Abstract
Intellectual disability (ID) affects 2–3% of the population. In the past, many genetic causes of ID remained unidentified due to its vast heterogeneity. Recently, whole exome sequencing (WES) studies have shown that de novo variants underlie a significant portion of sporadic cases of ID. Applying WES to patients with ID or global developmental delay at different centers, we identified three individuals with distinct de novo variants in HIVEP2 (human immunodeficiency virus type I enhancer binding protein), which belongs to a family of zinc-finger-containing transcriptional proteins involved in growth and development. Two of the variants were nonsense changes, and one was a 1 bp deletion resulting in a premature stop codon that was reported previously without clinical detail. In silico prediction programs suggest loss-of-function in the mutated allele leading to haploinsufficiency as a putative mechanism in all three individuals. All three patients presented with moderate-to-severe ID, minimal structural brain anomalies, hypotonia, and mild dysmorphic features. Growth parameters were in the normal range except for borderline microcephaly at birth in one patient. Two of the patients exhibited behavioral anomalies including hyperactivity and aggression. Published functional data suggest a neurodevelopmental role for HIVEP2, and several of the genes regulated by HIVEP2 are implicated in brain development, for example, SSTR-2, c-Myc, and genes of the NF-κB pathway. In addition, HIVEP2-knockout mice exhibit several working memory deficits, increased anxiety, and hyperactivity. On the basis of the genotype–phenotype correlation and existing functional data, we propose HIVEP2 as a causative ID gene.
Introduction
HIVEP2 (human immunodeficiency virus type I enhancer binding protein 2) is a large transcription factor with diverse roles in growth and development.1 It is responsible for regulating the activity of a number of different genes, many of which are implicated in brain development. These include somatostatin receptor II (SSTR-2),2 c-Myc,3 and genes in the NF-κB pathway.4 In addition to its involvement in the brain, HIVEP2 has a key role in systemic processes, including cellular immunity,5, 6, 7 bone remodeling,8 and adipogenesis.9
Only one de novo variant (c.5737del; p.Asp1913Metfs*15) in the gene encoding HIVEP2 has been reported to date in a whole exome sequencing (WES) study of 51 patients with non-syndromic sporadic severe intellectual disability (ID), with limited clinical information.10
In this report, we present two previously unpublished patients with HIVEP2 variants identified by WES, and detailed clinical information on the previously published patient. The three patients with different, likely pathogenic de novo variants in HIVEP2 all had global developmental delay (GDD) or ID, varying minimal structural brain anomalies including hypoplasia of corpus callosum or frontal atrophy, hypotonia, mild dysmorphic features, and in two instances behavioral abnormalities. We characterize the clinical features of these patients and suggest that variants in HIVEP2 gene are a cause of ID.
Materials and Methods
For Patient 1, her parents, and her unaffected brother, WES was performed by GeneDx Laboratory using the SureSelect XT2 All Exon V4 kit (Agilent, Santa Clara, CA, USA) and HiSeq 2000 system (Illumina, San Diego, CA, USA). The human reference genome (hg19, UCSC) was the reference sequence. Sequence changes in the patient were compared against other family members using XomeAnalyzer. All sequence variants were confirmed by Sanger sequencing.
For Patient 2, WES was performed by CeGaT Laboratory using the SureSelect XT V5 Exome kit (Agilent, Santa Clara, CA, USA) and HiSeq 2500 system (Illumina, San Diego, CA, USA) with 100 bp paired-end runs. The human reference genome (hg19, UCSC) was used as reference sequence. Confirmation of the variant in HIVEP2 (according to NM_006734.3; http://www.ncbi.nlm.nih.gov/nuccore/NM_006734) and its de novo origin was done by Sanger sequencing.
Patient 3 and her parents were analyzed as published previously10 using data generated with the SureSelect XT Human All Exon 50 Mb kits (Agilent) and HiSeq 2000 system (Illumina) with 100 bp paired-end runs. Presence of the sequence variant in the patient and its absence in the parents were confirmed by Sanger sequencing.
Genotype and phenotype data were submitted to Leiden Open Variation Database (http://www.lovd.nl/3.0/; patient IDs 00038503–00038505).
Clinical reports
Patient 1
Patient 1 (Figures 1b and c; Table 1) presented for neurodevelopmental evaluation when she was 3 years old. She is the second child of a non-consanguineous, healthy Caucasian couple with an unremarkable family history. Her prenatal course and newborn period were marked by various difficulties. The pregnancy was complicated by first-trimester vaginal bleeding treated with progesterone. Fetal movements, fetal ultrasound investigations, and amniotic fluid volume were normal. She was born at 37+5 weeks of gestation by normal spontaneous vaginal delivery, weighing 3150 g (other birth parameters unavailable). Immediately at birth, she had an asymmetric cry with accompanying weakness of her left lower lip and chin. In addition, she had physiological jaundice and left hip dysplasia requiring placement in a harness for several months.
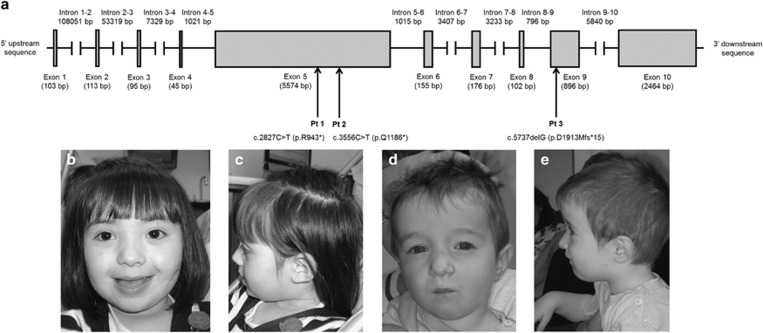
Figure 1.
Spectrum of HIVEP2 variants in the three patients. (a) Exon and intron region sizes are based on Ensembl (http://useast.ensembl.org/Homo_sapiens/Transcript/Exons?db=core;g=ENSG00000010818;r=6:142751467-142945201;t=ENST00000367603). (b) Portrait and (c) profile view of patient 1 at 4 years of age. (d) Portrait and (e) profile view of patient 2 at 26 months of age.
Table 1. Clinical characteristics of patients with HIVEP2 variants.
| Patient 1 | Patient 2 | Patient 3 | |
|---|---|---|---|
| Gender | Female | Male | Female |
| Age | 4 years 7 months | 3 years 9 months | 21 years |
| Variant | c.2827C>T (p.R943*) | c.3556C>T (p.Q1186*) | c.5737delG (p.D1913Mfs*15) |
| Inheritance | de novo | de novo | de novo |
| Growth | |||
| Birth parameters | Normal | Normal | Slight microcephaly |
| Height centile | 70th | 25th | 25–50th |
| Weight centile | 85th | 25–50th | 25–50th |
| OFC centile | 10th | 50th | 3rd |
| Neurological features | |||
| Motor development | Moderate delay; walked at 36 months | Moderate delay; walked at 30 months | Moderate delay; walked at 36 months |
| Cognitive | Moderate-severe ID | Moderate ID | Moderate-to-severe ID |
| Speech | Delayed; first words at 24 months | Delayed; first words at 24 months | Delayed; first words at 7.5 years |
| Behavioral anomalies | Hyperactivity, impulsivity, distractibility (reported later at 4 years 7 months of age) | — | Aggressive and impulsive behavior, self-stimulation, hyperactivity |
| Hypotonia | Mild | Moderate | Mild |
| Other neurological features | Frequent head tilt and leftward eye movements with preserved ability to fix/follow objects Subtle left facial weakness | Ataxic gait | |
| Head CT/brain MRI findings | Slightly thin corpus callosum | Hypoplasia of corpus callosum | Mild frontal atrophy |
| Dysmorphic features | |||
| Forehead | High forehead | ||
| Eyebrows | Medial eyebrow flare | Minor synophrys | |
| Eyes | Widely set eyes | Widely set eyes | Slightly upslanting palpebral fissures |
| Ears | Small ears with attached earlobes | ||
| Nose | Broad nasal root Slightly upturned nose | Broad nasal root | Prominent nose with high nasal bridge and columella extending below the alae nasi |
| Lips/Mouth | High-arched palate | Small mouth | Short philtrum Thin upper lip, overbite |
| Chin | Small chin | ||
| Extremities | Slightly tapering fingers | Tapering fingers Radial deviation of 4th fingers Wide feet with short toes | |
| Skin | Hirsutism | ||
| Systemic malformations | — | — | Minor pulmonary stenosis |
| Other features | Asymmetric cry Unilateral hip dysplasia Dysphagia | Hypermetropia | Chronic constipation Gingival swelling and bleeding High TSH levels Umbilical hernia |
Abbreviations: ID, intellectual disability; OFC, occipitofrontal circumference.
She exhibited global delays in development from an early age. She was hypotonic with poor head control at 4 months of age. She was not able to lift her neck up in the prone position for another 2 months. Eventually, she crawled at 22 months, cruised at 24 months, and walked independently at 36 months. She said her first word besides ‘mama' and ‘dada' and developed a vocabulary of about 10 words by 24 months of age. At the age of 4 years, the family expressed concerns for hyperactivity, impulsivity and distractibility possibly out of proportion to her developmental age.
When she was 2 years old, she was hospitalized for gastroparesis and dehydration after presenting with projectile vomiting, which resolved after treatment with erythromycin. Because of the concern for dysphagia, she received thickened liquids by mouth, but this requirement relaxed with time.
On examination at the age of 3 years 6 months, she had normal growth parameters: weight 17.1 kg (85th centile), height 100 cm (70th centile), and head circumference 48 cm (10th centile). She had mild craniofacial dysmorphism, including hypertelorism, broad nasal root, slightly upturned nose, and high-arched palate. The remainder of her general examination was normal. On neurological exam, she was social and affectionate. She had frequent head tilt and eye movement to the left but was able to fixate and follow objects without evidence for ocular motor apraxia and/or nystagmus. She had subtle weakness of her left chin manifesting as a very mild asymmetric smile. Muscle tone in her upper and lower extremities was decreased, but strength was full. Her gait was wide based. Developmental testing at the age of 3 years revealed a language age equivalent of 15 months (developmental quotient around 40%) and problem-solving age equivalent of 12 months (developmental quotient around 33%).
Brain MRI showed slightly thin corpus callosum. Routine electroencephalogram (EEG) was normal. Normal genetic testing included single-nucleotide polymorphism (SNP) array, methylation testing for Angelman syndrome, and MLL2 and MECP2 gene sequencing and multiplex ligation-dependent probe amplification (MLPA) analysis. Serum creatine kinase was slightly elevated (300 units/l) but repeat measurement was normal. Intestinal biopsy for disaccharidase levels was normal. Because of extensive unrevealing prior workup, she underwent WES.
Patient 2
Patient 2 (Figures 1d and e; Table 1) presented for genetic evaluation at the age of 17 months. He is the fourth child of a non-consanguineous, healthy German couple. The patient was born after 39 weeks of gestation with a length of 53 cm (75–90th centile), a weight of 3330 g (25–50th centile), and a head circumference of 35 cm (25–50th centile). No congenital anomalies were apparent, but the boy showed remarkable hypotonia and failure to thrive. One of his three older brothers has well-controlled seizures; otherwise, the family history is unremarkable.
After 6 months of age, delay in his psychomotor development became obvious. He learned to sit without support at the age of 13 months and walked independently at 30 months. He spoke his first words at the age of ~24 months, had an active vocabulary of 10 words at the age of 29 months and of 50 words at the age of 3 years and 9 months. He could communicate well through gesture and sign language. His receptive language skills were better than his expressive language skills. Because of hypermetropia, the patient required glasses, which he did not tolerate wearing.
On examination at the age of 17 months, his height was 80 cm (25th centile), his weight was 11 kg (25–50th centile) and his head circumference was 48 cm (50th centile). He had mild dysmorphism, such as a high forehead, medial eyebrow flare, widely set eyes, broad nasal root, small mouth, slightly tapering fingers, and flat feet. Moreover, he showed remarkable hypotonia along with significant salivation. Otherwise, the general examination was normal, and he was a very social and cheerful boy. At the age of 3 years and 9 months, he still had muscular hypotonia and a clumsy gait. Otherwise neurological examination was unremarkable.
Brain MRI showed hypoplasia of the corpus callosum. Before undergoing WES, he had an extensive workup that was unremarkable, including routine EEG, complete metabolic profile, karyotype, array comparative genomic hybridization, and ZFHX1B sequencing and MLPA analysis to evaluate for Mowat–Wilson syndrome.
Patient 3
Patient 3 (Table 1) presented for genetic evaluation at the age of 21 years. She is the third child of healthy non-consanguineous German parents with unremarkable family history. After an uneventful pregnancy, she was born at term by normal spontaneous vaginal delivery with low normal length (50 cm; 25th centile) and weight (2750 g; 3rd–10th centile), and microcephaly (32 cm; <3rd centile). A small umbilical hernia and mild pulmonary stenosis were diagnosed at the age of 1 and 6 months, respectively.
Global developmental delays became apparent at the age of ~1 year. The patient was able to sit at the age of 9 months and walk without support at the age of 3 years. Her gait was ataxic. Hypotonia and dystonia were diagnosed between 2 and 3 years of age. She uttered her first syllables at the age of ~6 years and spoke her first words at the age of 7 years after enrollment in a special needs school.
At the age of 21 years, she used longer sentences but had partially slurred articulation. She did not show regression in motor or speech development. Her ID was in the moderate range. She exhibited behavioral anomalies, such as aggression, impulsivity, self-stimulation, and hyperactivity. In addition to these neurodevelopmental features, she had constipation and hypertrophic gingiva with frequent bleeding.
On examination at the age of 21 years, height (164 cm; 25th–50th centile) and weight (59 kg; 25th–50th centile) were normal. She presented with microcephaly with a head circumference of 52.2 cm (3rd centile). She had minor dysmorphism, including upslanting palpebral fissures, mild synophrys, small ears with attached earlobes, a prominent nose with a high nasal bridge and a columella extending below the alae nasi, a short philtrum, a thin upper lip, an overbite, hypertrophic gingiva, tapering fingers, radial deviation of 4th fingers, wide feet with short toes, and hirsutism. Her feet were rotated inwards. Muscle tone in her upper and lower extremities was decreased. Her gait was clumsy.
Before undergoing WES, conventional karyotyping, MECP2 sequencing, and chromosomal microarray analysis using an Illumina HumanOmni1-Quad SNP array were normal. Head computed tomography at the age of 2 years showed mild frontal brain atrophy. EEGs were reportedly normal at different ages. Patient 3 was published previously without clinical details as BO63/11.10
Results
In these three patients, WES revealed two nonsense variants (Patients 1 and 2) and one frameshift variant (Patient 3) in HIVEP2 (NM_006734.3; http://www.ncbi.nlm.nih.gov/nuccore/NM_006734; Figure 1a). Patient 1 had a heterozygous de novo nonsense variant in exon 5 of HIVEP2: p.R943* (c.2827C>T). Patient 2 had a heterozygous de novo nonsense variant in exon 5 of HIVEP2: p.Q1186* (c.3556C>T). Patient 3 had a heterozygous de novo frameshift variant in exon 9 of HIVEP2 (c.5737delG) resulting in a preliminary stop codon at p.D1913Mfs*15.
In silico tools assessed the deleterious nature of these variants. MutationTaster (http://www.mutationtaster.org) classified all three variants as damaging and to cause nonsense mediated mRNA decay (NMD). SIFT (http://sift.jcvi.org) predicted a damaging effect and NMD for the frameshift variant in Patient 3. None of the variants was present in 2500 in-house control exomes or the Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org/). The Residual Variation Intolerance (RVI) score is a quantitative measure that ranks genes based on their intolerance to variation.11 The RVI score of HIVEP2 was −2.2 (1.37th percentile) and thus considerably lower than the average RVI score for genes known to cause developmental disorders (−0.56; 19.54th percentile).
Discussion
We report on three unrelated individuals with ID or GDD and loss-of-function variants of HIVEP2. We propose that HIVEP2 is a novel gene causing ID/GDD based on the following lines of evidence:
Individuals with de novo HIVEP2 loss-of-function variants share a non-specific and unrecognizable phenotype of ID, hypotonia, mild dysmorphic features, and variable minor structural brain anomalies
The three patients presented here represent the first detailed clinical descriptions of neurodevelopmental phenotypes associated with HIVEP2 variants. Clinical similarities between the three patients include global developmental delay and moderate ID. In addition, hypotonia may be a prominent part of the clinical presentation, leading to delays in achieving motoric milestones with clumsy or wide-based gait. All three patients showed a remarkable delay in speech development. However receptive language skills appeared to be less delayed. Growth parameters in these patients are within the normal range except a mild primary microcephaly in patient 3. Although congenital malformations and severe dysmorphisms are not part of the presentation, the three described patients share some minor dysmorphic features including hypertelorism, a broad nasal bridge and tapering fingers, seen in two out of the three patients. Nevertheless, these dysmorphic features do not constitute a recognizable phenotype. Hypoplasia of the corpus callosum was apparent on MRI in patients 1 and 2, whereas patient 3 showed mild frontal atrophy. Behavioral abnormalities like hyperactivity, impulsivity and aggression, seen in two out of the three patients, were a prominent feature. Because the phenotype is non-specific, it may be difficult for clinicians to decide to send targeted sequencing of HIVEP2. Instead, in cases like these with ID, a next-generation sequencing-based approach, ideally involving trios, may be advantageous.
HIVEP2 is a functionally convincing candidate gene
HIVEP2 (also known as Schnurri-2; MHC-binding protein 2/MBP2; C-Myc Intron Binding Protein 1; http://www.ncbi.nlm.nih.gov/gene/3097) belongs to a family of zinc-finger-containing transcriptional proteins involved in growth and development.1 In Drosophila, Schnurri is a downstream target of the decapentaplegic signal transduction pathway, which has an important role in embryonic patterning.12, 13 The vertebrate equivalent of decapentaplegic signaling is the transforming growth factor-β/bone morphogenetic protein (TGFβ/BMP) pathway, responsible for various developmental functions, including pituitary hormone production and bone and cartilage creation.14 Our patients were not examined for evidence of these features.
The mechanisms linking HIVEP2 to neurodevelopmental dysfunction are not entirely clear, but one possibility is a dysregulation of upstream and downstream signaling pathways. HIVEP2 regulates the activity of a number of different genes, many of which are implicated in brain development. HIVEP2 activates expression of SSTR-2.2 SSTR-2 is one of several G-protein-coupled receptors that mediate the effects of the regulatory peptide somatostatin throughout the central nervous system (CNS) and rest of the body, with effects on neurotransmission, endocrine function, and cell growth and secretion.15 Activation of SSTR-2 actually involves an additional component, transcription factor 4 (TCF4),2 defects in which are associated with Pitt–Hopkins syndrome. In fact, HIVEP2 and TCF4 are binding partners, and they are co-expressed during development in different parts of the brain important for learning, including the frontal cortex and hippocampus,2 overlapping with expression of SSTR-2. However, the clinical phenotype of Pitt–Hopkins syndrome does not show similarities with that of our patients except for ID/GDD. HIVEP2 represses transcription of c-Myc.3 c-Myc is a proto-oncogene that regulates aspects of cell growth, differentiation, and apoptosis,16 and Myc-regulated pathways are essential for normal brain growth and development.17 Finally, HIVEP2 represses transcription of genes in the NF-κB pathway.4 NF-κB is a ubiquitously expressed transcription factor that has a role in immunity, inflammation, and brain function, including synaptic transmission, neuronal plasticity, and neuronal development.18, 19 Apart from HIVEP2 affecting downstream pathways that mediate cell immunity and inflammation, there is evidence of more direct effects of HIVEP2 on these processes. HIVEP2 is important for activation and function of natural killer cells.7 In CD4+CD8+ double thymocytes, HIVEP2 subdues the mitochondrial death pathway in response to T-cell antigen receptor signals, thereby serving as an essential regulator of T-cell development.6 Survival of memory Th1/Th2 cells depends on the action of HIVEP2 on NF-κB.5
In the HIVEP2-knockout mouse model, there is evidence of brain inflammation, based on upregulation of genes involved in immune response and astrocyte activation in the hippocampus. Interestingly, treatment of the mice with anti-inflammatory agents both decreased astrocyte activation and improved some of the neurobehavioral features, including working memory.20 If an evaluation for astrocyte response and immune dysfunction in patients with HIVEP2 variants yielded evidence of CNS inflammation, the fact that animal models showed a positive response to anti-inflammatory drugs may open the window for investigation of therapeutic options in affected human patients.
The behavioral symptoms (particularly hyperactivity) present in our two older patients have been captured in related mouse models. When presented with novel situations, HIVEP2-knockout mice exhibit increased anxiety, accompanied by higher stress-induced corticosterone levels, compared with wild-type mice. In familiar environments, these mice also have greater locomotor activity than controls.21
In addition to anxiety and hyperactivity, HIVEP2-knockout mice show working memory deficits, reduced prepulse inhibition (PPI; in which the reaction induced by a startling stimulus is attenuated by a weak pre-stimulus), and impaired sociability, which are features seen in schizophrenia. In support of this, there is significant commonality in the expression profile of different genes in the medial prefrontal cortex (mPFC) of the HIVEP2-knockout mice compared with that of patients with schizophrenia. In addition, immunohistochemistry studies on the mutant mice show reduced expression of parvalbumin in the mPFC and area CA1 of the hippocampus, as well as reduced expression of glutamic acid decarboxylase 67 (GAD67) in the hippocampus. These alterations have been associated with schizophrenia in humans. Intriguingly, treatment of these mice with the antipsychotic haloperidol, used to treat schizophrenia, improved both the hyperactivity and PPI.20 This therapy has not been tried for our patients for the behavioral features, but given the animal model data it may be judicious to consider haloperidol in future trials of medications for aberrant behavior in humans with HIVEP2 variants.
On neuropathology, the HIVEP2-knockout mouse models show defects in neuronal maturation, neuronal growth, and dopaminergic receptor signaling that may explain some of the cognitive deficits seen in our patients. Specifically, the mutant mice demonstrate deficits in maturation of hippocampal dentate gyrus (DG) neurons, with increased numbers of immature neurons and decreased numbers of mature neurons. In addition, these mice have thinner cortices including the prelimbic cortex and primary visual cortex. Finally, in the DG, they exhibit decreased levels of D1 dopamine receptors.20 If such features are also true in patients with HIVEP2 variants, then a thin cortex, preponderance of immature neurons, and altered neurotransmitter signaling may serve as hypotheses for some of the etiological mechanisms of their ID and behavioral abnormalities. Consideration can be given toward CSF neurotransmitter profile analysis in patients with HIVEP2 variants. A thin prelimbic cortex would suggest dysregulated modulation of limbic functions, such as fear and stress, and may also account for some of the underlying behaviors in our patients, as dysregulated fear and stress can lead to impulsivity, hyperactivity, and aggression.
The HIVEP2 variants are likely deleterious
There are several lines of evidence suggesting that the two nonsense and one frameshift variant in the patients are deleterious. First, in silico tools predict these alterations to be damaging and to cause NMD, making haploinsufficiency the probable mechanism of action. Second, these changes are rare and de novo, and they have not been reported in in-house exomes from control patients or in public data sets such as the ExAC. Third, the RVI score of HIVEP2 is more negative than the average RVI score for a selection of genes associated with developmental disorders (−0.56; 19.54th percentile).11 On the basis of this, HIVEP2 seems to be even less resilient to deleterious changes than other genes implicated in developmental disorders.
Conclusions
In summary, we present three patients with de novo HIVEP2 variants, two nonsense and one frameshift, that are associated with moderate ID, minor brain abnormalities on neuroimaging, muscular hypotonia, and mild craniofacial dysmorphism. On the basis of the role that HIVEP2 has in the developing brain, and the deleterious nature and de novo occurrence of the variants, these genetic modifications are likely causative of the patients' clinical manifestations. Functional studies will be needed to confirm the pathophysiology of these variants as it relates to ID, behavioral abnormalities, and inflammation which may be addressed with rational therapeutics.
Acknowledgments
We thank the patients and their families for their patience and time. This work was supported in part by the German Ministry of Research and Education (grant numbers 01GS08164, 01GS08167, 01GS08163, German Mental Retardation Network/MRNET) as part of the National Genome Research Network.
The authors declare no conflict of interest.
References
- Wu L-C: ZAS: C2H2 zinc finger proteins involved in growth and development. Gene Expr 2002; 10: 137–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörflinger U, Pscherer A, Moser M, Rümmele P, Schüle R, Buettner R: Activation of somatostatin receptor II expression by transcription factors MIBP1 and SEF-2 in the murine brain. Mol Cell Biol 1999; 19: 3736–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S, Yamasaki Y, Iwaki T et al: Characterization of the biological functions of a transcription factor, c-myc intron binding protein 1 (MIBP1). J Biochem 2002; 131: 349–357. [DOI] [PubMed] [Google Scholar]
- Iwashita Y, Fukuchi N, Waki M, Hayashi K, Tahira T: Genome-wide repression of NF-κB target genes by transcription factor MIBP1 and Its modulation by O-Linked β-N-acetylglucosamine (O-GlcNAc) transferase. J Biol Chem 2012; 287: 9887–9900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Kimura MY: Memory Th1/Th2 cell generation controlled by Schnurri-2. Adv Exp Med Biol 2010; 684: 1–10. [DOI] [PubMed] [Google Scholar]
- Staton TL, Lazarevic V, Jones DC et al: Dampening of death pathways by schnurri-2 is essential for T-cell development. Nature 2011; 472: 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita J, Iwamura C, Mitsumori K et al: Murine Schnurri-2 controls natural killer cell function and lymphoma development. Leuk Lymphoma 2012; 53: 479–486. [DOI] [PubMed] [Google Scholar]
- Saita Y, Takagi T, Kitahara K et al: Lack of Schnurri-2 expression associates with reduced bone remodeling and osteopenia. J Biol Chem 2007; 282: 12907–12915. [DOI] [PubMed] [Google Scholar]
- Jin W, Takagi T, Kanesashi S et al: Schnurri-2 controls BMP-dependent adipogenesis via interaction with Smad proteins. Dev Cell 2006; 10: 461–471. [DOI] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E et al: Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 2012; 380: 1674–1682. [DOI] [PubMed] [Google Scholar]
- Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB: Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet 2013; 9: e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora K, Dai H, Kazuko SG et al: The Drosophila schnurri gene acts in the Dpp/TGF beta signaling pathway and encodes a transcription factor homologous to the human MBP family. Cell 1995; 81: 781–790. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton K, Laughon AS, Hoffmann FM: A Drosophila protein related to the human zinc finger transcription factor PRDII/MBPI/HIV-EP1 is required for dpp signaling. Dev Camb Engl 1995; 121: 3393–3403. [DOI] [PubMed] [Google Scholar]
- Kingsley DM: The TGF-beta superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev 1994; 8: 133–146. [DOI] [PubMed] [Google Scholar]
- Patel YC: Somatostatin and its receptor family. Front Neuroendocrinol 1999; 20: 157–198. [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan G: c-MYC: more than just a matter of life and death. Nat Rev Cancer 2002; 2: 764–776. [DOI] [PubMed] [Google Scholar]
- Wey A, Knoepfler PS: c-myc and N-myc promote active stem cell metabolism and cycling as architects of the developing brain. Oncotarget 2010; 1: 120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltschmidt B, Kaltschmidt C: NF-kappaB in the nervous system. Cold Spring Harb Perspect Biol 2009; 1: a001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LA, Kaltschmidt C: NF-kappa B a crucial transcription factor for glial and neuronal cell function. Trends Neurosci 1997; 20: 252–258. [DOI] [PubMed] [Google Scholar]
- Takao K, Kobayashi K, Hagihara H et al: Deficiency of schnurri-2, an MHC enhancer binding protein, induces mild chronic inflammation in the brain and confers molecular, neuronal, and behavioral phenotypes related to schizophrenia. Neuropsychopharmacolgy 2013; 38: 1409–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi T, Jin W, Taya K, Watanabe G, Mori K, Ishii S: Schnurri-2 mutant mice are hypersensitive to stress and hyperactive. Brain Res 2006; 1108: 88–97. [DOI] [PubMed] [Google Scholar]