Abstract
Fibronectin is a large vertebrate glycoprotein that is found in soluble and insoluble forms and involved in diverse processes. Protomeric fibronectin is a dimer of subunits, each of which comprises 29 to 31 modules—12 type I, two type II, and 15-17 type III. Plasma fibronectin is secreted by hepatocytes and circulates in a compact conformation before it binds to cell surfaces, converts to an extended conformation, and is assembled into fibronectin fibrils. Here we review biophysical and structural studies that have shed light on how plasma fibronectin transitions from the compact to the extended conformation. The three types of modules each have a well-organized secondary and tertiary structure as defined by NMR and crystallography and have been likened to “beads on a string”. There are flexible sequences in the N-terminal tail, between the fifth and sixth type I modules, between the first two and last two of the type III modules, and at the C-terminus. Several specific module-module interactions have been identified that likely maintain the compact quaternary structure of circulating fibronectin. The quaternary structure is perturbed in response to binding events, including binding of fibronectin to the surface of vertebrate cells for fibril assembly and to bacterial adhesins.
Keywords: fibronectin, integrin, plasma protein, bacterial adhesin, syndecan, heparan sulfate, fibronectin type I module, fibronectin type II module, fibronectin type III module
Introduction
Fibronectin (FN, human gene FN1, UniProt entry for human protein: http://www.uniprot.org/uniprot/P02751) is a 470- to 500-kDa glycoprotein of vertebrates that contributes to normal processes important for development, organogenesis, cell adhesion and migration, and hemostasis (George et al., 1993, Pankov and Yamada, 2002, Wang et al., 2014) and pathophysiologic processes such as angiogenesis (Zhou et al., 2008) and vascular remodeling (Chiang et al., 2009). FN has given its name to the three module types that constitute >90% of its sequence: FNI, FNII, and FNIII (Fig. 1). There are two general sources of FN: plasma FN that is synthesized by hepatocytes and secreted into blood where it circulates in a compact conformation and cellular FN that is secreted locally by cells (Tamkun and Hynes, 1983, Moretti et al., 2007). Many of the effects of FN require FN to be assembled into fibrils of the extracellular matrix. FN assembly has been studied mostly in cell culture and involves FN binding to molecules on cell surfaces, including syndecans and integrins, adopting a linear conformation, and recruiting additional FN molecules to form insoluble fibrils (Singh et al., 2010, Fruh et al., 2015). Assembly is initiated at sites of cell adhesion, and growing fibrils are translocated inwards (Pankov et al., 2000, Ohashi et al., 2002).
Fig. 1.
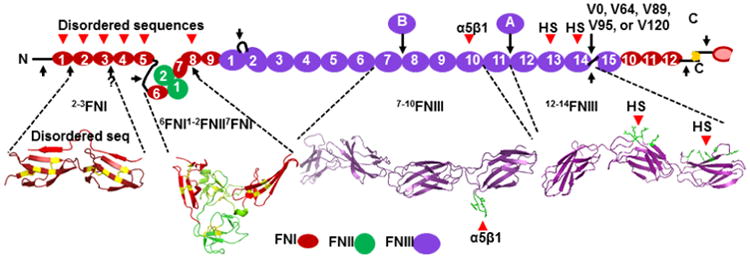
Model of FN subunit. One subunit of a FN dimer is displayed with FNI modules, FNII modules, and FNIII modules numbered as they are ordered from N- to C-termini. Locations of alternatively spliced AFNIII (A) and BFNIII (B) FNIII modules, when present, are indicated. Alternative splicing also results in five different sequences for the variable (V) region, labeled by the number of amino acids (V0, V64, V89, V95, or V120). Subunits of FN are connected by a brace of disulfide bonds at the extreme C-termini. Crystal structures of 2-3FNI (PDB 2RKZ), 6-7FNI (PDB 3MQL), 7-10FNIII (PDB 1FNF), and 12-14FNIII (PDB 1FNH) are shown below the display of the subunit. Structures have been manipulated by PyMOL (https://www.pymol.org) to emphasize features discussed in the review. The nomenclature used to designate arrays of modules in the figure is used throughout. Sites of interactions with disordered sequences of bacterial adhesins, α5β1 integrin, and heparan sulfate (HS) are indicated by arrowheads with sidechains interacting with α5β1 or heparan sulfate colored green. The crystal structure of 2-3FNI is shown with a peptide from a bacterial adhesion bound by anti-parallel β-strand addition. Likely stretches of disordered sequences (thick lines) within FN itself or sites of heightened flexibility between consecutive FN modules are indicated by arrows. A color version of the figure is available online in which type I, II, and III FN modules are red, green, and purple, respectively; disulfides are yellow; interacting side chains are green; and arrowheads are red.
Plasma FN circulates at 200 to 600 μg mL-1 (0.4-1.2 μM) in humans (Zerlauth and Wolf, 1984) and 100 to 400 μg mL-1 (0.2-0.8 μM) in mice (Tomasini-Johansson and Mosher, 2009). Remarkably, human plasma FN after purification and concentration is soluble in physiological saline at concentrations (15-20 mg mL1 or 30-40 μM) many-fold greater than that found in the circulation (Mosher and Johnson, 1983). Plasma FN is taken up by tissues and deposited in extracellular matrix fibrils alongside locally synthesized cellular FN (Oh et al., 1981, Moretti et al., 2007). The mechanism of in vivo deposition is presumed to be mimicked by assembly in vitro by cultured fibroblasts, which become competent to assemble plasma FN at low nM concentrations shortly after adhesion to culture dishes (Bae et al., 2004). Plasma FN is also assembled efficiently by adherent platelets in a flow chamber (Cho and Mosher, 2006c). Given the abundance of FN in the plasma, it has long been hypothesized that maintenance of plasma FN in a closed compact conformation is necessary to prevent aberrant interactions among FN protomers and between FN and cell surface receptors and other macromolecules (Pearlstein, 1978, Rocco et al., 1983, Johnson et al., 1999, Singh et al., 2010). Such interactions would be expected to lead to FN assembly in tissues and contribute to thrombus formation in the circulation.
Well before its primary structure was elucidated, purified plasma FN was characterized by a variety of biophysical techniques. The sedimentation velocity and diffusion coefficients were found to decrease as either ionic strength or pH is increased, suggesting that electrostatic interactions maintain FN in its compact conformation (Alexander et al., 1979, Williams et al., 1982, Tooney et al., 1983). Further, light scattering and intrinsic viscosity studies showed that the compact conformation can be perturbed easily inasmuch as the Stoke's radius of FN increased from ∼10.5 nm in ∼150 mM sodium chloride to ∼17.5 nm in ∼1 M sodium chloride (Rocco et al., 1983, Rocco et al., 1987). Rotary shadowing electron microscopy studies visualized FN in compact and V-shape conformations depending on the surface and conditions of adsorption (Odermatt and Engel, 1989). When studies were performed in high salt or acidic or basic conditions, FN had an extended V-shape conformation whereas studies in low salt showed a bent or “irregularly coiled compact” conformation (Tooney et al., 1983, Erickson and Carrell, 1983). On most surfaces, regardless of the conditions tested, there appeared to be a great deal of flexibility along the length of FN (Odermatt and Engel, 1989).
Based on such results and early primary sequence data demonstrating its modular nature, plasma FN was modeled as beads-on-a-string with differences along the string in the degrees that any two beads could bend or rotate relative to one another (Rocco et al., 1983). This model was further developed when the full sequence with its 58 modules (Fig. 1) was known (Rocco et al., 1987). Results of NMR, crystallography, and lower resolution studies are now available that characterize module-module interactions in FN and thus allow one to predict sites of flexibility between modules, shown as arrows in Fig. 1, and long distant interactions that determine quaternary structure of compact plasma FN (Fig. 2). Here, we work from these studies to arrive at models of the interactions that maintain plasma FN in a compact conformation and how these interactions may be broken and re-arranged in ways that are biological relevant. We pay particular attention to modulation of structure by segments of bacterial surface adhesins that interact specifically with FN.
Fig. 2.
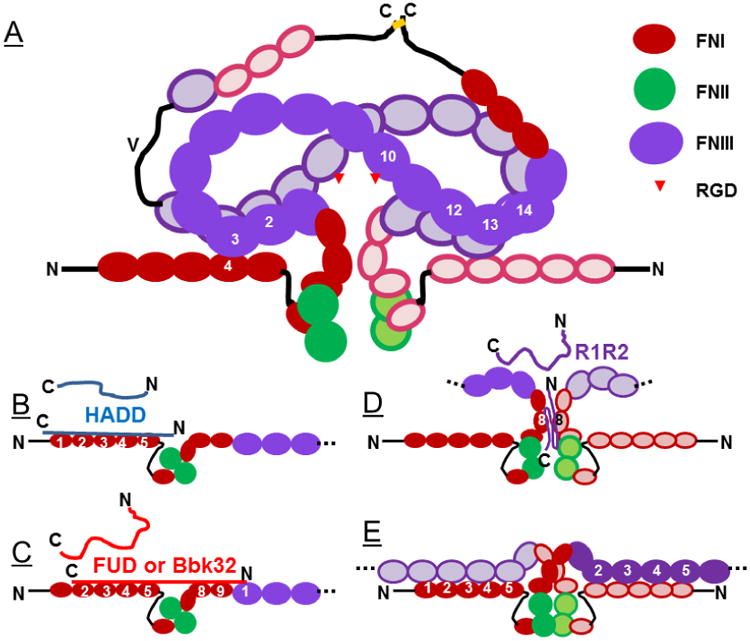
Models of the quaternary structure of globular dimeric plasma FN (A), how the structure may be altered by interaction with polypeptides based on the high affinity downstream domain (HADD) or functional upstream domain (FUD) of the F1 adhesin of S. pyogenes or the Bbk32 segment of BBK32 of B. burgorferi (B, C) or the R1R2 tandem repeats of S. equis subspecies equis (D), and how FN may interact to form fibrils in extracellular matrix (E). Color scheme and abbreviations are as for Fig. 1. Modules in one subunit are in solid whereas those of the opposite subunits are lighter and outlined. Arrowheads indicate the locations of RGD sequences in 10FNIII modules recognized by α5β1 integrin. As described in the text, the globular quaternary structure and cryptic state of 10FNIII are proposed to be favored by intra-subunit interactions between 4FNI and 3FNIII and inter-subunit interactions between 2-3FNIII and 12-14FNIII (A). The adhesin-derived polypeptides are depicted as being unstructured when unbound and interacting with specific FN modules by anti-parallel β-strand addition. Displacement of 3FNIII from 4FNI by HADD, FUD, or Bbk32 (B, C) is depicted as causing elongation of individual FN subunits whereas binding or R1R2 is favored by the compact conformation and ability to ligate both subunits. Panel E is highly schematic and should be compared to a similar schematic referenced in the text. A color version of the figure is available online in which type I, II, and III FN modules are red, green, and purple, respectively.
Modular make-up of FN and structures of FNI, FNII, and FNIII modules
FN is a dimer of 235-250-kDa subunits. Each subunit is composed of an array of 12 type I (FNI), two FNII, and 15-17 FNIII modules (Fig. 1). We use a nomenclature whereby module type is designated by a Roman number and the positions of the modules of each type, ordered from N- to C-terminus, are designated by superscript Arabic numbers. Dimerization is by a brace of disulfide bonds near the C-termini connecting the two subunits (An et al., 1992).
The variable number of type III modules is a result of differential mRNA splicing that determines inclusion of extra domain A (AFNIII) between 11FNIII and 12FNIII or extra domain B (BFNIII) between 7FNIII and 8FNIII (White et al., 2008, Astrof and Hynes, 2009) (Fig. 1). Additionally, differential splicing leads to five different sequences, ranging from 0 to 120 amino acids, that comprise the type III homology connecting segment or V region located between 14FNIII and 15FNIII (Muro et al., 2008, Petersen et al., 1989, Hynes, 1990). Random assortment of differential splicing of AFNIII, BFNIII, and the V region could yield 20 different versions of the human FN subunit and many more possible versions of homodimers and heterodimers. Plasma FN lacks AFNIII and BFNIII, and one subunit lacks residues in the V region, resulting in a simpler mix of dimers. Mice genetically manipulated to enforce the presence or absence of AFNIII and/or BFNIII in all FN subunits, including FN in plasma, manifest many abnormalities including changes in fibrosis and thrombosis (White et al., 2008, Astrof and Hynes, 2009). These observations along with biophysical studies of large FN fragments (Johnson et al., 1999) indicate that AFNIII and BFNIII have important effects on FN structure. However, purified intact FNs harboring AFNIII and/or BFNIII have been available in only limited quantities (e.g. (Guan et al., 1990, Manabe et al., 1997)) and have not been subjected to the extensive biophysical studies that have been carried out on plasma FN. Thus, this review concentrates on plasma FN alone.
The FNI module is unique to chordates, and the FN protein described above is only present in vertebrates (Tucker and Chiquet-Ehrismann, 2009, Adams et al., 2015). However, the FNI module is related structurally to a submodule within the von Willebrand factor-C module found widely in proteins of chordates and non-chordates, suggesting that FNI modules arose from a von Willebrand factor-C module (O'Leary et al., 2004). FN is the only vertebrate protein with tandem FNI modules. There are single FNI modules in tissue plasminogen activator (PLAT), blood coagulation factor XII (F12), and hepatocyte growth factor activator (HGFAC) (Downing et al., 1992, Smith et al., 1994, Maas et al., 2008). In FN, each FNI module is encoded by a single exon (Hynes, 1990). FNI modules are ∼45 amino acids in length and comprise minor AB and major CDE anti-parallel β-sheets, with conserved Cys1-Cys3 and Cys2-Cys4 disulfides connecting the A and D strands and D and E strands, respectively (Potts and Campbell, 1996). Ribbon structures of four FNI modules are shown in Fig. 1.
FNII modules are also encoded by individual exons (Hynes, 1990). The modules, both of which are shown in Fig. 1, are ∼60 amino acids in length and comprise two β-sheets that are perpendicular to each other. The majority of the sequence is in loops between strands. Two disulfides are in a Cys1-Cys3/Cys2-Cys4 pattern (Potts and Campbell, 1996, Pickford et al., 1997). Examples of FNII modules are found in matrix metalloproteinases MMP2 and MMP9 (Potts and Campbell, 1996) and other human proteins including F12, HGFAC, BSPH1, ELSPBP1, LY75, IGF2R, MRC1, MRC2, PLA2R1, and SEL1L (http://www.uniprot.org/uniprot/?query=domain%3A%22fibronectin+type+2+domain*%22+AND+human&sort=score).
Two exons encode the majority of FNIII modules of FN whereas single exons encode 9FNIII, AFNIII, and BFNIII (Muro et al., 2008, Hynes, 1990). FNIII modules are ∼90 amino acids long, fold into a seven-stranded β-sandwich, and lack disulfide bonds (Potts and Campbell, 1996). Ribbon structures of seven FNIII modules are shown in Fig. 1. FNIII modules are widespread in intracellular as well as extracellular proteins such that UniProt lists >7800 FNIII-module-containing proteins from diverse species, including 322 unique proteins in humans (http://www.uniprot.org/uniprot/?query=domain%3A%22fibronectin+type+3+domain*%22+human&sort=score). In many of these proteins, FNIII modules are present in long arrays, as is the case for FN.
To summarize, FN is a “vertebrate invention” and the only protein with tandem FNI modules and with FNI modules adjacent to a long array of FNIII modules. Other multi-modular extracellular matrix proteins such as laminins, tenascins, and fibrinogen/fibrin do not undergo large conformation changes. The uniqueness of plasma FN, therefore, offers a special challenge in discerning structure-function relationships that account for its ability to deposit in extracellular matrix seemingly at “a drop of a hat” and yet remain soluble at concentrations orders of magnitude greater than the concentrations at which it deposits into extracellular matrix or binds to cell surface receptors.
Flexible regions within FN
Approximately 90% of the FN sequence can be assigned to FNI, FNII, or FNIII modules. The rest of the sequence (depicted as black lines in Fig. 1) is likely unstructured and flexible (arrows in Fig. 1) as evidenced by NMR spectroscopy or susceptibility to proteolytic cleavage.
Human FN is synthesized with a 31-residue N-terminal “prepro” sequence ending in a consensus furin cleavage sequence (Petersen et al., 1989). The exact site of cleavage within this sequence by signal peptidase upon translocation into the endoplasmic reticulum and contribution of the remaining “pro” sequence to folding and secretion, to our knowledge, have not been approached experimentally. NMR of recombinant protein comprising the 18-residue N-terminal tail and 1FNI of mature FN indicated that the tail has a mobile random coil structure (Potts et al., 1995). This finding is important because near the tip of the tail are glutaminyl residues through which FN can be cross-linked to itself or other proteins via ε(γ-glutamyl)-lysine bonds catalyzed by activated blood coagulation Factor XIII and tissue transglutaminase (Hoffmann et al., 2011).
The 28-residue linker between 5FNI and 6FNI is much longer than the connectors between other pairs of FNI modules and contains six proline residues. The linking sequence is sensitive to limited proteolysis by trypsin, thrombin, plasmin, or thermolysin, yielding a N-terminal domain comprising five FNI modules (N-5FNI) (Pankov and Yamada, 2002). An NMR structure of the 1-2FNIII tandem construct showed that 2FNIII lacks the A-strand, and instead the A-strand is part of a ∼30-residue segment that has the potential to be a flexible connector between the G-strand of 1FNIII and the B-strand of 2FNIII (Vakonakis et al., 2007). Although this segment is sensitive to limited proteolysis by thermolysin (Borsi et al., 1986), “end-to-side” interactions between 1FNIII and 2FNIII may prevent the two ends of the connector from being mobilized (Vakonakis et al., 2007, Karuri et al., 2009).
The V region, when present, is sensitive to a variety of proteases (Hayashi and Yamada, 1981). Surprisingly, in a crystal structure of 12-15FNIII, none of the residues in 15FNIII were resolved (Sharma et al., 1999). However, 15FNIII has a buried free cysteine (Smith et al., 1982) and therefore is unlikely to be unstructured. It has been suggested that 15FNIII lacks residues needed for an A-strand and may use part of the V region for that purpose (Sharma et al., 1999). Finally, there are 32 residues between the C-terminal residue of 12FNI and the C-terminal inter-subunit disulfides that are likely flexible as evidenced by limited proteolysis experiments with trypsin, chymotrypsin, or plasmin, which produces a 3- to 6-kDa fragment at the C-terminus depending on the enzyme used(An et al., 1992, Pankov and Yamada, 2002).
Differential scanning calorimetry (DSC) has revealed a wide range of melting temperatures for FNIII modules (48°C-121°C) (Litvinovich and Ingham, 1995), and because FNIII modules lack constraining disulfide bonds, it has been hypothesized that strands of FNIII modules can be unfolded under certain conditions (Erickson, 1994). A study looking at exposure of buried cysteines placed by in vitro mutagenesis within FNIII modules indicated that in isolation 2FNIII, 3FNIII, 9FNIII, and 11FNIII unfold (Lemmon et al., 2011). Although FNIII modules in full length FN have not been shown to adopt an unfolded conformation in the circulation, there is substantial computational (Krammer et al., 1999, Paci and Karplus, 1999) and experimental (Lemmon et al., 2011, Baneyx et al., 2001, Baneyx et al., 2002, Smith et al., 2007) evidence that the structure of the FNIII modules can be altered by stretching forces, including those produced by cells during and after FN assembly. It has been suggested that unfolding of FNIII modules explains the observation that with force, the length of FN fibrils can increase 8-fold before 50% of the fibrils break (Ohashi et al., 1999, Klotzsch et al., 2009). Thinking about the additional mobility that could be afforded by unfolding of FNIII modules and how newly mobilized segments might interact with other regions of FN or with other molecules, especially by exchange of the β–strands (Erickson, 1994), is mind-boggling.
Anastellin, truncated 1FNIII lacking the A and B strands, offers an informative case study of the complexities of unravelling type III modules. This segment of FN was identified as a chymotryptic fragment that at low concentration partially inhibits deposition of FN into extracellular matrix (Morla and Ruoslahti, 1992) and subsequently studied as a small recombinant protein. The segment attracted considerable interest when it was found to cause aggregation of intact FN when added at high concentration (Morla et al., 1994) and thereby block tumor angiogenesis in mice (the name is derived from “anastello,” Greek for “inhibit” or “force a retreat”) (Yi and Ruoslahti, 2001, Yi et al., 2003) The truncation increases accessibility of the hydrophobic core of 1FNIII to solvent and causes some of the β–strands to acquire the potential to form backbone hydrogen bonds with β–strands of other proteins (Briknarova et al., 2003). Addition of anastellin to recombinant 3FNIII makes 3FNIII more sensitive to cleavage by thermolysin, suggesting that anastellin exposes sequences that are cryptic in intact 3FNIII (Ohashi et al., 2009). Interestingly, stopped-flow studies indicate that the rate constant for unfolding of 3FNIII is several orders of magnitude faster than the unfolding rate constants of other FNIII modules (Stine et al., 2015). The unusually fast rate is similar to the independently determined (Ohashi et al., 2009) rate of binding of anastellin to 3FNIII, consistent with a model in which 3FNIII has to unfold to interact with anastellin. The interaction of anastellin with 3FNIII, however, does not totally explain aggregation of FN induced by anastellin because only when 2FNIII, 3FNIII, and 11FNIII were stabilized by engineering disulfides into each was recombinant FN resistant to aggregation induced by anastellin (Ohashi and Erickson, 2011)
Interactions between adjacent modules
The beads-on-a-string model predicts differences in the degrees with which modules arrayed end-to-end can twist or tilt relative to one another (Rocco et al., 1983). Intrinsic viscosity measurements of plasma FN in its extended conformation at high ionic strength indicated a persistence length of <25 nm, i.e., that the string is quite flexible (Williams et al., 1982). Results of a number of NMR, crystallography, and lower resolution studies are now available that characterize many of the module-module interactions in FN and thus allow one to predict sites of flexibility between modules, shown as arrows in Fig. 1. Importantly, these studies identify regions of FN in which modules are not arrayed end-to-end.
An NMR structure of 1-2FNI showed no inter-module interactions suggesting that movement of these modules about the linker is minimally constrained (Potts et al., 1999, Pickford and Campbell, 2004). Two alternate crystal forms and an NMR structure of 2FNI-3FNI were solved that differed in inter-module orientation (Rudino-Pinera et al., 2007). The crystal structures showed buried interfaces between the two modules with the NMR structure being most similar to the crystal structure with the larger buried interface (Rudino-Pinera et al., 2007). A co-crystal of a peptide based on Staphylococcus aureus FN-binding adhesin FBPA binding by β-strand addition to consecutive E-strands of 2-3FNI (Fig. 1) contained no additional contacts between 2FNI and 3FNI (Bingham et al., 2008). There are both NMR and crystal structures of 4-5FNI (Bingham et al., 2008, Williams et al., 1994). The NMR structure demonstrated that movement between 4FNI and 5FNI is constrained by extensive contacts with 5FNI of a tryptophan in the C-D loop of 4FNI (Williams et al., 1994). The tryptophan interactions were also found in the crystal structure of 4-5FNI complexed by β-strand addition to a peptide based on FBPA (Bingham et al., 2008). Thus, three of the four potential interfaces between consecutive modules have been examined for the first five FNI modules and found to exhibit quite different flexibility from one another, large for 1-2FNI, limited for 2-3FNI, and none for 4-5FNI. We are not aware of information pertaining to inter-module contacts between 3FNI and 4FNI. The flexibility must allow the five N-terminal FNI modules to adopt a linear conformation with aligned E-strands as evidenced by high affinity binding of unstructured polypeptides based on bacterial adhesins from Streptococcus pyogenes that contain motifs for engaging consecutive E-strands of 1-5FNI by β-strand addition (Marjenberg et al., 2011, Norris et al., 2011, Maurer et al., 2012b). In addition, the spacing between the FNI modules is important for engagement with other proteins as evidenced by an assay in which stretching the inter-modular distances of 1-5FNI modules decreased the affinity of peptides from Streptococcus dysgalactiae or S. aureus to 1-2FNI or 4-5FNI modules (Chabria et al., 2010).
DSC, intrinsic fluorescence, and NMR indicate that all six modules of the gelatin-binding domain, 6-9FNI, are independently folded (Litvinovich et al., 1991). Consistently, NMR-deduced structures of 1FNII (Pickford et al., 1997), 2FNII (Sticht et al., 1998), and 7FNI (Baron et al., 1990) demonstrated the expected global fold for each. Further, an NMR structure of 1FNII-2FNII also revealed individual modules with canonical structures and no inter-modular interfaces (Smith et al., 2000). However, when 6FNI was added to yield 6FNI-2FNII, a hydrophobic interface between 6FNI and 2FNII was found that resulted in a compact triangular shape (Pickford et al., 2001). A similar interaction between 6FNI and 2FNII was found when 7FNI was added to yield 6-7FNI, i.e., a construct comprising 6FNI, 1-2FNII, and 7FNI; interactions also were noted between 7FNI and 1FNII-2FNII and between 1FNII and 2FNII (Erat et al., 2010) (Fig. 1). NMR of the same 6-7FNI construct complexed to a collagenous peptide demonstrated binding of the peptide to a groove on 2FNII and extending by β-strand addition to the E-strand of 7FNI (Erat et al., 2010). The link between 7FNI and 8FNI is likely flexible based on small angle x-ray scattering (SAXS) studies of 6-9FNI without and with binding of the collagenous peptide (Erat et al., 2013). The SAXS studies were interpreted as showing the non-ligated construct in multiple conformations with greater population of the ligated construct in a conformation that has a 90° kink between 7FNI and 8FNI modules (Erat et al., 2013). A co-crystal structure of 8FNI-9FNI with peptide based on the α(1) chain of type I collagen showed 8FNI and 9FNI as canonical type I modules in a tandem arrangement with a relatively small area buried and the collagenous peptide bound by β-strand addition to 8FNI (Erat et al., 2009). Putting together the structures of complexes of 6-7FNI or 8-9FNI with collagenous peptides, a model of collagen binding to 6-9FNI was proposed with the binding site comprising the 2FNII groove and E-strands of 7FNI and 8FNI (Erat et al., 2010).
A different structure was found when 6-9FNI was crystallized in presence of 30-50 mM Zn2+ (Graille et al., 2010). The part of the structure comprising 6FNI-2FNII is as described above (Pickford et al., 2001, Erat et al., 2010) and shown in Fig. 1. However, 8FNI lacks the FNI fold and instead is composed of two long strands that associate with 7FNI and 9FN1 to form a large β-sheet structure (Graille et al., 2010). The physiological relevance of the structure in Zn2+ is unclear; for instance, the concentrations of Zn2+ required for crystallization do not support binding of a collagenous peptide to the gelatin-binding domain (Graille et al., 2010). Further, binding to 8-9FNI of polypeptides derived from bacterial surface proteins was abolished in the presence of Zn2+ (Maurer et al., 2010, Ma et al., 2015a). However, the radically different structures of 8FNI within the whole gelatin-binding domain in Zn2+ compared to the tandem 8-9FNI construct indicate that this region in FN has the potential to undergo major conformational rearrangements.
The labile region seemingly extends into 9FNI1-2FNIII. Expression of the epitope of mouse monoclonal antibody L8 requires 9FNI and 1FNIII be contiguous (Chernousov et al., 1991). Although the L8 epitope was demonstrated to map to Gln690 of 1FNIII by in vitro mutagenesis (Maurer et al., 2010), the epitope is sensitive to reduction of disulfides in 9FNI (Chernousov et al., 1991). These findings indicate that 9FNI and 1FNIII share an interface through which the modules influence the structure of one another. 1FNIII is mechanically weak when subjected to atomic force microscopy compared to 1-2FNIII, suggesting that 1FNIII and 2FNIII also interact (Oberhauser et al., 2002). NMR of 2FNIII alone revealed that the A-strand is disordered, whereas NMR of 1-2FNIII was interpreted as showing that the C-terminal end of 1FNIII is “wedged” along the N-terminal to C-terminal axis of 2FNIII, creating ∼550-Å2 buried interface (Vakonakis et al., 2007). This arrangement involves a twist in the G-strand of 2FNIII allowed by the absence of a structured A-strand (Vakonakis et al., 2007). The residues from the missing A-strand of 2FNIII together with 17 residue linking 1FNIII and 2FNIII constitute a stretch of ∼30 residues that is unstructured. Consistent with an interaction between 1FNIII and 2FNIII, fluorescence resonance energy transfer studies indicated that opposite ends of 1FNIII and 2FNIII are within 50-60 Å instead of being extended linearly in opposite directions (Karuri et al., 2009). Mutagenesis studies of the tandem construct implicate a salt bridge between Lys672 in 1FNIII and Glu767 in 2FNIII with Lys669 playing a less critical role (Vakonakis et al., 2007, Karuri et al., 2009). It should be noted, however, that studies of a similar but not identical 1-2FNIII tandem construct failed to demonstrate the inter-module interaction or effects of mutations of putative salt bridge residues (Ohashi and Erickson, 2011). Further, the structure of 1-2FNIII in the context of neighboring modules, particularly 9FNI, is not known. Finally, it remains to be determined whether the ∼30 residues are disordered or structured in intact compact or extended FN,
A crystal structure of 7FNIII-10FNIII showed a buried interface of ∼550 Å2 between 7FNIII and 8FNIII or between 8FNIII and 9FNIII and a smaller buried interface of ∼300 Å2 between 9FNIII and 10FNIII (Leahy et al., 1996). The interface between 9FNIII and 10FNIII, which is of special interest because deletion of either the arginine-glycine-aspartic acid (RGD) cell adhesive sequence in 10FNIII or residues of the synergy site in 9FNIII results in >95% loss of integrin-associated cell adhesion (Obara et al., 1988, Aota et al., 1994), has been analyzed by NMR and other analyses for human (Spitzfaden et al., 1997) and mouse (Copie et al., 1998) 9-10FNIII. Most of the interactions between 9FNIII and 9FNIII were characterized as being of low energy (Spitzfaden et al., 1997). Despite this, correct orientation between the synergy site in 9FNIII and the RGD in 10FNIII is required to enhance the interactions of FN with integrins (Grant et al., 1997, Altroff et al., 2004). Addition of residues between 9FNIII and 10FNIII, which resulted in increased mobility of the modules as seen by NMR (Spitzfaden et al., 1997), was associated with a decrease in integrin-associated activities, including cell spreading (Grant et al., 1997). However, fixation of the orientation between 9FNIII and 10FNIII by introduction of a disulfide bond between the two modules also decreased integrin-associated activities (Altroff et al., 2004).
DSC and fluorescence denaturation experiments showed that 12FNIII, 13FNIII, 14FNIII, and 15FNIII all fold independently: 13FNIII was the least stable but appeared to be stabilized by 14FNIII (Novokhatny et al., 1992). A crystal structure of the V0 variant of 12-15FNIII revealed buried interfaces of ∼450 Å2 between 12FNIII and 13FNIII and ∼650 Å2 with several charge interactions between 13FNIII and 14FNIII (Sharma et al., 1999). None of the residues in 15FNIII were resolved. As described above, it has been suggested that 15FNIII lacks residues needed for an A-strand and when possible may use part of the V region for that purpose (Sharma et al., 1999).
With the exception of the three or four extended flexible sequences and the stretch of modules between 6FNI and 2FNIII, therefore, when examined the modules in a FN subunit have been arrayed head-to-tail with differences in the freedom of any two consecutive modules to tilt or twist relative to one another. A series of preferential module-module conformations could result in bending. However, it is difficult to predict such bending. For instance, the crystal structures of 7-10FNIII and 12-14FNIII shown in Fig. 1 exhibited a variety on tilts and twists such that one could envision the string of modules from 7FNIII to 14FNIII being anywhere from linear or circular (Sharma et al., 1999). For this reason, module-module interactions seem more likely to be facilitative rather than determinant in allowing plasma FN to adopt a distinct compact quaternary structure.
Long-range Interactions
Fig. 2 models how long-range interactions could drive plasma FN into a compact structure and when relieved could allow FN to open up and extend. It emphasizes interactions between 4FNI and 3FNIII of the same subunit and between 2-3FNIII and 12-14FNIII of the different subunits.
An NMR titration of FN fragments showed that recombinant 4-5FNI interacts with recombinant 3FNIII with a Kd of 0.15 mM (Vakonakis et al., 2009). This interaction was also studied in recombinant N-3FNIII. The interaction appeared to be electrostatic and involve Arg222 in 4FNI inasmuch as size exclusion chromatography showed that mutation of this residue to alanine caused an increase in the hydrodynamic radius of the recombinant protein (Vakonakis et al., 2009). The 4FNI-3FNIII interaction has possible significance in controlling the migration stimulatory activity of FN in collagen gels. This activity is lacking in intact plasma FN, and among naturally occurring FN splice variants lacking the bulk of the C-terminal modules a FN splice variant comprising N-1FNIII has activity whereas the splice variant comprising N-3FNIIIl does not (Vakonakis et al., 2009). When the R222A mutation was introduced into 4FNI of N-3FNIII, the construct gained the ability to stimulate fibroblast migration (Vakonakis et al., 2009).
Evidence for interactions involving 12-14FNIII comes from studies comparing behavior of solutions with low ionic strength of recombinant FN in which 12-14FNIII was deleted or replaced with FNIII domains A1 thru A3 from tenascin C or of large recombinant FN fragments (Johnson et al., 1999). For FN with deletion or replacement of 12-14FNIII, lower sedimentation velocities were found than when 12-14FNIII was present (Johnson et al., 1999). This finding suggests that 12-14FNIII mediates specific interactions that are necessary to maintain FN in a more compact conformation. Velocity sedimentation experiments using N- and C-terminal truncations of 2-14FNIII demonstrated dimerization only in the full-length construct, leading to the conclusion that 2-3FNIII from one subunit of FN interacts with 12-14FNIII from the other subunit (Johnson et al., 1999).
Fig. 2A, therefore, places 3FNIII next to 4FNI within the same subunit and 12-14FNIII in one subunit next to 2-3FNIII in the opposite subunit, creating two hubs that could drive plasma FN into a distinct compact conformation. The model is supported by immunoassays in which soluble plasma FN is asked to compete for binding of monoclonal antibody III-10 (mAbIII-10) (Ugarova et al., 1995) to FN adsorbed to microtiter plate plastic. mAbIII-10 recognizes 10FNIII, which contains the RGD cell adhesive sequence for α5β1 and is presumed to be fully exposed when FN is bound to plastic for cell adhesion assays. Competition by soluble plasma FN is poor at physiologic pH and ionic strength (Ugarova et al., 1995), consistent with the known inability of soluble plasma FN to prevent cell adhesion to adsorbed FN (Pearlstein, 1978). Thus, the RGD cell adhesive sequences in the 10FNIII are depicted as being cryptic in compact plasma FN (Fig. 2A). Removal of 1-5FNI from FN or ligation of 1-5FNI by β-strand addition greatly increases the recognition of plasma FN by mAbIII-10 (Ensenberger et al., 2004, Maurer et al., 2010, Maurer et al., 2012b). These results suggest that loss of the interaction of 4FNI with 3FNIII because 4FNI is missing or 3FNIII is displaced by the bacterial adhesin favors unravelling of the compact conformation responsible for the cryptic MAbIII-10 epitope. In addition, pre-incubation of soluble plasma FN with mABIII-10 or heparin increases binding of bacterial adhesins. This finding is consistent with an allosteric network linking binding and conformational change at 10FNIII or 12-14FNIII to binding and conformation of 1-5FNI (Maurer et al., 2012b). Rotary shadowing EM images indicated that mAbIII-10 employs its two FAB binding sites to bridge and engage the two subunits of extended adsorbed FN (Ugarova et al., 1995). The EM images raise the possibility that the antigenic epitopes in compact soluble FN are cryptic for two reasons: being buried within a compact conformation and not being separated by a distance that is optimal for bridging. In either case, the manipulations that improve the interaction of mAbIII-10 with intact soluble FN are compatible with unraveling and extension in response to loss of the interactions shown in Fig. 2A.
Binding of the N-terminal FNI modules to bacteria and disruption of the compact structure of FN
Bacteria have had ∼500 million years to adapt to FN since its “invention” in early vertebrates and seem to have done so with vigor, such that more than 100 different FN-binding bacterial proteins have been identified (Henderson et al., 2011). Many of these belong to the MSCRAMM (microbial surface components recognizing adhesive matrix molecules) family of proteins. MSCRAMMs are common in Gram-positive cocci including Staphylococcus aureus and Streptococcus pyogenes as well as spirochetes such as Borrelia burgdorferi, which causes Lyme disease (Schwarz-Linek et al., 2006). In general these proteins are thought to be involved in bacterial invasion of non-phagocytic host cells (Dziewanowska et al., 1998, Ozeri et al., 1998). MSCRAMMs contain variable numbers of FN binding repeats (FNBRs) with or without non-repetitive upstream or downstream sequences (Schwarz-Linek et al., 2006). Crystallographic and NMR studies indicate that FNBRs transition from a random coil to bind to the E-strands of consecutive FNI modules via anti-parallel tandem β-strand addition, also called β-zipper formation (Bingham et al., 2008, Schwarz-Linek et al., 2003). An example of such binding for a peptide based on a FNBR that interacts with 2-3FNI is shown in Fig. 1. The FNBR of the F1 adhesin (allelic variant SfbI) of S. pyogenes binds to 2-5FNI whereas the adjacent downstream non-repetitive domain binds to 1FNI (Bingham et al., 2008, Schwarz-Linek et al., 2003, Schwarz-Linek et al., 2004). Thus, the binding site for the recombinant polypeptide that we have dubbed HADD (for “high affinity downstream domain”) maps to 1-5FNI (Maurer et al., 2012b) (Fig. 2B). The adjacent upstream non-repetitive domain binds to 8-9FNI within the gelatin-binding domain of FN (Fig. 2C) (Maurer et al., 2010, Atkin et al., 2010). Mapping and homology modeling studies indicated that binding of FUD (“functional upstream domain”), a 49-amino acid polypeptide comprising the upstream domain and adjacent FNBR of the F1 adhesin, show that FUD binding requires modules 6-7FNI to “loop-out,” i.e., the binding site on fibronectin is discontinuous, jumping from 2-5FNI to 8FNI (Maurer et al., 2010) (Fig. 2C). B. burgdorferi, which is genetically distant from S. pyogenes, encodes the BBK32 adhesin that has a single FN-binding sequence and very different protein organization than the F1 adhesin (Kim et al., 2004). Remarkably, the binding sequence, which we have dubbed Bbk32, also binds to 2-5FNI and 8FNI (Harris et al., 2014) (Fig. 2C). As mentioned above, peptides from adhesins in S. dysgalactiae and S. aureus bind FN less well after FN is stretched (Chabria et al., 2010). This finding suggests that in order to engage the FNBR consecutive FNI modules must have favorable spacing.
Fluorescence polarization stopped-flow assays demonstrated that FUD and Bbk32 bind more rapidly to N-9FNI than to FN, likely due to competition for binding from the intramolecular interaction between 4FNI and 3FNIII shown in Fig. 2A (Ma et al., 2015b). The on-rates for binding of FUD and Bbk32 to plasma FN were similar, 3.9 × 105 M-1 sec-1 and 2.9 × 105M-1 sec-1, respectively, and ∼10-fold slower than binding of the two probes to N-9FNI. The off-rates to either FN or N-9FNI, however, were very different, ∼3 × 10-2 sec-1 for Bbk32 and ≪10-3 sec-1 for FUD. FUD and Bbk32 differ in the spacing between sequences that interact with 3FNI and 4FNI or with 5FNI and 8FNI. Thus, it has been suggested that these results indicate a “folding-after-binding” process after initial association of certain polypeptide sequences to FN that results in formation of complexes of variable stability and is a function of sequences between the engagement sites, and perhaps flexibility within the polypeptide-FN complex (Ma et al., 2015b). Thus, even though SfbI and BBK32 adhesins target the same extended binding site on plasma FN, the kinetics of binding are specific for the two adhesins. It has been suggested that bacterial pathogenicity may be determined by stability of adhesin-FN complexes, long-lasting in case of S. pyogenes that forms a biofilm in the pharynx and transient for B. burgdorferi, which is migratory (Ma et al., 2015b).
Isothermal calorimetric analysis of binding of FN to polypeptides with one or several FNBRs showed that binding of one FN to one FNBR influences FN binding to the adjacent FNBR, i.e., binding of FN to adhesins with multiple FNBRs is cooperative (Marjenberg et al., 2011). It was hypothesized that this binding paradigm would allow a single adhesin to capture and change the conformations of multiple FN dimers, thus exposing multiple RGD motifs that cause integrins to cluster and facilitate cellular invasion by bacteria (Marjenberg et al., 2011). Of note, strains of S. aureus cultured from blood of patients with infected cardiac devices have mutations in the FNBRs of the FNBPA adhesin that increase the strength of binding to FN (Messina et al., 2016)
Binding of plasma FN to collagen
Deposition of type I and type III collagen into the extracellular matrix in cell culture is dependent on the formation of a FN matrix (Velling et al., 2002, Sottile and Hocking, 2002, Sottile et al., 2007). How FN and collagen interact to form collagen fibrils and the relative importance of this interaction for in vivo collagen fibrillogenesis, however, are obscure (Kadler et al., 2008, Moriya et al., 2011). In fibroblast cultures, FN and type I collagen are found initially in ∼10-nm diameter aperiodic fibrils and later, especially after treatment of cells with ascorbate, in 40-nm diameter fibrils on which FN is present with 70-nm periodicity (Furcht et al., 1980). Thus, two separate types of interactions may take place, first of freshly secreted collagen molecules binding to a polymerized FN fibril, perhaps aligning collagen molecules (Singh et al., 2010), and second of freshly secreted FN molecules associating with sites displayed on fibrils of hydroxylated and polymerized collagen.
As described above, a model combining structural analyses of binding of collagenous peptides to 6-7FNI and 8-9FNI defines a binding pocket in FN extending from 2FNII to 9FNI that is dependent on intramolecular interactions among all six modules (6-9FNI) of the collagen-binding domain (Erat et al., 2010). The model suffers from the fact that the structural studies employed the same sequence from the “3/4 site” of type I or II collagen to probe binding to both 2FNII-7FNI and 8-9FNI (Erat et al., 2009, Erat et al., 2010). However, a longer peptide based on residues 70-118 at the “1/10-site” of the α2(I) chain of type I collagen was subsequently demonstrated to use its C-terminal and N-terminal halves specifically to engage 2FNII-7FNI and 8-9FNI and increase binding affinity cooperatively (Erat et al., 2013). Thus, type I collagen, with its two α1(I) chains and single α2(I) chain, is proposed to contain multiple sites that bind to the 6-9FNI collagen-binding domain with approximately equal micromolar affinity (Erat et al., 2013). One can imagine these sites displayed in collagen fibrils and mediating the periodic binding of FN.
Studies of the R1R2 polypeptide from SFS protein of Streptococcus equi, which is a strong inhibitor of FN-collagen binding, revealed a possible mechanism to achieve low nanomolar binding of collagen to FN. R1R2 contains two identical short GxxGE collagen-like repeats and binds to plasma fibronectin via modules 8-9FNI in a two-step reaction involving both fibronectin subunits (Ma et al., 2015a). Fig. 2D depicts R1R2 as folding back on itself so as to engage both 8FNI modules of the FN dimer by anti-parallel β-strand addition. Thus, unlike binding of FUD, HADD, or Bbk32, which is better when isolated N-9FNI is targeted (Maurer et al., 2010, Maurer et al., 2012b, Harris et al., 2014), R1R2 binds more tightly to dimeric FN. The two α1(I) and single α2(I) chains of type I collagen are in register and aligned N- to C-terminus (Brodsky and Persikov, 2005). In a soluble system, FN can be cross-linked efficiently to triple-helical type I collagen at 37°C but not at lower temperature (Mosher et al., 1979). At this temperature, there is partial unfolding of collagen at the “3/4 site” in proximity to the GxxGE motif where there is no proline over a stretch of 6 Gxx triplets (Stultz, 2002, Leikina et al., 2002, An et al., 2014). FN may mimic binding to the two repeats of R1R2 by associating with locally denatured sequences of two of the three subunits of type I collagen in a high affinity interaction (Ma et al., 2015a). A key consequence of this model is that for assembled FN to bind to, and possibly facilitate assembly of, nascent type I collagen, the two 8FNI modules must remain in proximity after incorporation of FN into extracellular matrix (Fig. 2D). Consistent with this notion, imaging studies employing fluorescence probes as an indicator of strain exerted on FN fibrils indicate that collagen associates preferentially with relaxed fibrils, as freshly elaborated FN fibrils are known to be (Kubow et al., 2015)
Assembly of plasma FN
Assembly of plasma FN requires that the protein becomes extended. There are two proposed ways to initiate extension: interaction of the N-terminal 70-kDa region of FN comprising N-9FNI with the cell surface (Tomasini-Johansson et al., 2006) and interaction of the integrin-binding RGD motif in 10FNIII with cell surface integrins (Geiger et al., 2001). Several pieces of evidence support initiation of FN assembly by binding of N-9FNI to cell-surface molecules. The presence of N-5FNI is essential for FN assembly (Schwarzbauer, 1991, Takahashi et al., 2007), isolated N-9FNI binds to the cell-surface with the same kinetics as full-length FN (Zhang et al., 1994, Cho and Mosher, 2006a), N-9FNI is a dominant negative inhibitor of FN assembly (McKeown-Longo and Mosher, 1985), and polypeptides such as FUD that bind to the 70K region block FN assembly (Tomasini-Johansson et al., 2001, Cho and Mosher, 2006b).
Cell-surface receptors for N-9FNI remain unknown. There are potential integrin-binding sites in N-9FNI, namely NGR motifs that spontaneously convert to integrin binding isoDGR motifs (Curnis et al., 2006), and IGD motifs, which stimulate fibroblast migration into type I collagen gels (Schor et al., 1999, Millard et al., 2007). However, neither the NGR motifs nor IGD motifs are required for binding of N-9FNI to the cell surface sites of assembly (Xu et al., 2010, Maurer et al., 2012a). N-9FNI appears to bind to the cell surface via multiple FNI modules as evidenced by studies showing that deletion of single FNI modules in N-5FNI destroys binding of N-9FNI to the cell surface (Sottile et al., 1991). Attempts to identify cell-surface binding partners of N-9FNI by photo-affinity cross-linking have identified only very large molecules (Zhang and Mosher, 1996). Such attempts need to be re-assessed knowing that N-9FNI can be engaged by rapid-on, rapid-off ligands such as Bbk32. When interactors of N-9FNI in plasma or platelet lysates were isolated by IFAST (immiscible filtration assisted by surface tension) in which bound molecules are separated from free by rapid passage through an immiscible barrier so as to capture transient binding, 36 interactors were identified by mass spectrometry, 31 in plasma and five in platelet lysate, of which only eight were previously known to interact with FN (Moussavi-Harami et al., 2013). The interactions were all specific as defined by blocking with FUD and therefore likely involve N-9FNI binding by β-strand addition to an intrinsically disordered region of the interactor. Disordered regions are common throughout the proteome and well-known mediators of protein-protein interactions (e.g. (Tompa et al., 2015)), and it seems likely that segments of multiple cell surface molecules mimic Bbk32 and interact transiently with the N-9FNI region of FN. Whether such interactions are productive of assembly may depend on whether the interactions coincide with other transient interactions such as unfolding of 3FNIII (Stine et al., 2015) or binding of α5β1 integrin to 10FNIII (Takagi et al., 2003).
To understand FN assembly further, one would like to know the molecular architecture of FN in fibrils, i.e., which parts of FN interact with one another. Fig. 2E is based on the concluding figure in a recent examination of native fibronectin fibrils by site-specific protein labelling and single-molecule localization by stepwise photobleaching or direct stochastic optical reconstruction microscopy (Fruh et al., 2015). Single end-labelled fibronectin molecules had an average end-to-end distance of 133 nm, and antibody epitopes displayed periodic punctate label patterns with ∼95 nm repeats and alternating N- and C-terminal regions. These measurements suggested an antiparallel 30-40 nm overlap between N-termini. Thus, the first five type I modules are depicted as binding type III modules of the adjacent molecule, in a sense reversing the interactions in compact FN proposed in Fig. 2A. However, exactly which modules interact with which and the nature of the interactions—β-strand addition, β-strand invasion and exchange, complementary surfaces of folded modules finding one another—remain obscure.
Concluding remarks
Research over the last several decades has greatly increased our understanding of the structures of the FNI, FNII, and FNIII modules that compose FN and of how certain of these modules array themselves within plasma FN. Here, we work from this progress to understand plasma FN itself. The models of plasma FN in its compact form and how the compact form can open up, although based on a coherent set of experimental observations, are speculative and offered to catalyze further thinking. Much needs to be learned. For instance, when and how does plasma FN adopt the compact form during biosynthesis? Is the propeptide important for adopting the form? How does FN avoid binding to the many potential interacting molecules in the secretory pathway, i.e., are there specific chaperones? Most of this review concentrates on the N-terminal part of the FN subunit. Do the FNI and FNIII modules near the C-terminus have similar susceptibility to conformational change and what is their structure and function? The review, to maintain focus, does not discuss the structural and functional consequences of introducing AFNIII and BFNIII into FN. These consequences are important and need to be understood. Advances in proteomics, imaging, and single molecule analysis should make it possible to address such issues and questions.
Acknowledgments
We thank our many colleagues for stimulating conversations and input.
Footnotes
Declaration of interest: The paper was written by ourselves. We report no conflicts of interest.
References
- Adams JC, Chiquet-Ehrismann R, Tucker RP. The evolution of tenascins and fibronectin. Cell Adh Migr. 2015;9:22–33. doi: 10.4161/19336918.2014.970030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SS, JR, Colonna G, Edelhoch H. The structure and stability of human plasma cold-insoluble globulin. The Journal of biological chemistry. 1979;254:1501–5. [PubMed] [Google Scholar]
- Altroff H, Schlinkert R, Van Der Walle CF, Bernini A, Campbell ID, Werner JM, Mardon HJ. Interdomain tilt angle determines integrin-dependent function of the ninth and tenth FIII domains of human fibronectin. The Journal of biological chemistry. 2004;279:55995–6003. doi: 10.1074/jbc.M406976200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An B, Abbonante V, Yigit S, Balduini A, Kaplan DL, Brodsky B. Definition of the native and denatured type II collagen binding site for fibronectin using a recombinant collagen system. The Journal of biological chemistry. 2014;289:4941–51. doi: 10.1074/jbc.M113.530808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An SS, Jimenez-Barbero J, Petersen TE, Llinas M. The two polypeptide chains in fibronectin are joined in antiparallel fashion: NMR structural characterization. Biochemistry. 1992;31:9927–33. doi: 10.1021/bi00156a010. [DOI] [PubMed] [Google Scholar]
- Aota S, Nomizu M, Yamada KM. The short amino acid sequence Pro-His-Ser-Arg-Asn in human fibronectin enhances cell-adhesive function. The Journal of biological chemistry. 1994;269:24756–61. [PubMed] [Google Scholar]
- Astrof S, Hynes RO. Fibronectins in vascular morphogenesis. Angiogenesis. 2009;12:165–75. doi: 10.1007/s10456-009-9136-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin KE, Brentnall AS, Harris G, Bingham RJ, Erat MC, Millard CJ, Schwarz-Linek U, Staunton D, Vakonakis I, Campbell ID, Potts JR. The streptococcal binding site in the gelatin-binding domain of fibronectin is consistent with a non-linear arrangement of modules. The Journal of biological chemistry. 2010;285:36977–83. doi: 10.1074/jbc.M110.156935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae E, Sakai T, Mosher DF. Assembly of exogenous fibronectin by fibronectin-null cells is dependent on the adhesive substrate. J Biol Chem. 2004;279:35749–59. doi: 10.1074/jbc.M406283200. [DOI] [PubMed] [Google Scholar]
- Baneyx G, Baugh L, Vogel V. Coexisting conformations of fibronectin in cell culture imaged using fluorescence resonance energy transfer. Proc Natl Acad Sci USA. 2001;98:14464–8. doi: 10.1073/pnas.251422998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneyx G, Baugh L, Vogel V. Fibronectin extension and unfolding within cell matrix fibrils controlled by cytoskeletal tension. Proc Natl Acad Sci USA. 2002;99:5139–43. doi: 10.1073/pnas.072650799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M, Norman D, Willis A, Campbell LD. Structure of the fibronectin type 1 module. Nature. 1990;345:642–646. doi: 10.1038/345642a0. [DOI] [PubMed] [Google Scholar]
- Bingham RJ, Rudino-Pinera E, Meenan NA, Schwarz-Linek U, Turkenburg JP, Hook M, Garman EF, Potts JR. Crystal structures of fibronectin-binding sites from Staphylococcus aureus FnBPA in complex with fibronectin domains. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12254–8. doi: 10.1073/pnas.0803556105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsi L, Castellani P, Balza E, Siri A, Pellecchia C, De Scalzi F, Zardi L. Large-scale procedure for the purification of fibronectin domains. Analytical biochemistry. 1986;155:335–45. doi: 10.1016/0003-2697(86)90443-4. [DOI] [PubMed] [Google Scholar]
- Briknarova K, Akerman ME, Hoyt DW, Ruoslahti E, Ely KR. Anastellin, an FN3 fragment with fibronectin polymerization activity, resembles amyloid fibril precursors. J Mol Biol. 2003;332:205–15. doi: 10.1016/s0022-2836(03)00890-8. [DOI] [PubMed] [Google Scholar]
- Brodsky B, Persikov AV. Molecular structure of the collagen triple helix. Advances in protein chemistry. 2005;70:301–39. doi: 10.1016/S0065-3233(05)70009-7. [DOI] [PubMed] [Google Scholar]
- Chabria M, Hertig S, Smith ML, Vogel V. Stretching fibronectin fibres disrupts binding of bacterial adhesins by physically destroying an epitope. Nature communications. 2010;1:135. doi: 10.1038/ncomms1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernousov MA, Fogerty FJ, Koteliansky VE, Mosher DF. Role of the I-9 and III-1 modules of fibronectin in formation of an extracellular fibronectin matrix. J Biol Chem. 1991;266:10851–10858. [PubMed] [Google Scholar]
- Chiang HY, Korshunov VA, Serour A, Shi F, Sottile J. Fibronectin is an important regulator of flow-induced vascular remodeling. Arterioscler Thromb Vasc Biol. 2009;29:1074–9. doi: 10.1161/ATVBAHA.108.181081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Mosher DF. Characterization of fibronectin assembly by platelets adherent to adsorbed laminin-111. J Thromb Haemost. 2006a;4:943–51. doi: 10.1111/j.1538-7836.2006.01862.x. [DOI] [PubMed] [Google Scholar]
- Cho J, Mosher DF. Enhancement of thrombogenesis by plasma fibronectin cross-linked to fibrin and assembled in platelet thrombi. Blood. 2006b;107:3555–63. doi: 10.1182/blood-2005-10-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Mosher DF. Role of fibronectin assembly in platelet thrombus formation. J Thromb Haemost. 2006c;4:1461–9. doi: 10.1111/j.1538-7836.2006.01943.x. [DOI] [PubMed] [Google Scholar]
- Copie V, Tomita Y, Akiyama SK, Aota SI, Yamada KM, Venable RM, Pastor RW, Krueger S, Torchia DA. Solution structure and dynamics of linked cell attachment modules of mouse fibronectin containing the RGD and synergy regions: comparison with the human fibronectin crystal structure. J Mol Biol. 1998;277:663–682. doi: 10.1006/jmbi.1998.1616. [DOI] [PubMed] [Google Scholar]
- Curnis F, Longhi R, Crippa L, Cattaneo A, Dondossola E, Bachi A, Corti A. Spontaneous formation of L-isoaspartate and gain of function in fibronectin. J Biol Chem. 2006;281:36466–76. doi: 10.1074/jbc.M604812200. [DOI] [PubMed] [Google Scholar]
- Downing AK, Driscoll PC, Harvey TS, Dudgeon TJ, Smith BO, Baron M, Campbell ID. Solution structure of the fibrin binding finger domain of tissue-type plasminogen activator determined by 1H nuclear magnetic resonance. J Mol Biol. 1992;225:821–33. doi: 10.1016/0022-2836(92)90403-7. [DOI] [PubMed] [Google Scholar]
- Dziewanowska K, Patti JM, Deobald CF, Bayles KW, Trumble WR, Bohach GA. Fibronectin Binding Protein and Host Cell Tyrosine Kinase are Required for Internalization of Staphylococcus aureus by Epithelial Cells. Infect Immun. 1998;67:4673–4678. doi: 10.1128/iai.67.9.4673-4678.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensenberger MG, Annis DS, Mosher DF. Actions of the functional upstream domain of protein F1 of Streptococcus pyogenes on the conformation of fibronectin. Biophys Chem. 2004;112:201–7. doi: 10.1016/j.bpc.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Erat MC, Schwarz-Linek U, Pickford AR, Farndale RW, Campbell ID, Vakonakis I. Implications for collagen binding from the crystallographic structure of fibronectin 6FnI1-2FnII7FnI. The Journal of biological chemistry. 2010;285:33764–70. doi: 10.1074/jbc.M110.139394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erat MC, Sladek B, Campbell ID, Vakonakis I. Structural analysis of collagen type I interactions with human fibronectin reveals a cooperative binding mode. The Journal of biological chemistry. 2013;288:17441–50. doi: 10.1074/jbc.M113.469841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erat MC, Slatter DA, Lowe ED, Millard CJ, Farndale RW, Campbell ID, Vakonakis I. Identification and structural analysis of type I collagen sites in complex with fibronectin fragments. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4195–200. doi: 10.1073/pnas.0812516106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP. Reversible unfolding of fibronectin type III and immunoglobulin domains provides the structural basis for stretch and elasticity of titin and fibronectin. Proc Natl Acad Sci USA. 1994;91:10114–10118. doi: 10.1073/pnas.91.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Carrell NA. Fibronectin in extended and compact conformations. Electron microscopy and sedimentation analysis. J Biol Chem. 1983;258:14539–44. [PubMed] [Google Scholar]
- Fruh SM, Schoen I, Ries J, Vogel V. Molecular architecture of native fibronectin fibrils. Nature communications. 2015;6:7275. doi: 10.1038/ncomms8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furcht LT, Smith D, Wendelschafer-Crabb G, Mosher DF, Foidart JM. Fibronectin presence in native collagen fibrils of human fibroblasts: Immunoperoxidase and immunoferritin localization. J Histo Cyto. 1980;28:1319–1333. doi: 10.1177/28.12.7014712. [DOI] [PubMed] [Google Scholar]
- Geiger B, Bershadsky A, Pankov R, Yamada KM. Transmembrane crosstalk between the extracellular matrix--cytoskeleton crosstalk. Nat Rev Mol Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- George EL, Georges-Labouesse EN, Patel-King RS, Rayburn H, Hynes RO. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development. 1993;119:1079–1091. doi: 10.1242/dev.119.4.1079. [DOI] [PubMed] [Google Scholar]
- Graille M, Pagano M, Rose T, Ravaux MR, Van Tilbeurgh H. Zinc induces structural reorganization of gelatin binding domain from human fibronectin and affects collagen binding. Structure. 2010;18:710–8. doi: 10.1016/j.str.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Grant RP, Spitzfaden C, Altroff H, Campbell ID, Mardon HJ. Structural requirements for biological activity of the ninth and tenth FIII domains of human fibronectin. The Journal of biological chemistry. 1997;272:6159–66. doi: 10.1074/jbc.272.10.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JL, Trevithick JE, Hynes RO. Retroviral expression of alternatively spliced forms of rat fibronectin. J Cell Biol. 1990;110:833–847. doi: 10.1083/jcb.110.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris G, Ma W, Maurer LM, Potts JR, Mosher DF. Borrelia burgdorferi protein BBK32 binds to soluble fibronectin via the N-terminal 70-kDa region, causing fibronectin to undergo conformational extension. The Journal of biological chemistry. 2014;289:22490–9. doi: 10.1074/jbc.M114.578419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Yamada KM. Differences in domain structures between plasma and cellular fibronectins. J Biol Chem. 1981;256:11292–300. [PubMed] [Google Scholar]
- Henderson B, Nair S, Pallas J, Williams MA. Fibronectin: a multidomain host adhesin targeted by bacterial fibronectin-binding proteins. FEMS microbiology reviews. 2011;35:147–200. doi: 10.1111/j.1574-6976.2010.00243.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann BR, Annis DS, Mosher DF. Reactivity of the N-terminal region of fibronectin protein to transglutaminase 2 and factor XIIIA. The Journal of biological chemistry. 2011;286:32220–30. doi: 10.1074/jbc.M111.255562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO. Fibronectins. New York: Springer-Verlag, Inc; 1990. [Google Scholar]
- Johnson KJ, Sage H, Briscoe G, Erickson HP. The compact conformation of fibronectin is determined by intramolecular ionic interactions. J Biol Chem. 1999;274:15473–15479. doi: 10.1074/jbc.274.22.15473. [DOI] [PubMed] [Google Scholar]
- Kadler KE, Hill A, Canty-Laird EG. Collagen fibrillogenesis: fibronectin, integrins, and minor collagens as organizers and nucleators. Current opinion in cell biology. 2008;20:495–501. doi: 10.1016/j.ceb.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karuri NW, Lin Z, Rye HS, Schwarzbauer JE. Probing the conformation of the fibronectin III1-2 domain by fluorescence resonance energy transfer. The Journal of biological chemistry. 2009;284:3445–52. doi: 10.1074/jbc.M805025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Singvall J, Schwarz-Linek U, Johnson BJ, Potts JR, Hook M. BBK32, a fibronectin binding MSCRAMM from Borrelia burgdorferi, contains a disordered region that undergoes a conformational change on ligand binding. J Biol Chem. 2004;279:41706–14. doi: 10.1074/jbc.M401691200. [DOI] [PubMed] [Google Scholar]
- Klotzsch E, Smith ML, Kubow KE, Muntwyler S, Little WC, Beyeler F, Gourdon D, Nelson BJ, Vogel V. Fibronectin forms the most extensible biological fibers displaying switchable force-exposed cryptic binding sites. Proc Natl Acad Sci USA. 2009;106:18267–72. doi: 10.1073/pnas.0907518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer A, Lu H, Isralewitz B, Schulten K, Vogel V. Forced unfolding of the fibronectin type III module reveals a tensile molecular recognition switch. Proc Natl Acad Sci USA. 1999;96:1351–6. doi: 10.1073/pnas.96.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubow KE, Vukmirovic R, Zhe L, Klotzsch E, Smith ML, Gourdon D, Luna S, Vogel V. Mechanical forces regulate the interactions of fibronectin and collagen I in extracellular matrix. Nature communications. 2015;6:8026. doi: 10.1038/ncomms9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy DJ, Aukhil I, Erickson HP. 2.0 Å crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- Leikina E, Mertts MV, Kuznetsova N, Leikin S. Type I collagen is thermally unstable at body temperature. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:1314–8. doi: 10.1073/pnas.032307099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon CA, Ohashi T, Erickson HP. Probing the folded state of fibronectin type III domains in stretched fibrils by measuring buried cysteine accessibility. J Biol Chem. 2011;286:26375–82. doi: 10.1074/jbc.M111.240028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvinovich SV, Ingham KD. Interactions between type III domains in the 110 kDa cell-binding fragment of fibronectin. J Mol Biol. 1995;248:611–626. doi: 10.1006/jmbi.1995.0246. [DOI] [PubMed] [Google Scholar]
- Litvinovich SV, Strickland DK, Medved LV, Ingham KC. Domain structure and interactions of the type I and type II modules in the gelatin-binding region of fibronectin. J Mol Biol. 1991;217:563–575. doi: 10.1016/0022-2836(91)90758-x. [DOI] [PubMed] [Google Scholar]
- Ma W, Ma H, Fogerty FJ, Mosher DF. Bivalent ligation of the collagen-binding modules of fibronectin by SFS, a non-anchored bacterial protein of Streptococcus equi. The Journal of biological chemistry. 2015a;290:4866–76. doi: 10.1074/jbc.M114.612259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Ma H, Mosher DF. On-Off Kinetics of Engagement of FNI Modules of Soluble Fibronectin by beta-Strand Addition. PloS one. 2015b;10:e0124941. doi: 10.1371/journal.pone.0124941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas C, Schiks B, Strangi RD, Hackeng TM, Bouma BN, Gebbink MF, Bouma B. Identification of fibronectin type I domains as amyloid-binding modules on tissue-type plasminogen activator and three homologs. Amyloid. 2008;15:166–80. doi: 10.1080/13506120802193498. [DOI] [PubMed] [Google Scholar]
- Manabe R, Ohe N, Maeda T, Fukuda T, Sekiguchi K. Modulation of cell-adhesive activity of fibronectin by the alternatively spliced EDA segment. The Journal of cell biology. 1997;139:295–307. doi: 10.1083/jcb.139.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjenberg ZR, Ellis IR, Hagan RM, Prabhakaran S, Hook M, Talay SR, Potts JR, Staunton D, Schwarz-Linek U. Cooperative binding and activation of fibronectin by a bacterial surface protein. J Biol Chem. 2011;286:1884–94. doi: 10.1074/jbc.M110.183053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer LM, Annis DS, Mosher DF. IGD motifs, which are required for migration stimulatory activity of fibronectin type I modules, do not mediate binding in matrix assembly. PloS one. 2012a;7:e30615. doi: 10.1371/journal.pone.0030615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer LM, Ma W, Eickstaedt NL, Johnson IA, Tomasini-Johansson BR, Annis DS, Mosher DF. Ligation of the fibrin-binding domain by beta-strand addition is sufficient for expansion of soluble fibronectin. The Journal of biological chemistry. 2012b;287:13303–12. doi: 10.1074/jbc.M111.294041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer LM, Tomasini-Johansson BR, Ma W, Annis DS, Eickstaedt NL, Ensenberger MG, Satyshur KA, Mosher DF. Extended binding site on fibronectin for the functional upstream domain of protein F1 of Streptococcus pyogenes. J Biol Chem. 2010;285:41087–99. doi: 10.1074/jbc.M110.153692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckeown-Longo PJ, Mosher DF. Interaction of the 70,000-mol-wt amino-terminal fragment of fibronectin with the matrix-assembly receptor of fibroblasts. J Cell Biol. 1985;100:364–374. doi: 10.1083/jcb.100.2.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina JA, Thaden JT, Sharma-Kuinkel BK, Fowler VG., JR Impact of Bacterial and Human Genetic Variation on Staphylococcus aureus Infections. PLoS pathogens. 2016;12:e1005330. doi: 10.1371/journal.ppat.1005330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard CJ, Ellis IR, Pickford AR, Schor AM, Schor SL, Campbell ID. The role of the fibronectin IGD motif in stimulating fibroblast migration. J Biol Chem. 2007;282:35530–5. doi: 10.1074/jbc.M707532200. [DOI] [PubMed] [Google Scholar]
- Moretti FA, Chauhan AK, Iaconcig A, Porro F, Baralle FE, Muro AF. A major fraction of fibronectin present in the extracellular matrix of tissues is plasma-derived. J Biol Chem. 2007;282:28057–62. doi: 10.1074/jbc.M611315200. [DOI] [PubMed] [Google Scholar]
- Moriya K, Bae E, Honda K, Sakai K, Sakaguchi T, Tsujimoto I, Kamisoyama H, Keene DR, Sasaki T, Sakai T. A fibronectin-independent mechanism of collagen fibrillogenesis in adult liver remodeling. Gastroenterology. 2011;140:1653–63. doi: 10.1053/j.gastro.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morla A, Ruoslahti E. A fibronectin self-assembly site involved in fibronectin matrix assembly: reconstruction in a synthetic peptide. J Cell Biol. 1992;118:421–429. doi: 10.1083/jcb.118.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morla A, Zhang Z, Ruoslahti E. Superfibronectin is a functionally distinct form of fibronectin. Nature. 1994;367:193–196. doi: 10.1038/367193a0. [DOI] [PubMed] [Google Scholar]
- Mosher DF, Johnson RB. In vitro formation of disulfide-bonded fibronectin multimers. J Biol Chem. 1983;10:6595–6601. [PubMed] [Google Scholar]
- Mosher DF, Schad PE, Kleinman HK. Cross-linking of fibronectin to collagen by blood coagulation factor XIIIa. J Clin Invest. 1979;64:781–787. doi: 10.1172/JCI109524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussavi-Harami SF, Annis DS, Ma W, Berry SM, Coughlin EE, Strotman LN, Maurer LM, Westphall MS, Coon JJ, Mosher DF, Beebe DJ. Characterization of molecules binding to the 70K N-terminal region of fibronectin by IFAST purification coupled with mass spectrometry. Journal of proteome research. 2013;12:3393–404. doi: 10.1021/pr400225p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro AF, Moretti FA, Moore BB, Yan M, Atrasz RG, Wilke CA, Flaherty KR, Martinez FJ, Tsui JL, Sheppard D, Baralle FE, Toews GB, White ES. An essential role for fibronectin extra type III domain A in pulmonary fibrosis. Am J Respir Crit Care Med. 2008;177:638–45. doi: 10.1164/rccm.200708-1291OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris NC, Bingham RJ, Harris G, Speakman A, Jones RP, Leech A, Turkenburg JP, Potts JR. Structural and functional analysis of the tandem beta-zipper interaction of a Streptococcal protein with human fibronectin. The Journal of biological chemistry. 2011;286:38311–20. doi: 10.1074/jbc.M111.276592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novokhatny V, Schwarz F, Atha D, Ingham K. Domain structure and domain-domain interactions in the carboxy-terminal heparin binding region of fibronectin. Journal of molecular biology. 1992;227:1182–91. doi: 10.1016/0022-2836(92)90530-w. [DOI] [PubMed] [Google Scholar]
- O'leary JM, Hamilton JM, Deane CM, Valeyev NV, Sandell LJ, Downing AK. Solution structure and dynamics of a prototypical chordin-like cysteine-rich repeat (von Willebrand Factor type C module) from collagen IIA. J Biol Chem. 2004;279:53857–66. doi: 10.1074/jbc.M409225200. [DOI] [PubMed] [Google Scholar]
- Obara M, Kang MS, Yamada KM. Site-directed mutagenesis of the cell-binding domain of human fibronectin: separable, synergistic sites mediate adhesive function. Cell. 1988;53:649–657. doi: 10.1016/0092-8674(88)90580-6. [DOI] [PubMed] [Google Scholar]
- Oberhauser AF, Badilla-Fernandez C, Carrion-Vazquez M, Fernandez JM. The mechanical hierarchies of fibronectin observed with single-molecule AFM. Journal of molecular biology. 2002;319:433–47. doi: 10.1016/S0022-2836(02)00306-6. [DOI] [PubMed] [Google Scholar]
- Odermatt E, Engel J. Physical properties of fibronectin. In: Mosher DF, editor. Fibronectin. New York: Academic Press; 1989. [Google Scholar]
- Oh E, Pierschbacher M, Ruoslahti E. Deposition of plasma fibronectin in tissues. Proc Natl Acad Sci USA. 1981;78:3218–3221. doi: 10.1073/pnas.78.5.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi T, Augustus AM, Erickson HP. Transient opening of fibronectin type III (FNIII) domains: the interaction of the third FNIII domain of FN with anastellin. Biochemistry. 2009;48:4189–97. doi: 10.1021/bi900001g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi T, Erickson HP. Fibronectin aggregation and assembly: the unfolding of the second fibronectin type III domain. The Journal of biological chemistry. 2011;286:39188–99. doi: 10.1074/jbc.M111.262337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi T, Kiehart DP, Erickson HP. Dynamics and elasticity of the fibronectin matrix in living cell culture visualized by fibronectin-green fluorescent protein. Proc Natl Acad Sci U S A. 1999;96:2153–2158. doi: 10.1073/pnas.96.5.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi T, Kiehart DP, Erickson HP. Dual labeling of the fibronectin matrix and actin cytoskeleton with green fluorescent protein variants. J Cell Sci. 2002;115:1221–9. doi: 10.1242/jcs.115.6.1221. [DOI] [PubMed] [Google Scholar]
- Ozeri V, Rosenshine I, Mosher DF, Fassler R, Hanski E. Roles of integrins and fibronectin in the entry of Streptococcus pyogenes into cells via protein F1. Mol Microbiol. 1998;30:625–37. doi: 10.1046/j.1365-2958.1998.01097.x. [DOI] [PubMed] [Google Scholar]
- Paci E, Karplus M. Forced unfolding of fibronectin type 3 modules: an analysis by biased molecular dynamics simulations. J Mol Biol. 1999;288:441–59. doi: 10.1006/jmbi.1999.2670. [DOI] [PubMed] [Google Scholar]
- Pankov R, Cukierman E, Katz BZ, Matsumoto K, Lin DC, Lin S, Hahn C, Yamada KM. Integrin dynamics and matrix assembly: tensin-dependent translocation of alpha(5)beta(1) integrins promotes early fibronectin fibrillogenesis. J Cell Biol. 2000;148:1075–90. doi: 10.1083/jcb.148.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankov R, Yamada KM. Fibronectin at a glance. J Cell Sci. 2002;115:3861–3. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- Pearlstein E. Substrate activation of cell adhesion factor as a prerequisite for cell attachment. Int J Cancer. 1978;22:32–35. doi: 10.1002/ijc.2910220108. [DOI] [PubMed] [Google Scholar]
- Petersen TE, Skorstengaard K, Vibe-Pedersen K. Primary structure of fibronectin. In: Mosher DF, editor. Fibronectin. New York: Academic Press; 1989. [Google Scholar]
- Pickford AR, Campbell ID. NMR studies of modular protein structures and their interactions. Chemical reviews. 2004;104:3557–66. doi: 10.1021/cr0304018. [DOI] [PubMed] [Google Scholar]
- Pickford AR, Potts JR, Bright JR, Phan I, Campbell LD. Solution structure of a type 2 module from fibronectin: implications for the structure and function of the gelatin-binding domain. Structure. 1997;5:359–370. doi: 10.1016/s0969-2126(97)00193-7. [DOI] [PubMed] [Google Scholar]
- Pickford AR, Smith SP, Staunton D, Boyd J, Campbell ID. The hairpin structure of the 6F11F22F2 fragment from human fibronectin enhances gelatin binding. EMBO J. 2001;20:1519–1529. doi: 10.1093/emboj/20.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts JR, Bright JR, Bolton D, Pickford AR, Campbell ID. Solution structure of the N-terminal F1 module pair from human fibronectin. Biochemistry. 1999;38:8304–8312. doi: 10.1021/bi990202b. [DOI] [PubMed] [Google Scholar]
- Potts JR, Campbell ID. Structure and function of fibronectin modules. Matrix Biol. 1996;15:313–320. doi: 10.1016/s0945-053x(96)90133-x. [DOI] [PubMed] [Google Scholar]
- Potts JR, Phan I, Williams MJ, Campbell ID. High-resolution structural studies of the factor XIIIa crosslinking site and the first type 1 module of fibronectin. Nature Struct Biol. 1995;2:946–950. doi: 10.1038/nsb1195-946. [DOI] [PubMed] [Google Scholar]
- Rocco M, Carson M, Hantgan R, Mcdonagh J, Hermans J. Dependence of the shape of the plasma fibronectin molecule on solvent composition. Ionic strength and glycerol content. J Biol Chem. 1983;258:14545–9. [PubMed] [Google Scholar]
- Rocco M, Infusini E, Daga MG, Gogioso L, Cuniberti C. Models for fibronectin. EMBO J. 1987;6:2343–2349. doi: 10.1002/j.1460-2075.1987.tb02510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudino-Pinera E, Ravelli RB, Sheldrick GM, Nanao MH, Korostelev VV, Werner JM, Schwarz-Linek U, Potts JR, Garman EF. The solution and crystal structures of a module pair from the Staphylococcus aureus-binding site of human fibronectin--a tale with a twist. Journal of molecular biology. 2007;368:833–44. doi: 10.1016/j.jmb.2007.02.061. [DOI] [PubMed] [Google Scholar]
- Schor SL, Ellis I, Banyard J, Schor AM. Motogenic activity of IGD-containing synthetic peptides. J Cell Sci. 1999;112(Pt 22):3879–88. doi: 10.1242/jcs.112.22.3879. [DOI] [PubMed] [Google Scholar]
- Schwarz-Linek U, Hook M, Potts JR. Fibronectin-binding proteins of gram-positive cocci. Microbes and infection / Institut Pasteur. 2006;8:2291–8. doi: 10.1016/j.micinf.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Schwarz-Linek U, Pilka ES, Pickford AR, Kim JH, Hook M, Campbell ID, Potts JR. High affinity streptococcal binding to human fibronectin requires specific recognition of sequential F1 modules. J Biol Chem. 2004;279:39017–25. doi: 10.1074/jbc.M405083200. [DOI] [PubMed] [Google Scholar]
- Schwarz-Linek U, Werner JM, Pickford AR, Gurusiddappa S, Kim JH, Pilka ES, Briggs JA, Gough TS, Hook M, Campbell ID, Potts JR. Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper. Nature. 2003;423:177–81. doi: 10.1038/nature01589. [DOI] [PubMed] [Google Scholar]
- Schwarzbauer JE. Identification of the fibronectin sequences required for assembly of a fibrillar matrix. J Cell Biol. 1991;113:1463–1473. doi: 10.1083/jcb.113.6.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Askari JA, Humphries MJ, Jones EY, Stuart DI. Crystal structure of a heparin- and integrin-binding segment of human fibronectin. EMBO J. 1999;18:1468–1479. doi: 10.1093/emboj/18.6.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Carraher C, Schwarzbauer JE. Assembly of fibronectin extracellular matrix. Annu Rev Cell Dev Biol. 2010;26:397–419. doi: 10.1146/annurev-cellbio-100109-104020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BO, Downing AK, Dudgeon TJ, Cunningham M, Driscoll PC, Campbell ID. Secondary structure of fibronectin type 1 and epidermal growth factor modules from tissue-type plasminogen activator by nuclear magnetic resonance. Biochemistry. 1994;33:2422–9. doi: 10.1021/bi00175a010. [DOI] [PubMed] [Google Scholar]
- Smith DE, Mosher DF, Johnson RB, Furcht LT. Immunological identification of two sulfhydryl-containing fragments of human plasma fibronectin. J Biol Chem. 1982;257:5831–5838. [PubMed] [Google Scholar]
- Smith ML, Gourdon D, Little WC, Kubow KE, Eguiluz RA, Luna-Morris S, Vogel V. Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol. 2007;5:e268. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SP, Hashimoto Y, Pickford AR, Campbell ID, Werner JM. Interface characterization of the type II module pair from fibronectin. Biochemistry. 2000;39:8374–81. doi: 10.1021/bi000427i. [DOI] [PubMed] [Google Scholar]
- Sottile J, Hocking DC. Fibronectin polymerization regulates the composition and stability of extracellular matrix fibrils and cell-matrix adhesions. Mol Biol Cell. 2002;13:3546–59. doi: 10.1091/mbc.E02-01-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottile J, Schwarzbauer J, Selegue J, Mosher DF. Five type I modules of fibronectin form a functional unit that binds to fibroblasts and Staphylococcus aureus. J Biol Chem. 1991;266:12840–12843. [PubMed] [Google Scholar]
- Sottile J, Shi F, Rublyevska I, Chiang HY, Lust J, Chandler J. Fibronectin-dependent collagen I deposition modulates the cell response to fibronectin. American journal of physiology Cell physiology. 2007;293:C1934–46. doi: 10.1152/ajpcell.00130.2007. [DOI] [PubMed] [Google Scholar]
- Spitzfaden C, Grant RP, Mardon HJ, Campbell ID. Module-module interactions in the cell binding region of fibronectin: stability, flexibility and specificity. Journal of molecular biology. 1997;265:565–79. doi: 10.1006/jmbi.1996.0736. [DOI] [PubMed] [Google Scholar]
- Sticht H, Pickford AR, Potts JR, Campbell ID. Solution structure of the glycosylated second type 2 module of fibronectin. Journal of molecular biology. 1998;276:177–87. doi: 10.1006/jmbi.1997.1528. [DOI] [PubMed] [Google Scholar]
- Stine JM, Sun Y, Armstrong G, Bowler BE, Briknarova K. Structure and unfolding of the third type III domain from human fibronectin. Biochemistry. 2015;54:6724–33. doi: 10.1021/acs.biochem.5b00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stultz CM. Localized unfolding of collagen explains collagenase cleavage near imino-poor sites. Journal of molecular biology. 2002;319:997–1003. doi: 10.1016/S0022-2836(02)00421-7. [DOI] [PubMed] [Google Scholar]
- Takagi J, Strokovich K, Springer TA, Walz T. Structure of integrin alpha5beta1 in complex with fibronectin. The EMBO journal. 2003;22:4607–15. doi: 10.1093/emboj/cdg445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Leiss M, Moser M, Ohashi T, Kitao T, Heckmann D, Pfeifer A, Kessler H, Takagi J, Erickson HP, Fassler R. The RGD motif in fibronectin is essential for development but dispensable for fibril assembly. The Journal of cell biology. 2007;178:167–78. doi: 10.1083/jcb.200703021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun JW, Hynes RO. Plasma fibronectin is synthesized and secreted by hepatocytes. The Journal of biological chemistry. 1983;258:4641–7. [PubMed] [Google Scholar]
- Tomasini-Johansson B, Mosher DF. Plasma fibronectin concentration in inbred mouse strains. Thrombosis and haemostasis. 2009;102:1278–80. doi: 10.1160/TH09-03-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini-Johansson BR, Annis DS, Mosher DF. The N-terminal 70-kDa fragment of fibronectin binds to cell surface fibronectin assembly sites in the absence of intact fibronectin. Matrix Biol. 2006;25:282–93. doi: 10.1016/j.matbio.2006.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini-Johansson BR, Kaufman NR, Ensenberger MG, Ozeri V, Hanski E, Mosher DF. A 49-residue peptide from adhesin F1 of Streptococcus pyogenes inhibits fibronectin. J Biol Chem. 2001;276:23430–23439. doi: 10.1074/jbc.M103467200. [DOI] [PubMed] [Google Scholar]
- Tompa P, Schad E, Tantos A, Kalmar L. Intrinsically disordered proteins: emerging interaction specialists. Current opinion in structural biology. 2015;35:49–59. doi: 10.1016/j.sbi.2015.08.009. [DOI] [PubMed] [Google Scholar]
- Tooney NM, Mosesson MW, Amrani DL, Hainfeld JF, Wall JS. Solution and surface effects on plasma fibronectin structure. J Cell Biol. 1983;97:1686–92. doi: 10.1083/jcb.97.6.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RP, Chiquet-Ehrismann R. Evidence for the evolution of tenascin and fibronectin early in the chordate lineage. Int J Biochem Cell Biol. 2009;41:424–34. doi: 10.1016/j.biocel.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Ugarova TP, Zamarron C, Veklich Y, Bowditch RD, Ginsberg MH, Weisel JW, Plow EF. Conformational transitions in the cell binding domain of fibronectin. Biochemistry. 1995;34:4457–4466. doi: 10.1021/bi00013a039. [DOI] [PubMed] [Google Scholar]
- Vakonakis I, Staunton D, Ellis IR, Sarkies P, Flanagan A, Schor AM, Schor SL, Campbell ID. Motogenic sites in human fibronectin are masked by long range interactions. J Biol Chem. 2009;284:15668–75. doi: 10.1074/jbc.M109.003673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakonakis I, Staunton D, Rooney LM, Campbell ID. Interdomain association in fibronectin: insight into cryptic sites and fibrillogenesis. The EMBO journal. 2007;26:2575–83. doi: 10.1038/sj.emboj.7601694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velling T, Risteli J, Wennerberg K, Mosher DF, Johansson S. Polymerization of type I and III collagens is dependent on fibronectin and enhanced by integrins alpha 11beta 1 and alpha 2beta 1. J Biol Chem. 2002;277:37377–81. doi: 10.1074/jbc.M206286200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Reheman A, Spring CM, Kalantari J, Marshall AH, Wolberg AS, Gross PL, Weitz JI, Rand ML, Mosher DF, Freedman J, Ni H. Plasma fibronectin supports hemostasis and regulates thrombosis. J Clin Invest. 2014;124:4281–93. doi: 10.1172/JCI74630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White ES, Baralle FE, Muro AF. New insights into form and function of fibronectin splice variants. J Pathol. 2008;216:1–14. doi: 10.1002/path.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams EC, Janmey PA, Ferry JD, Mosher DF. Conformational states of fibronectin. Effects of pH, ionic strength, and collagen binding. J Biol Chem. 1982;257:14973–14978. [PubMed] [Google Scholar]
- Williams MJ, Phan I, Harvey TS, Rostagno A, Gold L, Campbell ID. Solution structure of a pair of fibronectin type 1 modules with fibrin binding activity. J Mol Biol. 1994;235:1302–1311. doi: 10.1006/jmbi.1994.1083. [DOI] [PubMed] [Google Scholar]
- Xu J, Maurer LM, Hoffmann BR, Annis DS, Mosher DF. iso-DGR sequences do not mediate binding of fibronectin N-terminal modules to adherent fibronectin-null fibroblasts. J Biol Chem. 2010;285:8563–71. doi: 10.1074/jbc.M109.062646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Ruoslahti E. A fibronectin fragment inhibits tumor growth, angiogenesis, and metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:620–4. doi: 10.1073/pnas.98.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Sakai T, Fassler R, Ruoslahti E. Antiangiogenic proteins require plasma fibronectin or vitronectin for in vivo activity. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11435–8. doi: 10.1073/pnas.1635112100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerlauth G, Wolf G. Plasma fibronectin as a marker for cancer and other diseases. Am J Med. 1984;77:685–9. doi: 10.1016/0002-9343(84)90366-8. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Checovich WJ, Peters DM, Albrecht RM, Mosher DF. Modulation of cell surface fibronectin assembly sites by lysophosphatidic acid. J Cell Biol. 1994;127:1447–1459. doi: 10.1083/jcb.127.5.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Mosher DF. Crosslinking of the N-terminal region of fibronectin to molecules of large apparent molecular mass (LAMMs): Characterization of fibronectin assembly sites induced by the treatment of fibroblasts with lysophosphatidic acid. J Biol Chem. 1996;271:33284–33292. doi: 10.1074/jbc.271.52.33284. [DOI] [PubMed] [Google Scholar]
- Zhou X, Rowe RG, Hiraoka N, George JP, Wirtz D, Mosher DF, Virtanen I, Chernousov MA, Weiss SJ. Fibronectin fibrillogenesis regulates three-dimensional neovessel formation. Genes & development. 2008;22:1231–43. doi: 10.1101/gad.1643308. [DOI] [PMC free article] [PubMed] [Google Scholar]


