Abstract
Variants in CIB2 can underlie either Usher syndrome type I (USH1J) or nonsyndromic hearing impairment (NSHI) (DFNB48). Here, a novel homozygous missense variant c.196C>T and compound heterozygous variants, c.[97C>T][196C>T], were found, respectively, in two unrelated families of Dutch origin. Besides, the previously reported c.272 T>C functional missense variant in CIB2 was identified in two families of Pakistani origin. The missense variants are demonstrated not to affect subcellular localization of CIB2 in vestibular hair cells in ex vivo expression experiments. Furthermore, these variants do not affect the ATP-induced calcium responses in COS-7 cells. However, based on the residues affected, the variants are suggested to alter αIIβ integrin binding. HI was nonsyndromic in all four families. However, deafness segregating with the c.272T>C variant in one Pakistani family is remarkably less severe than that in all other families with this mutation. Our results contribute to the insight in genotype–phenotype correlations of CIB2 mutations.
Introduction
Genetic defects are important in the etiology of sensorineural hearing impairment (HI) especially in the early onset cases.1 Hereditary sensorineural HI is nonsyndromic (NSHI) in the majority of early onset cases, but part of these will be diagnosed with syndromic HI later in life. Usher syndrome is the most common sensorineural HI syndrome with a prevalence of 3–6.2 per 100 000 and the first symptoms of retinitis pigmentosa most often in the first or second decade of life.2 Vestibular defects occur in part of the subjects with Usher syndrome and lead to delayed motor development.2 To date, there are 13 loci and 10 genes known to be involved in the three clinical types of Usher syndrome.3, 4, 5, 6 Mutations in six of these genes, MYO7A,7, 8, 9, 10 CDH23,11, 12 PCDH15,13, 14 WHRN,15, 16 SANS17 and CIB2,4 can lead to either Usher syndrome or autosomal recessive NSHI (arNSHI). Mutations in USH2A can lead to either Usher syndrome or nonsyndromic retinitis pigmentosa.18, 19
CIB2 belongs to a family of calcium and integrin-binding proteins. CIB2 contains three predicted EF hand domains, of which only the two most C-terminal EF hands are able to bind Ca2+.20, 21 Upon Ca2+ binding, conformational changes occur that create a hydrophobic pocket that mediates the interaction with the C-terminal tail of integrin.22, 23 CIB2 has a broad expression pattern that includes the inner ear where it is immunohistochemically detected in supporting cells and in stereocilia and cuticular plate of hair cells in the cochlea and the vestibular system.4 Based on the functional impairment of hair cells in Cib2 zebrafish morphants, it has been suggested that Cib2 is important for Ca2+ homeostasis in sensory hair cells, and that loss of Cib2 function might affect processes like mechanotransduction, adaptation, electromotility and synaptic transmission.4, 24 Similarly, through functional studies in Drosophila, CIB2 was found to be essential for adequate phototransduction and maintenance of photoreceptor cells.4 Furthermore, CIB2 has been demonstrated to be integrated into the Usher interactome via the association with whirlin and myosin VIIa.4
In this study, we present molecular identity of functional variants in CIB2 in NSHI-affected families of Dutch and Pakistani origin (Figure 1). One is the recurrent c.272T>C CIB2 missense variant4 present homozygously in two Pakistani families with moderate-to-severe and profound prelingual arNSHI, respectively. The second is the novel c.196C>T missense variant found homozygously and in compound heterozygous state with the c.97C>T nonsense variant in two families of Dutch origin with profound arNSHI. We analyzed the effect of these deafness-associated alleles on the expression, subcellular localization and calcium binding abilities of CIB2.
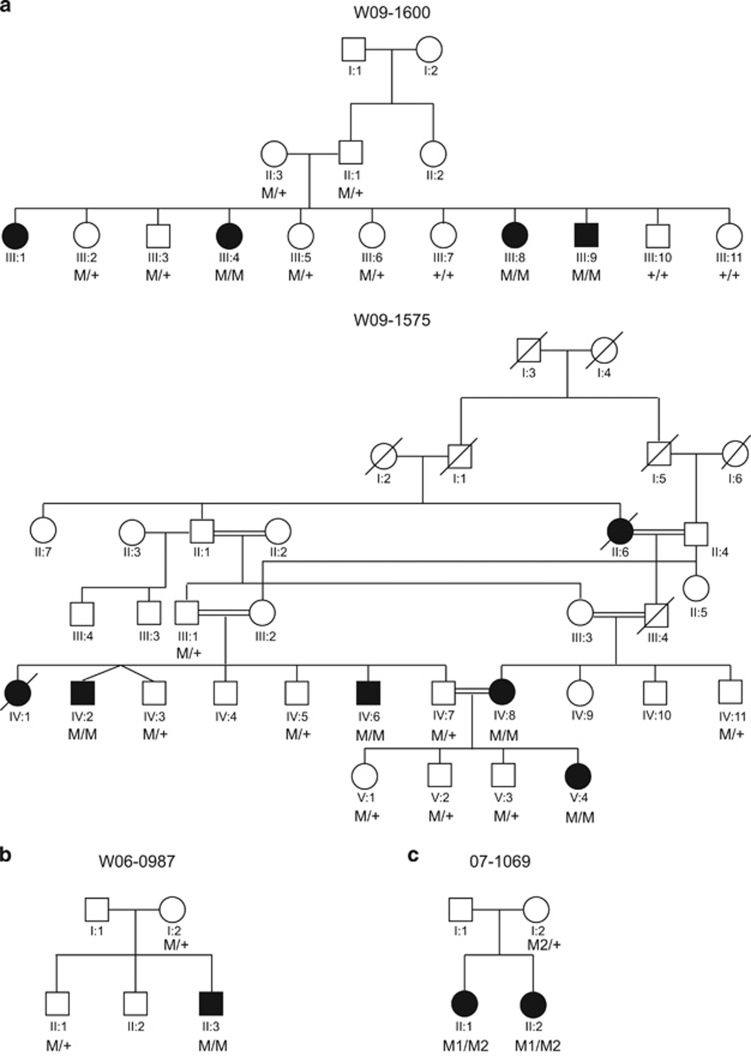
Figure 1.
Pedigrees and segregation of CIB2 variants (a) Pedigrees and segregation of the c.272T>C variant in the consanguineous Pakistani families W09-1575 and W09-1600. ‘M' represents the mutated allele c.272C and ‘+' the wild-type allele, c.272T. (b) Pedigree and segregation of c.196C>T variant in family W06-0987 of Dutch origin. ‘M' represents the mutated allele c.196T and ‘+' the wild-type allele, c.196C. (c) Pedigree and segregation of c.97C>T, indicated as M1, and c.196C>T, indicated as M2, variants in family 07-1069 of Dutch origin. The ‘+' represents the wild-type allele, c.97C. Individual II.1 underwent WES.
Materials and methods
Patients and their clinical and audiological evaluation
The current study conformed to the tenets of the Declaration of Helsinki Principles. Informed consent was obtained from all participating subjects of two families from Pakistan W09-1575 and W09-1600 (Figure 1a), and from the Dutch families W06-0987 and 07-1069 (Figures 1b and c). This study was approved by the Department of Biosciences Ethics Review Board of the COMSATS Institute of Information Technology, Islamabad, Pakistan, and the local ethics committee of the Radboud university medical center, Nijmegen, The Netherlands.
In all four families, other possible causes of HI, previous ear surgery and external ear deformities were excluded by a general ENT examination. In addition, a computed tomography (CT) scan of the temporal bone was performed for the affected individual of family W06-0987 and one affected member (II.1) of family 07-1069 in order to exclude possible anatomical causes of HI. Pure tone audiometry was performed to determine hearing thresholds at 0.25, 0.5, 1, 2, 4 and 8 kHz. Both air- and bone-conduction thresholds were obtained to exclude a conductive hearing loss. Brainstem-evoked response audiometry (BERA) was performed for the affected individual of family W06-0987 and individual II.1 of family 07-1069 according to current standards. Visual reinforcement audiometry (VRA) was performed before the age of 3 years in the affected individual of family W06-0987.25 GraphPad Prism 5.00 (GraphPad, San Diego, CA, USA) was employed for linear regression analysis to evaluate whether or not HI in family W06-0987 was progressive. Classification of the HI is in accordance with the GENDEAF guidelines (Hereditary Hearing Loss Homepage). For families W06-0987 and 07-1069 vestibular function was assessed by electronystagmography, rotatory tests and calorisation as previously described.26 To evaluate retinal involvement in the disease, fundoscopy was performed. In addition, renal, liver and thyroid functions were assessed in family W06-0987.
Genetic studies and functional evaluation of variants
Genomic DNA was isolated from peripheral blood lymphocytes by standard procedures. DNA of individual II.3 from family W06-0987 was genotyped with the Affymetrix GeneChip Human Mapping 250 K Nsp SNP array according to the manufacturer's protocol. Genotype calling and calculation of the regions of homozygosity was performed with the Genotyping Console software (Affymetrix, Santa Clara, CA, USA) and default settings. For families W09-1575 (IV:2, IV:6, IV:8 and V:4) and W09-1600 (III:1, III:4, III:8 and III:9), genotyping with Illumina HumanOmniExpress 700 K arrays was performed (San Diego, CA, USA). Homozygous regions were determined using the online tool Homozygosity Mapper with default settings. All exons of CIB2 including exon–intron boundaries were analyzed for the presence of variants as previously described,4 employing NM_006383.2 as reference sequence. Mutation analysis of TRIOBP was performed by Sanger sequencing.27 Primer sequences are available upon request. As a reference sequence NM_001039141.2 and NM_138632.2 were employed.
For affected member II.1 of family 07-1069, mutations and the common deletion in the GJB2 associated with DFNB1 have been excluded by Sanger sequencing and allele-specific PCR. Other known mutations in genes associated with severe congenital hearing loss, that is, CDH23, MYO7A, PCDH15, USH1C, USH1G, USH2A, GPR98, USH3A and DFNB31, were excluded by using the Asper Biotech array (version 6.0 with 612 variants) for Usher syndrome (Asper Biotech, Tartu, Estonia). Subsequently, variants in a set of 104 deafness genes (URL Gene list) were identified by whole-exome sequencing followed by data filtering and interpretation. Library preparation was performed using 3 μg of purified DNA of affected member II.1 of family 07-1069. For enrichment, the SOLiD optimized Sure Select All Human Exon Kit (50 Mb; Agilent Technologies, Santa Clara, CA, USA) was employed, followed by sequencing on 5500XL sequencers (Life Technologies, Carlsbad, CA, USA). Sequence reads were aligned to the human genome (hg19) using Lifescope v2.1 (Life Technologies), followed by variant calling on the aligned sequence. Variants were annotated using a custom analysis pipeline.28 The criteria to filter and interpret variants in the set of 104 genes known to be associated with deafness was as described previously.28, 29 An in-house-developed graphical user interface was used for data visualization and filtering within the respective gene set.
Molecular modeling, expression of CIB2 in hair cells by Helios gene gun transfection and calcium response assays of DsRed-tagged CIB2Phe91Ser, CIB2Arg66Trp and CIB2WT were performed as previously described.4
All variants identified in this study were submitted to the Leiden Open Variant Database with the following IDs: c.97C>T, #05182; c.196C>T, #05182; c.272T>C, #05182.
Results
Patient evaluation
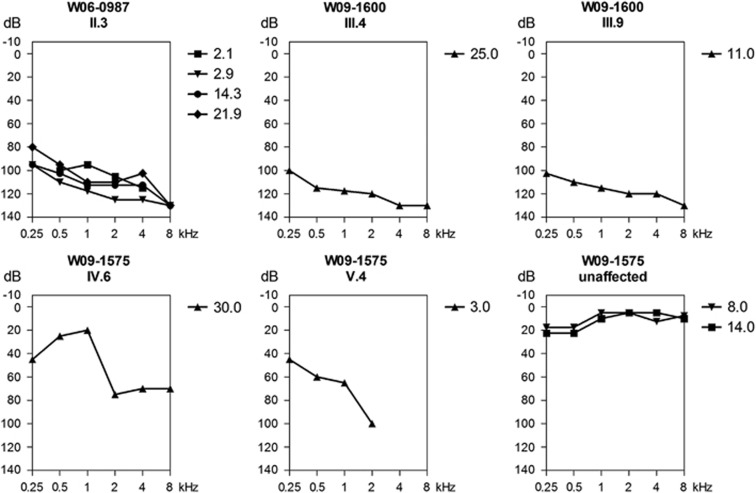
For none of the hearing-impaired individuals additional abnormalities were found during ENT examination. Affected individuals of Pakistani family W09-1575 (Figure 1a) were diagnosed with prelingual, bilateral, moderate-to-severe sensorineural HI (Figure 2). In another Pakistani family W09-1600 the HI was profound, prelingual, bilateral and sensorineural (Figure 2). There were no symptoms of retinal degeneration in the hearing-impaired individuals of these two families. The ages of the affected individuals at last visit were 5, 13, 24 and 29 years for family W09-1575 and 11, 15, 25 and 32 years for family W09-1600. However, no ophthalmologic examinations could be performed and therefore subclinical retinal abnormalities cannot be excluded. Individual II.3 of Dutch family W06-0987 (Figure 1b) presented with prelingual HI, probably congenital but diagnosed by a hearing test at 9 months of age. BERA performed before the age of 1 year demonstrated a response at 80 dB Hl. VRA and pure tone audiometry revealed bilateral, profound sensorineural HI with a stable character and a gently downsloping audiogram configuration (Figure 2). A CT scan did not show any structural abnormalities of the temporal bone. Renal, liver and thyroid function were found to be normal. Vestibular examination showed a slight hyperreflexia. Ophthalmologic examination including fundoscopy at the age of 24 years revealed no abnormalities except for bilateral epicanthic folds. Both individuals of Dutch family 07-1069 (Figure 1c) were diagnosed with congenital, profound sensorineural hearing loss as they both failed the neonatal hearing screening. During BERA no responses were found for both individuals. Vestibular examination in II.1 displayed excitable vestibules with normal responses on the right and an unilateral weakness at the left vestibule. This was examined after cochlear implantation at the left ear. A CT scan in individual II.1 displayed no temporal bone abnormalities. Also, ophthalmologic and renal examination in the same child revealed no abnormalities. Both hearing-impaired members of family 07-1069 received a cochlear implant with good results. At their last visit in the clinic at the ages of 7 (II.1) and 4 years (II.2), the affected sibs did not have any signs of night blindness as an early symptom of retinitis pigmentosa.
Figure 2.
Audiograms of affected individuals of families W06-0987, W09-1575 and W09-1600. Longitudinal binaural mean air-conduction pure tone thresholds are shown of the hearing-impaired member of family W06-0987. Binaural mean air-conduction pure tone thresholds are shown of affected members III.4 and III.9 of family W09-1600, individual V.4 and two unaffected members of family W09-1575 (IV:7 at 14 years of age and V:3 at 8 years of age). For individual IV.6 of W09-1575, the air-conduction thresholds of the left ear are depicted. Age (years) at which measurements are performed are indicated with a symbol key.
Genetic analyses
As a first step in the genetic analyses of the presented families, functional variants in GJB2 were excluded through Sanger sequencing, and allele-specific PCR was employed to exclude the common GJB2/GJB6 deletion. Homozygosity mapping was subsequently performed in families W06-0987, W09-1575 and W09-1600 as part of a larger project directed toward the identification of genetic defects underlying sensorineural HI in consanguineous Pakistani families and non-consanguineous families of The Netherlands.27, 30 In the only affected individual II.3 of family W06-0987, homozygosity mapping revealed nine homozygous regions larger than 1 Mb (Table 1). The second large homozygous region harbored TRIOBP, the gene associated with the known arNSHI type DFNB28.31, 32 No variants with a predicted functional effect were identified in TRIOBP by Sanger sequencing of the exons and exon–intron boundaries. The largest homozygous region of 16 Mb (SNP_A-1814998; SNP_A- A-1821537) contained the CIB2 gene, associated with DFNB48 and Usher syndrome type I (USH1J) by allelic functional variants.4 Genotyped affected individuals of the consanguineous family W09-1575 shared only one homozygous region larger than 3 Mb, which overlapped with the only shared homozygous region larger than 3 Mb in genotyped affected individuals of family W09-1600. CIB2 is located in the overlapping chromosomal segment (Table 1). In family W09-1575, an additional homozygous region of 4 Mb (rs1516570; rs12186082) was identified on chromosome 3. However, the affected individuals were homozygous for different alleles. Therefore this region was not further analyzed.
Table 1. Homozygosity mapping in families W06-0987, W09-1575 and W09-1600.
| No. family | Start SNP | End SNP | Chr. | Genome browser (February 2009) | Size (Mb) | Known arNSHI gene |
|---|---|---|---|---|---|---|
| W06-0987 | SNP_A-1814998 | SNP_A-1821537 | 15 | chr15:68,705,697-84,705,110 | 16.00 | CIB2 |
| SNP_A-1920225 | SNP_A-1945927 | 22 | chr22:36,758,849-40,089,893 | 3.33 | TRIOBP | |
| SNP_A-2032497 | SNP_A-2091266 | 2 | chr2:82,534,958-85,096,481 | 2.56 | — | |
| SNP_A-2033819 | SNP_A-2145805 | 14 | chr14:82,354,342-84,798,493 | 2.44 | — | |
| SNP_A-2210335 | SNP_A-1965012 | 2 | chr2:135,471,415-137,880,893 | 2.41 | — | |
| SNP_A-2277801 | SNP_A-1876469 | 9 | chr9:94,682,964-96,757,868 | 2.07 | — | |
| SNP_A-2029422 | SNP_A-4193502 | 1 | chr1:58,894,050-60,899,424 | 2.01 | — | |
| SNP_A-1856228 | SNP_A-2043311 | 8 | chr8:50,258,845-52,029,738 | 1.77 | — | |
| SNP_A-1934688 | SNP_A-4209616 | 3 | chr3:102,434,617-103,440,905 | 1.01 | — | |
| W09-1575 | rs4438265 | rs12912915 | 15 | chr15:70,826,268-92,013,118 | 21.19 | CIB2 |
| W09-1600 | rs8036698 | rs2732151 | 15 | chr15:68,716,776-84,203,685 | 15.49 | CIB2 |
Homozygous regions larger than 1 Mb in the affected individual of family W06-0987 and homozygous regions larger than 3 Mb shared by affected individuals in the consanguineous families W09-1575 and W09-1600.
Mutation analysis of CIB2 in the affected individual of family W06-0987 revealed a homozygous variant c.196C>T that is predicted to cause a substitution of a tryptophan for arginine (p.(Arg66Trp); Supplementary Figure 1A). The mother and unaffected sibling were carriers of this variant (Figure 1b). CIB2 c.196C>T affects all known CIB2 protein isoforms except isoform CIB2-006 as indicated in ENSEMBL (GRCh38) and in the Genome browser (GRCh37; Supplementary Figure 2).4 The c.196C>T variant is not present in the NCBI dbSNP138 database, the NHLBI Exome Sequencing Project (Exome Variant Server), the 1000 Genomes Project and the Nijmegen exome database (5031 exomes). In addition, c.196C>T is predicted to have a deleterious effect on protein function according to Polymorphism phenotyping version 2 with a score of 0.98 (range 0–1, 0 being benign and 1 being probably damaging), mutation taster with a score of 0.99 (range 0–1, 0 being benign and 1 being probably damaging) and Sorting Intolerant From Tolerant (SIFT) with a score of 0 (≤ 0.05 predicted to be damaging, >0.05 predicted to be tolerated). The c.196C>T missense variant affects the codon for amino-acid residue Arg66 that is moderately conserved with a score of 4 (9; conserved, 1; variable) according to the ConSeq online bioinformatic tool. The arginine residue at position 66 is conserved across the CIB2 orthologs and CIB4 paralogs in a wide range of species. In the paralogs CIB1 and CIB3 and several of the CIB2 orthologs, lysine is found at this position that is, like arginine, a positively charged amino acid (Supplementary figure 1B).
Sanger sequencing of CIB2 in families W09-1575 and W09-1600 revealed a homozygous missense variant c.272T>C (Supplementary figure 1A), which is predicted to substitute a serine for a phenylalanine. This functional variant was previously described to be a founder mutation and a common cause of arNSHI in the Pakistani population.4 However, in contrast to previously reported Pakistani families homozygous for the variant c.272T>C, the affected individuals of family W09-1575 had moderate-to-severe sensorineural HI (Figure 2).
For family 07-1069, DNA diagnostics was requested after the diagnosis of profound sensorineural HI. Known functional variants in genes for Usher syndrome were excluded. Subsequent analysis of 104 deafness genes in individual II.1 of family 07-1069 revealed only two likely functional variants: c.196C>T and c.97C>T in CIB2. The latter is a nonsense variant of the codon for Arg33 (p.(Arg33*)) and the former variant was already detected homozygously in family W06-0987). The presence of the variants was confirmed by Sanger sequencing in both II.1 and her affected sister, II.2. They inherited c.97C>T from their unaffected mother (Figure 1c), indicating that the variants in the two affected children are present in the compound heterozygous state (c.[97C>T][196C>T]). Variant c.97C>T is a rare SNP, rs201845656, found three times in 8586 alleles in the European American population (allele frequency of 0.03%) in EVS and eight times in the Nijmegen exome database (n=5031, allele frequency of 0.08%). The c.97C>T variant affects the CIB2 isoforms B and C but not isoforms A and CIB2-006 (Supplementary Figure 2).4 The c.97C>T variant is predicted to result in nonsense-mediated decay, as it introduces a premature termination codon more than 54 bp upstream of the last intron.33
The c.196C>T and c.272T>C variants do not affect targeting of CIB2 to the stereocilia tip of vestibular hair cells
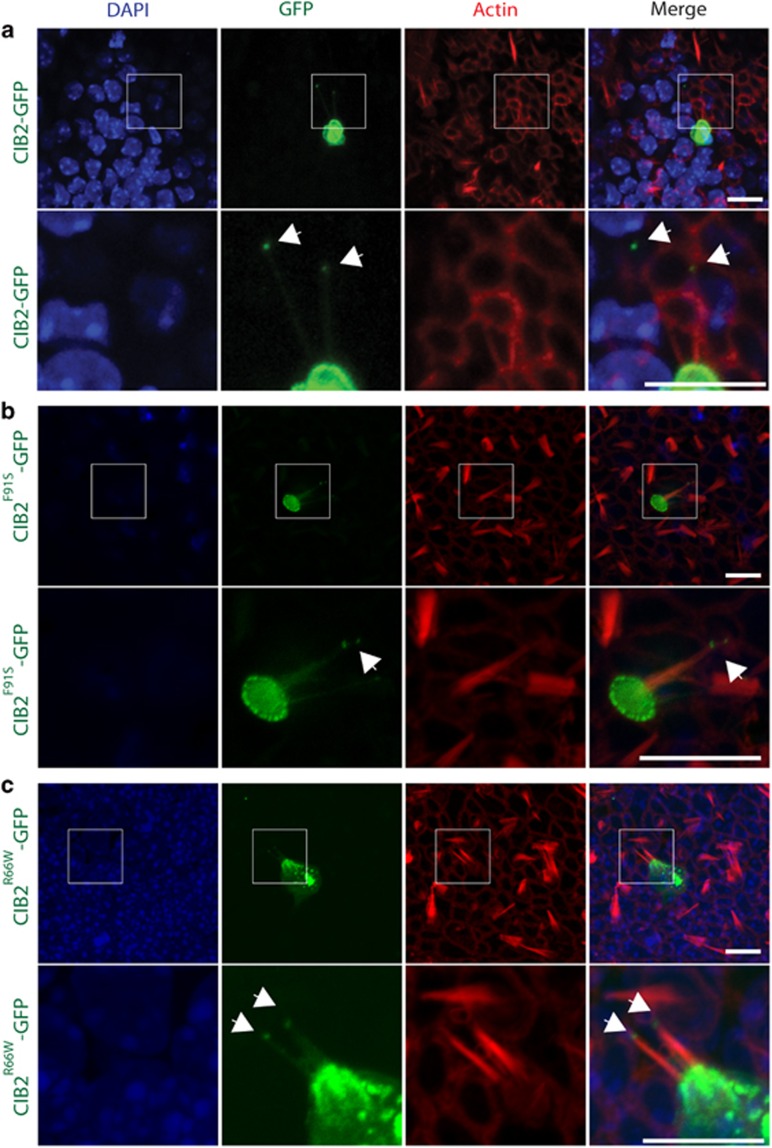
CIB2 is localized along the stereocilia of cochlear and vestibular hair cells, as well as at the stereocilia tips.4 The mechanism that permits the targeting of CIB2 to the region of the mechanotransduction machinery is still unclear. We transiently overexpressed a CIB2Arg66Trp-GFP and CIB2Phe91Ser-GFP constructs in P2 vestibular system explants in order to investigate their effect on CIB2 targeting. We found that, like wild-type control (CIB2WT-GFP), CIB2Arg66Trp-GFP, as well as CIB2Phe91Ser-GFP, accumulate in the cuticular plate, along and at the tip of the stereocilia of vestibular hair cells (Figure 3). These results suggest that, at least ex vivo, the arNSHI-associated variants do not affect CIB2 targeting in vestibular sensory hair cells (Figure 3).
Figure 3.
The c.196C>T variant does not affect the localization of CIB2 in hair cells of the vestibular system. (a–c) CIB2WT-GFP, CIB2Arg66Trp-GFP and CIB2Phe91Ser-GFP constructs were overexpressed in the P2 vestibular system of mouse using the gene gun technology. The CIB2WT-GFP variant is targeted to the tip of the stereocilia (green, arrows) and accumulates in the cuticular plate of sensory hair cells (a). The localization of CIB2 is not disrupted when c.196C>T and c.272T>C variants were introduced (b, c). Phalloidin labeling is in red and DAPI staining in blue. Scale bar: 10 μm.
The c.196C>T and c.272T>C variants do not affect the calcium-buffering property of CIB2
To test the calcium-buffering property of CIB2Arg66Trp variant, we performed a ratiometric Ca2+-imaging assay on transiently transfected COS-7 cells and measured ATP-induced inositol triphosphate-dependent calcium responses. Cells overexpressing DsRed-CIB2WT had a significant decrease in ATP-induced Ca2+ release as compared with mock-transfected cells (Figure 4c). The calcium responses of cells transfected with DsRed-CIB2Phe91Ser or DsRed-CIB2Arg66Trp constructs were comparable to cells transfected with a DsRed-CIB2WT construct showing that the amino-acid substitutions do not affect the calcium-buffering ability of CIB2 (Figure 4c).
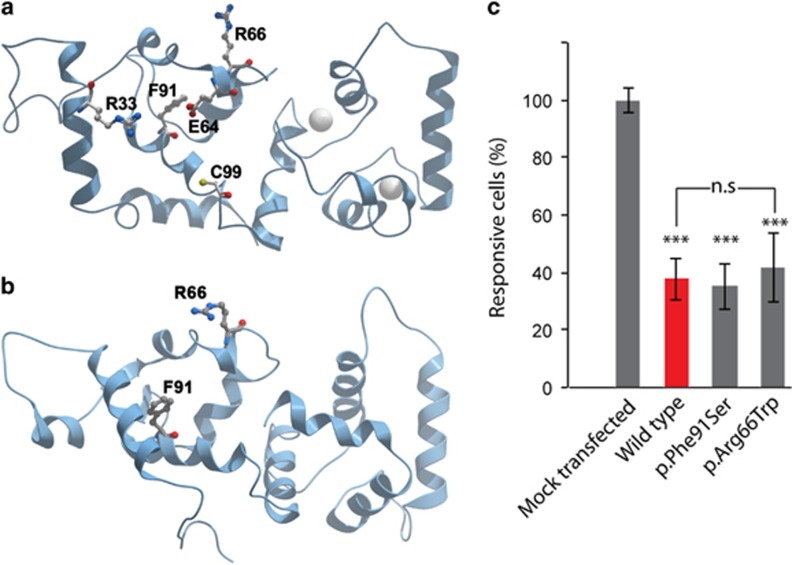
Figure 4.
(a, b) Molecular modeling of CIB2. The p.(Arg66Trp) and p.(Phe91Ser) missense mutations in CIB2, associated with arNSHI are predicted to alter the interaction of CIB2 with effector molecules. For molecular modeling, we used (a) the NMR structure of CIB1 bound to αIIβ integrin peptide and (b) the Protein Data Bank (PDB) 1XO5 crystal structure of Ca2+-CIB1. Two Ca2+ ions are represented by gray spheres. (c) The c.196C>T variant does not affect the calcium-buffering ability of CIB2. COS-7 cells were transfected with DsRed-CIB2WT, DsRed-CIB2Arg66Trp and DsRed-CIB2Phe91Ser constructs. The increase of cytosolic calcium concentration in response to the application of 1 μm of ATP was measured with ratiometric Fura-2 imaging as previously described.4 The ATP-evoked calcium responses decreased when DsRed-CIB2WT (red bar) is overexpressed due to the ability of CIB2 to bind and buffer Calcium. The p.(Phe91Ser) and p.(Arg66Trp) substitutions did not alter calcium-buffering ability of CIB2 (gray bars on the right). Data are normalized to the average response of mock-transfected cells and are shown as mean±SEM ***P<0.001; **P<0.01 and *P<0.05.
Molecular modeling suggests impaired interaction of CIB2 with effector molecules due to the c.196C>T and c.272T>C variants
When modeled using the known crystal structure of CIB1 as backbone4, 21, 34 in CIB2, the p.Arg66 maps to the N-terminal domain of CIB2 within a region that has been implicated in the interaction of CIB protein with the carboxy terminal, unstructured, negatively charged tail of αIIβ integrin. CIB1 is involved in the activation of integrin αIIβ3. It is thought that hydrophobic interactions between the αIIβ helix and the C-terminal domain of CIB1 primarily drive the interaction, with the N-domain involved in electrostatic stabilizing interactions with the negatively charged unstructured C-terminal tail of αIIβ (residues P998PLEEDDEEGQ1008). Implicated in this interaction are residues p.Arg33 and p.Lys65 of CIB1. By structure-based sequence alignment this corresponds to p.Arg33 and p.Arg66 of CIB2. Therefore, an amino acid change at p.Arg66 would likely destabilize the interaction with integrin and thus attenuate integrin activation (Figures 4a and b).22, 34 In consistence with our calcium-imaging data, Arg66 is located away from the Ca2+ binding pockets,20, 21 and thus the substitution of tryptophan for arginine at position 66 is predicted not to alter calcium-buffering abilities of CIB2. Similarly, the substitution of serine for phenylalanine at position 91 is also predicted to weaken the interaction of CIB2 with αIIβ integrin.4
Discussion
In this study we present a new homozygous functional variant, c.97C>T, and a compound heterozygous variants, c.[97C>T][196C>T] in CIB2 underlying DFNB48 arNSHI in an isolated case (W06-0987) and in a family (07-1609), respectively, of Dutch origin. Additionally, we report two consanguineous families of Pakistani origin (W09-1575 and W09-1600) with arNSHI carrying the previously described and recurrent CIB2 variant, c.272T>C. Due to this founder variant, defects in CIB2 are the major cause of arNSHI in Pakistan.4
DFNB48 and USH1J are caused by allelic CIB2 variants.4 Patients of the so far only family with USH1J present with prelingual, bilateral profound HI. Retinitis pigmentosa in the USH1J family progressed from mild to complete vision loss and was detected as early as 10 years of age. Besides that, vestibular dysfunction, later confirmed by ENG, was suspected as there was a delayed onset of ambulation.35 The DFNB48 phenotype found in three of the four families reported in the present study is consistent with the previously described DFNB48 phenotype,4, 36 severe to profound, prelingual bilateral HI without signs of retinitis pigmentosa and vestibular dysfunction. However, the HI in family W09-1575 with the recurrent homozygous variant, is less severe. The age at last evaluation of affected individuals in this family, who were diagnosed in early childhood, varies from 5 to 29 years and there are no signs of progression to profound HI. Genetic modifying factors might contribute to the less severe HI as compared with that in the previously described DFNB48 families.4 Therefore, it is worthwhile to perform more extensive genotyping in this family in order to identify candidate modifying DNA variants for CIB2 defects, which may provide insight into pathogenic mechanisms.
Interestingly, the c.196C>T variant associated with arNSHI is located very close to the c.192G>C variant associated with the USH1J phenotype.4 At their most recent evaluation at the ages of 24 years (family W06-0987) and 7 and 4 years (family 07-1069), the hearing-impaired individuals with the c.196C>T variant in the homozygous or compound heterozygous state did not have any symptoms of early retinitis pigmentosa. As electroretinography could not be performed, we cannot exclude an early stage of retinitis pigmentosa in these individuals. However, classical USH1J as diagnosed in the USH1J family with the c.192G>C variant in the homozygous state4, 35 is excluded in the present families because of the absence of vestibular areflexia, and for family W06-0987, in addition, because of lack of symptoms of retinal degeneration at 24 years of age.2 The difference in phenotypic effect of the c.192G>C (p.(Glu64Asp)) and c.196C>T (p.(Arg66Trp)) variants in CIB2 might be the result of a larger negative effect of the former mutation because of loss of a salt bridge between residues Arg33 and Glu64, which is formed in the absence of integrin.4 The associations of CIB2 with the Usher proteins myosin VIIa (USH1B) and whirlin (USH2D)16 are candidates for being affected differently by the two mutations but so far the region(s) of CIB2 that mediate(s) these direct or indirect interactions is still elusive. The identification of additional USH1J and DFNB48 families can shed light on the underlying reasons for the phenotypic differences, DFNB48 and USH1J, of mutations that affect neighboring amino-acid residues. Phenotypes varying from arNSHI, USH1J and HI combined with a variable age of onset of retinitis pigmentosa or asymptomatic retinitis pigmentosa-like findings has previously been described for functional variants in CDH23.37 Screening of CIB2 in more families in different populations will add more information on the frequency of functional variants of CIB2 and will contribute to define phenotype–genotype correlations of CIB2 variants.
Acknowledgments
We are thankful to the families for their participation in this study. We thank Saskia van der Velde-Visser for excellent technical assistance. This work was financially supported by grants from The Oticon Foundation (09-3742, to HK), ZonMW (40-00812-98-09047, to HK; 016.136.088, to MS; 016.136.091 to EvW and 40-42900-98-1006 to EvW). This study was partially sponsored by the National Institute on Deafness and Other Communication Disorders (NIDCD/NIH) research grants (R01 DC012564 to ZMA and R01 DC011803 to SR). Part of this work was supported by a core grant from the COMSATS Institute of Information Technology.
Web resources
1000 Genomes Project (http://www.1000genomes.org/). Conseq (http://conseq.tau.ac.il/). ENSEMBL (http://www.ensembl.org/Homo_sapiens/Info/Index). Exome variant server (EVS, http://evs.gs.washington.edu/EVS/). Genome browser (http://genome-euro.ucsc.edu). Gene list (https://www.radboudumc.nl/Informatievoorverwijzers/Genoomdiagnostiek/en/Pages/Hearingimpairment.aspx). Hereditary Hearing Loss Homepage (http://hereditaryhearingloss.org/). Homozygosity Mapper (www.homozygositymapper.org). Interpro (http://www.ebi.ac.uk/interpro/). LOVD (http://databases.lovd.nl/shared/variants/0000053133#05182; http://databases.lovd.nl/shared/variants/0000053131#05182; http://databases.lovd.nl/shared/variants/0000053132#05182). Mutation Taster (www.mutationtaster.org). Polymorphism phenotyping 2 (Polyphen2, http://genetics.bwh.harvard.edu/pph2/). Sorting Intolerant From Tolerant (SIFT, http://sift.jcvi.org/).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Mehl AL, Thomson V: The Colorado newborn hearing screening project, 1992-1999: on the threshold of effective population-based universal newborn hearing screening. Pediatrics 2002; 109: E7. [DOI] [PubMed] [Google Scholar]
- Cohen M, Bitner-Glindzicz M, Luxon L: The changing face of Usher syndrome: clinical implications. Int J Audiol 2007; 46: 82–93. [DOI] [PubMed] [Google Scholar]
- Millan JM, Aller E, Jaijo T, Blanco-Kelly F, Gimenez-Pardo A, Ayuso C: An update on the genetics of usher syndrome. J Ophthalmol 2011; 2011: 417217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S, Belyantseva IA, Giese AP et al: Alterations of the CIB2 calcium- and integrin-binding protein cause Usher syndrome type 1J and nonsyndromic deafness DFNB48. Nat Genet 2012; 44: 1265–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworek TJ, Bhatti R, Latief N, Khan SN, Riazuddin S, Ahmed ZM: USH1K, a novel locus for type I Usher syndrome, maps to chromosome 10p11.21-q21.1. J Hum Genet 2012; 57: 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D, Zallocchi M: Usher protein functions in hair cells and photoreceptors. Int J Biochem Cell Biol 2014; 46: 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil D, Blanchard S, Kaplan J et al: Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 1995; 374: 60–61. [DOI] [PubMed] [Google Scholar]
- Liu XZ, Walsh J, Mburu P et al: Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nat Genet 1997; 16: 188–190. [DOI] [PubMed] [Google Scholar]
- Weil D, Kussel P, Blanchard S et al: The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nat Genet 1997; 16: 191–193. [DOI] [PubMed] [Google Scholar]
- Liu XZ, Walsh J, Tamagawa Y et al: Autosomal dominant non-syndromic deafness caused by a mutation in the myosin VIIA gene. Nat Genet 1997; 17: 268–269. [DOI] [PubMed] [Google Scholar]
- Wayne S, Der Kaloustian VM, Schloss M et al: Localization of the Usher syndrome type ID gene (Ush1D to chromosome 10. Hum Mol Genet 1996; 5: 1689–1692. [DOI] [PubMed] [Google Scholar]
- Bork JM, Peters LM, Riazuddin S et al: Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet 2001; 68: 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Bernstein SL et al: Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet 2001; 69: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Ahmad J et al: PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum Mol Genet 2003; 12: 3215–3223. [DOI] [PubMed] [Google Scholar]
- Mburu P, Mustapha M, Varela A et al: Defects in whirlin, a PDZ domain molecule involved in stereocilia elongation, cause deafness in the whirler mouse and families with DFNB31. Nat Genet 2003; 34: 421–428. [DOI] [PubMed] [Google Scholar]
- Ebermann I, Scholl HP, Charbel Issa P et al: A novel gene for Usher syndrome type 2: mutations in the long isoform of whirlin are associated with retinitis pigmentosa and sensorineural hearing loss. Hum Genet 2007; 121: 203–211. [DOI] [PubMed] [Google Scholar]
- Oonk AM, van Huet RA, Leijendeckers JM et al: Nonsyndromic hearing loss caused by USH1G mutations: widening the USH1G disease spectrum. Ear Hear 2014; 36: 205–211. [DOI] [PubMed] [Google Scholar]
- Eudy JD, Weston MD, Yao S et al: Mutation of a gene encoding a protein with extracellular matrix motifs in Usher syndrome type IIa. Science 1998; 280: 1753–1757. [DOI] [PubMed] [Google Scholar]
- Rivolta C, Sweklo EA, Berson EL, Dryja TP: Missense mutation in the USH2A gene: association with recessive retinitis pigmentosa without hearing loss. Am J Hum Genet 2000; 66: 1975–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blamey CJ, Ceccarelli C, Naik UP, Bahnson BJ: The crystal structure of calcium- and integrin-binding protein 1: insights into redox regulated functions. Protein Sci 2005; 14: 1214–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry HR, Singer AU, Betts L et al: Structural and biochemical characterization of CIB1 delineates a new family of EF-hand-containing proteins. J Biol Chem 2005; 280: 8407–8415. [DOI] [PubMed] [Google Scholar]
- Huang H, Vogel HJ: Structural basis for the activation of platelet integrin alphaIIbbeta3 by calcium- and integrin-binding protein 1. J Am Chem Soc 2012; 134: 3864–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazejczyk M, Sobczak A, Debowska K et al: Biochemical characterization and expression analysis of a novel EF-hand Ca2+ binding protein calmyrin2 (Cib2) in brain indicates its function in NMDA receptor mediated Ca2+ signaling. Arch Biochem Biophys 2009; 487: 66–78. [DOI] [PubMed] [Google Scholar]
- Ahmed ZM, Frolenkov GI, Riazuddin S: Usher proteins in inner ear structure and function. Physiol Genomics 2013; 45: 987–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widen JE, Folsom RC, Cone-Wesson B et al: Identification of neonatal hearing impairment: hearing status at 8 to 12 months corrected age using a visual reinforcement audiometry protocol. Ear Hear 2000; 21: 471–487. [DOI] [PubMed] [Google Scholar]
- Theunissen EJ, Huygen PL, Folgering HT: Vestibular hyperreactivity and hyperventilation. Clin Otolaryngol Allied Sci 1986; 11: 161–169. [DOI] [PubMed] [Google Scholar]
- Schraders M, Lee K, Oostrik J et al: Homozygosity mapping reveals mutations of GRXCR1 as a cause of autosomal-recessive nonsyndromic hearing impairment. Am J Hum Genet 2010; 86: 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveling K, Feenstra I, Gilissen C et al: A post hoc comparison of the utility of sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum Mutat 2013; 34: 1721–1726. [DOI] [PubMed] [Google Scholar]
- de Ligt J, Willemsen MH, van Bon BW et al: Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 2012; 367: 1921–1929. [DOI] [PubMed] [Google Scholar]
- Shafique S, Siddiqi S, Schraders M et al: Genetic spectrum of autosomal recessive non-syndromic hearing loss in Pakistani families. PLoS One 2014; 9: e100146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S, Khan SN, Ahmed ZM et al: Mutations in TRIOBP, which encodes a putative cytoskeletal-organizing protein, are associated with nonsyndromic recessive deafness. Am J Hum Genet 2006; 78: 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahin H, Walsh T, Sobe T et al: Mutations in a novel isoform of TRIOBP that encodes a filamentous-actin binding protein are responsible for DFNB28 recessive nonsyndromic hearing loss. Am J Hum Genet 2006; 78: 144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E, Maquat LE: A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 1998; 23: 198–199. [DOI] [PubMed] [Google Scholar]
- Huang H, Ishida H, Yamniuk AP, Vogel HJ: Solution structures of Ca2+-CIB1 and Mg2+-CIB1 and their interactions with the platelet integrin alphaIIb cytoplasmic domain. J Biol Chem 2011; 286: 17181–17192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Khan SN, Friedman PL, Riazuddin S, Friedman TB: USH1H, a novel locus for type I Usher syndrome, maps to chromosome 15q22-23. Clin Genet 2009; 75: 86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad J, Khan SN, Khan SY et al: DFNB48, a new nonsyndromic recessive deafness locus, maps to chromosome 15q23-q25.1. Hum Genet 2005; 116: 407–412. [DOI] [PubMed] [Google Scholar]
- Astuto LM, Bork JM, Weston MD et al: CDH23 mutation and phenotype heterogeneity: a profile of 107 diverse families with Usher syndrome and nonsyndromic deafness. Am J Hum Genet 2002; 71: 262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.