Abstract
To evaluate the diagnostic value of single-nucleotide polymorphism (SNP) array testing in 1033 fetuses with ultrasound anomalies we investigated the prevalence and genetic nature of pathogenic findings. We reclassified all pathogenic findings into three categories: causative findings; unexpected diagnoses (UD); and susceptibility loci (SL) for neurodevelopmental disorders. After exclusion of trisomy 13, 18, 21, sex-chromosomal aneuploidy and triploidies, in 76/1033 (7.4%) fetuses a pathogenic chromosome abnormality was detected by genomic SNP array: in 19/1033 cases (1.8%) a microscopically detectable abnormality was found and in 57/1033 (5.5%) fetuses a pathogenic submicroscopic chromosome abnormality was detected. 58% (n=44) of all these pathogenic chromosome abnormalities involved a causative finding, 35% (n=27) a SL for neurodevelopmental disorder, and 6% (n=5) a UD of an early-onset untreatable disease. In 0.3% of parental samples an incidental pathogenic finding was encountered. Our results confirm that a genomic array should be the preferred first-tier technique in fetuses with ultrasound anomalies. All UDs involved early-onset diseases, which is beneficial for the patients to know. It also seems that UDs occur at a comparable frequency among microscopic and submicroscopic pathogenic findings. SL were more often detected than in pregnancies without ultrasound anomalies.
Introduction
The main goal of cytogenetic prenatal diagnosis is to inform prospective parents of the chromosomal status of their fetus, which if abnormal, usually causes an abnormal phenotype in childhood. Fetuses with ultrasound abnormalities are known to carry not only the highest percentage of microscopically visible chromosome abnormalities1, 2, 3 but also submicroscopic aberrations.4, 5, 6, 7, 8, 9, 10, 11 Before array implementation, these submicroscopic chromosome aberrations could only be detected if targeted testing was requested in case of specific ultrasound anomalies, for example, 22q11 deletion in fetuses with cardiac defects.12 However, based on prenatal phenotyping by ultrasound examination, it is not always easy to obtain a clinical diagnosis and to determine which locus/loci should be investigated. To increase the diagnostic yield genomic array testing is now recommended in case of fetal structural ultrasound anomalies.13
The aim of the present work was to study the prevalence and nature of pathogenic array findings and to evaluate the diagnostic value of prenatal genomic array testing in a large cohort of fetuses with ultrasound abnormalities. Unexpected diagnoses (UD) such as the detection of a late-onset disease or cancer risk factors are generally thought to be challenging for prenatal counseling.14 However, we hypothesized that UDs could be an additional advantage of array testing before birth, because the fetal phenotype is limited to ultrasound findings. To study this hypothesis and evaluate the diagnostic value of genomic array we reviewed all cases of fetal ultrasound anomalies tested with single-nucleotide polymorphism (SNP) array in our laboratory and we reclassified the pathogenic array findings as we suggested before.15 This study presents not only the prevalence of pathogenic array findings in fetuses with ultrasound anomalies but also the nature of the genetic abnormalities found. To our best knowledge this is the first report presenting the contribution of causative array findings (CAUs), UD and SL for neurodevelopmental disorders to the total amount of pathogenic array findings in a large prenatal cohort tested by a single laboratory and with the same array platform.
Materials and methods
Materials
After excluding the most common trisomies and triploidy by rapid aneuploidy detection (RAD), in 2009–2013, 1033 cases with fetal ultrasound anomalies were tested with SNP array. The cohort presented here includes cases published in our previous publications.6, 16 Since September 2012, SNP array testing is performed as a stand-alone first-tier test and therefore in only about 60 initial cases karyotyping was performed simultaneously to array. In some abnormal cases karyotyping was performed after array detected a pathogenic CNV to characterize the chromosome aberration and provide the risk for recurrence. Initially cases (~200 cases) were selected by both gynecologists and clinical geneticists,6 but since June 2011 array testing was performed in all cases of ultrasound anomalies (~800 cases) regardless of the severity of the phenotype. The fetal ultrasound anomalies included (a combination of) soft markers, non-structural anomalies (eg, such as intrauterine growth restriction or polyhydramnios), structural anomalies in a single or multiple organ systems and cases of nuchal translucency (NT) ≥3.5 mm. Cases of intrauterine fetal death (IUFD) (with or without structural anomalies) were also included. If RAD was not done and a common aneuploidy was found, such sample was excluded from the cohort presented in this paper.
The fetal material was obtained by chorionic villi (CV) biopsies (214/1033) and amniocentesis (801/1033) mainly in the first and second trimester, respectively. The remaining samples were 8 fetal blood samples and 10 skin fibroblasts that were also available after TOP. In most cases, parental blood was simultaneously sampled to determine inheritance of specific fetal CNVs (whole genome analysis was not performed).
Pre-test counseling
Initially we anticipated to deal with a relatively high number of unexpected diagnoses of late-onset diseases or cancer risk factors and therefore we offered patients a choice on the types of array findings they wished to be informed about.16 Since most of abnormalities found explained the fetal phenotype, and since there were no late-onset diseases found unexpectedly and we did not routinely release variants of unknown significance (VOUS), we realized that the extensive pre-test counseling system did not match the reality. We decided to simplify the pre-test counseling and stopped offering choices. Providing written information about array testing and a simplified pre-test counseling done by trained medical staff in the Department of Obstetrics and Gynaecology allowed the clinical geneticists to focus on rapid post-test counseling of cytogenetically abnormal cases and a more extensive pre-test genetic counseling in selected cases with ultrasound abnormalities (also to request other molecular testing). The following issues are addressed in the simplified pre-test counseling and the information leaflet: brief introduction of the technique as a method of higher detection potential than karyotyping, the need of parental testing in some cases, the chance of detecting abnormalities unrelated to the initial indication for invasive prenatal testing and the fact that only pathogenic array findings will be reported.
SNP array and reporting
In most cases, uncultured AF cells, mesenchymal core of CV or EDTA cord blood were used for automated DNA isolation and directly used for array. In all cases a backup culture was established. A total of 50–100 ng fetal DNA was tested with ~300 K Illumina HumanCytoSNP-12 (HCS) array (San Diego, CA, USA), as described before.6, 16 The array profiles were analyzed with a 0.15 Mb resolution in UCSC (Human Mar. 2006 (NCBI36/hg18) Assembly) by using initially Karyo Studio (Illumina), then Genome Studio (Illumina) and different versions of Nexus Copy Number (BioDiscovery: versions 5.0 and higher (Hawthorne, CA, USA)).6, 16 We did not use a filter, but in our experience events smaller that 150 kb most of the time occurred to be noisy and therefore we estimated the working resolution at 150 kb level. Fetal and parental DNA was simultaneously tested to determine the inheritance of CNVs, mainly to avoid subsequent testing, which delays the final reports.
The most important criterion for classification as pathogenic was association with a known abnormal phenotype. We followed the evidence-based approach of Riggs et al17 according to whom there is sufficient evidence for pathogenicity when there are at least three independent loss of function mutations or duplications in unrelated individuals with a similar phenotype published in the literature.
Array findings classification
We distinguish three main categories of array findings: pathogenic findings, VOUS and benign results. Of these only the pathogenic ones are routinely reported to clinical geneticists and gynecologists. For the purpose of this study all pathogenic array results of 1033 fetuses were reviewed and subclassified in the previously proposed categories:15 CAUs, unexpected diagnoses (UD) and susceptibility locus (SL) for neurodevelopmental disorders. A special category of pathogenic findings is formed by the incidental findings (IF) in parental samples that were discovered ‘by accident' during quality control of the array profiles. All pathogenic findings (including CAU, UD, SL and IF) were reported to patients.6, 18
For the purpose of this paper, we have categorized chromosome aberrations according to their size and depending on whether they were (or could be) detected by RAD. The following categories are recognized: RAD detectable, microscopic abnormalities and submicroscopic abnormalities.
RAD detectable aberrations are chromosome abnormalities found by RAD methods such as QF-PCR. These are the common aneuploidies (trisomy 13, 18, 21 and sex-chromosome aneuploidies) and triploidy, full blown as well as high mosaics (>30%). These were excluded from this cohort.
Microscopic chromosome abnormalities are aberrations that are potentially microscopically visible (with net imbalance >10 Mb). However we are aware that some abnormalities >10 Mb may be cryptic and not easily recognizable in the banded chromosome pattern.
Submicroscopic chromosome abnormalities are aberrations that are potentially missed with karyotyping since they are <10 Mb or since they involve cryptic unbalanced translocations or low level mosaicism.
The case details are presented in Supplementary Material and all pathogenic variants were submitted to ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/intro/) (accession numbers SCV000224042 - SCV000224117).
Genotype phenotype relationship
The causal character of array findings for particular ultrasound features was assessed based on cases published in the present literature. If a similar chromosomal imbalance was reported in association with comparable clinical features, the unbalanced chromosome aberration was assumed to be causal. Even when the fetus presented only some of the clinical features, the CNV was classified as causative because of the limitations of fetal phenotyping by ultrasound and phenotypic variability in many genetic syndromes. To show the actual frequency of susceptibility loci in a cohort of fetuses with ultrasound anomalies, we did not classify susceptibility loci (SL), that might have explained the fetal phenotype, as causative (eg, a 22q11 microduplication found in a fetuses with IUGR). In cases where the causal relationship was initially doubtful gene content was studied. Our classification is based on the prenatal phenotypes. The original classification was not changed after postnatal/postmortem examinations.
Results
Array testing was performed in all 1033 cases of fetal ultrasound anomalies. Most cases (n=736) were done in the past 2 years (2012 and 2013). Quality evaluation showed that in 30/736 (4%) cases array could not be performed on uncultured material and the reporting was delayed because of the need for cell culturing. Only 2/736 samples failed and no cytogenetic examination could be performed (one case of IUFD with >90% maternal cell contamination of the uncultured sample and no cell growth in the cultures and one case of <3 mg of CV with no backup culture).
Pathogenic array findings in 1033 fetuses
In 76/1033 (7.4%) fetuses a pathogenic array finding was detected by HumanCytoSNP-12 genomic array with testing resolution 0.15 Mb (Supplementary Table S1). In 4.3% (44/1033) a definitively causative abnormality explaining the abnormal fetal phenotype was found (Supplementary Table S1). In 0.5% (5/1033) an UD of a known syndrome was made that did not explain the abnormal ultrasound findings (based on the prenatally available phenotype) (Supplementary Table S2) and in 2.6% (27/1033) a SL for neurodevelopmental disorders was encountered (Supplementary Table S3). In 19/1033 (1.8%) cases a microscopically detectable abnormality was found. Submicroscopic abnormalities were found in 57/1033 (5.5%) cases (Table 1).
Table 1. The types of pathogenic array findings and their frequencies in 1033 fetuses with ultrasound anomalies after excluding aberrations detectable by RAD.
|
Number of cases with the particular type of pathogenic finding on the total number of tested fetuses (n=1033) (%) |
Percentage of the type of pathogenic finding in relation to all pathogenic findings in the same size category |
|||||
|---|---|---|---|---|---|---|
| Type of array findings | Microscopic findings | Submicroscopic findings | Both microscopic and submicroscopic | Microscopic findings total n=19 | Submicroscopic findings total n=57 | Both microscopic and submicroscopic total n=76 |
| CAU | 18 (1.74%) | 26 (2.52%) | 44 (4.26%) | 94.74% (18/19) | 47.37% (27/57) | 57.89% (44/76) |
| UD | 1 (0.1%) | 4 (0.39%) | 5 (0.48%) | 5.26% (1/19) | 7.01% (4/57) | 6.58% (5/76) |
| SL | 0 | 27 (2.61%) | 27 (2.61%) | 0 | 47.37% (27/57) | 35.53% (27/76) |
| Total | 19 (1.84%) | 57 (5.52%) | 76 (7.36%) | — | — | — |
Abbreviations: CAU, causative findings explaining the fetal phenotype; UD, unexpected diagnoses (all findings were early-onset untreatable disorders not related to the ultrasound findings); SL, susceptibility loci for a neurodevelopmental disorder.
This means that 58% (44/76) of all pathogenic array findings were causative for the phenotype, whereas 7% (5/76) involved an UD of an early-onset untreatable disease and 35% a SL for a neurodevelopmental disorder (Table 1). Moreover, 25% (19/76) of pathogenic findings were microscopically detectable and 75% (57/76) were submicroscopic aberrations.
In 31 out of 44 cases with causative findings, the chromosome aberrations were de novo and in 13 cases inherited. Eight out of 13 inherited cases involved unbalanced forms of a previously unknown familial translocation, four an inherited deletion associated with a variable phenotype and one an X-chromosome deletion in a male fetus inherited from the mother (see Supplementary Table S1). Unexpected findings were all de novo, except for one (case 5, Supplementary Table S2). In 8/27 cases of SL the inheritance was unknown, because the parents preferred not to be tested. In 32% (6/19) of cases the SL was de novo and in 68% (13/19) inherited from an apparently healthy parent.
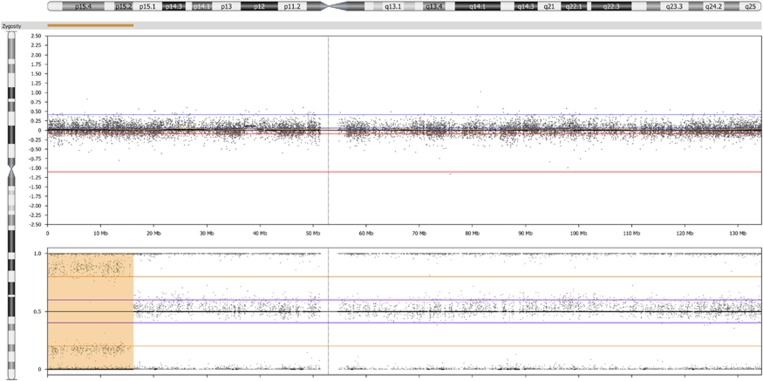
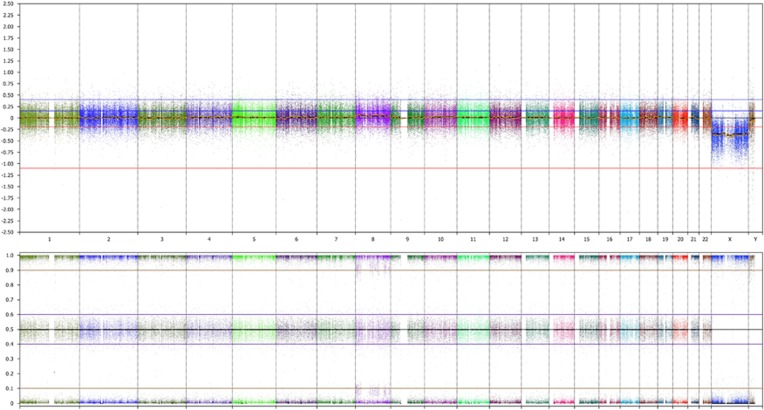
Two pathogenic findings (2/1033, 0.2%) were detected exclusively thanks to B-allelic frequency (BAF) plot analysis,16 which is 2.6% (2/76) of the pathogenic findings. These were the patients with a mosaic segmental uniparental isodisomy of 11p15 causing Beckwith–Wiedemann syndrome (Figure 1) and ~10% mosaic trisomy 8 (Figure 2).
Figure 1.
HumanCytoSNP-12 results on uncultured amniotic fluid showing the LogR ratio (upper panel) and B-allele frequency (BAF) (lower panel) along chromosome 11. A BAF of ~0.9 and ~0.1 (whereas one line at 0.5 is expected in case of two copies for heterozygous single-nucleotide polymorphisms) and a normal LogR ratio at 11p15 indicate a region of homozygosity in about 60% of the cells (a yellow bar along 11pter) fitting a mosaic segmental uniparental isodisomy of paternal origin associated with Beckwith–Wiedemann syndrome in a fetus with bilateral enlarged echogenic kidneys (case 25, Supplementary Table S1).
Figure 2.
HumanCytoSNP-12 results on uncultured amniotic fluid showing the LogR ratio (upper panel) and B-allele frequency (BAF) (lower panel) along the whole genome (different chromosomes are indicated in different colors). An extra BAF of ~0.9 and ~0.1 and a slightly elevated LogR ratio along chromosome 8 indicate ~10% mosaic trisomy 8 of meiotic origin in a fetus with ventriculomegaly (case 18, Supplementary Table S1).
Incidental findings in parental samples
Among 1826 parental samples, that were used for testing the inheritance of fetal CNVs, (from September 2010 to December 2013) we incidentally found six cases of a pathogenic finding during quality control of the parental arrays (Supplementary Table S4). Such a control allows only incidental detection of large anomalies and does not allow detection of smaller CNVs. This implicates that there is a risk of 0.3% for an incidental finding in parental samples, if parental and fetal samples are analyzed by using a SNP array and if a quality control requires a whole genome plot quality check. All these abnormal findings were communicated to the parents.
Discussion
The current paper describes the pathogenic findings in the biggest, so far published, cohort of fetuses with ultrasound anomalies, tested in one diagnostic center and analyzed with the same array platform. To our best knowledge this is the first report showing the actual ratio between causative and unexpected array findings in prenatal diagnosis. To evaluate the additional diagnostic value of SNP array testing in this group of fetuses, we discuss the hypothetical comparison with karyotyping and the nature of the pathogenic findings.
Higher diagnostic yield in comparison with karyotyping
Our results confirm that karyotyping as a stand-alone test is no longer an appropriate diagnostic approach to detect unbalanced chromosome aberrations in case of ultrasound anomalies. The increase in the detection of causative findings in our cohort supports this statement (from 1.7% microscopically visible to 4.3% in total Table 1). The frequency of pathogenic submicroscopic findings (5.5%) is similar to other published cohorts (6.8%).10 Since in 0.2% of all cases a chromosome aberration was involved that could only be detected by SNP array and since SNP array is very sensitive for the detection of maternal cell contamination,16 we strongly recommend SNP arrays instead of array CGH for prenatal diagnosis.
Genotype–phenotype relationship
To implement the proposed classification of array findings,15 a very close cooperation between clinical geneticists, laboratory specialists and obstetricians has to be established. In most cases the causal relation between the array finding and fetal phenotype is straightforward (eg, SIX3 deletion in a fetus with holoprosencephaly—case 1, Supplementary Table S1). The gene content of the array finding predetermines the characteristic phenotype associated with a particular syndrome. However genotype–phenotype correlations in prenatal diagnosis have several limitations.
Incompleteness of fetal phenotype
The fetal phenotype is often incomplete because of the limitations of the ultrasound examinations,19 which may complicate prenatal post-test counseling. For this reason, it may not always be easy to distinguish a causative finding (matching the fetal phenotype) from an unexpected diagnosis (pathogenic array finding that does not explain the fetal ultrasound anomalies, but leads to another disorder). Because of that, clinical validation in case of unexpected genetic findings is crucial for final classification. Subsequent (targeted) ultrasound examination, autopsy, postnatal examination, follow-up and family pedigree investigation may reveal additional information that may help in correlating the genetic findings with the fetal phenotype or even help in current clinical investigations in the family. In our cohort an Xq26.3-q28 deletion in a male fetus with megacystis and hydronephrosis (case 4, Supplementary Table S2) was prenatally assumed as unexpected. After birth, lower urinary tract obstruction and in addition anal atresia and underdevelopment of the legs were seen. Anal atresia was described before in a male patient with Fragile X syndrome and atypical obesity.20 Severe lower limb contractures were reported in another patient with a ~13 Mb deletion in Xq26.3-27.3.21 This chromosome aberration represents the only one in our cohort that could be reclassified as causative if considered after birth.
Phenotypic variability of syndromic disorders and incomplete penetrance
Phenotypic variability and incomplete penetrance are known for many genetic syndromes (eg, Turner syndrome, Greig syndrome, holoprosencephaly), which may complicate phenotype–genotype correlation in both prenatal and postnatal settings. Co-existing of other factors such as undetected mutations, polymorphisms and environmental influences may modulate the phenotype causing that one patient will present severe multiple congenital anomalies whereas others will be less affected. One of the prenatal factors that may play a significant role in determining the extent to which genetic risk factors are actually expressed, is prenatal nutrition.22 Even in syndromes such as Fragile X and Williams syndrome the relation between genotype and intellectual phenotype may be indirect and complex as illustrated by Karmiloff-Smith et al.22 In Down syndrome, the very well-known syndrome, there is great variability in phenotype, whereas all individuals have the same chromosome aberration.23 The phenotypic variability in known syndromes together with the incompleteness of fetal phenotype may cause that a fetus may present only atypical features that do not always directly indicate a particular syndrome. In our cohort (case 37, Supplementary Table S1) we showed a fetus with neural tube defect, single umbilical artery and clubfeet carrying a 22q11 microdeletion, which may be causative for NTD.24 Although the fetal phenotype was initially not suggestive for 22q11 microdeletion, based on the literature we classified this deletion as causative in this case.
Rarity of particular imbalances and postnatal bias
Rarity of abnormal cases and postnatal bias (most of the microdeletions are ascertained and described in postnatally tested patients/populations) also complicate the determination of a causal relationship between array finding and ultrasound features. Genetic syndromes are rare and emerging new postnatal cases dominate the literature. Present literature describing phenotypic effects of submicroscopic chromosome aberrations is most probably biased toward more severe phenotypes and genetic syndromes are mainly characterized by postnatal features including intellectual disability and dysmorphic features. Indeed, prenatal manifestations of microdeletion syndromes are mostly yet unknown.19 Therefore some cases may incorrectly be classified as unexpected diagnoses due to the lack of prenatal information in cases published so far.
Genotype–phenotype correlations are especially difficult in cases of larger CNVs, particularly in cases of unbalanced translocations, with similar chromosome imbalances hardly present in the literature. The only comparable cases often were not molecularly investigated and the precision of the breakpoints is limited to the cytogenetic nomenclature based on karyotyping or FISH. In such cases most often many genes are involved and the gene content may be different in cases that seem to have the same breakpoints based on karyotyping. Deletions and duplications of individual genes are so rare, that methodological epidemiological studies are often not possible. Because the phenotypic effect of large imbalances may result from an unique combination of deletion and/or duplication of several genes (either due to additive or multiplicative effect22), it is difficult to exclude that a particular ultrasound anomaly is not an effect of a large deletion/duplication even if a particular ultrasound feature was not seen in a patient with similar imbalance before. Although one could consider all large imbalances as causative, we carefully considered all individual cases.
Because of the limitations mentioned above, it is difficult to determine the actual ratio between clearly pathogenic causative findings and unexpected diagnoses in prenatal settings, which is reflected in the literature.25 Although unexpected findings are not new, there are only a few papers describing unexpected diagnoses in array testing.18, 26, 27
The frequencies of causative findings and unexpected diagnoses
Until now no paper specified the actual ratio between causative findings and UDs in prenatal array testing of fetuses with ultrasound anomalies. To assess the clinical utility of array testing, it is important not only to show the increase in causative findings, as we show in this paper, but also to evaluate the UDs that do not match the fetal phenotype or in general the indication of testing. The nature of UDs may influence the decision whether a particular test should be implemented in clinical practice. Because of the incompleteness of the fetal phenotype and of the fact that most submicroscopic pathogenic chromosome aberrations are associated with intellectual disability, we hypothesized that unexpected diagnoses of early-onset diseases may be of additional value in prenatal array testing.
We encountered UDs in ~0.5% of cases and these were ~7% of all pathogenic findings. Moreover, our results show that the UDs are also detected by karyotyping and that they represented about 5% (1/19) of pathogenic microscopic findings, which is comparable to the 7% (4/57) of pathogenic submicroscopic findings. The frequency of UDs in our cohort matches the frequency of pathogenic submicroscopic array findings in pregnancies without ultrasound abnormalities after excluding SL for neurodevelopmental diseases published in the literature.7, 9, 28, 29 Thus our results support the hypothesis that there is a background risk of ~0.5% for a clinically relevant submicroscopic chromosome abnormality (exclusive SL) in the general population.30 In our opinion, UDs found in our cohort are of additional value in routine prenatal genomic array testing as most (4/5) of them lead to (possibly) severe (including intellectual disability and/or a reduced life expectancy) early-onset diseases, which can be missed by ultrasound examination.
We did not unexpectedly detect any late-onset disease, which emphasizes the fact that we should not deny whole genome array testing due to the fear of detecting very rare late-onset diseases in prenatal diagnosis. However it is very important to set up a procedure and a policy for how to deal with such a finding. Therefore we recommend the formation of a broad multidisciplinary clinical team, including clinical geneticists, laboratory specialists, gynecologists and a psychologist that routinely discuss abnormal prenatal array cases to be able to accurately and rapidly deal with all kinds of pathogenic findings.
Incidentally pathogenic findings can also be encountered in parental samples (so called incidental findings).15 The frequency is dependent on the technique that has been used. In our cohort large chromosome aberrations were incidentally found during the quality control of the parental array profiles. It may be disputable whether such findings should be communicated to parents or whether we should better modify our protocol to avoid them. However, these IFs were clearly pathogenic and clinically actionable. Therefore our multidisciplinary team decided to communicate all findings summarized in Supplementary Table S4 to the patients. The individuals where a bone marrow malignancy was suspected remain under medical surveillance, the mosaic loss at chromosome 2 explained the recurrent miscarriages in one patient and the information on health issues such as increased risk of cardiovascular symptoms, endocrinological pathology and premature ovarian failure associated with Turner syndrome is clinically relevant.
SL for neurodevelopmental disorders
SL for neurodevelopmental phenotypes may be classified as pathogenic submicroscopic array findings in spite of their variable phenotypes and inheritance from (usually apparently) normal parents.15, 31, 32 It is well-established that the incidence of such CNVs among affected individuals is increased in comparison with the general population. Since these disorders of extreme phenotypic heterogeneity and/or of variable expressivity probably depend on the presence of one or more second-site variants to cause a disease,33, 34, 35 they should be classified as a separate category of findings even if they seem to match the indication.15 Moreover, at least some of the SL may modify abnormal phenotypes caused by well-known microdeletions/duplications resulting in more severe phenotypes than expected on the basis of the microdeletion/microduplication alone.34 Although there are already many cases of SL published, prenatal disclosure is still controversial because of the unquantifiable risks and unpredictable phenotypes after birth.36
In our cohort, SL were as often encountered as causative submicroscopic findings and therefore they need special clinical attention. Remarkably, our data show a statistically significant higher incidence of SL in fetuses with ultrasound abnormalities than in fetuses without ultrasound anomalies, 2.6% (27/1033) versus 1.35% (18/1330), respectively29 (odds ratio 1.956 with 95% CI 1.071, 3.572, P=0.01951) (Fisher exact). An increase in the prevalence of SL in affected pregnancies can also be observed in the data presented by Wapner et al (3.6 versus 0.8%) and by Scott et al (1.4 versus 0.55%).7, 37 If the phenotype of the SL involves a structural abnormality such as a heart anomaly in case of 1q21.1 duplication, an ultrasound investigation should be offered when the SL is encountered or when there is a family history of such a SL. Because of the higher prevalence of SL in fetuses with ultrasound abnormalities, it may seem to be defendable to offer a detailed second trimester ultrasound examination in all cases of SL or in families known to carry a SL. Moreover, long-term follow-up should also be done to provide more insight into the yet unquantifiable risk for an abnormal phenotype when a SL is prenatally detected in an uneventful pregnancy.
Conclusions
Our results confirm that a genomic SNP array should be the preferred first-tier technique to detect causative chromosome aberrations in fetuses with ultrasound anomalies. In addition to causative aberrations, UDs were found, but all involved early-onset diseases, which we considered to be beneficial for the patients to know. It also seems that UDs occur at a comparable frequency among microscopic and submicroscopic pathogenic findings. Cases of SL were more frequent than in cohorts of pregnancies without ultrasound anomalies and they were as common as causative submicroscopic findings in our cohort, therefore we believe they need clinical attention.
Acknowledgments
We thank all clinicians (gynecologists, sonographists and clinical geneticists) for referring patients and the prenatal laboratory technicians for their dedicated work and performing the tests.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Wladimiroff JW, Cohen-Overbeek TE, Ursem NT et al: Twenty years of experience in advanced ultrasound scanning for fetal anomalies in Rotterdam. Ned Tijdschr Geneeskd 2003; 147: 2106–2110. [PubMed] [Google Scholar]
- Caron L, Tihy F, Dallaire L: Frequencies of chromosomal abnormalities at amniocentesis: over 20 years of cytogenetic analyses in one laboratory. Am J Med Genet 1999; 82: 149–154. [DOI] [PubMed] [Google Scholar]
- Park SY, Kim JW, Kim YM et al: Frequencies of fetal chromosomal abnormalities at prenatal diagnosis: 10 years experiences in a single institution. J Korean Med Sci 2001; 16: 290–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman L, Fiegler H, Carter NP et al: Prenatal diagnosis by array-CGH. Eur J Med Genet 2005; 48: 232–240. [DOI] [PubMed] [Google Scholar]
- Hillman SC, Pretlove S, Coomarasamy A et al: Additional information from array comparative genomic hybridization technology over conventional karyotyping in prenatal diagnosis: a systematic review and meta-analysis. Ultrasound Obstet Gynecol 2011; 37: 6–14. [DOI] [PubMed] [Google Scholar]
- Srebniak MI, Boter M, Oudesluijs GO et al: Genomic SNP array as a gold standard for prenatal diagnosis of foetal ultrasound abnormalities. Mol Cytogenet 2012; 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapner RJ, Martin CL, Levy B et al: Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med 2012; 367: 2175–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman SC, McMullan DJ, Hall G et al: Prenatal chromosomal microarray use: a prospective cohort of fetuses and a systematic review and meta-analysis. Ultrasound Obstet Gynecol 2013; 41: 610–620. [DOI] [PubMed] [Google Scholar]
- Lee CN, Lin SY, Lin CH et al: Clinical utility of array comparative genomic hybridisation for prenatal diagnosis: a cohort study of 3171 pregnancies. BJOG 2012; 119: 614–625. [DOI] [PubMed] [Google Scholar]
- de Wit MC, Srebniak MI, Govaerts LC et al: The additional value of prenatal genomic array testing in fetuses with (isolated) structural ultrasound abnormalities and a normal karyotype: a systematic review of the literature. Ultrasound Obstet Gynecol 2014; 43: 139–146. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Rosenfeld JA, Dabell MP et al: Detection rates of clinically significant genomic alterations by microarray analysis for specific anomalies detected by ultrasound. Prenat Diagn 2012; 32: 986–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudjemline Y, Fermont L, Le Bidois J et al: Prevalence of 22q11 deletion in fetuses with conotruncal cardiac defects: a 6-year prospective study. J Pediatr 2001; 138: 520–524. [DOI] [PubMed] [Google Scholar]
- American College of Obstetricians and Gynecologists Committee on Genetics: ACOG Committee Opinion No. 581: the use of chromosomal microarray analysis in prenatal diagnosis. Obstet Gynecol 2013; 122: 1374–1377. [DOI] [PubMed] [Google Scholar]
- Vetro A, Bouman K, Hastings R et al: The introduction of arrays in prenatal diagnosis: a special challenge. Hum Mutat 2012; 33: 923–929. [DOI] [PubMed] [Google Scholar]
- Srebniak MI, Diderich KEM, Govaerts LCP et al: Types of array findings detectable in cytogenetic diagnosis: a proposal for a generic classification. Eur J Hum Genet 2014; 22: 856–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srebniak M, Boter M, Oudesluijs G et al: Application of SNP array for rapid prenatal diagnosis: implementation, genetic counselling and diagnostic flow. Eur J Hum Genet 2011; 19: 1230–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs ER, Church DM, Hanson K et al: Towards an evidence-based process for the clinical interpretation of copy number variation. Clin Genet 2012; 81: 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Opstal D, de Vries F, Govaerts L et al: Benefits and burdens of using a SNP array in pregnancies at increased risk for the common aneuploidies. Hum Mutat 2014; 36: 319–326. [DOI] [PubMed] [Google Scholar]
- Wapner RJ, Driscoll DA, Simpson JL: Integration of microarray technology into prenatal diagnosis: counselling issues generated during the NICHD clinical trial. Prenat Diagn 2012; 32: 396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan F, Zonana J, Gunter K et al: An atypical case of fragile X syndrome caused by a deletion that includes the FMR1 gene. Am J Hum Genet 1995; 56: 1042–1051. [PMC free article] [PubMed] [Google Scholar]
- Wolff DJ, Gustashaw KM, Zurcher V et al: Deletions in Xq26.3-q27.3 including FMR1 result in a severe phenotype in a male and variable phenotypes in females depending upon the X inactivation pattern. Hum Genet 1997; 100: 256–261. [DOI] [PubMed] [Google Scholar]
- Karmiloff-Smith A, Scerif G, Thomas M: Different approaches to relating genotype to phenotype in developmental disorders. Dev Psychobiol 2002; 40: 311–322. [DOI] [PubMed] [Google Scholar]
- Roper RJ, Reeves RH: Understanding the basis for Down syndrome phenotypes. PLoS Genet 2006; 2: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel RE, Pillers DA, Merkens M et al: Velo-cardio-facial syndrome and DiGeorge sequence with meningomyelocele and deletions of the 22q11 region. Am J Med Genet 1994; 52: 445–449. [DOI] [PubMed] [Google Scholar]
- Srebniak MI, Van Opstal D, Joosten M et al: Whole genome array as a first-line cytogenetic test in prenatal diagnosis. Ultrasound Obstet Gynecol 2014; 45: 363–372. [DOI] [PubMed] [Google Scholar]
- Cottrell CE, Prior TW, Pyatt R et al: Unexpected detection of dystrophin gene deletions by array comparative genomic hybridization. Am J Med Genet A 2010; 152A: 2301–2307. [DOI] [PubMed] [Google Scholar]
- Pichert G, Mohammed SN, Ahn JW et al: Unexpected findings in cancer predisposition genes detected by array comparative genomic hybridisation: what are the issues? J Med Genet 2011; 48: 535–539. [DOI] [PubMed] [Google Scholar]
- Fiorentino F, Napoletano S, Caiazzo F et al: Chromosomal microarray analysis as a first-line test in pregnancies with a priori low risk for the detection of submicroscopic chromosomal abnormalities. Eur J Hum Genet 2013; 21: 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Opstal D, de Vries F, Govaerts L et al: Benefits and burdens of using a SNP array in pregnancies at increased risk for the common aneuploidies. Hum Mutat 2015; 36: 319–326. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Dabell MP, Fisher AJ et al: Experience with microarray-based comparative genomic hybridization for prenatal diagnosis in over 5000 pregnancies. Prenat Diagn 2012; 32: 976–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky EB, Kaul V, Paschall J et al: An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet Med 2011; 13: 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S et al: A copy number variation morbidity map of developmental delay. Nat Genet 2011; 43: 838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman JA, Brunner HG: Understanding variable expressivity in microdeletion syndromes. Nat Genet 2010; 42: 192–193. [DOI] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Cooper GM et al: A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet 2010; 42: 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Coe BP et al: Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med 2012; 367: 1321–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld JA, Coe BP, Eichler EE et al: Estimates of penetrance for recurrent pathogenic copy-number variations. Genet Med 2013; 15: 478–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott F, Murphy K, Carey L et al: Prenatal diagnosis using combined quantitative fluorescent polymerase chain reaction and array comparative genomic hybridization analysis as a first-line test: results from over 1000 consecutive cases. Ultrasound Obstet Gynecol 2013; 41: 500–507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.