAGE Novex 4–12% Bis-Tris Protein Gels from Life Technologies. Samples were boiled for 5 minutes with NuPAGE LDS Sample Buffer (4X) supplemented with beta-mercaptoethanol (5% of total volume). Gels were run at 200 V for about 45 min. A pre-stained molecular weight marker was used to determine the end-point of the electrophoresis.
Gel fluorescent imaging
After electrophoresis, gels were removed from the cassette and briefly rinsed with water. Fluorescent pictures were taken using the blue laser (488 nm) of a Molecular Dynamics Typhoon 9410 Molecular Imager (GE Healthcare) with a 520 nm bandpass 30 filter for the detection of the fluorescent labeled proteins.
Western Blot
Western Blot was done using the iBlot Dry Blot System from Life Technologies. The pre-loaded program 3 was used according to manufacturer’s user manual. After completing transfer, the membrane was blocked in 5% Non Fat Dry Milk in TBS with 0.05% tween-20 for 1 hour. The membrane was incubated with the primary antibody for 18 hours at 4 °C. For the list of antibodies and dilutions see Supplementary Table 2. Positive bands were detected using SuperSignal West Pico Chemiluminescent Substrate (Pierce). Chemiluminescent pictures were taken with Alpha Innotech imager. Densitometry quantification was done with ImageJ.
ERBB2 and EGFR quantification
ERBB2 and EGFR quantification was carried out using DuoSet IC ELISA Development Systems from R&D Systems following the manufacturer’s protocol. The receptor concentration was calculated using GraphPad Prism, 4-parameter logistic fit function.
Small angle X-ray scattering (SAXS)
The empty NLP and ERBB2 NLP sample for SAXS were cell-free produced and nickel affinity purified as described above. After the nickel purification, both samples were dialyzed against 3 × 1 L 100 mM pH 7.5 HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffer. The dialyzed samples were then concentrated with Vivaspin 6 spin column (MWCO = 100 kD, Sartorius AG). The buffer in the concentrator flow through is used for subtraction. Each concentrated sample contained 0.23 mg/ml of total protein as measured by Nanodrop. SAXS data were collected at the SIBYLS beamline at the Advanced Light Source in Berkeley CA. Samples were collected as previously described22,23. Briefly, samples of 24 mL volume were placed 1.5 m away from a MAR165 detector and exposed to 12 keV X-rays with a flux of 1012 photons/second. Due to sample limitations and the relative nature of the study, empty NLPs relative to ERBB2 NLPs, only one concentration of sample was measured. Radiation damage was not observed. Data were analyzed using the ATSAS program package24. Some aggregation was observed in both samples with no Guinier region however the aggregation did not prevent the extraction of the primary spatial contribution to the SAXS.
Carbonate extraction
Two hundred μl of Superflow Ni-NTA resin (Qiagen) slurry was added to a 1 ml plastic column. Column was equilibrated with 1 ml native lysis buffer (50 mM NaH2PO4, 300 mM NaCl, pH 8.0). Cell-free reaction mixture (ERBB2-NLP or YopD-NLP, pre-labeled with FluoroTect™ GreenLys (Promega)) was added to the prepared Ni-NTA resin and mixed for 1 hour at 4C on a nutator. Columns were then washed with 2 × 200 μl of native lysis buffer with 10 mM imidazole, 2 × 200 μl of native lysis buffer with 20 mM imidazole. Samples were eluted with 2 × 200 μl of either 0.1 M sodium carbonate (pH 11.5), or native elution buffer, lysis buffer with 400 mM imidazole. Eluents were collected and concentrated using Vivaspin 500 spin column (MWCO = 10kD, Sartorius Stedim AG). The concentrated samples resolved by SDA-PAGE. Fluorescent pictures of the gels were taken using the blue laser (488 nm) of a Molecular Dynamics Typhoon 9410 Molecular Imager (GE Healthcare) with a filter 520 nm bandpass 30.
Phosphatase treatment and auto-phosphorylation assay
500 μl of cell-free reaction were set up for ERBB2-NLP expression. After 5 hours incubation, 1 unit of calf-intestinal alkaline phosphatase (CIP, NEB) was added to the reaction and incubated at room temperature for 1 hour. After that, the ERBB2-NLPs were Ni purified, dialyzed against 50 mM Tris-HCl (pH 7.5), and concentrated. The purified ERBB2-NLPs were then incubated with 15 mM MnCl2, 15 mM MgCl2, 2 mM DTT and 50 μM ATP for 30 min to allow for auto-phosphorylation.
Additional Information
How to cite this article: He, W. et al. Cell-free expression of functional receptor tyrosine kinases. Sci. Rep. 5, 12896; doi: 10.1038/srep12896 (2015).
Supplementary Material
Acknowledgments
This work was supported by funding from the NIH/NCI (RO1-CA155642-01A) and National Science Foundation grant through the Center for Biophotonics Science and Technology Center, managed by the University of California, Davis under Cooperative Agreement No. PHY 0120999. Work was also performed under the auspices of the U.S. Department of Energy under contract number DE-AC52-07NA27344, which includes IDAT funding through the US Department of Energy, Biological and Environmental Research.
Footnotes
Author Contributions W.H., C.T., K.L.C., P.T.H. and M.A.C. were involved in designing the research and discussion of the results and/or advised on the analyses; W.H., C.T., M.S., G.L.H. and S.H. conducted the experiments and performed the analyses; T.S., C.G. and J.A.T. provided critical scientific and technical expertise regarding protocols and reagents; and W.H. and M.A.C. wrote the manuscript with support and help from all the authors.
References
- Herbst R. S. Review of epidermal growth factor receptor biology. Int. J. Radiat. Oncol. Biol. Phys. 59, 21–26 (2004). [DOI] [PubMed] [Google Scholar]
- Pike L. J., Gallis B., Casnellie J. E., Bornstein P. & Krebs E. G. Epidermal growth factor stimulates the phosphorylation of synthetic tyrosine-containing peptides by A431 cell membranes. Proc. Natl. Acad. Sci. USA 79, 1443–1447 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamon D. J. et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235, 177–182 (1987). [DOI] [PubMed] [Google Scholar]
- Stephens P. et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature 431, 525–526 (2004). [DOI] [PubMed] [Google Scholar]
- Baselga J. & Swain S. M. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat. Rev. Cancer 9, 463–475(2009). [DOI] [PubMed] [Google Scholar]
- Yarden Y. & Sliwkowski M. X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2, 127–137 (2001). [DOI] [PubMed] [Google Scholar]
- Zhang X., Gureasko J., Shen K., Cole P. A. & Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125, 1137–1149(2006). [DOI] [PubMed] [Google Scholar]
- Bessman N. J. & Lemmon M. A. Finding the missing links in EGFR. Nat. Struct. Mol. Biol. 19, 1–3 (2012). [DOI] [PubMed] [Google Scholar]
- Endres N. F. et al. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell 152, 543–556 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraway K. L. 3rd & Cerione R. A. Fluorescent-labeled growth factor molecules serve as probes for receptor binding and endocytosis. Biochemistry 32, 12039–12045 (1993). [DOI] [PubMed] [Google Scholar]
- Mi L. Z. et al. Functional and structural stability of the epidermal growth factor receptor in detergent micelles and phospholipid nanodiscs. Biochemistry 47, 10314–10323 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson E. D., Gan R., Hodgman C. E. & Jewett M. C. Cell-free protein synthesis: applications come of age. Biotechnol Adv. 30, 1185–1194(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappuccio J. A. et al. Cell-free co-expression of functional membrane proteins and apolipoprotein, forming soluble nanolipoprotein particles. Mol. Cell Proteomics 7, 2246–2253(2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T. et al. Characterization of de novo synthesized GPCRs supported in nanolipoprotein discs. PLoS One 7, e44911 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T. et al. Characterizing diffusion dynamics of a membrane protein associated with nanolipoproteins using fluorescence correlation spectroscopy. Protein Sci. 20, 437–447 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W. et al. Controlling the diameter, monodispersity, and solubility of ApoA1 nanolipoprotein particles using telodendrimer chemistry. Protein Sci. 22, 1078–1086 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayburt T. H. & Sligar S. G. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 12, 2476–2481 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton M. R. et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl. Acad. Sci. USA 104, 7682–7687 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long A. R., O’Brien C. C. & Alder N. N. The Cell-Free Integration of a Polytopic Mitochondrial Membrane Protein into Liposomes Occurs Cotranslationally and in a Lipid-Dependent Manner. PLoS ONE 7, e46332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kynde S. A. et al. Small-angle scattering gives direct structural information about a membrane protein inside a lipid environment. Acta Crystallogr D Biol Crystallogr. 70, 371–383 (2014). [DOI] [PubMed] [Google Scholar]
- Hura G. L. et al. Comprehensive macromolecular conformations mapped by quantitative SAXS analyses. Nature Methods 10, 453–454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer K. N. et al. High-throughput SAXS for the characterization of biomolecules in solution: a practical approach. Methods Mol Biol. 1091, 245–258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura G. L. et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat Methods. 6, 606–612 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov M. V. & Svergun D. I. Applications of small-angle X-ray scattering to biomacromolecular solutions. Int J Biochem Cell Biol. 45, 429–437 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.