Abstract
X-linked congenital cerebellar ataxia is a heterogeneous nonprogressive neurodevelopmental disorder with onset in early childhood. We searched for a genetic cause of this condition, previously reported in a Buryat pedigree of Mongolian ancestry from southeastern Russia. Using whole-genome sequencing on Illumina HiSeq 2000 platform, we found a missense mutation in the ABCB7 (ABC-binding cassette transporter B7) gene, encoding a mitochondrial transporter, involved in heme synthesis and previously associated with sideroblastic anemia and ataxia. The mutation resulting in a substitution of a highly conserved glycine to serine in position 682 is apparently a major causative factor of the cerebellar hypoplasia/atrophy found in affected individuals of a Buryat family who had no evidence of sideroblastic anemia. Moreover, in these affected men we also found the genetic defects in two other genes closely linked to ABCB7 on chromosome X: a deletion of a genomic region harboring the second exon of copper-transporter gene (ATP7A) and a complete deletion of PGAM4 (phosphoglycerate mutase family member 4) retrogene located in the intronic region of the ATP7A gene. Despite the deletion, eliminating the first of six metal-binding domains in ATP7A, no signs for Menkes disease or occipital horn syndrome associated with ATP7A mutations were found in male carriers. The role of the PGAM4 gene has been previously implicated in human reproduction, but our data indicate that its complete loss does not disrupt male fertility. Our finding links cerebellar pathology to the genetic defect in ABCB7 and ATP7A structural variant inherited as X-linked trait, and further reveals the genetic heterogeneity of X-linked cerebellar disorders.
Introduction
X-linked congenital cerebellar ataxias
X-linked congenital cerebellar ataxia is a heterogeneous nonprogressive disease that occurs generally in the first years of life and is characterized by developmental delay and difficulties in coordination owing to hypoplasia/atrophy of the cerebellum. Most of the congenital cerebellar hypoplasia and atrophy cases, such as oligophrenin-1 syndrome (OMIM: 300486) or MICPCH/CASK syndrome (calcium/calmodulin-dependent serine protein kinase (membrane-associated guanylate kinase family (MAGUK) family); OMIM: 300749), are considered to be X-chromosome-associated intellectual disability pathologies.1, 2 Loss of function of oligophrenin-1 (OPHN1) gene leads to cerebellar hypoplasia and frontotemporal atrophy,1 and to hippocampal alterations3 by affecting Rho GTPase-dependent signaling required in cell migration and morphogenesis, synapse maturation and plasticity of neurons. Similarly, the dysfunction of CASK gene, which encodes the calcium/calmodulin-dependent serine protein kinase (MAGUK family), is associated with ataxia, mental retardation and microcephaly.2 The cerebellar dysfunctions are also linked to ion exchange abnormalities. In one familial case, the cause of the disease was a Ca2+ imbalance as a result of mutation in the ATP2B3 gene, an ATPase transmembrane transporter that moves Ca2+ ions out of the cell against concentration gradients.4 In another case, the X-linked spinocerebellar ataxia was linked to a nonsynonymous variant in the GJB1 gene encoding protein connexin 32, which supports ions and transfer of small molecules by forming gap junction channels between cells.5 Here, we report the case of inherited neurological pathology linked to genetic defects in genes for metal-ion transporters in a large Buryat pedigree (Figure 1).6
Figure 1.
Genealogy of the Buryat family. The whole-genome sequencing was performed for the patient designated by an arrow; solid symbols indicate affected individuals; dotted circles – obligate heterozygous carrier females; open symbols – unaffected individuals; slashed symbols – deceased subjects; asterisks – individuals who were used for genotyping.6
Clinical description
Magnetic resonance imaging (MRI) scans revealed hypoplasia of cerebellar hemispheres and vermis in affected males from the Buryat pedigree. Common neurological symptoms for all affected family members were development delay, difficulties in speech and coordination, limb and truncal ataxia and dysarthria (Supplementary Table 1). The patients examined were not able to sit without support any time before 15 months of age, or to walk independently before 7 years of age and to speak their first words before 4 years of age. For the majority of patients, nystagmus, ophthalmoplegia and increased tendon reflexes were observed. There were no signs of memory or cognitive impairment in all the patients from this pedigree.6
Features of sideroblastic anemia or copper disorder were not detected. Hematological tests performed for the patient (III-18) with X-linked ataxia from the Buryat pedigree showed no erythropoietic cell abnormalities or accumulation of iron granules. Hemoglobin values averaged 149 g/l (normal range, 130–180 g/l), color index averaged 1.0 (normal range, 0.80–1.05), the erythrocyte sedimentation test averaged 3 mm/h (normal range, 0–15 mm/h) and white blood cell count was generally normal. All biochemical tests were normal, except for a slightly elevated level of bilirubin, 14.5 mg/l (normal range, 5–12 mg/l), and the urine analysis was unremarkable.6
Materials and methods
Blood samples of all the subjects were collected previously with appropriate informed consent and these data have already been reported.6 No consanguinity was found to be present in the pedigree (Figure 1). A genomic library was made from 2 μg DNA sample of patient III-17 following the protocol for Paired-End DNA Sample Prep Kit (Illumina, San Diego, CA, USA). High-throughput genome sequencing was performed on Illumina HiSeq 2000 platform with at least 14-fold coverage depth of the genome GRCh37. The genome analysis was performed using 'ngs-pipeline' software designed by our group http://rogaevlab.ru/ngs-pipeline. The identified rare variants were submitted to NCBI ClinVar and are available at www.ncbi.nlm.nih.gov/clinvar/?LinkName=orgtrack_clinvar&from_uid=505407.
Paternity was determined by STR (short tandem repeat) analysis using homemade STR markers and PowerPlex 16 System (Promega, Madison, WI, USA).
Results
Whole-genome sequencing
Using pairwise linkage analysis and haplotype reconstruction, the gene responsible for the disorder was mapped to a 38-cM interval on chromosome Xp11.21-q24, flanked by the STR markers DXS991 and DXS1001. Multipoint linkage analysis showed the robust linkage (maximal lod score of 4.66) at the DXS1059 (Xq23).6 We next performed whole-genome sequencing and analysis of one male patient (III-17; Figure 2) from the Buryat family, with the described symptoms generating 542 million short reads using our bioinformatics pipeline (http://rogaevlab.ru/ngs-pipeline). The raw read sequences are freely available through the NCBI Sequence Read Archive under the accession number SRP049620 and from the website http://rogaevlab.ru/ataxia. Using these primary sequencing data, we reconstructed the complete mitochondrial genome of this person by assigning it to haplogroup C4b,7 which is commonly found in native populations of southern Siberia8, 9 (Supplementary Table 2). Assembly sequence of complete mitochondrial genome is available at the NCBI GenBank under the accession number KR153486. Analysis of Y-chromosome sequences linked it to haplogroup N1c1a1a*,10 a subgroup of N1c1 of Asian origin,11 which is frequent in Buryat and Mongolian populations12 (Supplementary Table 3). In total, 4.2 million variants, including 3.7 million single-nucleotide polymorphisms, were identified using GATK (The Genome Analysis Toolkit),13 and Pindel14 programs. To our knowledge, this is the first time that the whole-genome sequence of an individual from the ethnic Buryat of Mongolian ancestry has been presented.
Figure 2.
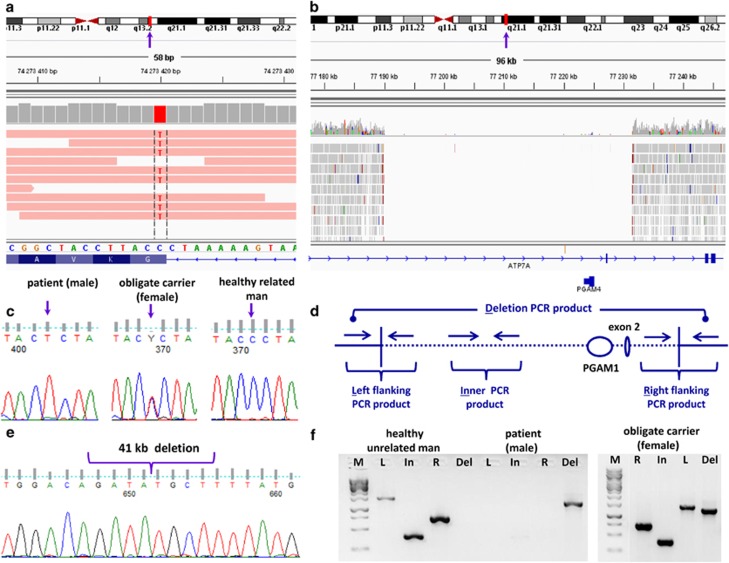
Identification of variants in ABCB7 and ATP7A genes. Representation of genomic region containing mutations using IGV (Integrative Genomics Viewer) tools: (a) exon 16 of ABCB7 gene (hg19 chrX:g.742734204C>T), (b) 41.4 kb deleted region of the ATP7A gene (hg19 chrX:g.77190006_77231471del), including exon 2; vertical arrows in (a) and (b) indicated the chromosomal loci for ABCB7 and ATP7A genes. (c) Sequencing verification of candidate mutations in exon 16 of ABCB7 gene. (d) Primers scheme for validation of deletion in ATP7A gene. (e) Sequencing validation by Sanger sequencing of deletion in ATP7A gene in the patients. (f) PCR products of the ATP7A gene deleted region: M, DNA ladder; R, upstream ‘right' flanking with deletion region; In, region inner deletion; L, downstream ‘left' flanking with deletion region (R, In, L – products are present if deletion is absent; Del – product is present if there is deletion in DNA sample).
We selected all rare variants (minor allele frequency (MAF)) <5% 1000 Genomes Project Consortium15) located in the specified region on Xp11.21-q24 locus, flanked by the STR markers DXS991 and DXS1001, and identified five rare or novel variants that can alter protein coding exon sequences (Supplementary Table 4). We narrowed down the candidate-gene list by excluding NHSL2 (Gene ID: 340527), AMER1 (Gene ID: 139285), PHKA1 (Gene ID: 5255) and RGAG1 (Gene ID: 57529) based on evolutionary analysis, gene expression profiles in tissues and protein topology position (Supplementary Figure 1). NHSL2 gene was excluded because there are no reports on association of rare variants in the NHSL2 gene, including the rs72630038 (SCV000221324) (MAF=0.045 in 1000 Genomes Project Consortium15) found in the Buryat patient, with any disorder; low expression of the gene in the cerebellum at prenatal period;16, 17 and that at least one mammalian species (hedgehog) bears the same rare amino-acid variant in NHSL2. Mutations in the AMER1 gene are known as genetic factors underlying osteopathia striata, cranial sclerosis and intellectual disability syndrome (OMIM: 300647 and 300373). We excluded AMER1 gene because of the clinical phenotype differences in our patients and location of the variant (SCV000221323) in evolutionarily non-conserved region of the protein. Mutations in the PHKA1 gene lead to X-linked muscle glycogenosis (OMIM: 311870 and OMIM 300559), which is also distant from the observed clinical symptoms in our patients. The amino-acid substitution arginine to glutamine (SCV000221325) in the PHKA1 protein occurred in non-conserved position. Variant in RGAG1 (retrotransposon gag domain containing 1 protein) gene (SCV000221326) is also localized in a non-conserved position. Ultimately, we identified the genetic defects in two closely linked genes, both essential for metal transport. We found that the missense mutation in exon 16 of the ABCB7 (ABC-binding cassette transporter B7) gene (SCV000221289), which is in linkage disequilibrium with a large 41.4 kb deletion in the ATP7A gene on X-chromosome (SCV000221290) in this family, is the most probable causative factor for the neurological disease of the Buryat patients (Figure 2).
ABCB7 gene
Nonsynonymous missense variant hg19 chrX:g.74273420C>T in exon 16 (AF241887) of ABCB7 gene (Gene ID: 22) leads to substitution of glycine to serine in position 682 of ATP-binding cassette member 7 protein (NP_001258625, UniProt O75027) (Figures 2a and c). As the identified variant is localized near an acceptor splice site within the first coding triplet of exon 16, we examined if the substitution could lead to the formation of a new strong splice site. The bioinformatic tools testing putative splicing sites based on nucleotide frequencies in each site position (from −20 to +3) for constitutive, cassette, inner acceptors and outer acceptors predicted no splicing alterations for the mutant allele18, 19 (Supplementary Table 5).
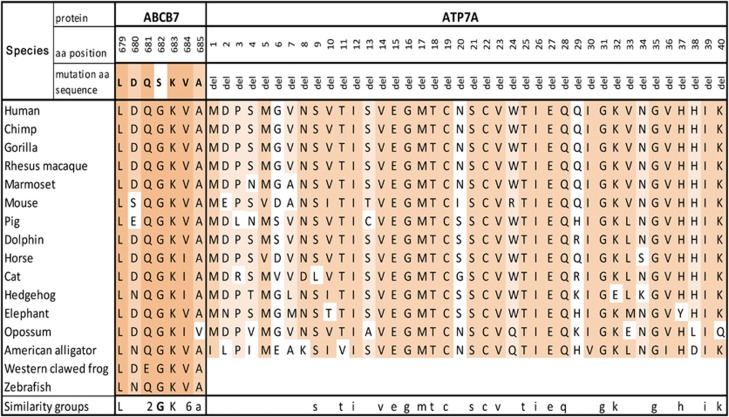
There are several known transcripts of ABCB7 gene encoding protein isoforms, and the identified variant affects all predicted protein isoforms. The structure and function of ABCB7 protein domains are not fully understood. The mutation is located in the nucleotide-binding domain (NBD) of ABCB7 protein (NP_001258625, UniProt O75027), and disruption of this region in ATM1 yeast ortholog leads to the loss of protein function and iron accumulation inside the mitochondrion (Supplementary Figure 3A).20 We studied structural alterations in the ABCB7 protein bearing mutant serine variant using the bioinformatics program Phyre2.21 Comparative analysis of ~1000 paralogous and orthologous amino-acid sequences of superfamily ABC transporter proteins shows a highly conserved glycine variant in position 682 (Figure 3). Serine substitution leads to rotation in the motionless protein side. Additional bioinformatic predictions for this mutation were performed. The mutant variant in ABCB7 gene was assessed as deleterious by SIFT22 and probably damaging (0.996) by PolyPhen.23 To date, four other mutations were found in the ABCB7 gene associated with sideroblastic anemia and cerebellar ataxia (Supplementary Figure 3A).
Figure 3.
Evolutionary analysis of mutation region. The missense mutation in ABCB7 and deletion in ATP7A genes change the amino-acid sequences that are highly conserved in vertebrates. Level of evolutionarily amino acid conservation illustrated by shading from white for non-conserved to dark gray for highly conserved. Alignments were obtained from the UCSC Genome Browser (Multiz Alignments of 100 Vertebrates),48 ClustalW.49
ATPase, Cu2+ transporting, alpha polypeptide (ATP7A) gene
We identified a deletion of region hg19 chrX:g.77190006_77231471del encompassing exon 2 (U27361), partially, introns 2 and 3 of the ATP7A gene (Gene ID: 538) and PGAM4 (phosphoglycerate mutase family member 4) retrogene (Gene ID: 441531) (Figures 2b, d and f). This deletion of the ATP7A gene is located at a distance of 2.8 Mb from ABCB7 gene and eliminates the original translation start site in exon 2. We suggest that an alternative start codon, for example, in exon 3 can be instead used for the same open reading frame (predicted by ATGpr24). The resulting protein will lack the first 68 amino-acid residuals, which constitute the first of six metal-binding domains (MBDs) (Supplementary Figure 3B). As the ATP7A protein (NP_000043.4, Q04656) has six isoforms,25 the deletion disrupts the open reading frame for the two short ones with unknown biological significance (Supplementary Figure 4). The ATP7A gene homologs are present in a broad range of organisms: from unicellular to complex multicellular organisms. The first MBD encoded by exon 2 is evolutionarily conserved in mammals, birds and reptiles. However, prokaryotes and lower eukaryotes have only one to three MDBs. We tested ATP7A gene expression in the adult cortex and cerebellum and found the presence of a transcript product corresponding to protein-encoding transcript sequence in the frontal cortex but not in the cerebellum (Supplementary Figure 5).
Mutations in the ATP7A gene have been linked to copper deficiency/Menkes disease (MD; OMIM: 309400), characterized by severe mental retardation, seizures, growth retardation, hypothermia, kinky or steely hair, laxity of skin and joints and early age mortality. More mild occipital horn syndrome (OHS) is also associated with mutations in the ATP7A gene (OMIM: 304150). Yet, neither of the clinical features of these diseases have been detected in Buryat patients lacking the second exon of the ATP7A gene. We assumed that the deletion may have a relatively modest, if any, effect on clinical phenotype in these patients, which is unrelated to previously described clinical symptoms associated with mutations in the ATP7A gene.
PGAM4 gene
Protein-coding retrogene PGAM4 (Gene ID: 441531) is localized in the first intron of the ATP7A gene and is transcribed from the opposite strand. PGAM4 is a functional retrocopy of PGAM1 gene. Phosphoglycerate mutase catalyzes the isomerization reactions of 3-phosphoglycerate to 2-phosphoglycerate in the glycolytic pathway. The gene for original functional enzyme PGAM1 is highly expressed in the brain (Illumina Human Body Map 2.0, ArrayExpress accession no. E-MTAB-513).26 PGAM4 retrogene has promoter-specific sequences of TATA boxes and CAAT box and is highly expressed in the testes. However, there is no evidence of activity of this gene in human cerebellum or cerebral cortical neurons27 (Supplementary Figure 6). This retrogene PGAM4 protein (NP_001025062, Q8N0Y7) has LxRHGExxxN motif specific for enzymatic activity of phosphoglycerate mutase family. The PGAM4 is present only in higher primates: human, chimpanzees and gorillas. The biological significance of the gene is unknown, but PGAM4 has been associated with male fertility28 and spermatogenesis.29 Interestingly, although our data indicate that the full sequence of PGAM4 retrogene was lost, the Buryat patient with cerebellar pathology and carrier of the above-described X-linked ABCB7-ATP7A-PGAM4 genetic variants was married and fathered three healthy sons. As anticipated, the genotyping of one of the sons available for this study revealed inheritance of X-chromosome wild-type allele from his mother. Given the potential role of the PGAM4 gene on male fertility, we verified genetic paternity for the patient and his sons. Using homemade and commercial informative STR markers, we confirmed the authentic biological paternity of the patient bearing the X-linked ABCB7-ATP7A-PGAM4 genetic variants. Thus, these results suggest that the naturally occurring knockout of PGAM4 does not disrupt the male fertility. However, given that all offsprings of the patient were males, the pathogenic effect of gene loss on generation or maintenance of X-chromosome-bearing spermatozoids cannot be ruled out.
Genetic heterogeneity
We found the mutation/deletion in ABCB7 and ATP7A genes in all four examined affected male relatives of this Buryat family with the same diagnosis and in four asymptomatic females relatives with the affected sons (Supplementary Table 6). The mutation/deletion was in the hemizygotic state in all examined affected males and in every female carrier it was heterozygous. All four examined healthy men from the same pedigree were hemizygous for wild-type alleles.
We further tested four unrelated families of patients with the very similar clinical manifestation and with diagnosis of congenital nonprogressive ataxia occurring exclusively in males (Supplementary Table 7). The analysis of all exons of the ABCB7 gene by direct sequencing (Supplementary Table 8) did not exhibit any differences when compared with human reference genome sequences. Similarly, we found no deletion of exon 2 of the ATP7A gene and retrogene PGAM4 in affected males in these families. The genotyping of these families for mutation NM_001001344.2:c.3321G>A (p.Gly1107Asp) in the ATP2B3 gene, which was previously described in cases of cerebellum ataxia,4 also revealed no difference from wild-type reference allele.
Discussion
The common feature among nonprogressive ataxias is a noticeable delay in early motor development and a disturbance of motor coordination during the life as a result of cerebellar hypoplasia/atrophy. Here, we report a novel mutation in ABCB7 that is a causative factor of cerebellar hypoplasia/atrophy and nonprogressive ataxia, thus further expanding the list of mutations in this gene linked to cerebellar ataxia. Previously, four different mutations found in ABCB7 were linked to cerebellar pathology with sideroblastic anemia. In sideroblastic anemia iron is not incorporated into hemoglobin as it is required to produce healthy erythrocytes by bone marrow. However, the remarkable feature of our case is that the patients have nonprogressive ataxia without the classic symptoms of sideroblastic anemia. It is tempting to speculate that this is because of a unique location of the mutation in ABCB7. Alternatively, the deletion of N-terminal part of the ATP7A may potentially have a role in modifying the genetic factor.
ABCB7 protein consists of six transmembrane domains and two intramitochondrial regions, forming NBD (Supplementary Figure 3A). It is localized in the mitochondrial inner membrane, as well as on the cytoplasmic membrane and in the cytoplasm.30 ABCB7 has an important role in maturation of cytosolic and mitochondrial Fe-S cluster proteins, affecting the function of cytochrome c, NADH dehydrogenase (nicotinamide adenine dinucleotide, the reduced form) and succinate dehydrogenase. It is involved in DNA repair and nucleotide excision repair, in oxidative DNA damage repair, in ribosome biogenesis and tRNA thiomodification.31 The mutation of ABCB7 protein in our patients is located in the domain facing the inner mitochondrial space. All other mutations described to date, causing refractory anemia and ataxia, are localized near or in the transmembrane domains.20, 32, 33, 34, 35, 36 As such, this mutation may affect its activity or binding with other co-factors, as ABCB7 controls mitochondrial iron transport and protoporphyrin binding during hemoglobin synthesis.20,32,33,36
The mental and neurological symptoms are varied in patients with described mutations in the ABCB7 gene. The depression, cognitive declines or intellectual disabilities and even schizophrenia were described for some patients (Supplementary Table 9). Neurological manifestations in the affected members of the Buryat family demonstrate some common features with the ABCB7 mutation – associated symptoms in other cases, such as cerebellar ataxia, lack of muscle weakness and deficiency of sensation (Supplementary Table 9). As shown by MRI in the Buryat family case, the brain atrophy was mostly localized in the cerebellar hemispheres and the vermis. However, we failed to detect abnormalities in other regions, including pons and medulla shown in other cases.34
The ABCB7 mutation in our patients is coupled with the deletion in ATPase copper-transporter gene (ATP7A) (Supplementary Figure 7). Surprisingly, the deletion in the evolutionarily conserved N-terminal part of ATP7A found in individuals from the Buryat family does not lead to disease manifestations described for carriers of ATP7A mutations. The disruption of this gene is linked to several clinically different disorders characterized by copper metabolic disturbance (OMIM: 300011) including MD (OMIM: 309400) and OHS (OMIM: 304150). Depending on the specific mutation, it can lead to early childhood lethality (before 3 years old), which is characterized by infantile-onset cerebral and cerebellar neurodegeneration, heavy mental retardation and arrest of development. A milder form of MD is not lethal and exhibits weaker clinical symptoms.37 The ATP7A gene mutations also lead to OHS, characterized by a connective tissue disorder and skeletal abnormalities often associated with mild mental retardation (OMIM: 304150), or to distal motor neuropathy, characterized by motor nerve degeneration in the anterior horn of the spinal cord and atrophy of lower and upper extremities (OMIM: 300489). Interestingly, the expression of ATP7A protein is inversely correlated with the severity of the disease phenotype.38 Despite the loss of the first of six MBDs of ATP7A, the patients from the Buryat family did not show any clinical signs of the ATP7A-associated disorders reported to date. Among the mutation spectrum described for ATP7A gene, the gross deletions were represented in ~15% of the MD patients.39 Thus, it is tempting to speculate that in males carrying the ATP7A deletion of the first MBD, the remaining five copper-binding domains are efficiently used to retain the function of ATP7A.
We cannot rule out that the dysfunction in ABCB7 and modestly modified activity of ATP7A may mutually affect homeostasis in some tissue cells, as ABCB7 impairment may cause an accumulation of iron ions in the mitochondria, whereas a mutation of ATP7A transporter may affect copper ion levels in the cell cytoplasm. The iron deficiency increases intestinal tissue expression of ATP7A40 and brain copper levels.41 Copper is known to regulate iron utilization in the bone marrow and in hemoglobin synthesis.42, 43, 44, 45 Importantly, both ABCB7 and ATP7A genes are expressed in the bone marrow26, 46 and in various tissues (Illumina Human Body Map 2.0),26, 46, 47 (Supplementary Figure 5). Given the mutual regulatory influence of the copper and iron uptake, the clinical phenotype described in the Buryat patients may depend on presumable interaction of genetic defects in two genes for the MBPs in some tissue cells, an interesting case of syndrome inherited as a X-linked monogenic trait. We, however, argue that the major cause of cerebellar hypoplasia and the clinical phenotype of ataxia in these patients is caused by this novel mutation in the ABCB7 gene.
Acknowledgments
This work was supported by the Government of the Russian Federation (No. 14.B25.31.0033). We thank Oleg Balanovsky and Michail Ignashkin for assistance with ancestry and paternity analysis and Walter J Lukiw, Arya Byragin and anonymous reviewers for providing helpful comments on the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Bergmann C, Zerres K, Senderek J et al: Oligophrenin 1 (OPHN1 gene mutation causes syndromic X-linked mental retardation with epilepsy, rostral ventricular enlargement and cerebellar hypoplasia. Brain 2003; 126: 1537–1544. [DOI] [PubMed] [Google Scholar]
- Najm J, Horn D, Wimplinger I et al: Mutations of CASK cause an X-linked brain malformation phenotype with microcephaly and hypoplasia of the brainstem and cerebellum. Nat Genet 2008; 40: 1065–1067. [DOI] [PubMed] [Google Scholar]
- Nadif Kasri N, Nakano-Kobayashi A, Malinow R, Li B, Van Aelst L: The Rho-linked mental retardation protein oligophrenin-1 controls synapse maturation and plasticity by stabilizing AMPA receptors. Genes Dev 2009; 23: 1289–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanni G, Calì T, Kalscheuer VM et al: Mutation of plasma membrane Ca2+ ATPase isoform 3 in a family with X-linked congenital cerebellar ataxia impairs Ca2+ homeostasis. Proc Natl Acad Sci USA 2012; 109: 14514–14519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramins M, Colebatch JG, Bainbridge MN et al: Exome sequencing identification of a GJB1 missense mutation in a kindred with X-linked spinocerebellar ataxia (SCA-X1). Hum Mol Genet 2013; 22: 4329–4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illarioshkin SN, Tanaka H, Markova ED, Nikolskaya NN, Ivanova-Smolenskaya IA, Tsuji S: X-linked nonprogressive congenital cerebellar hypoplasia: clinical description and mapping to chromosome Xq. Ann Neurol 1996; 40: 75–83. [DOI] [PubMed] [Google Scholar]
- Van Oven M, Kayser M: Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat 2009; 30: E386–E394. [DOI] [PubMed] [Google Scholar]
- Derenko M, Malyarchuk B, Grzybowski T et al: Origin and post-glacial dispersal of mitochondrial DNA haplogroups C and D in northern Asia. PLoS One 2010; 5: e15214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Lott MT, Procaccio V et al: An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res 2007; 35: D823–D828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rootsi S, Zhivotovsky LA, Baldovic M et al: A counter-clockwise northern route of the Y-chromosome haplogroup N from Southeast Asia towards Europe. Eur J Hum Genet 2007; 15: 204–211. [DOI] [PubMed] [Google Scholar]
- International Society of Genetic Genealogy Y-DNA Haplogroup Tree 2014, Version: 9.117, 9 November 2014. Available at: http://www.isogg.org/tree/ (accessed 13 November 2014).
- Kharkov VN, Khamina KV, Medvedeva OF, Simonova KV, Eremina ER, Stepanov VA: Gene pool of Buryats: clinal variability and territorial subdivision based on data of Y-chromosome markers. Russ J Genet 2014; 50: 180–190. [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E et al: The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z: Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009; 25: 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium, Abecasis GR, Auton A et al: An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding S-L, Sunkin SM et al: Transcriptional landscape of the prenatal human brain. Nature 2014; 508: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz M, Ng L, Feng D, Sunkin S, Szafer A, Dang C The Allen Brain Atlas; in: Kasabov N (ed): Springer Handbook of Bio-/Neuroinformatics. Berlin, Heidelberg, Germany: Springer, 2014, pp 1111–1126. [Google Scholar]
- Stamm S, Zhu J, Nakai K, Stoilov P, Stoss O, Zhang MQ: An alternative-exon database and its statistical analysis. DNA Cell Biol 2000; 19: 739–756. [DOI] [PubMed] [Google Scholar]
- Nurtdinov RN, Neverov AD, Mal'ko DB et al: EDAS, databases of alternatively spliced human genes. Biofizika 2006; 51: 589–592. [PubMed] [Google Scholar]
- Allikmets R, Raskind WH, Hutchinson A et al: Mutation of a putative mitochondrial iron transporter gene (ABC7 in X-linked sideroblastic anemia and ataxia (XLSA/A). Hum Mol Genet 1999; 8: 743–749. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJE: Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 2009; 4: 363–371. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC: Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P et al: A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamov AA, Nishikawa T, Swindells MB: Assessing protein coding region integrity in cDNA sequencing projects. Bioinformatics 1998; 14: 384–390. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium: Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res 2014; 42: D191–D198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapushesky M, Emam I, Holloway E et al: Gene expression atlas at the European bioinformatics institute. Nucleic Acids Res 2010; 38: D690–D698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulha HP, Crisci JL, Reshetov D et al: Human-specific histone methylation signatures at transcription start sites in prefrontal neurons. PLoS Biol 2012; 10: e1001427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda H, Tsujimura A, Irie S et al: A single nucleotide polymorphism within the novel sex-linked testis-specific retrotransposed PGAM4 gene influences human male fertility. PLoS One 2012; 7: e35195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q, Pan H, Wang B et al: The PGAM4 gene in non-obstructive azoospermia. Syst Biol Reprod Med 2013; 59: 179–183. [DOI] [PubMed] [Google Scholar]
- Zutz A, Gompf S, Schägger H, Tampé R: Mitochondrial ABC proteins in health and disease. Biochim Biophys Acta 2009; 1787: 681–690. [DOI] [PubMed] [Google Scholar]
- Hollenstein K, Dawson RJP, Locher KP: Structure and mechanism of ABC transporter proteins. Curr Opin Struct Biol 2007; 17: 412–418. [DOI] [PubMed] [Google Scholar]
- Bekri S, Kispal G, Lange H et al: Human ABC7 transporter: gene structure and mutation causing X-linked sideroblastic anemia with ataxia with disruption of cytosolic iron-sulfur protein maturation. Blood 2000; 96: 3256–3264. [PubMed] [Google Scholar]
- Maguire A, Hellier K, Hammans S, May A: X-linked cerebellar ataxia and sideroblastic anaemia associated with a missense mutation in the ABC7 gene predicting V411L. Br J Haematol 2001; 115: 910–917. [DOI] [PubMed] [Google Scholar]
- Hellier KD, Hatchwell E, Duncombe AS, Kew J, Hammans SR: X-linked sideroblastic anaemia with ataxia: another mitochondrial disease? J Neurol Neurosurg Psychiatr 2001; 70: 65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagon RA, Bird TD, Detter JC, Pierce I: Hereditary sideroblastic anaemia and ataxia: an X linked recessive disorder. J Med Genet 1985; 22: 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Hooghe M, Selleslag D, Mortier G et al: X-linked sideroblastic anemia and ataxia: a new family with identification of a fourth ABCB7 gene mutation. Eur J Paediatr Neurol 2012; 16: 730–735. [DOI] [PubMed] [Google Scholar]
- Tchan MC, Wilcken B, Christodoulou J: The mild form of Menkes disease: a 34 year progress report on the original case. JIMD Rep 2013; 9: 81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donsante A, Tang J, Godwin SC et al: Differences in ATP7A gene expression underlie intrafamilial variability in Menkes disease/occipital horn syndrome. J Med Genet 2007; 44: 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tümer Z, Birk Møller L, Horn N: Screening of 383 unrelated patients affected with Menkes disease and finding of 57 gross deletions in ATP7A. Hum Mutat 2003; 22: 457–464. [DOI] [PubMed] [Google Scholar]
- Jiang L, Ranganathan P, Lu Y, Kim C, Collins JF: Exploration of the copper-related compensatory response in the Belgrade rat model of genetic iron deficiency. Am J Physiol Gastrointest Liver Physiol 2011; 301: G877–G886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnot AD, Behl M, Ho S, Zheng W: Regulation of brain copper homeostasis by the brain barrier systems: effects of Fe-overload and Fe-deficiency. Toxicol Appl Pharmacol 2011; 256: 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyatskowit JW, Prohaska JR: Iron injection restores brain iron and hemoglobin deficits in perinatal copper-deficient rats. J Nutr 2008; 138: 1880–1886. [DOI] [PubMed] [Google Scholar]
- Reeves PG, DeMars LCS: Signs of iron deficiency in copper-deficient rats are not affected by iron supplements administered by diet or by injection. J Nutr Biochem 2006; 17: 635–642. [DOI] [PubMed] [Google Scholar]
- Pyatskowit JW, Prohaska JR: Copper deficient rats and mice both develop anemia but only rats have lower plasma and brain iron levels. Comp Biochem Physiol C 2008; 147: 316–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JF, Prohaska JR, Knutson MD: Metabolic crossroads of iron and copper. Nutr Rev 2010; 68: 133–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizio M, Harshbarger J, Shimoji H et al: Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol 2015; 16: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L, Hallström BM, Oksvold P et al: Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteom 2014; 13: 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS et al: The human genome browser at UCSC. Genome Res 2002; 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP et al: ClustalW and ClustalX version 2. Bioinformatics 2007; 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.