Abstract
The Brazilian population is considered to be highly admixed. The main contributing ancestral populations were European and African, with Amerindians contributing to a lesser extent. The aims of this study were to provide a resource for determining and quantifying individual continental ancestry using the smallest number of SNPs possible, thus allowing for a cost- and time-efficient strategy for genomic ancestry determination. We identified and validated a minimum set of 192 ancestry informative markers (AIMs) for the genetic ancestry determination of Brazilian populations. These markers were selected on the basis of their distribution throughout the human genome, and their capacity of being genotyped on widely available commercial platforms. We analyzed genotyping data from 6487 individuals belonging to three Brazilian cohorts. Estimates of individual admixture using this 192 AIM panels were highly correlated with estimates using ~370 000 genome-wide SNPs: 91%, 92%, and 74% of, respectively, African, European, and Native American ancestry components. Besides that, 192 AIMs are well distributed among populations from these ancestral continents, allowing greater freedom in future studies with this panel regarding the choice of reference populations. We also observed that genetic ancestry inferred by AIMs provides similar association results to the one obtained using ancestry inferred by genomic data (370 K SNPs) in a simple regression model with rs1426654, related to skin pigmentation, genotypes as dependent variable. In conclusion, these markers can be used to identify and accurately quantify ancestry of Latin Americans or US Hispanics/Latino individuals, in particular in the context of fine-mapping strategies that require the quantification of continental ancestry in thousands of individuals.
Introduction
The American continent as seen today is the result of the contribution of mainly three people: Native American (Amerindians), European, and African. The proportions of ancestral contributions vary according to the history of each American country.1, 2, 3 The Brazilian population is considered to be highly admixed. Colonization history shows that the main contributing ancestral populations were European and African, with Amerindians contributing to a lesser extent.4
Along the five geographical regions of Brazil (North, Northeast, Center-West, Southeast, and South), following a North to South gradient, European ancestry increases in all urban populations, achieving the highest values in populations from the South. The populations in the North have a significant proportion of Native American ancestry, which is approximately twice as high as that of African ancestry. Conversely, in the Northeast, Center-West, and Southeast, the African contribution is the second most important.5, 6
A study performed by Pena et al,7 analyzing a group of 40 validated insertion–deletion DNA polymorphisms in 934 Brazilians resident in the four most populous Brazilian regions, unraveled a high level of individual admixture in Brazilians across all regions.
The first Brazilian genome-wide survey of 1129 individuals (Brazilians and HapMap III) with 365 116 SNPs was performed by Giolo et al.8 The results showed that the Brazilian population was even more admixed than expected, bringing unique possibilities to the search of relevant genetic markers via admixture mapping.
The characterization of the patterns of genetic variation has contributed to a better understanding of the relationship between one's genetic makeup and race/ethnicity classification, as well as improved the design and analysis of genetic association studies conducted in samples from mixed populations. It is well known that the difference in allele frequencies between continental populations potentially creates a great confounding problem in interpreting association studies,9, 10 this is especially relevant in admixed populations. This confounding effect because of population stratification will become particularly evident, and important, as more genetic association studies are conducted among multiethnic samples.11, 12
In genome-wide association studies, this is not an unsurpassable problem as there is a high number of SNPs available, which can be used to obtain the ancestral components of population structure for each participant. However, controlling for stratification is just as important in replication studies in independent samples, which, usually, focus on only a small number of polymorphic markers. Similarly, candidate gene association studies, as well as fine-mapping and sequencing studies, also require knowledge of individual ancestry. Because a small number of candidate markers may not be sufficiently informative for ancestry and genotyping a large number of SNPs is expensive for thousands of samples, there is the need to define a minimum set of ancestry informative markers (AIMs) able to measure and, therefore, assess differences in continental population structure at the individual level. In addition, resequencing and exome studies are biased towards coding regions of the genome and the overall ancestry components of sequenced individuals may not be correctly derived from these data.
Many AIM sets have been developed to estimate admixture proportions and continental structure in admixed Latin American samples. However, at least two main difficulties are imposed in applying these panels in a new study sample, (1) AIMs such as insertion–deletion5, 6, 7 or microsatellite markers13 are difficult to multiplex, need primer design by the developers and standardization of PCR conditions, hindering the access and reproduction in large samples. If the AIMs are SNPs, another difficulty (also present in the first case) is (2) the adequacy of reference ancestral populations used in AIMs selection and its effect on ancestry estimates for the specific population to be studied. This occurs, in general, when local/regional samples are part of the used reference population panel.6, 14, 15
The aim of this study is to define a minimum set of SNPs (AIMs) for determining ancestry and quantifying differences in contributions of continental populations by a cost- and time-efficient strategy.
Methods
Population samples
EpiGen-Brazil Project is a consortium studying 6487 individuals belonging to three Brazilian population-based cohorts with at least 10 years of follow-up, with small losses and monitored over time. The cohorts consist of: (1) birth cohort of Pelotas, Rio Grande do Sul, Southern Brazil (3736 individuals16); (2) cohort of Bambuí, Minas Gerais, Southeast Brazil (1442 individuals17); and (3) cohort of Salvador, Bahia, Northeast Brazil (1309 individuals18). We used DNA samples from these individuals to perform the genetic ancestry analysis.
The 1982 Pelotas birth cohort study was conducted in Pelotas, a city in the extreme South of Brazil, near the Uruguay border, with 214 000 urban inhabitants in 1982. Throughout 1982, the three maternity hospitals in the city were visited daily and births were recorded, corresponding to 99.2% of all births in the city. The 5914 live-born infants whose families lived in the urban area constituted the original cohort. At age 23 years, 3736 participants categorized their color/race according to five groups: White, Brown, Black, Amerindian, and Yellow. Further details are shown in a previous publication.16
The Bambuí cohort study of aging is ongoing in Bambuí, a city of ~15 000 inhabitants, in Minas Gerais State in Southeast Brazil. The population eligible for the cohort study consisted of all residents aged 60 years and over on 1 January 1997, who were identified from a complete census in the city. Of a total of 1742 older residents, 1606 constituted the original cohort. At baseline, 1442 participants categorized their color/race into the above-mentioned color groups; no individuals categorized themselves as Amerindian or Yellow. Further details of the Bambuí study can be seen elsewhere.17
The Salvador-SCAALA project is a longitudinal study involving a sample of 1445 children aged 4–11 years in 2005, living in Salvador, a city of 2.7 million inhabitants in Northeast Brazil. The population is part of an earlier observational study that evaluated the impact of sanitation on diarrhea in 24 small sentinel areas selected to represent the population without sanitation in Salvador. In the 2013 follow-up, 879 participants categorized their color/race according to the previous mentioned groups, and were included in the present analysis. Further details can be seen elsewhere.18
Regarding skin color distribution, in this population study self-reported White individuals predominated in Pelotas (77.5%), followed by Bambuí (60.6%) and Salvador (7.4%); individuals self-reported Black were predominant in Salvador (49.3%), followed by Pelotas (16.6%) and Bambuí (2.5%); and self-reported Brown individuals predominated in Salvador (43.3%), followed by Bambuí (36.9%) and Pelotas (5.8%).
This study was developed under the approval of the National Committee of Ethics in Research (CONEP).
We assumed as reference ancestral populations individuals from the Human Genome Diversity Project (HGDP): Pima, Maya as Amerindians and from the HapMap project, Africans: YRI (Yoruba in Ibadan, Nigeria), LWK (Luhya in Webuye, Kenya), ASW (Americans of African Ancestry in SW, USA); European: CEU (Utah Residents (CEPH) with Northern and Western European ancestry) and TSI (Tuscan in Italia).
Genotyping data
Individuals from the EpiGen-Brazil project were genotyped using the Omni2.5 Illumina array by the Illumina Laboratory (San Diego, CA, USA). To analyze genetic ancestry, we used the shared SNPs between the reference populations and EpiGen-Brazil project samples, totaling 370 539 common SNPs. Genotyping data has been deposited at the European Genome-phenome Archive (EGA, https://www.ebi.ac.uk/ega/), which is hosted by the EBI, under the accession number EGAS00001001245 (more details about EpiGen-Brazil Project are also available at https://epigen.grude.ufmg.br/).
Statistical analysis
We performed principal component analysis (PCA) on individual genotypes, which is a dimensionality reduction technique19 to analyze the data. PCA allows reducing the data size by obtaining variation axes (eigenvalues and corresponding eigenvectors), which contain most of the data variability, these axes are the principal components (PCs). The first PC is the linear combination of variables that explains the largest proportion of the total data variability, the second PC explains the second largest proportion of the total variability, and so on. Plots of the eigenvectors associated with the largest eigenvalues were then used to investigate the ancestral structure of the study sample.
The global ancestry analysis was performed using the Admixture program.20 Admixture is a software tool for maximum-likelihood estimation of individual ancestries from multilocus SNP genotype data sets. Specifically, Admixture uses a block relaxation approach to alternately update allele frequency and ancestry fraction. This software estimates parameter standard errors using bootstrapping. As the contributions of different ancestral genomes have previously been described by our group, as well as others, we used a supervised approach for ancestry determination. We used 200 bootstrap replicates (default) and k=3 (number of populations assumed for the analysis). This analysis was carried out with the data set containing the genotyping of 370 539 common SNPs for EpiGen-Brazil plus reference populations.
Selection of the minimum set of AIMs
For selection of a minimum set of AIMs, we constructed several panels using two different methodologies: PCA by EINGENSTRAT, and the AIMs algorithm selector,19 implemented in a Python script by Galanter et al.15 We inferred genetic individual ancestry with each panel and calculated the linear correlation with the inferred ancestry using all common 370 539 SNPs. The following section details the used methodologies.
Selection by PCA
For this selection, we used the PC load scores obtained from the analysis with the 370 539 common SNPs. We conducted stepwise reductions in the set of SNPs to obtain the smallest set of SNPs able to get the proportions of ancestries more correlated with all SNPs. We formed 19 sets spanning from 3000 to 30 most influential SNPs for the first, second, and third PCs and 19 sets spanning from 2000 to 20 SNPs for the first and second PCs (Table 1). For each predefined set, we determined the ancestry using Admixture and calculated the Spearman's correlation between each panel and the ancestries determined using the total set of common SNPs.
Table 1. Spearman's rank correlation (ρ) between inferred ancestry of EpiGen individual by ~300 K common SNPs and ancestry inferred by 192 AIMsa.
| ~370 K SNPs common | |||
|---|---|---|---|
| 192 AIMs Ref. pop. | African ancestry | European ancestry | Amerindian ancestry |
| Original pop | 0.889384 | 0.912636 | 0.703948 |
| Group 1 | 0.888627 | 0.912070 | 0.702670 |
| Group 2 | 0.889038 | 0.912053 | 0.703259 |
| Group 3 | 0.888894 | 0.912882 | 0.704362 |
| Group 4 | 0.888158 | 0.911343 | 0.702319 |
| Group 5 | 0.888487 | 0.912286 | 0.704278 |
| Group 6 | 0.887827 | 0.912126 | 0.703009 |
| Group 7 | 0.887272 | 0.911433 | 0.703291 |
| Group 8 | 0.887519 | 0.892294 | 0.647826 |
| Group 9 | 0.889163 | 0.912375 | 0.703697 |
Considering different groups of reference populations in analysis by 192 AIMs.
Selection by the Galanter algorithm generator of AIMs
For this selection, we applied to our data an algorithm generator of AIMs implemented as a Python script described by Galanter et al.15 For each SNP, for each parental group, we used the PLINK software (Boston, MA, USA)21 to calculate allele frequencies. For each marker, the algorithm calculates the statistics of informativeness, including delta, Fst,22 and Rosenberg's informativeness for assignment statistic,23 for each pair of ancestral populations (African/European, European/Amerindian, and African/Amerindian), based on reference allele frequencies. Locus-specific branch length (LSBL)24 statistics were estimated for each population and each statistic of informativeness to translate the pairwise metrics into a population-specific statistic. At each stage, the algorithm selected the polymorphism with the highest LSBL for the population with the lowest cumulative LSBL that met the inclusion criteria. The criteria are detailed below.
The algorithm excludes polymorphisms if they are in linkage disequilibrium (r2≥0.1) or within a predefined physical distance (≤500 kb pairs) of previously selected AIMs. This ensures maximum independent informativeness and a fair distribution of AIMs throughout the genome. In addition, in order for potential AIMs to be applicable to all sub-populations within a continental group, potential AIMs are also excluded if there is evidence of significant allele frequency heterogeneity between the samples representing each ancestral group (X2 P-value ≤0.01).
By the algorithm, we can directly choose the final number of AIMs. We chose 25 panels spanning between 2000 and 20 AIMs (Supplementary Table S1). For each panel formed, we calculated the ancestry using of Admixture and calculated the Spearman's correlation coefficients between the ancestry estimate with each of them and with the total set of ~370 K shared SNPs.
Adequacy assessment of the minimum set of AIMs in respect to different reference populations
On genotype public databases one has several populations that are used as reference in studies that infer proportion of genetic ancestry in admixed populations. This analysis seeks to evaluate how much the minimum set selected is stable when using different groups of people, from those used to obtain the panel, as reference. The populations used in this analysis belong to HapMap, HGDP, and 1000 Genomes projects. Supplementary Table S2 specifies all used reference samples.
The population groups were created by stages of substitutions of a population by another from the same continent. First, we substituted one continental reference, then we substituted two continental references and finally the three continental references were substituted. Supplementary Table S3 shows the group compositions highlighting substitutions in bold. In group 1, we substituted only African references, in group 2 only European references, in group 3 only Amerindian references, in group 4 we substituted African and European references, in group 5 European and Amerindian references, in group 6 African and Amerindian references, in group 7 all reference populations were substituted (African, European, and Amerindian), and groups 8 and 9 were formed with a reduced number of reference populations.
To evaluate the stability of the panel on changes in the reference populations, we formed 10 data sets (1 original reference – used to create the panel, and 9 reference populations groups) with each group and EpiGen individuals containing the minimum set. For each data set, we ran Admixture analysis to infer genetic ancestry for EpiGen individuals. For each result of Admixture, we calculated the Spearman's correlation with inferences using the ~370K SNPs.
We also calculated the Spearman's correlation between the ancestry inferred by the original reference populations and the ancestry inferred by the groups formed using different reference populations.
Relation between genetic ancestry and allele associated with skin pigmentation
The SNP rs1426654, within the SLC24A5 gene, results in a nonsynonymous amino-acid substitution strongly associated with skin pigmentation.25, 26 The G allele is more frequent in African populations and the A allele is more frequent in European populations. Here, we used this SNP as a proxy for a trait in which the association with ancestry is strong and well established (skin color). In this scenario, we want to investigate if ancestry inferred by the minimum set predicts rs1426654 genotypes, as well as ancestry inferred by genomic data (370 K SNPs). We performed a linear regression of ancestry on rs1426654 genotype status (coded as 0, 1 or 2 copies of the allele).
Results
Structure and global ancestry
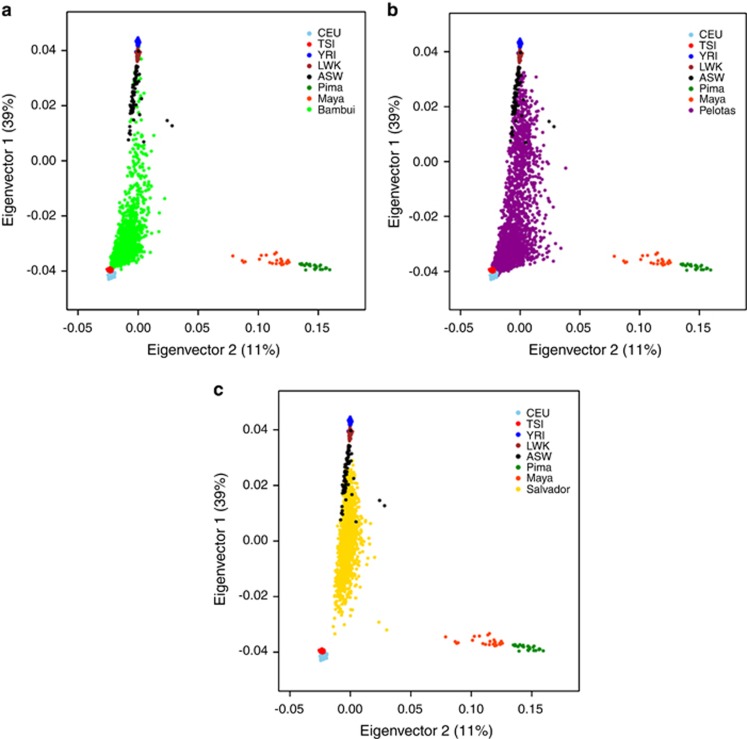
PCA analysis showed pronounced patterns of genetic variation within and among the cohorts. As expected, most variation (PC1) occurs between European and African populations (39% of the total variation), the two principal ancestry contributions to Brazil. The second largest source of variation (PC2) occurs between European and Amerindian populations (11% of the total variation). To visualize these patterns graphically, we plotted, by cohort, the first two PCs and their respective percentage of total variance (Figure 1).
Figure 1.
PCA results. PCA plot projecting 6487 individual of Brazilian cohorts (Bambui (a), Pelotas (b) and Salvador (c)) and reference populations (HapMap and HGDP) on their first and second axes variation (PCs).
The Brazilian population formed a continuum between Europeans and Africans. The Southeast and Southern Brazilian cohorts (Bambuí and Pelotas) are closer to European reference samples compared with the Northeast cohort (Salvador), in accordance with previous studies,8 and consistent with the higher European contribution in the Southeast and Southern cohorts and the higher African contribution in the Northeast cohort. Population ancestries estimated by Admixture (the mean of individual ancestries) using the ~370 K SNPs were, for European, African, and Amerindian, respectively: 0.77, 0.15, and 0.08 for the Pelotas cohort; 0.79, 0.14, and 0.07 for the Bambuí cohort; and 0.43, 0.50, and 0.07 for the Salvador cohort (Figure 2).
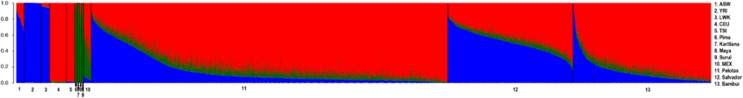
Figure 2.
Admixture proportions by individual. Barplot Admixture analysis shows the individual ancestry of Brazilian cohorts and potentially parents population. Red, green, and blue represent the proportion of inferred ancestry from European, Amerindian and African parental population. 1. ASW, 2. YRI, 3. LWK 4. CEU, 5. TSI, 6. Pima, 8. Maya, 10. MEX, 11. Pelotas, 12. Salvador, and 13. Bambuí.
Minimum set of AIMs
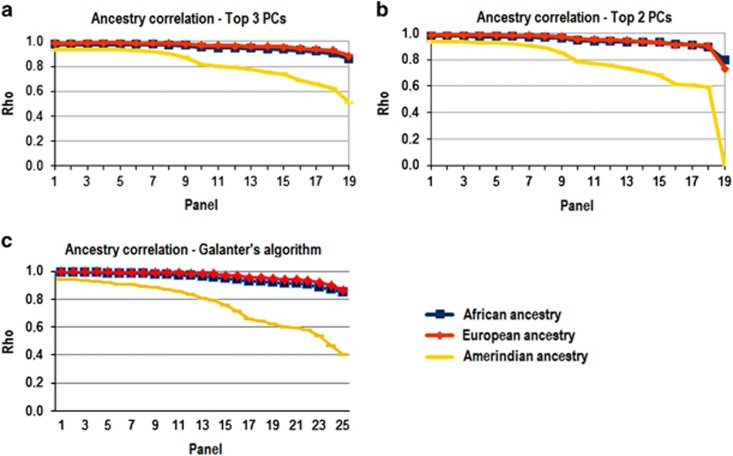
Defining small sets of AIMs have been the focus of several studies.14, 27 Figure 3 shows the correlation between European, African, and Native American ancestry, estimated by our 19 reduced panels of SNPs and ancestry inferences based on 370 K SNP. The three parts of the figure correspond to the different strategies of AIMs selection: from maximizing load scores of 1–2 PCs (Figure 3a), from maximizing load scores of 1–3 PCs (Figure 3b), and using the Galanter algorithm, which is based on population genetic statistics (Figure 3c). Although the correlations were high for African and European ancestry across all the spectrum of panels, correlations are lower for Amerindian ancestry, and using only the two largest PCs to select AIMs reduce the correlation even more, particularly when <300 AIMs are used (Figure 3b).
Figure 3.
Ancestry correlations in process of panel selection. Graphic representation of linear correlation between genetic ancestry inferred by each AIM panel and gold standard (~370 K SNPs), considering the panels obtained by the three selection methods presented in the Methods section. (a) Selection by 3 first PCs; (b) selection by first 2 PCs; and (c) selection by Galanter's algorithm.
To maximize the information regarding Amerindian ancestry, we manually conducted a second round of selection on the selection made by the Galanter's algorithm. This algorithm identifies which SNPs are more informative for each ancestral component. At the end, we identified a set of 192 AIMs combined manually, hereafter called the minimum set of AIMs. This reduced set is able to capture 91%, 92%, and 74% of, respectively, African, European, and Native American ancestry components. Tables 1 and 2 present further analysis results exploring the correlations between the ancestries inferred by ~370 K SNPs and the ancestry inferred by 192 AIMs using other reference population groups. These results are presented with more details below. The complete description of 192 AIMs and their respective statistics are in Supplementary Table S4.
Table 2. Spearman's rank correlation (ρ) between inferred ancestries of EpiGen individuals by 192 AIMs using original reference populations and using different groups of reference populations.
| 192 AIMs – original populations | |||
|---|---|---|---|
| 192 AIMs Ref. pop. | African ancestry | European ancestry | Amerindian ancestry |
| Group 1 | 0.999719 | 0.999832 | 0.999768 |
| Group 2 | 0.999852 | 0.999917 | 0.999594 |
| Group 3 | 0.999435 | 0.999378 | 0.995298 |
| Group 4 | 0.999676 | 0.999763 | 0.999691 |
| Group 5 | 0.999437 | 0.999362 | 0.995802 |
| Group 6 | 0.999014 | 0.999314 | 0.994651 |
| Group 7 | 0.999030 | 0.999298 | 0.995476 |
| Group 8 | 0.999484 | 0.985098 | 0.975782 |
| Group 9 | 0.999925 | 0.999965 | 0.999820 |
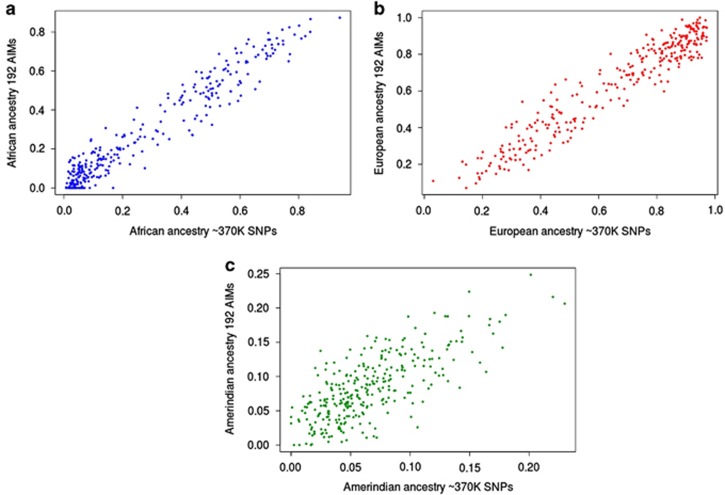
We also show that the correlation between the ancestries inferred by 192 SNPs and the ancestries inferred by ~370 K SNPs is good at the individual level. For this, we randomly selected 100 individual from each cohort and plotted the proportions of ancestry inferred by both SNP sets (Figure 4).
Figure 4.
Comparison between the genetic ancestry inferred by ~370 K SNPs and genetic ancestry inferred by 192 SNPs at the individual level. Graphic representations comparing genetic ancestry inferred by AIM panel or by the ‘gold standard' (~370 K SNPs). For this representation, we randomly selected a subset of 100 individuals from each cohort. (a) Correlation structure for African ancestry; (b) correlation structure for European ancestry; and (c) correlation structure for Amerindian ancestry.
Adequacy of AIMs to different reference populations
Table 1 shows the correlations between inferred ancestry by each reference group (using 192 AIMs) and the inferred ancestry by all common SNPs. All groups presented practically the same correlations for African, European, and Amerindian ancestry, except in the case of Group 8 for Amerindian ancestry. However, this reduced correlation is expected because the Group 8 does not include any Native American population as the reference, but only an admixed Mexican sample as a proxy. Table 2 shows the correlations between inferred ancestry by each new reference group and inferred ancestry by the original reference group, but in this case, only the minimum panel of 192 SNPs was used to infer ancestry. Results with all groups showed almost 100% of correlation with results with the original group. These results suggest that our strategy of AIMs selection and our minimum panel produce very stable ancestry estimations, regardless of the reference population used.
Relation between genetic ancestry and rs1426654 alleles, associated with skin pigmentation
Table 3 shows the proportions of genetic ancestry and frequencies of the rs1426654G allele by cohort. Bambuí and Pelotas have the smallest proportions of African ancestry, and also the smallest frequency of allele G, 0.18 and 0.20, respectively. Salvador has 50% of African ancestry and a 0.46 frequency for allele G. As expected, African ancestry had a positive association with allele G and European ancestry had a negative association, in all cohorts. Amerindian ancestry had positive association in Bambuí and Pelotas cohorts, and negative association in Salvador cohort, but the β-coefficients values are very low (−0.004 and −0.002). Most importantly, however, the association estimates were almost identical when using panel- derived ancestries (Table 4).
Table 3. Proportion of genetic ancestry (using 370 K SNPs) and allele frequency of rs1426654 (skin color) in EpiGen cohorts.
| Genetic ancestry proportion | Allele frequency (G) | |||
|---|---|---|---|---|
| Population | African | European | Amerindian | Rs1426654 |
| Salvador | 0.50 | 0.43 | 0.07 | 0.4614 |
| Bambui | 0.14 | 0.79 | 0.07 | 0.1813 |
| Pelotas | 0.15 | 0.77 | 0.07 | 0.2009 |
| EpiGen | 0.22 | 0.70 | 0.08 | 0.2647 |
Table 4. β-Coefficient of association between genetic ancestry and rs1426654 (allele G) in EpiGen cohorts.
| African ancestry | European ancestry | Amerindian ancestry | ||||
|---|---|---|---|---|---|---|
| Population | Genomic | AIMs | Genomic | AIMs | Genomic | AIMs |
| Salvador | 0.085* | 0.107* | −0.081* | −0.0104* | −0.004* | −0.002 |
| Bambui | 0.129* | 0.133* | −0.151* | −0.151* | 0.023* | 0.018* |
| Pelotas | 0.198* | 0.200* | −0.214* | −0.212* | 0.016* | 0.013* |
| EpiGen | 0.212* | 0.216* | −0.220* | −0.221* | 0.008* | 0.005* |
*P-value<0.001
Discussion
The current study was undertaken using a large sample of Brazilian individuals from different parts of the country to provide researchers with a set of AIMs to estimate accurately the continental ancestral components (African, European, and Amerindian) of contemporary American admixed samples, especially the Brazilian population.
Previous studies identified AIMs that exhibit large differences in allele frequencies across populations of European, Amerindian, and African descent, and therefore confer increased power for detecting levels of Latino American population stratification at the continental level.1, 2, 3, 14, 15 However, few Brazilian populations were included in those studies. Here, we present results based on three large Brazilian population-based cohorts from different parts of the country, with different demographic and admixture history.
Many populations in contemporary Latin American result from three main ancestry contributions: Native American (Amerindians), European, and African. The colonization history of each country is the most important predictor of the current mean proportions of ancestral contributions. African immigrants in Brazil, and in other Latin American countries where such ancestral component is found, were mainly from the slave trade. European immigrants were mainly from Spain, for Spanish-speaking countries, and Portugal, for Brazil. The Amerindian contribution was made by the natives of each region of America where today the countries are, and, in Brazil, these were isolated groups.28 Mexico, Central America, and South America (with the exception of Brazil) have about 40–48% Amerindian ancestry. Cuba, Dominican Republic, and Puerto Rico have about 6 to 13% of Amerindian ancestry.3 By our results, urban Brazilian population has about 7% of Amerindian ancestry contribution. This ancestral component is the most important factor in explaining the differences in the ancestry structure between Latin American countries, because its origins are very heterogeneous. In addition, the great variability in the reference populations used as a proxy of Amerindian ancestry (in general, investigators use local native groups as their reference samples for Native American) is also one of the main drivers of ‘incompatibility' between different proposed AIMs panels for Latin American populations. Future work on the understanding of the specific ancestral contribution of different Native American populations are warranted and should, in the future, increase the robustness of Amerindian genomic ancestry inference.
PCA shows extensive admixture between individuals of African and European descent. This occurred since the beginning of colonization and persists until the present day (Figure 1). Thus, the genomes of Brazilian individuals consist of chromosomal segments of distinct ancestries mainly derived from European and African-related admixture.
Figure 1 shows the ancestral structure by cohort. Bambuí (Figure 1a) and Pelotas (Figure 1b) cohorts are closer to European ancestral references comapred with the Salvador cohort (Figure 1c). These observations are concordant with the colonization history of these regions. These structures are consistent with the percentages of self-reported race information from each cohort.
There was strong correlation between ancestry estimates obtained from the AIMs panel and those obtained from ~370 K common SNPs between EpiGen Project and populations from public databases, providing strong support for the use of the proposed AIMs panel for the accurate estimation of ancestry. The correlation was lower for the Amerindian component, the minor ancestral component.
The correlations were calculated using the whole study sample, but in Figure 4 it is possible to evaluate the correlations at the individual level for a subset of studied individuals. Thus, we suggest that the derived panel of AIMs can be applied both at the individual and populational levels.
We also observed that genetic ancestry inferred by the small number of AIMs (around 200) provides almost the same results than the ancestry inferred by genomic data (370 K SNPs) in a simple association test. The SNP chosen for this test was the rs1426654, an SNP related to skin pigmentation. The allele A has a frequency of 98–100% in Europeans, whereas the allele G has a frequency of 97–100% in Africans.25 In our Brazilian sample, the frequencies ranged from 0.18 to 0.46. The association results between genetic ancestry and allele G were almost the same for genomic and AIM- defined ancestry.
The proposed AIMs panel will have important implications for the correct design and analytical planning of studies exploring complex traits in this population. This study uses data from South, Southeast, and Northeast Brazilian cohorts and was able to capture and incorporate into the derived panel the overall admixture variability that has been previously described for the Brazilian population.
For the typing of this AIM's panel, flexible, PCR-based SNP multiplexing platforms could be used. The cost of this kind of genotyping is determined by the size of the PCR reaction volume. As fewer reagents are needed for smaller volumes, platforms that perform the genotyping using low volume are most suitable to get a more cost-effective result. Indeed, we have designed a TaqMan SNP genotyping chip containing the 192 AIMs using the OpenArray Real-Time PCR platform (Applied Biosystems, Foster City, CA, USA). This genotyping platform is able to perform PCR reactions for a large number of samples using a very low volume, thus typing can be carried out quickly and efficiently with low cost. The assays are already commercially available (assay IDs and more information are available upon request). We also remember that each of the SNPs is also part of the Omni2.5 Illumina array.
Finally, our results provide strong reassurance that these 192 AIMs can be used for characterizing sample sets from diverse admixed population groups. These markers can be applied either to identify and quantify ancestry of individuals from a particular study, including individuals from Latin American or from Hispanic/Latino US populations, or to adjust for genetic ancestry in association studies, reducing population ancestry heterogeneity that may also correspond to reducing genetic heterogeneity for specific traits. The use of our minimum panel also contributes to minimize the probability of obtaining false-positive results by adjusting for genetic population structure.
Acknowledgments
We thank the following funding agencies: Department of Science and Technology (DECIT/ SCTIE) and National Fund for Scientific and Technological Development (FNDCT), Ministry of Health, Brazil; Funding of Studies and Projects (FINEP), Ministry of Science and Technology, Brazil; Coordination of Improvement of Higher Education Personnel (CAPES), Ministry of Education, Brazil. HCS is supported by a grant from the São Paulo Research Foundation (FAPESP).
Author contributions
HCS performed the data analysis and interpretation, and wrote the manuscript. AVRH, FR-S, MLB, BLH, MFL-C, MG, MM, TMS, JMS, and NE participated in analysis. WCSM, MRR, and FSGK participated in analysis and revised the manuscript. ET-S participated in data collection and revised the manuscript. ACP participated in the concept and design of the study, provided support for data analysis and interpretation, and revised the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Shtir CJ, Marjoram P, Azen S et al: Variation in genetic admixture and population structure among Latinos: the Los Angeles Latino eye study (LALES). BMC Genet 2009; 10: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hildesheim A, Wang SS et al: Genetic admixture and population substructure in Guanacaste Costa Rica. PLoS One 2010; 5: e13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Palmas W, Rodriguez CJ et al: Population structure of Hispanics in the United States: the multi-ethnic study of atherosclerosis. PLoS Genet 2012; 8: e1002640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves VF, Carvalho CMB, Bortolini MC, Bydlowski SP, Pena SDJ: The phylogeography of African Brazilians. Hum Hered 2008; 65: 23–32. [DOI] [PubMed] [Google Scholar]
- Saloum de Neves Manta F, Pereira R, Vianna R et al: Revisiting the genetic ancestry of Brazilians using autosomal AIM-Indels. PLoS One 2013; 8: e75145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos NPC, Ribeiro-Rodrigues EM, Ribeiro-Dos-Santos AKC et al: Assessing individual interethnic admixture and population substructure using a 48-insertion-deletion (INSEL) ancestry-informative marker (AIM) panel. Hum Mutat 2010; 31: 184–190. [DOI] [PubMed] [Google Scholar]
- Pena SDJ, Di Pietro G, Fuchshuber-Moraes M et al: The genomic ancestry of individuals from different geographical regions of Brazil is more uniform than expected. PLoS One 2011; 6: e17063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giolo SR, Soler JMP, Greenway SC et al: Brazilian urban population genetic structure reveals a high degree of admixture. Eur J Hum Genet 2012; 20: 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Butler J, Patterson N et al: Discerning the ancestry of European Americans in genetic association studies. PLoS Genet 2008; 4: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Plenge RM, Ransom M et al: Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet 2008; 4: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper RS, Tayo B, Zhu X: Genome-wide association studies: implications for multiethnic samples. Hum Mol Genet 2008; 17: R151–R155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayo BO, Teil M, Tong L et al: Genetic background of patients from a university medical center in Manhattan: implications for personalized medicine. PLoS One 2011; 6: e19166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C, Fernandez-Formoso L, Gelabert-Besada M et al: Development of a novel forensic STR multiplex for ancestry analysis and extended identity testing. Electrophoresis 2013; 34: 1151–1162. [DOI] [PubMed] [Google Scholar]
- Kosoy R, Nassir R, Tian C, White P: Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat 2009; 30: 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanter JM, Fernandez-Lopez JC, Gignoux CR et al: Development of a panel of genome-wide ancestry informative markers to study admixture throughout the Americas. PLoS Genet 2012; 8: e1002554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora CG, Barros FC: Cohort profile: the 1982 Pelotas (Brazil) birth cohort study. Int J Epidemiol 2006; 35: 237–242. [DOI] [PubMed] [Google Scholar]
- Lima-Costa MF, Firmo JOA, Uchoa E: Cohort profile: the Bambui (Brazil) Cohort Study of Ageing. Int J Epidemiol 2011; 40: 862–867. [DOI] [PubMed] [Google Scholar]
- Barreto ML, Cunha SS, Alcântara-Neves N et al: Risk factors and immunological pathways for asthma and other allergic diseases in children: background and methodology of a longitudinal study in a large urban center in Northeastern Brazil (Salvador-SCAALA study). BMC Pulm Med 2006; 6: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick N a, Reich D: Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909. [DOI] [PubMed] [Google Scholar]
- Alexander D, Novembre J, Lange K: Fast model-based estimation of ancestry in unrelated individuals. Genome Res 2009; 19: 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger KE, Weir BS: Genetics in geographically structured populations: defining, estimating and interpreting F(ST). Nat Rev Genet 2009; 10: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N a, Li LM, Ward R, Pritchard JK: Informativeness of genetic markers for inference of ancestry. Am J Hum Genet 2003; 73: 1402–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver M, Kennedy G, Parra E: The genomic distribution of population substructure in four populations using 8525 autosomal SNPs. Hum genomics 2004; 1: 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason RL, Mohideen M-APK, Mest JR et al: SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 2005; 310: 1782–1786. [DOI] [PubMed] [Google Scholar]
- Beleza S, Johnson NA, Candille SI et al: Genetic architecture of skin and eye color in an African-European admixed population. PLoS Genet 2013; 9: e1003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Li H, Criswell LA et al: Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Hum Genet 2005; 118: 382–392. [DOI] [PubMed] [Google Scholar]
- Kuhn PC, Horimoto ARVR, Sanches JM et al: Genome-wide analysis in Brazilian Xavante Indians reveals low degree of admixture. PLoS One 2012; 7: e42702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.