Abstract
The relationship between genetic and linguistic diversification in human populations has been often explored to interpret some specific issues in human history. The Albanian-speaking minorities of Sicily and Southern Italy (Arbereshe) constitute an important portion of the ethnolinguistic variability of Italy. Their linguistic isolation from neighboring Italian populations and their documented migration history, make such minorities particularly effective for investigating the interplay between cultural, geographic and historical factors. Nevertheless, the extent of Arbereshe genetic relationships with the Balkan homeland and the Italian recipient populations has been only partially investigated. In the present study we address the genetic history of Arbereshe people by combining highly resolved analyses of Y-chromosome lineages and extensive computer simulations. A large set of slow- and fast-evolving molecular markers was typed in different Arbereshe communities from Sicily and Southern Italy (Calabria), as well as in both the putative Balkan source and Italian sink populations. Our results revealed that the considered Arbereshe groups, despite speaking closely related languages and sharing common cultural features, actually experienced diverging genetic histories. The estimated proportions of genetic admixture confirm the tight relationship of Calabrian Arbereshe with modern Albanian populations, in accordance with linguistic hypotheses. On the other hand, population stratification and/or an increased permeability of linguistic and geographic barriers may be hypothesized for Sicilian groups, to account for their partial similarity with Greek populations and their higher levels of local admixture. These processes ultimately resulted in the differential acquisition or preservation of specific paternal lineages by the present-day Arbereshe communities.
Introduction
The term ‘linguistic minority' generally refers to a group of people speaking a linguistic variety, which is markedly different from the official language of the surrounding geographic area.1 Usually, these populations originated from a restricted number of founders and remained at least partially isolated from the subsequent events of admixture.2 Compared with open populations, linguistic minorities can therefore offer a simplified model of investigation to test hypotheses concerning the interplay between geographic and cultural factors, reducing the confounding noise otherwise ascribable to the multilayered history of a population. In addition, their condition of linguistic isolation (potentially acting as a genetic barrier) may have helped these communities to preserve a more direct genetic link with their original background.3
Among the several ethnolinguistic groups currently established in the Italian territory,1, 3 the Albanian-speaking Arbereshe represent one of the largest (88 727 inhabitants).4 Unlike most of the other Italian ethnolinguistic minorities, the Arbereshe case is well documented. The available historical–demographic data provide quite a precise understanding of their place and time of origin, as well as of the main geographic and chronological patterns of diffusion in Southern Italy (including Sicily). Their origins are generally referred to massive movements of Albanians taking place between the fifteenth and sixteenth centuries, mainly to escape the invasion of the Balkans by the Ottoman Empire.5 However, their presence in Italy was actually the result of several migratory events, either originated directly in Toskeria (Southern Albania) or arrived in Italy after stopovers in Greece (particularly in the Peloponnese). Present-day Arbereshe people survive in 41 municipalities distributed across seven Southern Italian regions (Abruzzo, Molise, Campania, Puglia, Basilicata, Calabria and Sicily).6
The Arbereshe of Calabria persist in 19 municipalities of the province of Cosenza (38 018 inhabitants, 42.8% of the entire Italian Arbereshe people),4, 6 scattered either around the highland area of the Pollino massif or along the Crati River valley. Besides a common geographical marginality, these communities have also demonstrated a deep sense of group identity, maintaining social, religious (Greek Orthodox) and linguistic (Arberisht) original traits at least until the first half of the twentieth century.6 Previous biodemographic studies demonstrated that such a strong cultural identity helped these communities to preserve a clear differentiation from the surrounding non-Arbereshe villages.7, 8 Accordingly, our first genetic surveys, besides confirming the discontinuity of Calabrian Arbereshe with the Italian genetic background, revealed a shared genetic ancestry with modern Balkan populations.3, 9
The Arbereshe of Sicily survive today in only three municipalities of the province of Palermo (9057 inhabitants, 10,2%).4, 6 They are characterized by lower geographic isolation and smaller population size, as well as by higher levels of linguistic erosion. Historical evidence also suggests a more intricate formative process, which involved intermediate steps in the Balkans and Italy, as well as subsequent repeopling events from Greece.10 Our first investigation of the uniparental genetic structure of Sicilian Arbereshe confirmed stronger paternal links with Greek populations and higher similarities with Sicilians from the maternal perspective.3
Building on these studies, here we present a new high-resolution analysis of Y-chromosome genetic variation in the Arbereshe population by (i) increasing the sampling coverage for both Sicilian and Calabrian Arbereshe; (ii) deepening the resolution of Y-chromosome analysis; (iii) providing new data for the putative source populations; and (iv) testing alternative demographic models through extensive forward-in-time computer simulations. In particular, we seek to explore potential signals of Arbereshe dispersion in Sicily and Southern Italy, and to assess the amounts of their genetic continuity or local admixture, respectively, with Balkan source and Italian recipient populations.
Materials and methods
Population samples and DNA isolation
The data set primarily consists of unpublished Y-chromosome data generated for 150 unrelated individuals coming from 13 Arbereshe villages of the province of Cosenza (Calabria) and 2 Arbereshe communities of the province of Palermo (Sicily). We will refer to the former as Calabrian Arbereshe (ARB_CAL) and to the latter as Sicilian Arbereshe (ARB_SIC). Blood samples or buccal swabs were collected from healthy male volunteers, selected according to the self-declared affiliation to Arbereshe people and the patrilineal residence in an Arbereshe village for at least three generations. On the basis of previous biodemographic researches,7, 8 the 13 Calabrian Arbereshe communities were grouped into three internally homogeneous clusters: (i) populations located in the Crati River valley (VAL_CRA; n=46); (ii) populations located on the Southwestern side of the Pollino area (POL_SW; n=36); (iii) populations located in the highland area of the Pollino massif (POL_AREA; n=24). Y-chromosome data (12 Y-STRs and 30 Y-SNPs) of 40 ARB_CAL individuals were previously published in Boattini et al.9 The two Sicilian Arbereshe communities of Contessa Entellina (CON_ENT; n=26) and Piana degli Albanesi (PIA_ALB; n=18) were analyzed separately by considering the postulated complexity in their evolutionary histories.10
The same set of Y-chromosome markers was also typed for 223 unrelated Albanian males, collected from the two major dialect groups of Albania, Gheg from the North (N=119) and Tosk from the South (N=104). All the participants identified themselves as members of the given ethnic group, with at least two generations of unrelated paternal ancestry in their region of birth. Twelve Y-STRs and 18 Y-SNPs data for these populations were previously published in Ferri et al.11
We finally included 263 males from Sicily (N=216) and Southern Italy (N=47)12, 13 as local reference population and 209 unpublished Greek samples from Korinth (N=113) and Euboea (N=96)14 as additional source groups, reaching a total of 845 Y-chromosomes (Figure 1 and Supplementary Table S1). The additional Greek comparisons were introduced to account for two important aspects concerning the historical links among Albania, Greece and Southern Italy. Euboea has been considered the source area of many Greek colonies in Sicily and Southern Italy (Magna Grecia) during the Hellenic colonization in the Archaic Age.14 Korinth was settled by southern Albanians between the thirteenth and sixteenth centuries, during population movements that took place in parallel with or as a part of those migratory events later arrived in Sicily and Southern Italy.15
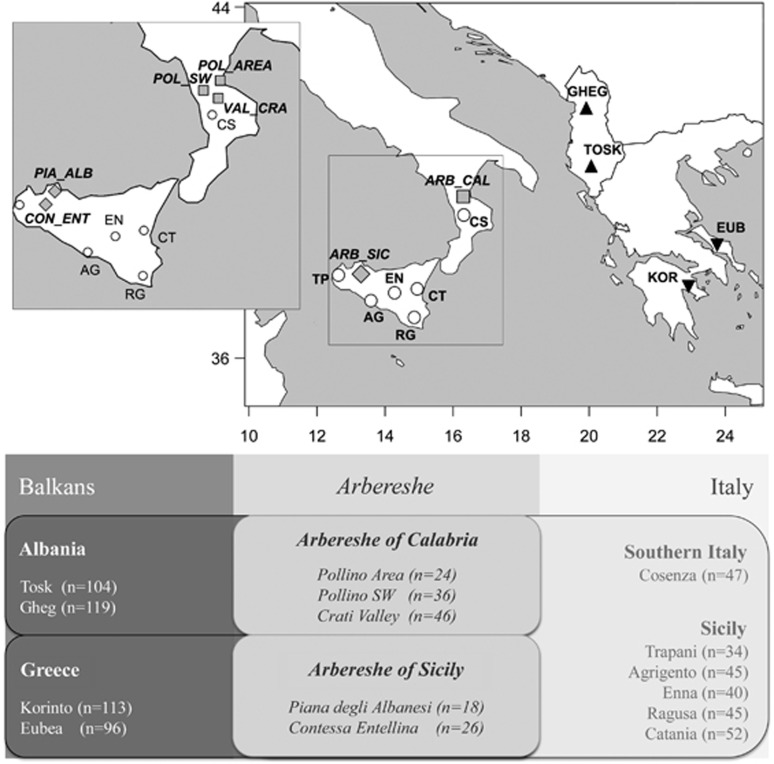
Figure 1.
Sampling map showing the location of Arbershe groups, Balkan source and Italian recipient populations. The enlarged box at the top-left details the communities decomposition for each of the two Arbereshe groups. The table at the bottom of the plot outlines the sampling structure and the number of samples analyzed for each population. Population codes as in Supplementary Table S1.
All donors provided a written informed consent to the data treatment and project objectives, according to the ethical standards of the involved institutions. The Ethics Committee of the Azienda Ospedaliero-Universitaria Policlinico S.Orsola-Malpighi in Bologna (Italy) approved the procedure. Genomic DNA was extracted by using a modified salting out protocol.16
Y-chromosome genotyping
Y-chromosome haplogroups (HGs) were assigned by typing 44 bi-allelic markers as described previously.13 Y-SNP information are published by Karafet et al.17 and by the International Society of Genetic Genealogy (www.isogg.org/tree/). All samples were additionally typed for the 17 Y-STRs implemented in the AmpFlSTR Yfiler PCR Amplification Kit (Applied Biosystems, Foster City, CA, USA). PCR and locus information can be found on Mulero et al.18 and on the Y-STR Haplotype Reference Database (YHRD, www.yhrd.org). As Yfiler kit amplifies DYS385a/b simultaneously, avoiding the determination of each of the two alleles, this locus was excluded from all the analyses performed. The locus DYS389b was obtained by subtracting DYS389I from DYS389II.
Newly-generated Y-chromosome data have been submitted to the YHRD database with accession numbers YA003636 (Albanian Tosk), YA003637 (Albanian Gheg), YA004046 (Sicilian Arbereshe) and YA004047 (Calabrian Arbereshe).
Statistical analyses
Descriptive analyses
Standard measures of genetic diversity and the analysis of molecular variance (AMOVA) were calculated with Arlequin software 3.5.1.2.19 Haplogroup frequencies were estimated by direct counting. Fisher exact tests were performed on HG frequencies to determine significantly over- or under-represented lineages in any of the considered populations. To reach a common level of Y-chromosome resolution, sub-lineages were combined in 36 HGs.
Multivariate analyses
To explore the relationship of Arbereshe ethnolinguistic minorities with both Balkan and Italian comparison populations, a correspondence analysis (CA) based on HG frequencies was performed by using the function dudi.coa in R software (package ade4).20, 21 Genetic distances based on STRs22, 23 were used to generate a multidimensional scaling (MDS) plot with the function isoMDS in R (library MASS)24 as well as to perform an admixture-like analysis13 as follows: (i) a nonhierarchical method based on Gaussian mixture models (R package mclust25, 26) was first exploited to find population clusters; (ii) the posterior membership probabilities (for each population to belong at each identified cluster) were then inferred by using DAPC (Discriminant Analysis of Principal Components; R package adegenet27) and represented with barplots.
Forward-in-time computer simulations
A forward-in-time approach was preferred to backward simulations (based on coalescent modeling) for the following reasons: (i) our case study is centered upon reproductively closed communities whose Ne is supposed to be of the same order of their relative sample size; (ii) observed values can be directly tested against parameter distributions modeled upon known demographic data. The respective contribution of source and recipient populations to present-day Arbereshe communities was formally estimated via a hypothesis testing approach. The variation through time of DHS—a molecular distance based on the extent and type of shared haplotypes between pairs of diverging pools28—was calculated at Y-STR and Y-SNP variants. The closer the divergence between pools of haplotypes the lower the value of DHS, ranging from 0 (all haplotypes shared) to 1 (no shared haplotypes).
Different sets of parameters (see Supplementary Table S2) were modeled under a stochastic Markov Chain Monte Carlo (MCMC) method, as implemented in the software ASHES (codeplex.ashes.com). As starting pools we used three putative source samples (Tosks, Ghegs and a 50% random blend of Greeks and Tosks) and two sink non-Arbereshe groups (Sicilians and Calabrians). For each simulation model we considered two populations coming into contact at time t0 and exchanging M=Nem haplotypes from the source to the sink pool. From time t1, the two populations were allowed to evolve independently for 20 generations—which correspond to the time elapsed since the oldest evidence of Arbereshe migrations (~560 YBP) by considering 28 years per generation (best guess for Y-chromosome estimated in translocated historical groups29). For each model, 500 iterations were performed and summary statistics of DHS values were calculated. To take into account the effect of demographic dynamics since contact event, realistic prior parameters were used (Supplementary Table S2). Varying parameters were the number of exchanged haplotypes M (5, 10, 20, 30 and 50% contribution of local haplotypes to the final pool), the mutation rates (2.49 × 10−3 (95% confidence interval (CI): 9.18 × 10−4–5.48 × 10−3) mut/site/gen for STRs, obtained by averaging single-locus germline mutation rates as estimated by Ballantyne et al.;30 and 3.0 × 10−8 (95% CI: 8.9 × 10−9–7.0 × 10−8) mut/site/gen for SNPs31) and the increment rates (0.00 for the stationary model; 0.07 for the growth model). The starting haplotype pools of source and local populations were built on n-time reiterations of the real data, with a final Ne corresponding to one-sixth of the respective averaged present-day census size. According to the standard definition of ‘effective size', as the number of breeding individuals in a panmictic population, a good estimate of Ne in modern human groups is around one-third of the census size. Assuming an equal sex ratio and little size fluctuation over time, a rough estimate of Ne for Y-haplotypes is thus one-sixth of the census size. Doubling or halving Ne and recalculating the increment rate accordingly did not affect appreciably simulation outcomes (data not shown). The distributions of simulated DHS values were compared with empirical values calculated for each pair of samples. The data were considered to fit the model when observed DHS values fell within two SD by the mean of the simulated distribution.
Coalescence analysis
The phylogenetic relationships between haplotypes within the most important HGs were inferred through Median Joining network applied on the output of a Reduced Median network, as implemented in the Network software 4.5.32, 33 STR loci were weighted according to the inverse of their variance.34 Time to the Most Recent Common Ancestor (TMRCA) was calculated for population-specific clusters of haplotypes. These clusters were identified within phylogenetic networks of the most important HGs following Balanovsky et al.35 and by selecting only those clusters appearing to have specifically evolved within particular population groups (frequency higher than 70%). TMRCAs were estimated by means of the SD estimator36 and the 95% CI were calculated based on the SE. Since time estimates based on variance are sensitive to the presence of outliers, all Y-chromosome age estimates were corrected for the presence of outlier STR loci as in Boattini et al.12 As events involving the Arbereshe population are relatively recent (~500 YBP) time estimates were computed based on all STR loci (minus DYS385a/b), by averaging locus-specific mutation rates as estimated by Ballantyne et al.30 A generation time of 28 years has been considered.
Results
Y-STR and Y-SNP genotyping results for the newly typed Arbereshe (n=150) and Albanian (n=223) individuals are provided in Supplementary Table S3. The corresponding frequencies of Y-chromosome HGs are detailed in Supplementary Table S4. The geographic distribution of Y-chromosome main lineages in the studied populations is shown in Supplementary Figure S1.
Within ethnic-group genetic variability
Intrapopulation genetic diversity of Arbereshe communities (Supplementary Table S5) exhibits values comparable to those reported in previous studies on Arbereshe Y-chromosome variability.3, 9 On the whole, both haplotype and haplogroup diversity in Arbereshe communities do not significantly differ from the range of values observed among Italian and Balkan comparison populations (Mann–Whitney U-test: W=10.5, P-value=0.075 for STR data and W=12, P-value=0.113 for SNP data). Nevertheless, departures from the observed range of variability have been detected in the Sicilian Arbereshe community of Contessa Entellina as well as in the Albanian Gheg population (Supplementary Table S5), showing the lowest values for Y-STR (0.9785±0.0166) and Y-SNP (0.7695±0.0251) markers respectively.
Among populations comparison
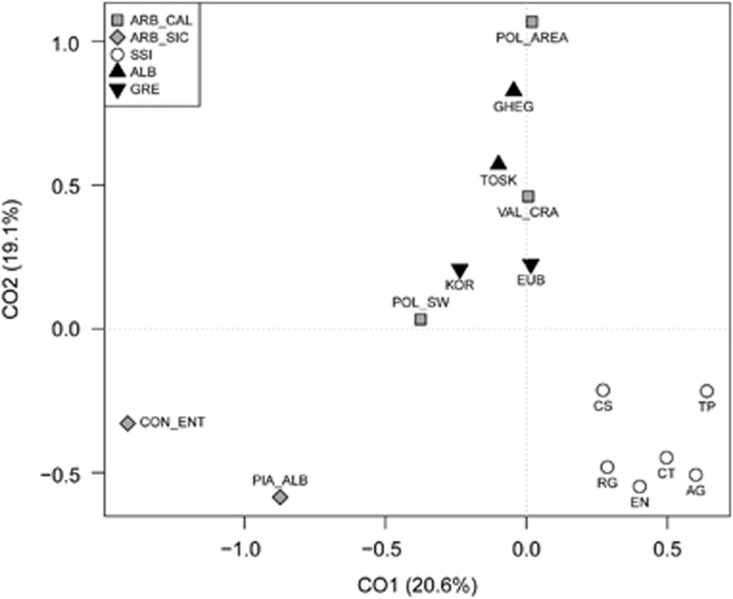
To visualize the genetic relationships of Arbereshe communities with Balkan and Italian populations, a correspondence analysis was performed on HG frequencies (Figure 2). The first two components, together accounting for ~40% of the total variance, suggest different patterns of genetic similarities for Calabrian and Sicilian Arbereshe, from both a geographical and linguistic point of view (see also AMOVA results, Supplementary Table S6). Although Calabrian Arbereshe clearly fall within the genetic space occupied by Balkan populations, the Sicilian Arbereshe reveal more complex patterns. In fact, the first component (20.6%) locates these samples at the opposite side of the autochthonous Sicilian populations, whereas the second component (19.1%) separates all the Sicilian groups (regardless of their linguistic affiliation) from the rest of analyzed populations. Consistently with correspondence analysis results, the MDS plot based on STR genetic distances confirms the heterogeneity observed among the two Arbereshe ethnolinguistic groups (Supplementary Figure S2).
Figure 2.
Correspondence Analysis (CA) based on Y-chromosome haplogroup frequencies. Population codes as in Supplementary Table S1. Symbols and color codes as in the legends at the top-left. Abbreviations: ARB_CAL, Arbereshe of Calabria; ARB_SIC, Arbereshe of Sicily; SSI, Sicilians and Southern-Italians; ALB, Albanians; GRE, Greeks.
Paternal contributions to the Arbereshe genetic pool
The admixture contributions of source (Balkan) and recipient (Italian) populations to the present-day Arbereshe genetic pool were estimated by performing forward-in-time DHS-based simulations, using the MCMC method implemented in the program ASHES.28
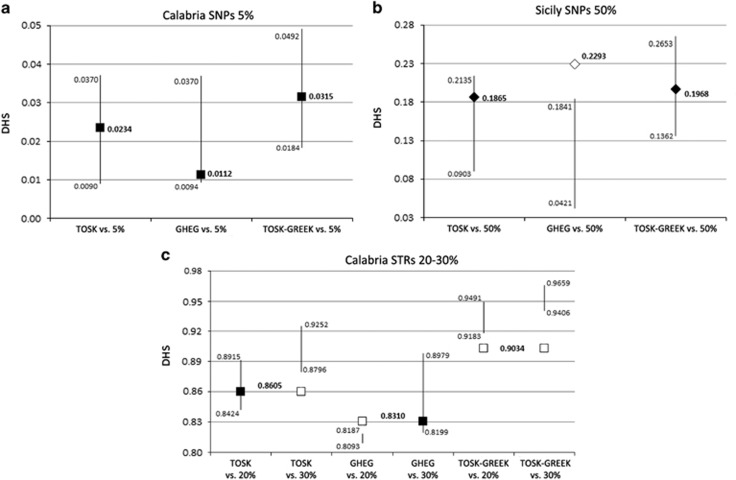
By comprehensively considering STR- and SNP-based results under a stationary model, the scenario that best approximates empirical data in the case of ARB_CAL is a migration from Toskeria to Calabria by 200–900 migrants and a local contribution to the genetic pool of the new settlements comprised between 5 and 20% (Table 1, Figure 4). Evolutionary scenarios with Gheg as parental population fit models with a local contribution of 5–30% and 100–900 migrants. Nevertheless, admixture with Gheg should be considered less likely since the observed DHS values match the lower tails of the expected distributions (Figure 4).
Table 1. Fitting of observed and expected values of the DHS statistics for STR and SNP genetic systems under a stationary model.
| % Local contribution | |||||||
|---|---|---|---|---|---|---|---|
| Marker | Source pop | 5 | 10 | 20 | 30 | 50 | Obs DHS |
| Calabria | |||||||
| SNP | Tosks | 0.0370 | 0.0809 | 0.1957 | 0.3054 | 0.4993 | 0.0234 |
| 0.0090 | 0.0291 | 0.0828 | 0.1441 | 0.2527 | |||
| Ghegs | 0.0370 | 0.0687 | 0.1459 | 0.2165 | 0.4209 | 0.0112 | |
| 0.0094 | 0.0255 | 0.0702 | 0.0626 | 0.1348 | |||
| Tosks–Greek | 0.0492 | 0.1187 | 0.2441 | 0.3664 | 0.5592 | 0.0315 | |
| 0.0184 | 0.0524 | 0.1239 | 0.1988 | 0.3409 | |||
| STR | Tosks | 0.6751 | 0.7967 | 0.8915 | 0.9252 | 0.9594 | 0.8605 |
| 0.5726 | 0.7483 | 0.8424 | 0.8796 | 0.9140 | |||
| Ghegs | 0.6211 | 0.7448 | 0.8187 | 0.8979 | 0.9379 | 0.8310 | |
| 0.5468 | 0.6845 | 0.8093 | 0.8199 | 0.8629 | |||
| Tosks–Greek | 0.8415 | 0.8973 | 0.9491 | 0.9659 | 0.9815 | 0.9034 | |
| 0.7988 | 0.8624 | 0.9183 | 0.9406 | 0.9619 | |||
| Sicily | |||||||
| SNP | Tosks | 0.0060 | 0.0236 | 0.0681 | 0.1079 | 0.2135 | 0.1865 |
| 0.0000 | 0.0023 | 0.0188 | 0.0288 | 0.0903 | |||
| Ghegs | 0.0105 | 0.0267 | 0.0652 | 0.0773 | 0.1841 | 0.2293 | |
| 0.0010 | 0.0055 | 0.0229 | 0.0313 | 0.0421 | |||
| Tosks–Greek | 0.0140 | 0.0337 | 0.0845 | 0.1401 | 0.2653 | 0.1968 | |
| 0.0021 | 0.0116 | 0.0356 | 0.0564 | 0.1362 | |||
| STR | Tosks | 0.4861 | 0.6105 | 0.7649 | 0.8316 | 0.8987 | 0.9825 |
| 0.3988 | 0.5591 | 0.7088 | 0.7833 | 0.8504 | |||
| Ghegs | 0.4379 | 0.5810 | 0.7038 | 0.7760 | 0.8537 | 1.0000 | |
| 0.3128 | 0.5319 | 0.6475 | 0.7123 | 0.7831 | |||
| Tosks–Greek | 0.6732 | 0.7805 | 0.8810 | 0.9188 | 0.9490 | 0.9770 | |
| 0.5562 | 0.7428 | 0.8472 | 0.8889 | 0.9224 | |||
A percent contribution of local versus putative source population was accepted as the most likely model when the observed value fell within a two SD interval (in bold) of the simulated distribution at time t=20 generations.
Evolutionary scenarios for ARB_SIC fit only models based on SNP data and seem unlikely unless extensive gene flows between source and local groups or complete population displacements are invoked. In fact, 120 (or fewer) migrants from a Tosk or a Tosk/Greek source population (Figure 4) and a local contribution >50% (Table 1) were estimated. In the case of ARB_SIC, evolutionary scenarios with Gheg as parental population do not fit any model (Table 1, Figure 4). In general, variation of the increment rate (I=0.07, growth model) did not appreciably affect the fitting of the considered models (data not shown).
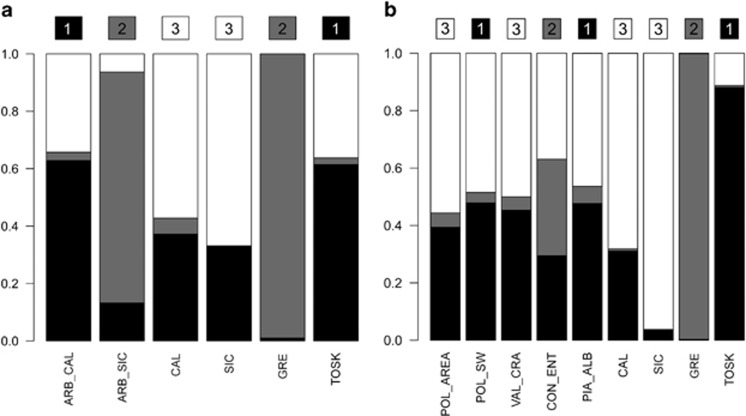
Consistently with simulation results, admixture-like plots reveal ARB_CAL as conserving clearer signals of descent from Albanian populations (black component, cluster 1, Figure 3a), whereas ARB_SIC exhibit a more complex genetic pattern involving links with Greek populations (gray component, cluster 2, Figure 3a). In particular, CON_ENT seems affected by a relevant Greek contribution (gray component, Figure 3b). It is also worth noting the clustering of POL_SW with the Sicilian Arbereshe population of PIA_ALB (cluster 1, Figure 3b), rather than with the other Calabrian Arbereshe communities of POL_AREA and VAL_CRA.
Figure 3.
Admixture-like barplots for the Arbereshe groups (a) and the single-Arbereshe communities (b). The inferred cluster affiliation of each population (mclust algorithm) and the corresponding cluster's color code (1: black; 2: gray; 3: white) are represented by numbers (within colored squares) at the top of each bar. The probability (DAPC-based posterior membership probabilities) of each population belonging to the inferred clusters is represented by vertical bars in the plot. Abbreviations: ARB_CAL, Arbereshe of Calabria; ARB_SIC, Arbereshe of Sicily; CAL, Italians of Calabria; SIC, Italians of Sicily; GRE, Greeks; TOSK, Tosks. Codes for single-Arbereshe communities as in Supplementary Table S1.
Haplogroup composition and haplotype clusters
Five haplogroups were found to particularly affect the genetic variability within and between the two Arbereshe groups (Supplementary Figure S1,Supplementary Table S4). The two most frequent HGs in ARB_CAL (E-V13, 16.9% I-M223, 14.2%) do not match those found in ARB_SIC (I-P215, 20.5% E-M123, 18.2%). By contrast, the third most frequent HG (R-SRY10831.2) is found at comparatively high frequencies in both Calabrian (9.43%) and Sicilian (11.35%) Arbereshe (Supplementary Table S4).
Haplotype networks of both E-V13 (Supplementary Figure S3) and I-P215* (Supplementary Figure S4) show fairly compact star-like structures centered around Albanian (mainly Tosk) haplotypes, yet highlighting the differential presence of E-V13 ARB_CAL (77.8% E-V13α, Supplementary Figure S3) and I-P215 ARB_SIC (85.7% I-P215α, Supplementary Figure S4) specific clusters. Both of these clusters reveal in their star-like structures signals of recent expansions and their SD-based age estimates (469±118 and 649±170 YBP) are consistent with times of Arbereshe migrations in Sicily and Southern Italy. Non-Arbereshe haplotypes from these clusters were autonomously recognized as ‘outliers' by the jackknife-like omitting procedure of TMRCA estimation, and excluded from the age calculation.
R-SRY10831.2 haplotype network (Supplementary Figure S5), despite a relatively complex structure, reveals the presence of a shared Arbereshe-specific cluster (R-SRY10831.2α) dating at 556±146 YBP.
Consistently with their presumptive Balkan origin, Y-STR haplotype diversities for each of these three lineages is higher in the considered Balkan populations (0.990–0.995 for E-V13; 0.985–0.995 for I-P215; and 0.990–1.000 for R-SRY10831.2) than among ARB_CAL (0.948±0.039 for E-V13), ARB_SIC (0.962±0.064 for I-P215*) or the whole Arbereshe people (0.980±0.013 for R-SRY10831.2). By contrast, I-M223 and E-M123 appear significantly over-represented in the Arbereshe ethnolinguistic minorities (P-value<0.001 and P-value=0.0015, respectively) with respect to Southern Italy and the Balkans. In addition, these lineages show differential genetic links between the Sicilian Arbereshe and the Calabrian sample of POL_SW (E-M123: 18.2% and 22.2%, respectively), compared with the other Calabrian Arbereshe communities of VAL_CRA and POL_AREA (I-M223: 17.4% and 25%, respectively). Accordingly, whereas the I-M223 network (Supplementary Figure S6) mainly reflects its specificity for ARB_CAL (being cluster I-M223α completely absent in ARB_SIC), the E-M123 network shows the co-existence of two distinct Arbereshe-specific clusters (Supplementary Figure S7): a 100% ARB_CAL (E-M123α) one and a 100% ARB_SIC (E-M123β) one. Age estimates are similar for both E-M123 clusters (1133±314 and 883±228 YBP), yet predating the time of Arbereshe settlement in Southern Italy. Analogously, the I-M223α cluster dates at 2122±548 YBP.
Discussion
In this study, we addressed some specific questions concerning the population history and genetic structure of Arbereshe people by combining high-resolution investigations of Y-chromosome lineages with extensive computer simulations.
With the partial exception of Contessa Entellina (showing lower values for STR data), all the analyzed Arbereshe communities do not exhibit any significant reduction of gene diversity. When Arbereshe were compared with Italian and Balkan groups, our results (Figure 2 and Supplementary Figure S2) revealed the presence of both geographic and cultural clustering patterns (see also AMOVA results, Supplementary Table S6). At a broader geographic scale, Southern Italy and the Balkans were previously proven to form quite a consistent homogeneous group in the Mediterranean genetic landscape.13 When adopting a microgeographical perspective, they however show significant degrees of cultural and genetic complexity. Cultural factors were confirmed to have played an important role for Arbereshe people to preserve their original genetic background. However, our simulation-based approach suggests markedly different genetic histories between Calabrian and Sicilian groups. The Arbereshe of Calabria attest a tight relationship with modern Albanian populations—the most likely candidate source being Tosks—and show relatively low levels of admixture with Italian local groups (Table 1 and Figure 4). As for Sicilian Arbereshe, composite founder events (involving population replacements) and/or lower pressures of linguistic and geographic barriers may be hypothesized to account for partial similarities with Greek populations as well as for the higher levels of local admixture.
Figure 4.
Plots of observed and expected DHS values for fitting scenarios under a stationary model. Evolutionary scenarios with the three putative source populations (Tosks, Ghegs and a 50% random blend of Tosks and Greeks) were compared for those models (bold in Table 1) showing at least one fit in the simulations results: (a) Calabria SNPs 5%, (b) Sicily SNPs 50% and (c) Calabria STRs 20–30%. The SD intervals of the simulated distributions are represented by vertical bars. Observed DHS values are represented by symbols filled in black or white depending on whether or not they match the simulated intervals. Squares and diamonds stand for Calabrian and Sicilian Arbereshe, respectively.
In particular, Contessa Entellina (CON_ENT) reveals the strongest genetic affinity with Greeks (Figure 3b). Interestingly, historical research showed that, at around 1500 CE, Contessa Entellina was repopulated by one hundred families coming from the island of Andros, in the Peloponnese.10 On the contrary, the other Arbereshe sample from Sicily (PIA_ALB), similarly to all the three Calabrian Arbereshe groups (VAL_CRA, POL_SW, POL_AREA), confirm a considerable Tosk contribution from Albania (Figure 3b). Such patterns are however complicated by the fact that POL_SW seem to be somehow related with Sicilian Arbereshe, rather than clustering with the other Calabrian Arbereshe communities.
In accordance with admixture analyses, the Y-chromosome lineage dissection highlights different patterns of HG composition between and within the two Arbereshe groups (Supplementary Figure S1 and Supplementary Table S4). The high proportion of Balkan lineages (E-V13, I-P215* and R1a-SRY10831.2) observed in both Sicilian and Calabrian Arbereshe, coupled with the corresponding loss of Y-STR variability relative to putative founding populations, support the described patterns of Arbereshe diffusion in Sicily and Southern Italy from the Balkans. However, whereas some lineages (R-SRY10831.2) proved not to significantly differ between Calabrian and Sicilian Arbereshe (9.43% and 11.35%), other haplogroups proved to be differentially present in ARB_CAL (E-V13) and ARB_SIC (I-P215) or to differentially link some communities with other ones (E-M123 and I-M223). This might suggest that the Albanian source populations were at least in part already differentiated before their migration to Italy.
Further evidence is provided by our dating experiments. The ages estimated for clusters of haplotypes within the most frequent Balkan HGs (E-V13, I-P215*, R-SRY10831.2) yielded dates that are consistent with historical evidence of Arbereshe migrations (469±118, 649±170, 556±146 YBP). Nevertheless, as E-V13 and I-P215* show Calabrian-specific (E-V13α) and Sicilian-specific (I-P215α) haploype clusters, we hypothesize that their expansions may have been the result of two contemporary but independent events. Finally, the time estimates of the two remaining major Arbereshe lineages (E-M123 and I-M223) are referable to time periods (~1000–2000 YBP) which largely predate the settlement of Arbereshe people in Italy. These facts possibly reflect events of genetic differentiation occurred in the Balkans earlier than the Arbereshe migrations took place.
In summary, our results suggest that the considered Arbereshe groups, despite the shared origins in the Balkan Peninsula and the common cultural features, may have been generated by two parallel but independent peopling processes, from already differentiated source populations. One of these events have possibly led to the formation of present-day Arbereshe communities settled in the Calabrian areas of the Pollino massif (POL_AREA) and of the Crati River Valley (VAL_CRA). The other one is presumptively related with the origin of Sicilian Arbereshe, but involving also part of the Calabrian communities of POL_SW. The two founding populations may have diverged from each other for two possible reasons: (a) the presence of genetic sub-structuring in the Middle Ages Southern Balkans; (b) episodes of admixture with other populations (eg Greece, Sicily and Calabria) during and after the migratory processes.
Future research, facilitated by analyses of complementary genetic systems will enable us to achieve an even more detailed picture of the Arbereshe genetic history and population variability, within a multi-disciplinary effort to compare and integrate these genetic overviews with the results offered by other disciplines, such as linguistics.
Acknowledgments
This study was supported by the European Research Council ERC-2011-AdG 295733 grant (Langelin) and by the MIUR-PRIN grants to DP and DL. CC and FB are supported by the British Academy (BARDA-47870 ‘The Greeks in the West'). SS and EB are supported by the European Research Council ERC-2011-AdG 295733 grant (Langelin). Special thanks are due to Dr A. Ales and Dr L. Màtesi for their invaluable help in selecting the populations and in performing the sampling campaign of the Sicilian Arbereshe communities of Piana degli Albanesi and Contessa Entellina. We are indebted to all the Calabrian and Sicilian Arbereshe communities involved in the project and we gratefully acknowledge all the volunteers who kindly accepted to participate in this study. We thank Marilisa Carta, Vincenzo Motta and Serena Tucci for their assistance in the lab analysis. We would like to thank the anonymous reviewers for their insightful comments that improved the quality of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Toso F: The study of language islands: an interdisciplinary approach. J Anthropol Sci 2014; 92: 1–5. [DOI] [PubMed] [Google Scholar]
- Destro-Bisol G, Anagnostou P, Batini C et al: Italian isolates today: geographic and linguistic factors shaping human biodiversity. J Anthropol Sci 2008; 86: 179–188. [PubMed] [Google Scholar]
- Capocasa M, Anagnostou P, Bachis V et al: Linguistic, geographic and genetic isolation: a collaborative study on Italian populations. J Anthropol Sci 2014; 92: 1–32. [DOI] [PubMed] [Google Scholar]
- ISTATXIV Censimento Generale delle Popolazioni e delle Abitazioni. Roma: Istituto Centrale di Statistica Editore, 2003. [Google Scholar]
- Zangari D: Le Colonie Italo-Albanesi di Calabria. Naples: Casella Editore, 1941. [Google Scholar]
- Tagarelli A: Studio antropologico della comunità Arbëreshe della provinciaf di Torino, (eds) Librare, 2004, pp 47–66. [Google Scholar]
- Fiorini S, Tagarelli G, Boattini A, Luiselli D, Piro A, Tagarelli A, Pettener D: Ethnicity and evolution of the biodemographic structure of Arbereshe and Italian populations of the Pollino area, Southern Italy (1820–1984). Amer Anthropol 2007; 109: 735–746. [Google Scholar]
- Tagarelli G, Fiorini S, Piro A, Luiselli D, Tagarelli A, Pettener D: Ethnicity and biodemographic structure in the Arbëreshe of the province of Cosenza, southern Italy, in the XIX century. Coll Antropol 2007; 31: 331–338. [PubMed] [Google Scholar]
- Boattini A, Luiselli D, Sazzini M, Useli A, Tagarelli G, Pettener D: Linking Italy and the Balkans. A Y-chromosome perspective from the Arbereshe of Calabria. Ann Hum Biol. 2010; 38: 59–68. [DOI] [PubMed] [Google Scholar]
- Giunta F, Mandalà M: Albanesi in Sicilia. Palermo: Mirror, 2003; p. 25. [Google Scholar]
- Ferri G, Tofanelli S, Alù M et al: Y-STR variation in Albanian populations: implications on the match probabilities and the genetic legacy of the minority claiming an Egyptian descent. Int J Legal Med 2010; 124: 363–370. [DOI] [PubMed] [Google Scholar]
- Boattini A, Martinez-Cruz B, Sarno S et al: Uniparental markers in Italy reveal a sex-biased genetic structure and different historical strata. PLoS One 2013; 8: e65441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarno S, Boattini A, Carta M et al: An ancient Mediterranean melting pot: investigating the uniparental genetic structure and population history of Sicily and Southern Italy. 2014. PLoS One 2014; 9: e96074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofanelli S, Brisighelli F, Anagnostou P et al: The Greeks in the West: genetic signatures of the Hellenic colonization in southern Italy and Sicily. Eur J Hum Genet 2015. (Submitted). [DOI] [PMC free article] [PubMed]
- Hall JM: Ethnic Identity in Greek Antiquity. Cambridge: Cambridge University Press, 2000. [Google Scholar]
- Miller SA, Dykes DD, Polesky HF: A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF: New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res 2008; 18: 830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero JJ, Chang CW, Calandro LM, Green RL, Li Y, Johnson CL, Hennessy LK: Development and validation of the AmpFlSTR Yfiler PCR amplification kit: a male specific, single amplification 17 Y-STR multiplex system. J Forensic Sci 2006; 51: 64–75. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S: Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 2007; 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- Dray S, Dufour AB, Chessel D: The ade4 package-II: two-table and K-table methods. R News 2007; 7: 47–52. [Google Scholar]
- Dray S, Dufour AB: The ade4 package: implementing the duality diagram for ecologists. Journal of Statistical Software 2007; 22: 1–20. [Google Scholar]
- Reynolds JB, Weir BS, Cockerham CC: Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics 1983; 105: 767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval G, SanCristobal M, Chevalet C: Measuring genetic distances between breeds: use of some distances in various short term evolution models. Genet Sel Evol 2002; 34: 481–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN, Ripley BD: Modern Applied Statistics with S, 4th edn. New York: Springer, 2002. [Google Scholar]
- Fraley C, Raftery AE: Model-based clustering, discriminant analysis, and density estimation. J Am Stat Assoc 2002; 97: 611–631. [Google Scholar]
- Fraley C, Raftery AE: Bayesian regularization for normal mixture estimation and model-based clustering. J Classif 2007; 24: 155–181. [Google Scholar]
- Jombart T, Devillard S, Balloux F: Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 2010; 11: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofanelli S, Bertoncini S, Castrì L, Luiselli D, Calafell F, Donati G, Paoli G: On the origins and admixture of Malagasy: new evidence from high-resolution analyses of paternal and maternal lineages. Mol Biol Evol 2009; 26: 2109–2124. [DOI] [PubMed] [Google Scholar]
- Bonné-Tamir B, Korostishevsky M, Redd AJ, Pel-Or Y, Kaplan ME, Hammer MF: Maternal and paternal lineages of the Samaritan isolate: mutation rates and time to most recent common male ancestor. Ann Hum Genet 2003; 67: 153–164. [DOI] [PubMed] [Google Scholar]
- Ballantyne KN, Goedbloed M, Fang R et al: Mutability of Y-chromosomal microsatellites: rates, characteristics, molecular bases, and forensic implications. Am J Hum Genet 2010; 87: 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wang Q, Long Q et al: Human Y chromosome base-substitution mutation rate measured by direct sequencing in a deep-rooting pedigree. Curr Biol 2009; 19: 1453–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A: Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 1999; 16: 37–48. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB: Mitochondrial portraits of human populations using median networks. Genetics 1995; 141: 743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Weiss G, von Haeseler A: Pattern of nucleotide substitution and rate heterogeneity in the hypervariable regions I and II of human mtDNA. Genetics 1999; 152: 1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanovsky O, Dibirova K, Dybo A et al: Parallel evolution of genes and languages in the Caucasus region. Mol Biol Evol 2011; 28: 2905–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Zhivotovsky LA, King R et al: Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet 2006; 78: 202–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.