Abstract
Diarrhea continues to be a leading cause of death in children younger than 5 years in developing countries. Enterotoxigenic Escherichia coli (ETEC) is a leading bacterial cause of children’s diarrhea and travelers’ diarrhea. ETEC bacteria initiate diarrheal disease by attaching to host receptors at epithelial cells and colonizing in small intestine. Therefore, preventing ETEC attachment has been considered the first line of defense against ETEC diarrhea. However, developing vaccines effectively against ETEC bacterial attachment encounters challenge because ETEC strains produce over 23 immunologically heterogeneous adhesins. In this study, we applied MEFA (multiepitope fusion antigen) approach to integrate epitopes from adhesin tips or adhesive subunits of CFA/I, CS1, CS2, CS3, CS4, CS5, CS6, CS21 and EtpA adhesins and to construct an adhesin tip MEFA peptide. We then examined immunogenicity of this tip MEFA in mouse immunization, and assessed potential application of this tip MEFA for ETEC vaccine development. Data showed that mice intraperitoneally immunized with this adhesin tip MEFA developed IgG antibody responses to all nine ETEC adhesins. Moreover, ETEC and E. coli bacteria expressing these nine adhesins, after incubation with serum of the immunized mice, exhibited significant reduction in attachment to Caco-2 cells. These results indicated that anti-adhesin antibodies induced by this adhesin tip MEFA blocked adherence of the most important ETEC adhesins, suggesting this multivalent tip MEFA may be useful for developing a broadly protective anti-adhesin vaccine against ETEC diarrhea.
Keywords: ETEC (enterotoxigenic Escherichia coli), MEFA (multiepitope fusion antigen), adhesin tip, vaccine, diarrhea
Introduction
Diarrhea remains a leading cause of death in children younger than 5 years who live in developing countries particularly of South Asia, South America and sub-Saharan Africa [1–8]. Enterotoxigenic Escherichia coli (ETEC; a heterogeneous group of E. coli strains producing enterotoxins) is a top bacterial cause of children’s diarrhea [1, 9–11]. In addition, ETEC is a common cause of travelers’ diarrhea among children above 5 years and adults travelling from developed countries to low-income countries, as well as military and civil personnel deployed at ETEC endemic regions [12–16].
Key virulence factors of ETEC strains are bacterial adhesins and enterotoxins [17–19]. Adhesins including colonization factor antigens (CFAs) and coli surface antigens (CSs) initiate bacterial attachment to host cell receptors and colonization in host small intestines. Attachment and colonization bring ETEC bacteria in close proximity to host cells. That allows bacteria to effectively deliver enterotoxins into host small intestinal epithelial cells to disrupt fluid homeostasis, leading to fluid hyper-secretion and watery diarrhea. Without attachment to host receptors and colonization in small intestines, ETEC bacteria are unable to cause diarrheal disease in young pigs [20–22]. Thus, bacterial attachment is the initial and essential step of ETEC infection; developing a vaccine to prevent ETEC bacterial attachment and colonization has long been considered the first line of defense against ETEC diarrhea [19, 23].
However, developing an effective vaccine against ETEC bacterial attachment and colonization remains to be difficult. The main challenge is heterogeneity of ETEC strains. ETEC strains produce 23 or more immunologically heterogeneous CFA adhesins [24–27]. Since ETEC strains producing any one or two CFA adhesins (with heat-labile toxin - LT, heat-stable toxin - STa, or both enterotoxins) can cause diarrhea, an effective ETEC vaccine needs to protect against all or a majority of these adhesins. But protecting against 23 or more adhesins is not feasible with current technology. An alternative approach is to target the most important adhesins (instead of all adhesins). This approach is preferred indeed in current ETEC vaccine development. Among the characterized ETEC CFA adhesins, CFA/I, CFA/II (CS1, CS2, CS3) and CFA/IV (CS4, CS5, CS6) are found often more prevalent than others among ETEC strains. ETEC strains producing these seven CFA adhesins were estimated to cause about 70% − 80% of the ETEC diarrhea cases (caused by ETEC strains with known adhesins) and also moderate to severe diarrhea cases [19, 28–31]. Recent studies indicated that Longus pilus (CS21) and outer-member protein adhesin EtpA are also more frequently detected among ETEC strains associated with children’s diarrhea and travelers’ diarrhea [32–37]. A vaccine blocking attachment of these prevalent adhesins likely will be effective against ETEC diarrhea [23, 31, 38–40].
To develop a vaccine preventing attachment and colonization from CFA/I, CS1-CS6, CS21 and EtpA ETEC adhesins, in this study we in silico identified antigenic B-cell epitopes from adhesin tips or adhesive subunits of these nine adhesins, and applied MEFA (multiepitope fusion antigen) approach [41] to integrate the most antigenic epitope predicted from each of these nine adhesin tips or adhesive subunits into a single MEFA protein. This adhesin tip MEFA was then used to immunize mice and examined for anti-adhesin antigenicity, and was assessed for potential application in ETEC vaccine development.
Materials and Methods
Bacterial strains and plasmids
E. coli strains listed in Table 1 were used for PCR amplification of adhesin tip or adhesive subunit genes of CFA/I, CS1 - CS6, CS21 and EtpA adhesins, and for in vitro antibody adherence inhibition assays. Recombinant E. coli strains expressing each adhesin tip, adhesive subunit protein, or the tip MEFA protein were also included in Table 1. Expression vector pET28α (Novagen, Madison, WI) and E. coli strain BL21 (GE Healthcare, Piscataway, NJ) were used to express adhesin tips, adhesive subunits, and the adhesin tip MEFA protein.
Table 1.
A list of Escherichia coli strains used in this study. E. coli and ETEC strains were used for PCR amplification of adhesin tip and adhesive subunit genes and also for antibody adherence inhibition assays. Recombinant E. coli strains expressing each adhesin tip or adhesive subunit constructed in this study were also included.
| Strain | Relevant characteristics | Source |
|---|---|---|
| H10407 | O78:H11; CFA/I, LT, STa | Johns Hopkins University |
| THK38/pEU405 | CS1 | Emory University (47) |
| DH5α/pEU588 | CS2 | Emory University (48) |
| E116 (E19446) | CS3, LT, STa | University of Gothenburg |
| E106 (E11881/9) | CS4/CS6, LT, STa | University of Gothenburg |
| UM 75688 | CS5/CS6, LT, STa | Johns Hopkins University |
| JF2423 ETP98066 | CS6, LT, STa | Washington University |
| JF2101 | CS21, EtpA, STa | Washington University |
| JF2318 ETP050008 | EtpA, STa | Washington University |
| CFA/I knockout | ΔCFA/I H10407, EtpA, LT, STa | University of Maryland (46) |
| 9473 | CfaE (CFA/I) in pET28α/ BL21 Kan+ | this study |
| 9474 | CooD (CS1) in pET28α/ BL21 Kan+ | this study |
| 9475 | CotD (CS2) in pET28α/BL21 Kan+ | this study |
| 9505 | CstH (CS3) in pET28α/BL21 Kan+ | this study |
| 9504 | CsaE (CS4) in pET28α/BL21 Kan+ | this study |
| 9533 | CsfD (CS5) in pET28α/BL21 Kan+ | this study |
| 9506 | CssB (CS6) in pET28α/BL21 Kan+ | this study |
| 9468 | LngA (CS21) in pET28α/BL21 Kan+ | this study |
| 9507 | EtpA in pET28α/BL21 Kan+ | this study |
| 9450 | adhesin tip MEFA in pET28α/BL21 Kan+ | this study |
Cloning and expression of adhesin tips, adhesive subunits and adhesin tip MEFA
PCR primers used to amplify the adhesin tip genes of CFA/I, CS1, CS2, CS3, CS4 and CS5, and adhesive subunit (major structural subunit involving in adhesion) genes of CS6 and CS21, and the 5’ end of EtpA gene are listed in Table 2. PCR-amplified adhesin tip or adhesive subunit gene products were digested with restriction enzymes EagI and NcoI or NheI (New England BioLabs Inc., Ipswich, MA). Digested products were cloned into vector pET28α and expressed in E. coli strain BL21.
Table 2.
PCR primers used to amplify adhesin tip or adhesive subunits in this study. Restriction sites, NheI or NcoI in forward primers and EagI in reverse primers, were underlined.
| Primer | Sequence (5’ → 3’) |
|---|---|
| CfaE-F | CTAGCTAGCGATAAAAATCCCGGAAGTG |
| CfaE-R | GATCGGCCGCTAGAGTGTTTGACTACTTG |
| CS1 (CooD)-F | CTAGCTAGCGGGCGATACCCGGAAACTACAG |
| CS1 (CooD)-R | GATCGGCCGTCATAAATTTTCGACACTGG |
| CS2 (CotD)-F | CTAGCTAGCCAATCATGGCATACGAACGTAG |
| CS2 (CotD)-R | GATCGGCCGTTACAGACTTGAACTACTAGGAG |
| CS3 (CstH)-F | AGTTACATCCATGGGCACTCTAACCAAAGAACTGGCATTAAATGTGC |
| CS3 (CstH)-R | TACATGATCGGCCGTTAATTACCTGAAACTAAATGTTCGTTACC |
| CS4 (CsaE)-F | CTAGCTAGCGATAAAATTCCCGGAGATGAAAG |
| CS4 (CsaE)-R | GATCGGCCGCTAGAGTGTTTGACTACTTGGTGTG |
| CS5 (CsfD)-F | TTTTCCATGGTTATGGTTCAGGCTGCTACA |
| CS5 (CsfD)-R | AGATCGGCCGTTATTTATTGTAACATTTCC |
| CS6 (CssB)-F | AGTTACATCCATGGGCTGGCAATATAAATCTCTGGATGTAAATG |
| CS6 (CssB)-R | ATGTAGATCGGCCGTTAAGTCAAATTTCCTGCATAAGTACCAGAC |
| CS21 (LngA)-F | CTAGCTAGCATGAGCCTGCTGGAAGTTATCATTG |
| CS21 (LngA)-R | GATCGGCCGTTAACGGCTACCTAAAGTAATTG |
| EtpA-F | CTAGCTAGCGGCGTGGGTAATGCAAAAGCCACG |
| EtpA-F | ATGTAGATCGGCCGTTAGCTGAAGGTGTAACGACGGTTCATG |
To construct adhesin tip MEFA gene, we first used web-based B-cell epitope prediction software to in silico identify B-cell epitopes from the adhesin tips of six adhesins (CFA/I, CS1-CS5) and adhesive subunits of three adhesins (CS6, CS21, EtpA), as previously described [41]. We then selected CFA/I tip CfaE gene (cfaE) the backbone, truncated eight nucleotide fragments coding surface-exposed but less antigenic peptides from the cfaE gene, and replaced them with nucleotide fragments coding the most antigenic epitope of the other eight adhesin tips or adhesive subunits. After in silico optimization for placing the substitutional epitopes in the backbone gene (for similar antigenic propensity of the native backbone protein), this adhesin tip MEFA gene was synthesized (Genewiz, South Plainfield, NJ). The synthetic gene was digested with enzymes NcoI and EagI, and was cloned into vector pET28α and expressed in E. coli BL21.
A single colony from each subunit recombinant strain or the tip MEFA strain was cultured in 5 ml Lysogeny broth (LB) supplemented with kanamycin (30μg/ml) at 37°C overnight. Overnight-grown culture was added to 200 ml 2x YT medium (2x Yeast Extract Tryptone). After optical density (OD) reached 0.6, bacteria were induced with 25 – 100 μM Isopropyl β-D-1-thiogalactopyranoside (IPTG; Sigma, St. Louis, MO) and grew for four more hours. Bacteria were collected with centrifugation of 12,000 – 13,000 rpm for 15 minutes. Pellets were suspended in 10 ml bacterial protein extraction reagent (B-PER; Pierce, Rockford, IL). After 30 min at room temperature, bacterial lysates were centrifuged at 12,000 – 13,000 rpm for 15 min at 4°C. Pellets were suspended in 10 – 20 ml B-PER with vortex and pipetting, mixed with 200 –400 μl freshly prepared lysozyme (10 mg/ml, in B-PER), and incubated at room temperature for 20 min. Suspensions were centrifuged; resultant pellets were suspended in B-PER supplemented with lysozyme again. After centrifugation, pellets were suspended in 100 ml 1:10 diluted B-PER, vortexed, and centrifuged. Pellets were washed three times with 100 ml PBS, vortexed vigorously, centrifuged at 12,000 rpm for 15 min at 4°C. Final pellets were dissolved in 10 ml PBS.
Extracted proteins were then refolded using a protein refolding kit by following manufacturer’s protocol (Novagen). Briefly, extracted proteins were mixed with 1xIB solubilization buffer (50 mM CAPs) supplemented with 0.3% N-lauroylsarcosine and 1mM EDTA. After incubation at room temperature for 15 min, suspension was centrifuged (12,000 rpm for 20 min at room temperature) to collect soluble proteins. Solubilized proteins were transferred to molecular porous membrane tubing (Spectrum Laboratories, Inc., Rancho Dominquez, CA) and refolded using Dialysis Buffer (20 mM Tris-HCl, pH 8.5) supplemented with 0.1 mM DTT at 4°C. After 3 – 4 h, protein samples (in tubing) were moved to Dialysis Buffer without DTT, and followed by two more buffer changes. Refolded adhesin tip, adhesive subunit or adhesin tip MEFA proteins were collected with centrifugation at 12,000 rpm for 10 min at 4°C, measured for protein concentration, aliquoted, and stored at −80°C.
Mouse immunization with adhesin tip MEFA
Before being used for mouse immunization, refolded adhesin tip MEFA protein sample, 10 or 15 μl (1 mg/ml), was examined in 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Mouse anti-CFA/I antiserum (1:3000) and IRDye-labeled goat anti-mouse IgG (1:5000; LI-COR, Lincoln, NE) were used as the primary and secondary antibody, respectively. LI-COR Odyssey premium infrared gel imaging system was used to visualize bound proteins. In addition, Coomassie blue staining was used to assess purity of the refolded adhesin tip MEFA protein.
Eight seven-week-old female BALB/c mice (Charlies River Laboratories International, Inc., Wilmington, MA) were each intraperitoneally immunized with 150 μg (in 200 μl PBS) adhesin tip MEFA protein and 1μg dmLT adjuvant (LTR192G/L211A; provided by Walter Reed Army Institute of Research, Silver Spring, MD). Immunized mice received two booster injections intraperitoneally at the same dose of the primary, in an interval of two weeks. Immunized mice were sacrificed two weeks after the second booster. Eight mice without immunization were used as the control.
Serum samples collected from each mouse before the primary and two weeks after the second booster were stored at -80°C until use. Mouse immunization study complied with the Animal Welfare Act by following the 1996 National Research Council guidance and was approved by the Kansas State University‘s Institutional Animal Care and Use Committee.
Mouse anti-CFA IgG antibody titration
Refolded recombinant adhesin tip and adhesive subunit proteins (about 1 mg/ml; a yield of 25–50 mg per liter culture) were examined for purity in SDS-PAGE with Coomassie blue staining, and used as coating antigens in ELISAs to titrate mouse serum anti-CfaE (CFA/I), -CooD (CS1), -CotD (CS2), -CstH (CS3), -CsaE (CS4), -CsfD (CS5), -CssB (CS6), anti-LngA (CS21) and anti-EtpA IgG antibodies, respectively. Serum samples collected from each mouse were initially diluted in 1:400 and then two-folded diluted till to 1:51,200. For serum of the control mice, dilutions of 1:200 were also included in ELISAs. Each adhesin tip or subunit recombinant protein (400 ng per well), in 100 μl coating buffer [41], was added to wells of Immulon 2HB 96-well plates (Thermo Fisher Scientific, Rochester, NY). Plates incubated at 37°C for 1 h and then 4°C overnight were washed three times with PBS-0.05% Tween 20 (PBST). After incubation with 5% nonfat milk at 37°C for 1 h to block uncoated sites, wells were washed three times with PBST and incubated with mouse serum twofold dilutions at 37°C for 1 h, then followed by three washes with PBST and incubation with horseradish peroxidase (HRP) conjugated goat-anti-mouse IgG antibodies (1:3000; Sigma) at 37°c for 1 h. After three washes with PBST and two washes with PBS, each well was incubated with 100 μl 3,3’,5,5’-tetramethylbezidine (TMB) Microwell Peroxidase Substrate (KPL, Gaithersburg, MD) at room temperature for 30 min and measured for OD650. The highest dilution that gave an OD reading above 0.3 after subtraction of background readings was calculated (OD x dilution) for antibody titer, and antibody titers were presented in log10 as described previously [41–44].
Mouse serum antibody adherence inhibition assay
Mouse serum in vitro antibody adherence inhibition assay was carried out as described previously [41, 44, 45]. Briefly, 30 μl serum pooled from mice in the immunization group or the control group was added to ETEC or E. coli bacteria (4.5x106 CFUs, in 150 μl PBS; pre-treated with 10% mannose) that express CFA/I, CS1, CS2, CS3, CS4/CS6, CS5/CS6, CS6, CS21, or EtpA. Incubated at room temperature for 1 h on a shaker (50 rpm), each serum/bacteria mixture was brought to 300 μl with PBS and added to Caco-2 cells (3x105 cells, in 700 μl DMEM-10% FBS; at a multiplicity-of-infection ratio of 15 bacteria to 1 cell). After incubation at 37°C for 1 h in a CO2 incubator (5% CO2), Caco-2 cells were gently washed three times with PBS to remove non-adherent bacteria, and dislodged with 0.5% triton X (300 μl per well; 37°C for 20 min at room temperature). Adherent bacteria (and dislodged Caco-2 cells) were collected with centrifugation, suspended in 1 ml PBS, serially diluted, and plated on LB plates. Bacteria on plates cultured at 37°C overnight were counted (CFUs).
Statistical analysis
Data of antibody adherence inhibition assay were analyzed using the mixed procedure (SAS for windows, version 8; SAS Institute, Cary, NC). Results were expressed in means ± standard deviations. Student’s t-test was used to compare differences between the immunization group and the control group. Differences were considered significant if p < 0.05 when treatments were compared at two-tailed distribution and two-sample unequal variance.
Results
Adhesin tip MEFA protein carried epitopes of nine adhesin tips or adhesive subunits
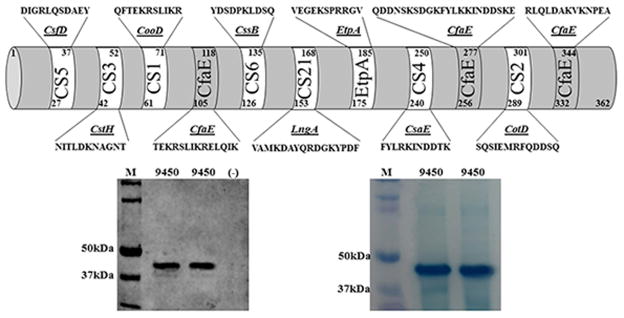
Epitopes ‘QFTEKRSLIKR’, ‘SQSIEMRFQDDSQ’, ‘NITLDKNAGNT’, ‘FYLRKINDDTK’, ‘DIGRLQSDAEY’, ‘YDSDPKLDSQ’, ‘VAMKDAYQRDGKYPDF’, and ‘VEGEKSPRRGV’ were in silico predicted from CooD of CS1, CotD of CS2, CstH of CS3, CsaE of CS4, CsfD of CS5, CssB of CS6, LngA of CS21 and EtpA of EtpA , respectively. Three epitopes from CfaE of CFA/I: ‘TEKRSLIKRELQIK’, ‘QDDNSKSDGKFYLKKINDDSKE’ and ‘RLQLDAKVKNPEA’ were retained in the tip MEFA backbone (Fig. 1). This 362-amino acid polypeptide (25 – 50 mg per liter culture) was expressed, extracted, refolded, and recognized by mouse anti-CFA/I antiserum (Fig 1).
Figure 1.
Schematic illustration of the constructed ETEC adhesin tip MEFA protein. Amino acid sequences of epitopes representing each adhesin tip or adhesive subunit were showed in the top panel. Bottom left panel showed results of Western blot with mouse anti-CFA/I antiserum. 9450 was the adhesin tip MEFA protein (10 and 15 μl), and (−) was the total proteins from BL21 E. coli. Bottom right panel showed Coomassie blue staining of refolded solubilized adhesin tip MEFA proteins (lane 1: protein marker; lane 2: 10 μl tip MEFA protein; lane 3: 15 μl tip MEFA protein).
Mice intraperitoneally immunized with the adhesin tip MEFA developed immune responses to all nine adhesins
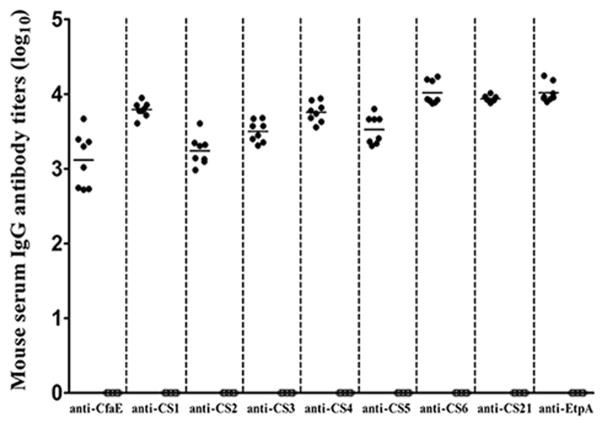
All eight mice intraperitoneally immunized with the adhesin tip MEFA recombinant protein developed antibody responses to nine adhesin tips or adhesive subunits (Fig. 2). ELISAs using refolded recombinant adhesin subunit proteins detected anti-CfaE, -CooD, -CotD, -CstH, -CsaE, -CsfD, -CssB, and -LngA and anti-EtpA IgG antibody titers at 3.1 ± 0.36, 3.8 ± 0.10, 3.2 ± 0.19, 3.5 ± 0.14, 3.8 ± 0.13, 3.5 ± 0.19, 4.0 ± 0.16, 3.9 ± 0.04 and 4.0 ± 0.13 (in log10) from the serum samples of the immunized mice. No antibody responses to these adhesin tip or subunit antigens were detected from the serum samples of the control mice or serum collected before the primary immunization.
Figure 2.
Mouse serum anti-CfaE (CFA/I), -CooD (CS1), -CotD (CS2), -CstH (CS3), -CsaE (CS4), -CsfD (CS5), -CssB (CS6), anti-LngA (CS21) and anti-EtpA (EtpA) IgG antibody titers (log10). Solid circles (●) were titers of mice intraperitoneally immunized with the adhesin tip MEFA protein; empty circles (○) were of titers of the control mice. Each circle represented the IgG titer from an individual mouse. Bars indicated the mean titers of the group specific to each adhesin.
Mouse serum antibodies inhibited adherence of ETEC or E. coli bacteria expressing these nine adhesins
ETEC bacteria expressing CFA/I, CS3, CS4/CS6, CS5/CS6, CS6, CS21 or EtpA, or H10407 CfaE knock out mutant which expresses EtpA [46], and recombinant E. coli strains expressing CS1 or CS2 [47, 48] incubated with pooled serum sample of the immunized mice showed significantly reduction in adherence to Caco-2 cells, compared to the same bacteria incubated with pooled serum from the control mice (Table 3).
Table 3.
In vitro antibody adherence inhibition assay using E. coli or ETEC strains expressing one or two adhesins and mouse serum samples pooled from the immunization group or the control group. Adherent bacteria were expressed in means and standard deviations. P values were calculated from Student t-test.
| E .coli strains used in bacterial adherence assays | Number (x103) of bacteria adherent to Caco-2 cells | ||
|---|---|---|---|
| pooled serum of immunized micea | pooled serum of control mice | p-values | |
| H10407 - CFA/I | 152.8 ± 10.9 | 407.8 ± 36.8 | p < 0.01 |
| THK38/pEU405 – CS1 | 139.2 ± 13.5 | 605 ± 29.2 | p < 0.01 |
| DH5α/pEU588 – CS2 | 62 ± 7.4 | 284.4 ± 31.7 | p < 0.01 |
| E116 (E19446) – CS3 | 165.8 ± 22.2 | 392 ± 48.2 | p < 0.01 |
| E106 (E11881/9) – CS4/CS6 | 159.6 ± 17.6 | 426.6 ± 44.5 | p < 0.01 |
| UM 75688 – CS5/CS6 | 40.4 ± 16.8 | 172.8 ± 12.1 | p < 0.01 |
| 2423 ETP98066 – CS6 | 136.2 ± 18.9 | 394.6 ± 21.3 | p < 0.01 |
| JF2101- CS21 | 138 ± 13.4 | 309.6 ± 25.5 | p < 0.01 |
| JF2318 ETP050008 - EtpA | 183.2 ± 21.4 | 540 ± 66.2 | p < 0.01 |
: pooled serum sample from mice immunized with multi-tip adhesin epitope and adjuvant dmLT.
Discussion
The single adhesin tip MEFA protein created from this study carried epitopes from nine different ETEC adhesins and induced neutralizing antibodies against each of these adhesins. That may rejuvenate the epitope vaccine concept of which multiple epitopes can be linked together to develop a broadly protective vaccine against heterogeneous pathogens. However, different from the epitope vaccine that stacks several epitopes into a linear peptide antigen and often results in inferior immunogenicity and protection, this MEFA (multiepitope fusion antigen) approach uses a backbone protein and substitutes surface-exposed peptides of the backbone protein with epitopes of foreign antigens. This MEFA approach tends to lead to proteins with similar structure of the backbone protein and better presentation of epitopes of interested. Future protein structure studies including crystal chromatography will help to characterize better the structure of this tip MEFA protein.
We demonstrated previously that ETEC CFA MEFA carrying epitopes of the major subunits of CFA/I and CS1 to CS6 adhesins induced antibodies broadly protecting against adherence of bacteria expressing these seven adhesins [41, 44]. That CFA MEFA was generated by using CFA/I major subunit CfaB as the backbone and replacing six CfaB surface exposed peptides with the most antigenic epitopes predicted from the major structural subunits of CS1 to CS6 adhesins. Given a typical E. coli fimbrial adhesin consists of hundreds of copies of a major structural subunit and a single or a few copies of minor subunits, adhesin vaccines derived from major structural subunits are expected effective. Abundance of major subunits makes it easy for derived antibodies to target against adhesins, leading to effective protection against bacterial adherence and ETEC diarrhea. But recent studies suggested that adhesin tips play a key role in bacteria attaching to host receptors, and antibodies against tips are thought critical for protection against ETEC adherence and colonization [49, 50]. Data from this study indicated that antibodies derived from ETEC adhesin tips inhibited adherence of E. coli or ETEC bacteria expressing CFA/I, CS1-CS6, CS21 and EtpA to Caco-2 cells. Whether antibodies derived from adhesin tips are more effective against ETEC bacterial adherence than antibodies derived from major structural subunits remained undetermined momently. Future comparative studies of adherence inhibition activities from antibodies induced by a major structural subunit or an adhesin tip antigen may help to understand better interaction between bacteria adhesins and host cells, and likely provide guidance for ETEC vaccine development.
Epitopes selection can affect antigenicity of MEFA proteins. In this study, we selected an epitope from CssB subunit to represent CS6 adhesin for the tip MEFA. Both CssA and CssB are major structural subunits and play roles in adherence by CS6 adhesin [51, 52]. CssA epitope ‘72QVTVYPV78’ was found to induce antibodies inhibiting CS6 adhesin from adherence to Caco-2 cells [41]. Data from the current study showed that CssB epitope ‘YDSDPKLDSQ’ also induced antibodies that inhibited CS6 adherence in vitro. In contrast to CS6 which has two major structural subunits, CS3 adhesin has CstH the major structural subunit and also the adhesive subunit. CstH epitope ‘NITLDKNAGNT’ in this tip MEFA induced strong anti-CstH antibody response. Comparison of antibody titers derived from the previously constructed major subunit MEFA versus this tip MEFA may not be much informative since different coating antigens were used in antibody titration ELISAs. In vitro antibody adherence inhibition assays suggested antibodies derived against these two CstH epitopes showed similar levels at inhibiting adherence of CS3 adhesin to Caco-2 (50% from anti-CFA MEFA antibodies vs. 58% from anti-adhesin tip MEFA antibodies). Future studies to immunize mice with equivalent molecule copies of this adhesin tip MEPA or the major subunit CFA MEFA may help to identify optimal CS6 and CS3 epitopes and to improve vaccine candidacy.
Results from this study showed that an adhesin tip MEFA carried adhesin tip or adhesive subunit epitopes induced antibody responses to nine ETEC adhesins, and derived antibodies inhibited adherence of E. coli and ETEC bacteria expressing these nine adhesins to Caco-2 cells. It needs to point out that the current study only used IP route and examined IgG antibody responses. Future studies using other parenteral routes (ID, IM, SC) or oral immunization to examine both systemic and mucosal IgG and IgA antibody responses and eventually challenge studies to assess protective efficacy will evaluate better antigenicity of this tip MEFA and potential in ETEC vaccine development.
Highlight.
An ETEC adhesin tip MEFA (multiepitope fusion antigen) carrying antigenic elements of 9 ETEC adhesins was constructed.
This adhesin tip MEFA induces antibody responses to all nine most important ETEC adhesins.
Induced anti-adhesin antibodies inhibited adherence of ETEC or E. coli bacteria expressing any of these nine adhesins.
This adhesin tip MEFA can be used for broadly protective vaccines against ETEC associated children’s diarrhea and travelers’ diarrhea.
Acknowledgments
The authors thank Drs. Ann-Mari Svennerholm (University of Gothenburg, Sweden), June Scott (Emory University), Eileen Barry (University of Maryland), and James Fleckenstein (Washington University) for providing ETEC or recombinant E. coli strains, and Walter Reed Army Institute of Research for supplying dmLT adjuvant. Financial support for this study was provided by NIH (AI109209-02) and research funds from Kansas State University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Black RE, Cousens S, Johnson HL, Lawn JE, Rudan I, Bassani DG, et al. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet. 2010;375(9730):1969–87. doi: 10.1016/S0140-6736(10)60549-1. [DOI] [PubMed] [Google Scholar]
- 2.Fischer Walker CL, Sack D, Black RE. Etiology of diarrhea in older children, adolescents and adults: a systematic review. PLoS Negl Trop Dis. 2010;4(8):e768. doi: 10.1371/journal.pntd.0000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamberti LM, Fischer Walker CL, Black RE. Systematic review of diarrhea duration and severity in children and adults in low- and middle-income countries. BMC public health. 2012;12:276. doi: 10.1186/1471-2458-12-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine MM, Kotloff KL, Nataro JP, Muhsen K. The Global Enteric Multicenter Study (GEMS): impetus, rationale, and genesis. Clin Infect Dis. 2012;55(Suppl 4):S215–24. doi: 10.1093/cid/cis761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L, Johnson HL, Cousens S, Perin J, Scott S, Lawn JE, et al. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet. 2012;379(9832):2151–61. doi: 10.1016/S0140-6736(12)60560-1. [DOI] [PubMed] [Google Scholar]
- 6.Fischer Walker CL, Perin J, Aryee MJ, Boschi-Pinto C, Black RE. Diarrhea incidence in low- and middle-income countries in 1990 and 2010: a systematic review. BMC public health. 2012;12:220. doi: 10.1186/1471-2458-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanata CF, Fischer-Walker CL, Olascoaga AC, Torres CX, Aryee MJ, Black RE, et al. Global causes of diarrheal disease mortality in children <5 years of age: a systematic review. PLoS One. 2013;8(9):e72788. doi: 10.1371/journal.pone.0072788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamberti LM, Bourgeois AL, Fischer Walker CL, Black RE, Sack D. Estimating diarrheal illness and deaths attributable to Shigellae and enterotoxigenic Escherichia coli among older children, adolescents, and adults in South Asia and Africa. PLoS Negl Trop Dis. 2014;8(2):e2705. doi: 10.1371/journal.pntd.0002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO. Future directions for research on enterotoxigenic Escherichia coli vaccines for developing countries. Wkly Epidemiol Rec. 2006;81:97–107. [PubMed] [Google Scholar]
- 10.Kotloff KL, Blackwelder WC, Nasrin D, Nataro JP, Farag TH, van Eijk A, et al. The Global Enteric Multicenter Study (GEMS) of diarrheal disease in infants and young children in developing countries: epidemiologic and clinical methods of the case/control study. Clin Infect Dis. 2012;55(Suppl 4):S232–45. doi: 10.1093/cid/cis753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382(9888):209–22. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 12.Sack DA, Shimko J, Torres O, Bourgeois AL, Francia DS, Gustafsson B, et al. Randomised, double-blind, safety and efficacy of a killed oral vaccine for enterotoxigenic E. Coli diarrhoea of travellers to Guatemala and Mexico. Vaccine. 2007;25(22):4392–400. doi: 10.1016/j.vaccine.2007.03.034. Epub 2007/04/24. [DOI] [PubMed] [Google Scholar]
- 13.Sack RB. The epidemiology of diarrhea due to enterotoxigenic Escherichia coli. J Infect Dis. 1978;137(5):639–40. doi: 10.1093/infdis/137.5.639. Epub 1978/05/01. [DOI] [PubMed] [Google Scholar]
- 14.Sanders JW, Putnam SD, Riddle MS, Tribble DR. Military importance of diarrhea: lessons from the Middle East. Curr Opin Gastroenterol. 2005;21(1):9–14. Epub 2005/02/03. [PubMed] [Google Scholar]
- 15.Black RE. Epidemiology of travelers' diarrhea and relative importance of various pathogens. Rev Infect Dis. 1990;12(Suppl 1):S73–9. doi: 10.1093/clinids/12.supplement_1.s73. Epub 1990/01/01. [DOI] [PubMed] [Google Scholar]
- 16.Hill DRBN. Travelers' diarrhea. Curr Opin Infect Dis. 2010;23(5):481–7. doi: 10.1097/QCO.0b013e32833dfca5. [DOI] [PubMed] [Google Scholar]
- 17.Smith HW, Linggood MA. Observations on the pathogenic properties of the K88, Hly and Ent plasmids of Escherichia coli with particular reference to porcine diarrhoea. J Med Microbiol. 1971;4(4):467–85. doi: 10.1099/00222615-4-4-467. Epub 1971/11/01. [DOI] [PubMed] [Google Scholar]
- 18.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11(1):142–201. doi: 10.1128/cmr.11.1.142. Epub 1998/02/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Svennerholm AM, Tobias J. Vaccines against enterotoxigenic Escherichia coli. Expert Rev Vaccines. 2008;7(6):795–804. doi: 10.1586/14760584.7.6.795. Epub 2008/07/31. [DOI] [PubMed] [Google Scholar]
- 20.Rutter JM, Burrows MR, Sellwood R, Gibbons RA. A genetic basis for resistance to enteric disease caused by E. coli. Nature. 1975;257(5522):135–6. doi: 10.1038/257135a0. [DOI] [PubMed] [Google Scholar]
- 21.Gibbons RA, Sellwood R, Burrows M, Hunter PA. Inheritance of resistance to neonatal E. coli diarrhoea in the pig: examination of the genetic system. TAG Theoretical and applied genetics. Theoretische und angewandte. Genetik. 1977;51(2):65–70. doi: 10.1007/BF00299479. [DOI] [PubMed] [Google Scholar]
- 22.Sellwood R. Escherichia coli diarrhoea in pigs with or without the K88 receptor. Vet Rec. 1979;105(10):228–30. doi: 10.1136/vr.105.10.228. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W, Sack DA. Progress and hurdles in the development of vaccines against enterotoxigenic Escherichia coli in humans. Expert Rev Vaccines. 2012;11(6):677– 84. doi: 10.1586/erv.12.37. [DOI] [PubMed] [Google Scholar]
- 24.Gaastra W, Svennerholm AM. Colonization factors of human enterotoxigenic Escherichia coli (ETEC) Trends Microbiol. 1996;4(11):444–52. doi: 10.1016/0966-842x(96)10068-8. [DOI] [PubMed] [Google Scholar]
- 25.Wolf MK. Occurrence, distribution, and associations of O and H serogroups, colonization factor antigens, and toxins of enterotoxigenic Escherichia coli. Clin Microbiol Rev. 1997;10(4):569–84. doi: 10.1128/cmr.10.4.569. Epub 1997/10/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qadri F, Svennerholm A-M, Faruque ASG, Sack RB. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin Microbiol Rev. 2005;18(3):465–83. doi: 10.1128/CMR.18.3.465-483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W. Perspective of use of vaccines for preventing enterotoxigenic Escherichia coli diarrhea in humans. In: Morabito S, editor. Pathogenic Escherichia coli: molecular and cellular microbiology. Norfolk, UK: Caister Academic Press; 2014. pp. 273–302. [Google Scholar]
- 28.Qadri F, Das SK, Faruque AS, Fuchs GJ, Albert MJ, Sack RB, et al. Prevalence of toxin types and colonization factors in enterotoxigenic Escherichia coli isolated during a 2-year period from diarrheal patients in Bangladesh. J Clin Microbiol. 2000;38(1):27–31. doi: 10.1128/jcm.38.1.27-31.2000. Epub 2000/01/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svennerholm AM, Lundgren A. Recent progress toward an enterotoxigenic Escherichia coli vaccine. Expert Rev Vaccines. 2012;11(4):495–507. doi: 10.1586/erv.12.12. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W, Sack DA. Progress and hurdles in the development of vaccines against enterotoxigenic Escherichia coli in humans. Expert Rev Vaccines. 2012;11(6):677–94. doi: 10.1586/erv.12.37. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W, Sack DA. Current progress in developing subunit vaccines against enterotoxigenic Escherichia coli (ETEC) associated diarrhea. Clin Vaccine Immunol. 2015 doi: 10.1128/CVI.00224-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giron JA, Levine MM, Kaper JB. Longus: a long pilus ultrastructure produced by human enterotoxigenic Escherichia coli. Mol Microbiol. 1994;12(1):71–82. doi: 10.1111/j.1365-2958.1994.tb00996.x. [DOI] [PubMed] [Google Scholar]
- 33.Giron JA, Viboud GI, Sperandio V, Gomez-Duarte OG, Maneval DR, Albert MJ, et al. Prevalence and association of the longus pilus structural gene (lngA) with colonization factor antigens, enterotoxin types, and serotypes of enterotoxigenic Escherichia coli. Infection and immunity. 1995;63(10):4195–8. doi: 10.1128/iai.63.10.4195-4198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pichel MG, Binsztein N, Qadri F, Giron JA. Type IV longus pilus of enterotoxigenic Escherichia coli: occurrence and association with toxin types and colonization factors among strains isolated in Argentina. J Clin Microbiol. 2002;40(2):694–7. doi: 10.1128/JCM.40.2.694-697.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fleckenstein JM, Roy K, Fischer JF, Burkitt M. Identification of a two-partner secretion locus of enterotoxigenic Escherichia coli. Infect Immun. 2006;74(4):2245–58. doi: 10.1128/IAI.74.4.2245-2258.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isidean SD, Riddle MS, Savarino SJ, Porter CK. A systematic review of ETEC epidemiology focusing on colonization factor and toxin expression. Vaccine. 2011;29(37):6167–78. doi: 10.1016/j.vaccine.2011.06.084. [DOI] [PubMed] [Google Scholar]
- 37.Luo Q, Qadri F, Kansal R, Rasko DA, Sheikh A, Fleckenstein JM. Conservation and immunogenicity of novel antigens in diverse isolates of enterotoxigenic Escherichia coli. PLoS Negl Trop Dis. 2015;9(1):e0003446. doi: 10.1371/journal.pntd.0003446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fleckenstein JM, Sheikh A. Designing vaccines to neutralize effective toxin delivery by enterotoxigenic Escherichia coli. Toxins (Basel) 2014;6(6):1799–812. doi: 10.3390/toxins6061799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svennerholm AM. From cholera to enterotoxigenic Escherichia coli (ETEC) vaccine development. Indian J Med Res. 2011;133(2):188–96. Epub 2011/03/19. [PMC free article] [PubMed] [Google Scholar]
- 40.Walker RI. An assessment of enterotoxigenic Escherichia coli and Shigella vaccine candidates for infants and children. Vaccine. 2015;33(8):954–65. doi: 10.1016/j.vaccine.2014.11.049. [DOI] [PubMed] [Google Scholar]
- 41.Ruan X, Knudsen DE, Wollenberg KM, Sack DA, Zhang W. Multiepitope Fusion Antigen Induces Broadly Protective Antibodies That Prevent Adherence of Escherichia coli Strains Expressing Colonization Factor Antigen I (CFA/I), CFA/II, and CFA/IV. Clin Vaccine Immunol. 2014;21(2):243–9. doi: 10.1128/CVI.00652-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang W, Zhang C, Francis DH, Fang Y, Knudsen D, Nataro JP, et al. Genetic fusions of heat-labile (LT) and heat-stable (ST) toxoids of porcine enterotoxigenic Escherichia coli elicit neutralizing anti-LT and anti-STa antibodies. Infect Immun. 2010;78(1):316–25. doi: 10.1128/IAI.00497-09. Epub 2009/10/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu M, Ruan X, Zhang C, Lawson SR, Knudsen DE, Nataro JP, et al. Heat-labile- and heat-stable-toxoid fusions (LTR(1)(9)(2)G-STaP(1)(3)F) of human enterotoxigenic Escherichia coli elicit neutralizing antitoxin antibodies. Infection and immunity. 2011;79(10):4002–9. doi: 10.1128/IAI.00165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruan X, Sack DA, Zhang W. Genetic fusions of a CFA/I/II/IV MEFA (multiepitope fusion antigen) and a toxoid fusion of heat-stable toxin (STa) and heat-labile toxin (LT) of enterotoxigenic Escherichia coli (ETEC) retain broad anti-CFA and antitoxin antigenicity. PLoS ONE. 2015 doi: 10.1371/journal.pone.0121623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruan X, Liu M, Casey TA, Zhang W. A tripartite fusion, FaeG-FedF-LT(192)A2:B, of enterotoxigenic Escherichia coli (ETEC) elicits antibodies that neutralize cholera toxin, inhibit adherence of K88 (F4) and F18 fimbriae, and protect pigs against K88ac/heat-labile toxin infection. Clin Vaccine Immunol. 2011;18(10):1593–9. doi: 10.1128/CVI.05120-11. Epub 2011/08/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baker KK, Levine MM, Morison J, Phillips A, Barry EM. CfaE tip mutations in enterotoxigenic Escherichia coli CFA/I fimbriae define critical human intestinal binding sites. Cell Microbiol. 2009;11(5):742–54. doi: 10.1111/j.1462-5822.2009.01287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perez-Casal J, Swartley JS, Scott JR. Gene encoding the major subunit of CS1 pili of human enterotoxigenic Escherichia coli. Infect Immun. 1990;58(11):3594–600. doi: 10.1128/iai.58.11.3594-3600.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Froehlich BJ, Karakashian A, Sakellaris H, Scott JR. Genes for CS2 pili of enterotoxigenic Escherichia coli and their interchangeability with those for CS1 pili. Infect Immun. 1995;63(12):4849–56. doi: 10.1128/iai.63.12.4849-4856.1995. Epub 1995/12/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sincock SA, Hall ER, Woods CM, O'Dowd A, Poole ST, McVeigh AL, et al. Immunogenicity of a prototype enterotoxigenic Escherichia coli adhesin vaccine in mice and nonhuman primates. Vaccine. 2016;34(2):284–91. doi: 10.1016/j.vaccine.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 50.Luiz WB, Rodrigues JF, Crabb JH, Savarino SJ, Ferreira LC. Maternal Vaccination with a Fimbrial Tip Adhesin and Passive Protection of Neonatal Mice against Lethal Human Enterotoxigenic Escherichia coli Challenge. Infection and immunity. 2015;83(12):4555–64. doi: 10.1128/IAI.00858-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tobias J, Lebens M, Kallgard S, Nicklasson M, Svennerholm AM. Role of different genes in the CS6 operon for surface expression of Enterotoxigenic Escherichia coli colonization factor CS6. Vaccine. 2008;26(42):5373–80. doi: 10.1016/j.vaccine.2008.07.091. [DOI] [PubMed] [Google Scholar]
- 52.Wajima T, Sabui S, Fukumoto M, Kano S, Ramamurthy T, Chatterjee NS, et al. Enterotoxigenic Escherichia coli CS6 gene products and their roles in CS6 structural protein assembly and cellular adherence. Microb Pathog. 2011;51(4):243–9. doi: 10.1016/j.micpath.2011.06.004. [DOI] [PubMed] [Google Scholar]