Abstract
Cerebral visual impairment (CVI) is a major cause of low vision in children due to impairment in projection and/or interpretation of the visual input in the brain. Although acquired causes for CVI are well known, genetic causes underlying CVI are largely unidentified. DNAs of 25 patients with CVI and intellectual disability, but without acquired (eg, perinatal) damage, were investigated by whole-exome sequencing. The data were analyzed for de novo, autosomal-recessive, and X-linked variants, and subsequently classified into known, candidate, or unlikely to be associated with CVI. This classification was based on the Online Mendelian Inheritance in Man database, literature reports, variant characteristics, and functional relevance of the gene. After classification, variants in four genes known to be associated with CVI (AHDC1, NGLY1, NR2F1, PGAP1) in 5 patients (20%) were identified, establishing a conclusive genetic diagnosis for CVI. In addition, in 11 patients (44%) with CVI, variants in one or more candidate genes were identified (ACP6, AMOT, ARHGEF10L, ATP6V1A, DCAF6, DLG4, GABRB2, GRIN1, GRIN2B, KCNQ3, KCTD19, RERE, SLC1A1, SLC25A16, SLC35A2, SOX5, UFSP2, UHMK1, ZFP30). Our findings show that diverse genetic causes underlie CVI, some of which will provide insight into the biology underlying this disease process.
Introduction
Cerebral visual impairment (CVI) is one of the major causes of visual impairment in Western countries, as it accounts for 27% of low vision in childhood.1 CVI is a collective term of visual disorders, resulting from damage or malfunctioning of cerebral parts of the visual system, such as the optic tracts, optic radiations, and the visual cortex. It is diagnosed when no ocular abnormality can explain the impairment in vision, which can consist of a reduced visual acuity and/or visual field defects.2 In addition, abnormal visual behavior, such as staring into light or delayed fixation, can be present. Deficits in higher perceptual functions, for example, difficulties with recognition of objects and faces, or visio-spatial disorders can occur and are sometimes the only features of CVI.3, 4, 5 CVI can occur in isolation, but more often additional features are present, such as intellectual disability (ID), epilepsy and/or deafness.6, 7, 8 An important cause of CVI is acquired damage to the brain, mainly the result of perinatal problems (eg, cerebral hemorrhage or periventricular leukomalacia), but also other types of acquired damage, such as congenital infection, hypoglycemia, meningitis, or head trauma, can be causal.2 Furthermore, West syndrome and hydrocephalus can result in CVI.9, 10 So far, less attention has been paid to genetic causes of CVI, although associations with several neurodegenerative causes and chromosomal aberrations have been described.11 Recently, we reported in 7% of CVI patients associations with copy number variants, among others trisomy 21, 1p36 deletion, and 22q13.3 deletion (Phelan–McDermid syndrome).12 Moreover, CVI was recently shown to be caused by de novo variants in NR2F1, leading to the Bosch–Boonstra–Schaaf optic atrophy syndrome (#615722, http://www.NR2F1gene.com).13 In other neurological disorders, such as ID, epileptic encephalopathies, or autism, a high rate of (probably) disease-causing de novo variants were identified by whole-exome sequencing (WES) by using a child–parents trio approach.14, 15, 16, 17, 18, 19, 20 In addition, WES has also shown to be a powerful tool for identifying autosomal-recessive and X-linked variants in persons with ID.19, 20, 21, 22, 23, 24 Here we used WES to identify underlying genetic causes for CVI.
Subjects and methods
Twenty-five patients with CVI and a visual acuity ≤0.3 were included, and WES was performed in the patients and their parents (detailed methods are presented in the Supplementary Methods). After performing quality filtering, the common variants (>1%) were excluded, and the data were analyzed for variants following a de novo, X-linked, and autosomal-recessive inheritance pattern. Truncating variants and variants predicted to affect function were validated by Sanger sequencing. The genes in which the variants were identified were further classified partly based on the method reported by de Ligt et al15 and Gilissen et al.25 (Figure 1). A second, more stringent filtering was used for the de novo and X-linked variants (frequency ≤0.1% in controls) and truncating variants (frequency of truncating variants in controls across the whole gene ≤0.1 or ≤1.0% (autosomal-recessive) in controls). This study was approved by the Ethics Committee of the Radboud University Medical Center (Commissie Mensgebonden Onderzoek, regio Arnhem-Nijmegen), and written informed consent was obtained for all enrolled subjects. Three patients (12, 13, and 23) were part of previous reports.13, 26 The variants identified have been submitted to the Leiden Open Variation Databases (LOVDs) (http://databases.lovd.nl/, patient IDs #00025011 and #00039389–#000394012).
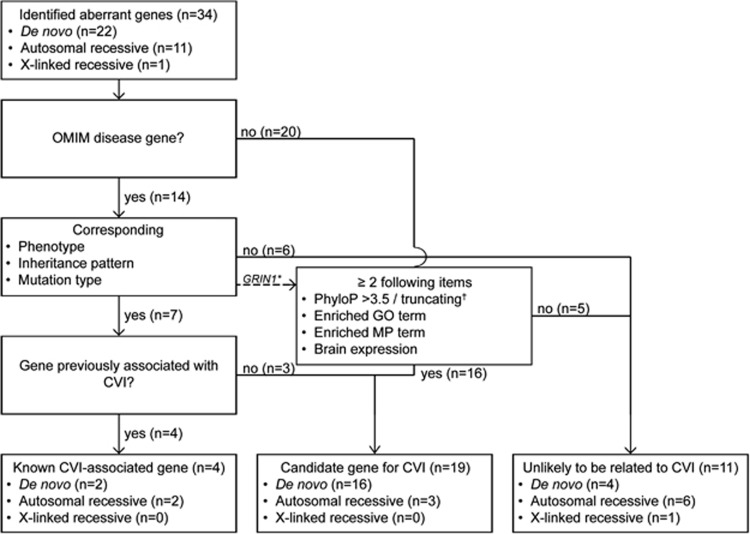
Figure 1.
Flow chart of gene classification. *Inheritance does not fit with the reported OMIM disease (autosomal-recessive instead of the reported de novo autosomal dominant). However, this variant is further classified according to de Ligt et al15 and Gilissen et al25; see Results section. †Frameshift, nonsense or splice site variant.
Results
The clinical characteristics of the 25 patients analyzed were as described in Table 1 and in more detail per patient in Supplementary Table S1. The mean age was 12 years (range 1–33 years) and one patient had a visual acuity <0.05, which is defined as blindness by the WHO (http://apps.who.int/classifications/icd10/browse/2015/en). In addition to CVI, all patients had ID, ranging from mild to severe. In all patients, WES was performed by trio approach with an average read depth of 120 × (Illumina Hiseq 2000, Illumina, San Diego, CA, USA), 115 × (Illumina HiSeq, Illumina), or 72 × (Solid 5500XL, Life Technologies, Carlsbad, CA, USA). A de novo ratio could not be established, because only protein truncating variants and missense variants predicted to affect function were validated. After prioritization and validation of the identified variants in a patient and its parents, further segregation analysis for the autosomal-recessive and X-linked variants was performed in the families for whom this was possible. The variants in one autosomal-recessive gene, ITPRIPL1, and five variants in X-linked genes, CACNA1F, CNGA2, FLNA, PCDH11X, and ZMAT1, could be discarded because of their presence in healthy (male) family members (Supplementary Table S2).
Table 1. Cohort characteristics.
| Characteristic | Number of patients |
|---|---|
| Gender | |
| Male | 17 |
| Female | 8 |
| Age group | |
| <4 years | 4 |
| 4–10 years | 9 |
| 11–20 years | 6 |
| >20 years | 6 |
| Visual acuity | |
| ≤0.3 and ≥0.05 | 24 |
| <0.05 | 1 |
| Fixation abnormalities | |
| Yes | 13 |
| No | 12 |
| Visual field defects | |
| Yes | 17 |
| No | 8 |
| Developmental delay and/or intellectual disability | |
| Yes | 25 |
| No | 0 |
| Able to walk independently | |
| Yes | 15 |
| No | 10 |
| Speak words | |
| Yes | 12 |
| No | 13 |
| Microcephaly (<3rd centile) | |
| Yes | 4 |
| No | 20 |
| Unknown | 1 |
| Macrocephaly (>97th centile) | |
| Yes | 3 |
| No | 21 |
| Unknown | 1 |
| Abnormality on brain MRI | |
| Yes | 14 |
| No | 8 |
| Not assessed | 3 |
| Epilepsy | |
| Yes | 9 |
| No | 16 |
| Hearing loss | |
| Yes | 1 |
| No | 23 |
| Unknown | 1 |
In total, 52 variants fulfilling the prioritization criteria in 45 genes were identified: 28 de novo variants in 27 genes, 19 autosomal-recessive variants in 13 genes, and 5 X-linked recessive variants in 5 genes (Supplementary Tables S3 and S4). Two of the de novo variants were, based on the exome data and Sanger sequencing results, probably mosaic variants: one stop variant in PPFIA4 and one missense variant in SLC1A1. In addition, five de novo frameshift variants were identified in AHDC1, AKAP9, DLG4, RAB11FIP1, and TRIOBP, one de novo variant in AMOT with a possible splice effect and 20 de novo missense variants. The autosomal-recessive variants consisted of 7 homozygous and 12 compound heterozygous variants of several variant types. The X-linked recessive variants consisted of four missense and one variant in the translation-initiating Methionine. The latter variant was identified in ALAS2, and other transcripts of this gene have alternate start codons (Biosoftware Alamut, version 2.3 rev2, Interactive Biosoftware, Rouen, France).
All 45 genes were classified, but 11 genes (AKAP9, ALAS2, FAM166B, KAL1, MAP3K15, MUT, POF1B, PPFIA4, SLC6A13, SPTBN5, and TRIOBP), which were excluded based on the more stringent criteria (Supplementary Table S4), are not further discussed. Of the remaining 34 genes, 14 have previously been indicated in OMIM diseases (Figure 1).
For seven genes, the phenotype of the patients was in line with the reported phenotype: CVI was reported previously in four genes, AHDC1, NGLY1, NR2F1, and PGAP1, whereas the other three genes were classified as candidate genes for CVI, GRIN2B, KCNQ3, and SLC35A2.13, 27, 28, 29
For five genes, HSPG2, PHKB, SRP72, SYNE1, and TENM3, the reported phenotype in literature was not in line with the phenotype of the patient, and those variants were classified as unlikely to be causative for CVI in those patients (#224410, #255800, #261750, #614675, #612998, #610743, #615145). In APOPT1, the phenotype was also not in line with the reported phenotype (#220110). Moreover, we identified a de novo heterozygous variant in this gene, whereas the related OMIM disease has an autosomal-recessive inheritance pattern and no second variant could be identified in the raw exome data. For another OMIM disease gene, GRIN1 (#614254), de novo heterozygous variants in this gene have been reported to cause ID; however, we identified a homozygous variant. Therefore, this variant was further classified according to the method by de Ligt et al15 and Gilissen et al25 as a candidate gene for CVI. This method was also used for the non-OMIM disease-related genes (n=20) and consisted of the assessment of the variant characteristics (PhyloP/truncating variant) in combination with the functional relevance of the gene (GO-terms, MP-terms, and brain expression), leading to a classification of 15 candidate genes for CVI and 5 genes as unlikely to be related with CVI (Figure 1).
In total, 4 known CVI-associated genes, 19 candidate CVI genes, and 11 genes unlikely to be related to CVI were identified (Table 2 and Supplementary Table S3), and in five of the 25 patients (20%), a genetic diagnosis for the CVI could be established. In another 11 patients (44%), one or more candidate genes for CVI could be identified. Photographs of the patients in whom variants in known or candidate genes were identified are shown in Figure 2 and their clinical features are summarized in Table 1 and Supplementary Table S1. Pictures of patients 12, 13, and 23 have previously been published.13, 26
Table 2. Identified known and candidate genes for CVI per patient.
| Patient | Dominant de novo | Autosomal recessive |
|---|---|---|
| 1 | DLG4 | |
| 2 | NGLY1a | |
| 3 | ||
| 4 | ||
| 5 | ||
| 6 | SLC35A2 | |
| 7 | AHDC1a | |
| 8 | GABRB2, ARHGEF10L | |
| 9 | AMOT | |
| 10 | UHMK1 | |
| 11 | SLC25A16 | GRIN1, DCAF6 |
| 12 | PGAP1a | |
| 13 | NR2F1a | |
| 14 | ||
| 15 | ||
| 16 | ||
| 17 | SOX5, KCTD19 | |
| 18 | ||
| 19 | GRIN2B, ZFP30 | |
| 20 | ATP6V1A, UFSP2 | |
| 21 | ||
| 22 | RERE, SLC1A1b | |
| 23 | NR2F1a | ACP6 |
| 24 | KCNQ3 | |
| 25 |
Identified known CVI-associated gene.
Probably mosaic mutation.
Figure 2.
Photographs of the patients in whom known or candidate genes for CVI were identified, except for patient 11 (consent to publish could not be obtained).
Discussion
WES was performed in 25 patients with CVI and a visual acuity of ≤0.3, without acquired risk factors for CVI nor pathogenic copy number variants. We identified variants in four known CVI-associated genes, namely AHDC1, NGLY1, NR2F1, and PGAP1, and 19 candidate genes for CVI. In some patients, multiple variants in more than one gene were found. In addition, de novo variants in NR2F1 were identified in two patients (13 and 23). The identification of variants in known CVI-associated genes strengthened our hypothesis that WES is the right approach to identify the underlying genetic causes in patients with CVI. Moreover, several identified candidate genes have a functional link with genes known to be associated with CVI.
Three genes, in which variants were identified, have been implicated in glycosylation: NGLY1, SLC35A2, and PGAP1. Previously, CVI has been reported as part of congenital disorders of glycosylation (CDG) type 1a (PMM2), type 1q (SRD5A3), and type 1v (NGLY1).8, 28, 30, 31 The phenotype of patient 2 with NGLY1 variants is similar to the previously reported patients, including the microcephaly, hypotonia, movement disorder, and alacrima.28, 32, 33 Variants in SLC35A2 lead to CDG type 2m, featured by ID, epilepsy, facial dysmorphisms, and transient abnormalities in transferin testing.34, 35, 36 In the seven reported patients with CDG type 2m, CVI has not been mentioned, but other features, including the facial dysmorphism, epilepsy and severe ID were present in patient 6. The third glycosylation gene in which a variant was identified, PGAP1 (patient 12, reported elsewhere), is important in the GPI-anchor synthesis pathway.26 Several other genes, PIGA, PIGN, and PIGT, implicated in this pathway are known to be implicated in CVI and ID as well,29, 37, 38, 39, 40 and recently, also PGAP1 variants were found to be associated with CVI and ID.29, 41
RERE is another gene with a functional link with a gene known to be aberrant in CVI. A de novo variant in this arginine-glutamic acid repeats-encoding gene was identified in patient 22. RERE binds directly to NR2F1, which has recently been identified to be aberrant in Bosch–Boonstra–Schaaf optic atrophy syndrome, of which one of the features is CVI.13, 42 In addition, RERE null mice show severe central nervous system abnormalities and defects of the optic vesicles.43 Furthermore, RERE forms a complex with NR2F2 and EP300 and positively regulates retinoic acid signaling in mice.44 Retinoic acid signaling induces optic vesicle and brain development and Nr2f1 and Nr2f2 transcription in mice stem cells.45 These findings indicate that RERE is a likely candidate gene for CVI (http://www.REREgene.com).
Several other candidate genes, GRIN2B, GRIN1, KCNQ3, GABRB2, and SOX5, have been implicated in neurological diseases other than CVI. In GRIN2B, a de novo missense variant was identified in patient 19. Variants in GRIN2B have previously been found in individuals with ID.46 GRIN2B encodes the subunit NR2B of the NMDA receptor, which is present during development. In the first decade, during the critical period of developing cerebral visual cortex, NR2B is replaced by NR2A, encoded by GRIN2A.47 For GRIN2A, one 4-year-old girl has been reported with low vision.48 So it might be expected that variants in GRIN2B can lead to a disturbed development and subsequently CVI. So far, 18 patients with GRIN2B variants have been reported.17, 18, 46, 49, 50, 51, 52 In two patients with West syndrome, poor eye contact was reported,50 whereas in the other patients no assessment for CVI or visual acuity measurement was mentioned.
In patient 11, a homozygous missense variant in GRIN1 was identified. GRIN1 encodes for NR1, which, together with NR2B or NR2A, forms the NMDA receptor. NR1 is an essential subunit for the NMDA receptor, and full Nr1 knockout mice are not viable.53 So far, only four patients with de novo heterozygous variants in GRIN1 have been reported with ID with or without epilepsy.18, 54, 55 Those variants are located in or nearby the transmembrane domains,54 in contrast to the homozygous variant identified in patient 11, which is situated in the extracellular ligand-binding domain of GRIN1 (http://www.ebi.ac.uk/interpro).56 Whether the here identified variant might be considered as a hypomorphic variant, giving rise to an autosomal-recessive disorder, awaits functional proof.
In patient 24, with ID and absence seizures, a de novo missense variant in KNCQ3 was identified. The same variant was previously reported in a patient with severe ID and multifocal abnormalities on EEG.16 KNCQ3 encodes a potassium channel subunit and has been implicated in benign epileptic seizures. Several families have been reported with seizures that resolve before the age of 6 years without CVI or ID.57, 58, 59, 60, 61, 62, 63, 64, 65 However, recently two additional families with seizures and variants in KCNQ3 have been reported in which family members had various IQ levels from severe ID to normal.66, 67 Therefore, the phenotypic spectrum of KCNQ3 variants appeared to be broader than benign epilepsy only and might well include CVI.
A de novo GABRB2 variant in the transmembrane domain was identified in patient 8. In addition to CVI and ID, by EEG he had continuous spike and wave during slow wave sleep epilepsy, a severe epileptic encephalopathy, from the age of 6 years. One de novo GABRB2 variant, in the N-terminal extracellular domain implicated in GABA-binding, has been reported in a patient with ID and febrile seizures, tonic clonic convulsions, and partial seizures.68 Variants in other genes encoding GABA type A-receptor subunits have been identified in different epilepsy syndromes, making GABRB2 a likely explanation for the phenotype in our patient.
Finally, in patient 17 a de novo missense variant in SOX5 was identified, located in the HMG-domain, which is important for DNA and protein binding. Intragenic deletions in SOX5 have been reported as a cause for ID.69, 70, 71 In one patient, optic nerve hypoplasia was reported, which is in agreement with the slightly pale optic discs in our patient. However, several intragenic deletions are reported in healthy individuals in the Database of Genomic Variants (http://dgv.tcag.ca/dgv/app/home).72 Without functional assays, it is difficult to ascertain whether the here identified missense variant leads to loss or gain of function.
In literature, several genes, for example, PAX6 and SOX2, are reported to influence the structural development of eye and brain.73 These genes may affect other parts of the visual system as well. However, whether aberrant genes lead to a vision disorder exclusively owing to structural eye defects, or whether an additional cerebral component is present, is difficult to distinguish. In the here presented patients without structural eye abnormalities, no rare variants have been identified in these genes.
In total, in 5 out of the 25 patients (20%) a genetic diagnosis for the CVI could be established. The proportion of genetically solved cases is lower than previously reported for ID, but this is probably due to the fact that only a few CVI genes are yet known.15, 16, 19, 20 In another 11 patients (44%), variants in one or more candidate genes for CVI could be identified with several showing a functional link with known genes for CVI. In addition, several candidate genes have been implicated in a neurological disorder, such as ID and epilepsy. In those reported patients, ophthalmological investigation has not always been performed or mentioned, and especially in patients with ID CVI can easily remain unnoticed.74 In addition, for some disorders only few patients have been reported, and the full clinical spectrum and its variability is probably not yet clear. Nevertheless, for the identified candidate genes reported here it is of importance to find more patients with CVI and a variant in the same gene to establish a causal relationship. In combination with additional functional studies, this will increase our insight into the development of the visual system. So far, CVI has been mainly investigated in the light of acquired brain damage. Previously, we associated several chromosomal aberrations and NR2F1 with CVI.12, 13 Here we show the importance of monogenetic disorders in the pathogenesis of CVI and the necessity to test for genetic defects using genome-wide diagnostic tools.
Acknowledgments
We are grateful to the individuals involved and their families for their support and cooperation. We thank the technical ophthalmological assistant and orthoptists for their assistance during the ophthalmological examinations, with special thanks to Piet Rison. We thank Lisenka Vissers for the helpful discussions and support. This work has been supported by grants from Stichting ODAS (to FNB and FPMC), Vereniging Bartiméus-Sonneheerdt (5781251 to FNB and FPMC), and the Dutch Organisation for Health Research and Development (ZON-MW grants 917-86-319 and 912-12-109 to BBAdV). Additionally, this study was accomplished in part through the Center for Mendelian Genomics research effort funded by the National Institutes of Health and supported by the National Human Genome Research Institute grant U54HG006542 to the Baylor-Hopkins Center for Mendelian Genomics.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Boonstra N, Limburg H, Tijmes N, van Genderen M, Schuil J, van Nispen R: Changes in causes of low vision between 1988 and 2009 in a Dutch population of children. Acta Ophthalmol 2012; 90: 277–286. [DOI] [PubMed] [Google Scholar]
- Dutton GN, Jacobson LK: Cerebral visual impairment in children. Semin Neonatol 2001; 6: 477–485. [DOI] [PubMed] [Google Scholar]
- Good WV, Jan JE, DeSa L, Barkovich AJ, Groenveld M, Hoyt CS: Cortical visual impairment in children. Surv Ophthalmol 1994; 38: 351–364. [DOI] [PubMed] [Google Scholar]
- Ortibus E, Lagae L, Casteels I, Demaerel P, Stiers P: Assessment of cerebral visual impairment with the L94 visual perceptual battery: clinical value and correlation with MRI findings. Dev Med Child Neurol 2009; 51: 209–217. [DOI] [PubMed] [Google Scholar]
- Saidkasimova S, Bennett DM, Butler S, Dutton GN: Cognitive visual impairment with good visual acuity in children with posterior periventricular white matter injury: a series of 7 cases. J AAPOS 2007; 11: 426–430. [DOI] [PubMed] [Google Scholar]
- Fazzi E, Signorini SG, Bova SM et al: Spectrum of visual disorders in children with cerebral visual impairment. J Child Neurol 2007; 22: 294–301. [DOI] [PubMed] [Google Scholar]
- Khetpal V, Donahue SP: Cortical visual impairment: etiology, associated findings, and prognosis in a tertiary care setting. J AAPOS 2007; 11: 235–239. [DOI] [PubMed] [Google Scholar]
- Bosch DG, Boonstra FN, Willemsen MA, Cremers FP, de Vries BB: Low vision due to cerebral visual impairment: differentiating between acquired and genetic causes. BMC Ophthalmol 2014; 14: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin R, Garcia-Revillo J, Fitz C: Orbital hydrocephalus: a proven cause for optic atrophy. Pediatr Radiol 1998; 28: 995–997. [DOI] [PubMed] [Google Scholar]
- Castano G, Lyons CJ, Jan JE, Connolly M: Cortical visual impairment in children with infantile spasms. J AAPOS 2000; 4: 175–178. [PubMed] [Google Scholar]
- Afshari MA, Afshari NA, Fulton AB: Cortical visual impairment in infants and children. Int Ophthalmol Clin 2001; 41: 159–169. [DOI] [PubMed] [Google Scholar]
- Bosch DG, Boonstra FN, Reijnders MR, Pfundt R, Cremers FP, de Vries BB: Chromosomal aberrations in cerebral visual impairment. Eur J Paediatr Neurol 2014; 18: 677–684. [DOI] [PubMed] [Google Scholar]
- Bosch DG, Boonstra FN, Gonzaga-Jauregui C et al: NR2F1 mutations cause optic atrophy with intellectual disability. Am J Hum Genet 2014; 94: 303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, de Ligt J, Gilissen C et al: A de novo paradigm for mental retardation. Nat Genet 2010; 42: 1109–1112. [DOI] [PubMed] [Google Scholar]
- de Ligt J, Willemsen MH, van Bon BW et al: Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 2012; 367: 1921–1929. [DOI] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E et al: Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 2012; 380: 1674–1682. [DOI] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Girirajan S et al: Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nat New Biol 2012; 485: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epi4K Consortium, Epilepsy Phenome/Genome ProjectEpi4K Consortium, Epilepsy Phenome/Genome Project, Allen AS et al: De novo mutations in epileptic encephalopathies. Nature 2013; 501: 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CF, Fitzgerald TW, Jones WD et al: Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet 2015; 385: 1305–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Xia F et al: Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 2014; 312: 1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurs-Hoeijmakers JH, Vulto-van Silfhout AT, Vissers LE et al: Identification of pathogenic gene variants in small families with intellectually disabled siblings by exome sequencing. J Med Genet 2013; 50: 802–811. [DOI] [PubMed] [Google Scholar]
- Tarpey PS, Smith R, Pleasance E et al: A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet 2009; 41: 535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najmabadi H, Hu H, Garshasbi M et al: Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 2011; 478: 57–63. [DOI] [PubMed] [Google Scholar]
- Alazami AM, Patel N, Shamseldin HE et al: Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep 2015; 10: 148–161. [DOI] [PubMed] [Google Scholar]
- Gilissen C, Hehir-Kwa JY, Thung DT et al: Genome sequencing identifies major causes of severe intellectual disability. Nature 2014; 511: 344–347. [DOI] [PubMed] [Google Scholar]
- Bosch DG, Boonstra FN, Kinoshita T et al: Cerebral visual impairment and intellectual disability caused by PGAP1 variants. Eur J Hum Genet 2015, e-pub ahead of print 25 March 2015 doi:10.1038/ejhg.2015.42. [DOI] [PMC free article] [PubMed]
- Xia F, Bainbridge MN, Tan TY et al: De novo truncating mutations in AHDC1 in individuals with syndromic expressive language delay, hypotonia, and sleep apnea. Am J Hum Genet 2014; 94: 784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enns GM, Shashi V, Bainbridge M et al: Mutations in NGLY1 cause an inherited disorder of the endoplasmic reticulum-associated degradation pathway. Genet Med 2014; 16: 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, Jiang YH, Shashi V et al: Additional evidence that PGAP1 loss of function causes autosomal recessive global developmental delay and encephalopathy. Clin Genet 2015, e-pub ahead of print 30 March 2015 doi:10.1111/cge.12581. [DOI] [PubMed]
- Jensen H, Kjaergaard S, Klie F, Moller HU: Ophthalmic manifestations of congenital disorder of glycosylation type 1a. Ophthalmic Genet 2003; 24: 81–88. [DOI] [PubMed] [Google Scholar]
- Morava E, Wevers RA, Cantagrel V et al: A novel cerebello-ocular syndrome with abnormal glycosylation due to abnormalities in dolichol metabolism. Brain 2010; 133: 3210–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Shashi V, Hitomi Y et al: Mutations in NGLY1 gene linked with new genetic disorder: parents' reports of children's symptoms help facilitate the discovery. Am J Med Genet A 2014; 164: viii–vi. [DOI] [PubMed] [Google Scholar]
- Caglayan AO, Comu S, Baranoski JF et al: NGLY1 mutation causes neuromotor impairment, intellectual disability, and neuropathy. Eur J Med Genet 2015; 58: 39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodera H, Nakamura K, Osaka H et al: De novo mutations in SLC35A2 encoding a UDP-galactose transporter cause early-onset epileptic encephalopathy. Hum Mutat 2013; 34: 1708–1714. [DOI] [PubMed] [Google Scholar]
- Ng BG, Buckingham KJ, Raymond K et al: Mosaicism of the UDP-galactose transporter SLC35A2 causes a congenital disorder of glycosylation. Am J Hum Genet 2013; 92: 632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorre K, Olczak M, Wada Y et al: A new case of UDP-galactose transporter deficiency (SLC35A2-CDG): molecular basis, clinical phenotype, and therapeutic approach. J Inherit Metab Dis 2015, e-pub ahead of print 17 March 2015. [DOI] [PubMed]
- Swoboda KJ, Margraf RL, Carey JC et al: A novel germline PIGA mutation in Ferro-Cerebro-Cutaneous syndrome: a neurodegenerative X-linked epileptic encephalopathy with systemic iron-overload. Am J Med Genet A 2014; 164A: 17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maydan G, Noyman I, Har-Zahav A et al: Multiple congenital anomalies-hypotonia-seizures syndrome is caused by a mutation in PIGN. J Med Genet 2011; 48: 383–389. [DOI] [PubMed] [Google Scholar]
- Kvarnung M, Nilsson D, Lindstrand A et al: A novel intellectual disability syndrome caused by GPI anchor deficiency due to homozygous mutations in PIGT. J Med Genet 2013; 50: 521–528. [DOI] [PubMed] [Google Scholar]
- Nakashima M, Kashii H, Murakami Y et al: Novel compound heterozygous PIGT mutations caused multiple congenital anomalies-hypotonia-seizures syndrome 3. Neurogenetics 2014; 15: 193–200. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Tawamie H, Maeda Y et al: Null mutation in PGAP1 impairing Gpi-anchor maturation in patients with intellectual disability and encephalopathy. PLoS Genet 2014; 10: e1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Rajan H, Pitman JL, McKeown M, Tsai CC: Histone deacetylase-associating Atrophin proteins are nuclear receptor corepressors. Genes Dev 2006; 20: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BJ, Zaveri HP, Shchelochkov OA et al: An allelic series of mice reveals a role for RERE in the development of multiple organs affected in chromosome 1p36 deletions. PLoS One 2013; 8: e57460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilhais-Neto GC, Maruhashi M, Smith KT et al: Rere controls retinoic acid signalling and somite bilateral symmetry. Nature 2010; 463: 953–957. [DOI] [PubMed] [Google Scholar]
- Laursen KB, Mongan NP, Zhuang Y, Ng MM, Benoit YD, Gudas LJ: Polycomb recruitment attenuates retinoic acid-induced transcription of the bivalent NR2F1 gene. Nucleic Acids Res 2013; 41: 6430–6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endele S, Rosenberger G, Geider K et al: Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet 2010; 42: 1021–1026. [DOI] [PubMed] [Google Scholar]
- Yashiro K, Philpot BD: Regulation of NMDA receptor subunit expression and its implications for LTD, LTP, and metaplasticity. Neuropharmacology 2008; 55: 1081–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswaran S, Myers KA, Smith AC et al: Whole-exome sequencing in an individual with severe global developmental delay and intractable epilepsy identifies a novel, de novo GRIN2A mutation. Epilepsia 2014; 55: e75–e79. [DOI] [PubMed] [Google Scholar]
- Freunscht I, Popp B, Blank R et al: Behavioral phenotype in five individuals with de novo mutations within the GRIN2B gene. Behav Brain Funct 2013; 9: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke JR, Hendrickx R, Geider K et al: GRIN2B mutations in West syndrome and intellectual disability with focal epilepsy. Ann Neurol 2014; 75: 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Fu W et al: Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 2012; 338: 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Deriziotis P, Lee C et al: Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet 2011; 43: 585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest D, Yuzaki M, Soares HD et al: Targeted disruption of NMDA receptor 1 gene abolishes NMDA response and results in neonatal death. Neuron 1994; 13: 325–338. [DOI] [PubMed] [Google Scholar]
- Hamdan FF, Gauthier J, Araki Y et al: Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet 2011; 88: 306–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redin C, Gerard B, Lauer J et al: Efficient strategy for the molecular diagnosis of intellectual disability using targeted high-throughput sequencing. J Med Genet 2014; 51: 724–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A, Chang HY, Daugherty L et al: The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res 2015; 43: D213–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier C, Singh NA, Ryan SG et al: A pore mutation in a novel KQT-like potassium channel gene in an idiopathic epilepsy family. Nat Genet 1998; 18: 53–55. [DOI] [PubMed] [Google Scholar]
- Fister P, Soltirovska-Salamon A, Debeljak M, Paro-Panjan D: Benign familial neonatal convulsions caused by mutation in KCNQ3, exon 6: a European case. Eur J Paediatr Neurol 2013; 17: 308–310. [DOI] [PubMed] [Google Scholar]
- Hirose S, Zenri F, Akiyoshi H et al: A novel mutation of KCNQ3 (c.925T—>C) in a Japanese family with benign familial neonatal convulsions. Ann Neurol 2000; 47: 822–826. [PubMed] [Google Scholar]
- Li H, Li N, Shen L et al: A novel mutation of KCNQ3 gene in a Chinese family with benign familial neonatal convulsions. Epilepsy Res 2008; 79: 1–5. [DOI] [PubMed] [Google Scholar]
- Zara F, Specchio N, Striano P et al: Genetic testing in benign familial epilepsies of the first year of life: clinical and diagnostic significance. Epilepsia 2013; 54: 425–436. [DOI] [PubMed] [Google Scholar]
- Neubauer BA, Waldegger S, Heinzinger J et al: KCNQ2 and KCNQ3 mutations contribute to different idiopathic epilepsy syndromes. Neurology 2008; 71: 177–183. [DOI] [PubMed] [Google Scholar]
- Singh NA, Westenskow P, Charlier C et al: KCNQ2 and KCNQ3 potassium channel genes in benign familial neonatal convulsions: expansion of the functional and mutation spectrum. Brain 2003; 126: 2726–2737. [DOI] [PubMed] [Google Scholar]
- Allen NM, Mannion M, Conroy J et al: The variable phenotypes of KCNQ-related epilepsy. Epilepsia 2014; 55: e99–105. [DOI] [PubMed] [Google Scholar]
- Fusco C, Frattini D, Bassi MT: A novel KCNQ3 gene mutation in a child with infantile convulsions and partial epilepsy with centrotemporal spikes. Eur J Paediatr Neurol 2015; 19: 102–103. [DOI] [PubMed] [Google Scholar]
- Soldovieri MV, Boutry-Kryza N, Milh M et al: Novel KCNQ2 and KCNQ3 mutations in a large cohort of families with benign neonatal epilepsy: first evidence for an altered channel regulation by syntaxin-1A. Hum Mutat 2014; 35: 356–367. [DOI] [PubMed] [Google Scholar]
- Miceli F, Striano P, Soldovieri MV et al: A novel KCNQ3 mutation in familial epilepsy with focal seizures and intellectual disability. Epilepsia 2015; 56: e15–e20. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Cohen J, Pevsner J et al: A novel variant in GABRB2 associated with intellectual disability and epilepsy. Am J Med Genet A 2014; 164A: 2914–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb AN, Rosenfeld JA, Neill NJ et al: Haploinsufficiency of SOX5 at 12p12.1 is associated with developmental delays with prominent language delay, behavior problems, and mild dysmorphic features. Hum Mutat 2012; 33: 728–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RW, Bodurtha J, Cohen J, Fatemi A, Batista D: Deletion 12p12 involving SOX5 in two children with developmental delay and dysmorphic features. Pediatr Neurol 2013; 48: 317–320. [DOI] [PubMed] [Google Scholar]
- Schanze I, Schanze D, Bacino CA, Douzgou S, Kerr B, Zenker M: Haploinsufficiency of SOX5, a member of the SOX (SRY-related HMG-box) family of transcription factors is a cause of intellectual disability. Eur J Med Genet 2013; 56: 108–113. [DOI] [PubMed] [Google Scholar]
- MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW: The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res 2014; 42: D986–D992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson KA, FitzPatrick DR: The genetic architecture of microphthalmia, anophthalmia and coloboma. Eur J Med Genet 2014; 57: 369–380. [DOI] [PubMed] [Google Scholar]
- Nielsen LS, Skov L, Jensen H: Visual dysfunctions and ocular disorders in children with developmental delay. I. prevalence, diagnoses and aetiology of visual impairment. Acta Ophthalmol Scand 2007; 85: 149–156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.