Abstract
The ends of each chromosome are capped by the telomere assembly to protect chromosomal integrity from telomere attrition and DNA damage. In response to DNA damage, DNA repair factors are enriched at damage sites by a sophisticated signaling and recruitment cascade. However, DNA damage response at telomeres is different from non-telomeric region of genomic DNA due to specialized sequences and structures of the telomeres. In the course of normal DNA replication or DNA damage repair, both the telomere shelterin protein complex and the condensed telomeric chromatin structure in mammalian cells are modified to protect telomeres from exposing free DNA ends which are subject to both telemere shortening and chromosome end fusion. Initiation of either homologous recombination or non-homologous end joint repair at telomeres requires disassembling and/or post-translational modifications of the shelterin complex and telomeric chromatin. In addition, cancer cells utilize distinct mechanisms to maintain telomere length and cell survival upon damage. In this review, we summarize current studies that focus on telomere end protection and telomere DNA repair using different methodologies to model telomere DNA damage and disruption. These include genetic ablation of sheltering proteins, targeting endonuclease to telomeres, and delivering oxidative damage directly. These different approaches, when combined, offer better understanding of the mechanistic differences in DNA damage response between telomeric and genomic DNA, which will provide new hope to identify potential cancer therapeutic targets to curtail cancer cell proliferation via induction of telomere dysfunctions.
Keywords: oxidative DNA damage, telomere, recombination, KillerRed, shelterin
Introduction
The integrity of the genome is crucial for sustained cell proliferation and hence plays a critical role in cancer development, treatment, and aging. Telomere oxidative damage and attrition leads to genomic instability and is a major threat to the maintenance of chromosomal integrity [1]. Telomeric DNA consists of long duplex telomeric DNA and single-strand DNA overhangs of TTAGGG repeats [2,3]. To avoid inappropriate DNA damage response at the end of chromosomes, the unique T-loop structure of telomeres is formed by protrusion of 3′ single-strand end into the DNA duplex strands to avoid the ends of the chromosome to be detected as a strand break. Meanwhile, telomere integrity is protected by the shelterin protein complex including TRF1, TRF2, RAP1, TIN2, POT1, and TPP1, which physically coats telomere repeating sequences [4,5]. Telomere region is frequently compromised by DNA damage, as evidenced by the frequent DNA damage foci observed at telomere sites during aging [6]. A most likely endogenous source is oxidation-mediated base modifications and associated strand breaks [7–9]. In addition, the densely compacted nature of telomere by shelterin protein complex may render telomere DNA damage more resistant to DNA repair [10]. If not repaired efficiently, damaged telomeres will suffer accelerated erosion and restrict the proliferative lifespan of cells [11]. To date, how the telomeric structure and the shelterin proteins protect telomere in the face of frequent DNA damage is an urgent issue for understanding the role of telomeres during oncogenesis. Furthermore, many cancer therapies involve DNA-targeting modalities that have a direct effect on telomeres. For example, 40% of the cancers are treated by radiation therapy, which introduces base oxidation, DNA strand breaks on both nuclear DNA and telomere DNA. Understanding how cells respond to and repair oxidation-induced damage is important for controlling cancer progression and response to DNA-targeting therapies.
DNA Damage Response Induced by Genetic Ablation of Shelterin Proteins
Several groups, including the de Lange laboratory, have made the original discovery that inappropriate engagement of DNA repair mechanisms at normal telomeres is avoided by the assembly of shelterin proteins onto telomere DNA structures [12,13]. Among the key shelterin proteins, TRF1 and TRF2 share protein structural resemblance and bind to double-strand telomeric sequence TTAGGG. However, each protein seems to offer a different type of telomere protection mechanism. Early studies showed that inhibition of endogenous TRF2 function via a dominant negative mutant resulted in loss of telomeric 3′-overhang and end-to-end chromosome fusion, which activated ATM- and p53-dependent cell cycle arrest and apoptosis, respectively [14,15]. Knockout of TRF2 further confirmed the presence of DNA damage response at chromosome ends and recapitulated the activation of ATM and end-to-end chromosome fusion phenotypes [16–18]. Several DNA repair factors, MRN, ATM, Ku70–80, ligase IV, and 53BP1 which are involved in non-homologous end joining (NHEJ) pathway and damage signaling, have been identified to participate in NHEJ-induced chromosome end fusions at telomeres [16,19–21]. However, inhibition of other shelterin components including TRF1 and POT1/Tpp1 in the presence of TRF2 activates Ataxia telangiectasia and Rad3 related signal but not ATM. Homologous recombination (HR) is triggered by the absence of Pot1 function, underlying the observed increase in telomere-sister chromatin exchanges [22–24]. Pot1 depletion in the absence of TRF1 and TRF2 unravels a new DNA repair pathway, alternative-NHEJ at telomeric DNA repair [13], indicating that protective functions of shelterin proteins on telomere are distinct yet partially redundant in regulating different damage repair pathways.
DNA damage-induced and aging-associated telomere shortening is often accompanied by insufficient protection of shelterin proteins [4]. The enrichment of DNA repair factors at genomic DNA double-strand breaks engages either the NHEJ or the HR pathway. NHEJ is an error-prone repair pathway and tends to introduce deletions or insertions both in nucleosomal and telomere DNA. Upon DNA damage, the shelterin proteins do not block the recruitment of a specific set of repair proteins; however, the existence of DNA damage per se does not seem to be sufficient to activate DNA repair in the presence of the intact shelterin–telomere structure [6]. Loss of telomere shelterin proteins in cells results in the initiation of NHEJ repair or HR repair. NHEJ-induced chromosome end fusion is proposed to be a major mechanism of telomere crisis-induced cell death and cancerous transformation [25,26]. The recruitment of NHEJ repair factor in the presence of shelterin proteins at telomere is more likely to confer a signal event for cell cycle arrest than the full deployment of the NHEJ process. Unlike the genomic DNA damage which arrests cells at G2/M phases, telomere DNA damage tends to trigger G1 phase arrest [27]. Moreover, interactions between shelterin proteins and DNA repair factors can mediate the feedback for DNA damage response at telomeres and regulate the spatial and temporal activation of checkpoint kinase [28,29]. The physiological significance of separation between the recruitment of NHEJ factors and the execution of NHEJ process at telomere needs to be further elucidated. Together, the fundamental difference between shelterin-based telomere structure and nucleosome structure entails distinct regulatory mechanisms for how DNA damage repair mechanisms are applied. The roles of each shelterin protein in the selective activation of DNA damage responses also warrant further studies.
HR and Alternative Lengthening of Telomeres Cells
Telomerase-deficient cells adapt to the alternative lengthening of telomeres (ALT) pathway to maintain their telomere length. The constitutive presence of HR factors at telomeres, identified as ALT-associated promyelocytic leukemia bodies, is employed to maintain telomere length and cellular immortalization by HR. Thus, ALT cells are characterized by the heterogeneous telomere length and constant localization of HR proteins at telomere. About 10%–15% cancer cells acquire the ALT mechanism to support the infinite proliferation and survival [30]. In addition to supporting ALT-based elongation of telomeres, HR functions as the major damage repair pathway since telomeres are intrinsically resistant to repair through the NHEJ pathway. Recently, TRF1-tagged FOK1, a sequence-independent DNA endonuclease, has emerged as a new approach to model DNA double-strand break repair at telomeres. The TRF1-Fok1 fusion protein induces double-strand breaks which exhibit enhanced telomere movement in ALT cells, presumably resulted from the increased homology searching from the HR-mediated damage repair [31]. It remains unclear as to what are the intrinsic determinants to control ALT cells to utilize HR as the first choice to repair double-strand breaks at telomeres. The special structure of single-strand G-rich overhang at telomeres might serve as a potential marker distinct from nucleosomal DNA. The single-strand portion of telomere sequence is coated by the shelterin protein POT1 and may potentially function to avoid HR initiation [17,22]. It is plausible that regulated POT1 removal or exposed single-strand telomeric DNA can serve as an initiating event to allow Rad51 binding and the subsequent homology search. Future studies on how HR is regulated by shelterin proteins with or without telomerase will help to develop mechanism-specific cancer therapy for ALT- and telomerase-positive cells.
Effects of Oxidative Damage on Telomeres, Cell Senescence, and Cancer
Oxidative DNA damage accumulates during aging. Reactive oxygen radicals in the forms of superoxide and other derivative forms are mainly originated from mitochondria metabolism, which is enhanced during elevated oncogenic metabolic stress. When the amplitude of reactive oxygen species (ROS) production exceeds the cellular capacity of available ROS scavengers and enzymatic capabilities, nuclear DNA will be inevitably subject to the overload of oxidative DNA damage. The crucial marker for aging is the gradual shortening of telomere, which can be induced by increased oxidative stress [1,32,33]. The consequence of increased formation of 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxoG) in the G-rich telomere repeat may saturate the base-excision repair capacity and contribute to telomere shortening [1,33,34]. As a result, the accumulation of oxidative DNA damage may lead to both telomere shortening and strand breaks from stalled telomere replication process [34]. However, testing these potential scenarios has proven to be technically challenging because modeling oxidative DNA damage response at the telomeres requires that induction of oxidative damage is specific to the telomeric DNA region.
Studies on telomere DNA damage response, for the most part, have been carried out by exposing cells globally to DNA damaging agents. In the case of oxidative DNA damage, peroxide treatment is the typical method of choice. These treatments induce global DNA damage, which affect gene expression that generates confounding effects on telomeres and the genome. Moreover, global cellular exposure to DNA damaging agents also incurs extensive cytoplasmic effects including mitochondrial dysfunction and metabolic disruptions, which also impacts nuclear functions and potentially telomere maintenance [35–37]. Additionally, because telomere sequences account for <0.5% of the chromosomal DNA in any mammalian genome [38], >99% of the damage will be on chromatin DNA rather than on telomeric DNA when exogenous damage is applied to the whole cell. Genetic manipulation of shelterin proteins or using TRF-localized endonuclease at telomeres, while capable of generating telomere-specific effects, does not fully recapitulate the primary and secondary effects of oxidative telomere DNA damage. The KillerRed (KR) protein is an engineered derivative of the red florescent protein [39]. Upon excitation by a visible light photon, it emits an oxygen atom with linear dose dependence. Our laboratory has generated a KR-TRF1 fusion in which the protein functions are preserved. When expressed in cells, the KR-TRF1 is incorporated into the shelterin complex at the telomeres. Upon microscopic visible light beam (550–580 nm) illumination, oxidative DNA damage can be introduced in a spatial and temporal manner with an extended linear dosage for damage delivery. It also allows that oxidative damage is selectively inflicted on telomeres specifically and simultaneously in a single cell. Moreover, the instantaneous damage formation makes it possible to conduct live cell imaging for kinetic studies of DNA repair factor recruitment and telomere end dynamics [34]. Compared with pan cellular exposure to damaging reagents, oxidative lesions inflicted by KR provide a well-defined platform for studying telomeric oxidative DNA damage response with far fewer confounding elements.
Using the KR-TEL system, our group has shown that telomeric oxidative DNA damage leads to a number of cellular responses including increased cellular senescence, cell death, telomere loss, and sister chromatid association [34]. These observations demonstrate the importance of oxidative lesion repair in telomeres and in chromosomal integrity. The phenotype of telomere oxidation in cells is different from what is observed from shelterin protein depletion which induces primarily chromosomal fusions in the case of TRF2. The distinct roles of shelterin proteins are necessary for protecting telomere DNA integrity. Shelterin protein disruption, as an experimental approach, is ideal for elucidating the telomere structure but not adequate for studying damage response under physiological conditions. Complete loss of capping and resolution of the T-loop structure is unlikely sustainable even for cancer cells. Meanwhile, oxidative DNA damage may be one of the main sources of telomere instability that threatens the minimum essential chromosomal stability of cancer cells. Therefore, oxidative DNA damage introduced to normal telomere structures should be an ideal model to reveal the physiological response of telomeres to aging and DNA damage. This approach is complementary to the TRF protein-guided endonuclease which allows modeling of DNA strand breaks occurred at telomere DNA. The combined use of both the KR-TEL and TRF-FOK1 systems will provide a more in-depth understanding of telomere biology during aging and cancer development.
The KR-TEL system generates oxidative telomeric DNA damage which ranges from base damages to double-strand breaks. Upon DNA damage, base-excision repair factors, single- and double-strand break repair proteins are detectable at the damaged telomeres, indicating that the same repair process is shared by nucleosomal and telomeric DNA [34]. To date, our knowledge is limited with regard to whether or how the same DNA repair machinery might operate differentially to counteract the primary and secondary effects of oxidative DNA damage between telomere and nucleosomal DNA. Compared with the substantial knowledge and advances in telomere structural biology, the understanding of how oxidative stress and oxidative damage are repaired at the telomeres is relatively far from enough. It will be important to further investigate how telomere integrity is maintained during oncogenesis with implications on tumor treatment.
In addition to aberrant signaling-induced cell senescence, telomere dysfunction also induces replicative senescence [40]. Although both types of senescence are characterized by the DNA damage foci at telomeres and activation of cell cycle checkpoint mechanisms [41,42], replicative senescence can actually be reversed by the introduction of active telomerase. Conversely, senescence triggered by accumulated and persistent DNA damage at telomeres, such as those induced by genotoxic and oxidative stresses, seems to be more frequently compounded by the fact that the telomere DNA base content and repetitive nature renders it more prone to DNA damage than nucleosomal DNA [7]. Therefore, endogenous DNA lesions are the prevalent elements in inducing cellular senescence regardless of telomere length or structural status. The elevated spontaneous damage at telomeres can be found in cells with low telomerase activity or in ALT cells with impaired binding capacity of shelterin proteins [6]. Future studies regarding the interplays among telomerase, shelterin proteins with or without oxidation of telomeres are expected to improve our understanding of this important aspect of telomere physiology.
Telomeric Chromatin and Chromosome Chromatin upon DNA Damage
Besides the shelterin complex, telomeres have a condensed chromatin structure as an additional protection against DNA damage. Genomic nucleosomes consist of core histones H2A, H2B, H3, and H4 that form the histone octamer which is wrapped by ~147 bp DNA. The nucleosomes are separated by linker DNAs associated with histone H1. Compared with the genomic nucleosomes, telomeric chromatin forms more compact nucleosomal structure with shorter nucleosomal linkers, highlighting the particular configuration of telomeric chromatin [43,44]. The telomere chromatin resembles heterochromatin, as evidenced by the presence of heterochromatin markers, such as high levels of trimethylation of H3K9 and H4K20 and low levels of acetylated H3 and H4 [45], a common pattern found in yeast, flies, and mammalian cells [46–48]. The telomeric chromatin is further assembled to form telomere structure by interacting with shelterin proteins [49,50]. Similar to the normal nucleosomal DNA, histone markers such as H3K9me3 and γH2AX serve as the DNA damage anchors at the telomeres to recruit DNA repair proteins. As a result, the condensed structure of telomere chromatin is altered by chromatin modifying enzymes such as histone methyltransferases [47,48,51], deacetylase [52,53], and histone code readers [46]. These concerted events create access for DNA repair factors and facilitate HR or lesion removal. However, the heterochromatin characteristics of telomere may help telomere to maintain a more compact state for the better concealment of DNA ends and to function synergistically with shelterin proteins to protect telomere from DNA damage signal recognition [49,50,54].
Regardless of the protective function of the telomere structure, telomeric DNA is inevitably exposed temporally during replication. Extensive studies have shown that uncapped telomeric DNA, either due to replication or due to loss of shelterin proteins, leads to altered telomeric chromatin modifications, resulting in de-condensation of telomeric chromatin and accumulation of DNA damage response proteins [55–57]. In addition, uncapping the telomere by removal of the shelterin complex induces strong DNA damage response and telomere fusions, but it keeps the telomeric DNA intact in the nucleosome [13]. However, how the removal of the shelterin complex leads to the changes in the modification of telomeric chromatin as well as the inner chromosomal structural changes is still unknown. Post-translational modifications of telomeric chromatin are important for both the maintenance of telomere DNA and the repair of telomere DNA damage [58], suggesting that histone modification-mediated chromatin remodeling is a crucial step in the creation of accessible DNA and in recruiting DNA damage response proteins. Moreover, shelterin proteins also serve as docking sites to recruit protein complex during damage repair. Both the heterochromatin structure and shelterin proteins play important roles in telomere protection. How the two protective mechanisms cooperate in protecting telomere integrity needs to be further investigated.
Conclusion
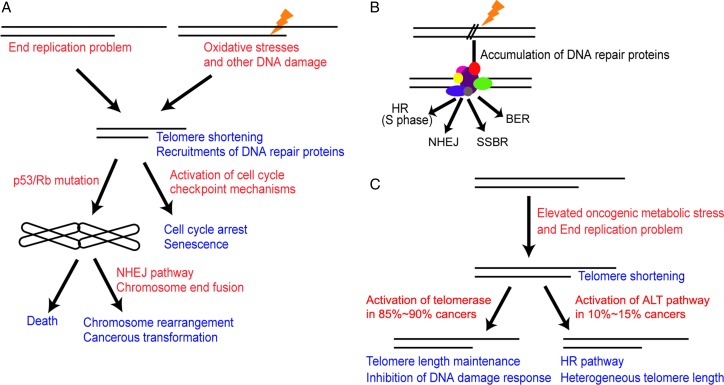
Telomeres provide a safeguard to the very end of the chromosomal DNA to protect it from DNA damage and erroneous processing. Factors, such as incomplete end replication, exposure to intracellular and extracellular DNA damage, and insufficient protection from shelterin proteins from aging or genetic deficiencies, will subject the telomeres to accelerated shortening. In normal cells, accumulation of DNA damage at telomeres serves as a signal to activate cell cycle checkpoint mechanisms rather than immediately initiate NHEJ- or HR-based repairs. However, elevated telomere DNA damage will activate the p53 and Rb pathways and arrest cell proliferation to enter the senescent state. Cells deficient in p53 or Rb are able to circumvent senescence and continue to proliferate with eroding telomeres. This will eventually induce chromosome end fusions and in most cases chromosomal catastrophe. Telomere crisis, characterized by telomere structural disintegration and chromosome fusion phenotypes, can result in cell death as well as gross chromosome instabilities [25]. Cells that survive the telomere crisis are highly predisposed to cancer development [26,59] (Fig. 1A). In contrast, non-telomeric chromosomal DNA damage activates the DNA damage signal cascade and initiates the immediate process by the DNA repair enzymes. DNA repair is carried out more efficiently in nucleosomal DNA which possesses a relatively simple structure than the telomere (Fig. 1B). In immortalized cancer cells, around 80%–90% cells have telomerase expression and 10%–15% cells depend on ALT for telomere elongation. Telomerase extends the telomere length and inhibits the DNA damage signaling. ALT cells utilize HR to maintain the functionality of telomeres, which often results in heterogeneous telomere size distribution (Fig. 1C). Given the essential role of telomeres in chromosomal stability and in human diseases, further understanding of DNA damage response of telomeres will better our understanding of oncogenesis and aging in general [60–62]. The availability of novel and effective methods of delivering defined DNA damage specifically at telomeres will significantly improve the understanding on this unique chromosomal structure, which is highly protective of the chromosome ends and also presents barriers to DNA damage removal. Future studies are required to unravel how cells resolve such conflict to maintain genome stability.
Figure 1.
DNA damage response at telomeres and chromosome sites in somatic cells and cancer cells (A) Telomeric DNA damage response in normal cells. Cells consistently lose telomeres with each replication cycle and/or with the exposure to endogenous and exogenous DNA damages. Upon DNA damage, repair proteins are recruited to damaged telomeres, which also serve as signals for the activation of the cell cycle checkpoint mechanism rather than initiation of impropriate repair leading to chromosomal fusions. The persistent cell cycle arrest transits the cells into senescence. The deficiencies of p53 or Rb genes antagonize cell cycle arrest, leading to cell death or cancerous transformation. Cells that undergo ‘telomere crisis’ exhibit short telomeres or chromosomal fusions. (B) DNA damage response at non-telomeric regions. A subsequent DNA repair pathways including base-excision repair (BER), single-strand break repair (SSBR), HR, and NHEJ and the sequential DNA damage signal cascade are activated at sites of DNA damage. (C) Telomere maintenance in cancer cells. About 80% cancer cells express telomerase and 15% cancer cells use ALT pathway for telomere maintenance. Telomerase catalyzes the elongation of the telomeres and inhibits the DNA damage signals. Cancer cells utilize ALT pathway to prevent telomere shortening characterized by heterogeneous telomere lengths. ALT cancer cells are resistant to NHEJ in response to telomeric DNA damage, but show frequent recruitments of HR proteins at telomeres.
Funding
This work was supported by the grant from the National Institutes of Health (NIH) (No. AG045545).
References
- 1.von Zglinicki T. Oxidative stress shortens telomeres. Trends Biochem Sci 2002, 27: 339–344. [DOI] [PubMed] [Google Scholar]
- 2.Henderson E, Hardin CC, Walk SK, Tinoco I Jr, Blackburn EH. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell 1987, 51: 899–908. [DOI] [PubMed] [Google Scholar]
- 3.Henderson ER, Blackburn EH. An overhanging 3′ terminus is a conserved feature of telomeres. Mol Cell Biol 1989, 9: 345–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 2005, 19: 2100–2110. [DOI] [PubMed] [Google Scholar]
- 5.Songyang Z, Liu D. Inside the mammalian telomere interactome: regulation and regulatory activities of telomeres. Crit Rev Eukary Gene Expr 2006, 16: 103–118. [DOI] [PubMed] [Google Scholar]
- 6.Cesare AJ, Kaul Z, Cohen SB, Napier CE, Pickett HA, Neumann AA, Reddel RR. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat Struct Mol Biol 2009, 16: 1244–1251. [DOI] [PubMed] [Google Scholar]
- 7.Hewitt G, Jurk D, Marques FD, Correia-Melo C, Hardy T, Gackowska A, Anderson R, et al. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat Commun 2012, 3: 708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen S, Saretzki G, von Zglinicki T. Preferential accumulation of single-stranded regions in telomeres of human fibroblasts. Exp Cell Res 1998, 239: 152–160. [DOI] [PubMed] [Google Scholar]
- 9.Rhee DB, Ghosh A, Lu J, Bohr VA, Liu Y. Factors that influence telomeric oxidative base damage and repair by DNA glycosylase OGG1. DNA Rep 2010, 10: 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fumagalli M, Rossiello F, Clerici M, Barozzi S, Cittaro D, Kaplunov JM, Bucci G, et al. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat Cell Biol 2012, 14: 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Rhee DB, Lu J, Bohr CT, Zhou F, Vallabhaneni H, de Souza-Pinto NC, et al. Characterization of oxidative guanine damage and repair in mammalian telomeres. PLoS Genet 2010, 6: e1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lange T. How telomeres solve the end-protection problem. Science 2009, 326: 948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science 2012, 336: 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell 1998, 92: 401–413. [DOI] [PubMed] [Google Scholar]
- 15.Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 1999, 283: 1321–1325. [DOI] [PubMed] [Google Scholar]
- 16.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol 2005, 7: 712–718. [DOI] [PubMed] [Google Scholar]
- 17.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 2007, 448: 1068–1071. [DOI] [PubMed] [Google Scholar]
- 18.Konishi A, de Lange T. Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes Dev 2008, 22: 1221–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng Y, Guo X, Ferguson DO, Chang S. Multiple roles for MRE11 at uncapped telomeres. Nature 2009, 460: 914–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol 2006, 8: 885–890. [DOI] [PubMed] [Google Scholar]
- 21.Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 2008, 456: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 2006, 126: 49–62. [DOI] [PubMed] [Google Scholar]
- 23.Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell 2006, 126: 63–77. [DOI] [PubMed] [Google Scholar]
- 24.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 2009, 138: 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi MT, Cesare AJ, Rivera T, Karlseder J. Cell death during crisis is mediated by mitotic telomere deprotection. Nature 2015, 522: 492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maciejowski J, Li Y, Bosco N, Campbell PJ, de Lange T. Chromothripsis and kataegis induced by telomere crisis. Cell 2015, 163: 1641–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cesare AJ, Hayashi MT, Crabbe L, Karlseder J. The telomere deprotection response is functionally distinct from the genomic DNA damage response. Mol Cell 2013, 51: 141–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR III, Denchi EL. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature 2013, 494: 502–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rai R, Li JM, Zheng H, Lok GT, Deng Y, Huen MS, Chen J, et al. The E3 ubiquitin ligase Rnf8 stabilizes Tpp1 to promote telomere end protection. Nat Struct Mol Biol 2011, 18: 1400–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med 1997, 3: 1271–1274. [DOI] [PubMed] [Google Scholar]
- 31.Cho NW, Dilley RL, Lampson MA, Greenberg RA. Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell 2014, 159: 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grahame TJ, Schlesinger RB. Oxidative stress-induced telomeric erosion as a mechanism underlying airborne particulate matter-related cardiovascular disease. Part Fibre Toxicol 2012, 9: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawanishi S, Oikawa S.. Mechanism of telomere shortening by oxidative stress. Ann NY Acad Sci 2004, 1019: 278–284. [DOI] [PubMed] [Google Scholar]
- 34.Sun L, Tan R, Xu J, LaFace J, Gao Y, Xiao Y, Attar M, et al. Targeted DNA damage at individual telomeres disrupts their integrity and triggers cell death. Nucleic Acids Res 2015, 43: 6334–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malinin NL, West XZ, Byzova TV. Oxidation as ‘the stress of life’. Aging 2011, 3: 906–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Passos JF, Saretzki G, Ahmed S, Nelson G, Richter T, Peters H, Wappler I, et al. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol 2007, 5: e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, Cooper M, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature 2011, 470: 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding Z, Mangino M, Aviv A, Spector T, Durbin R, Consortium UK. Estimating telomere length from whole genome sequence data. Nucleic Acids Res 2014, 42: e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulina ME, Lukyanov KA, Britanova OV, Onichtchouk D, Lukyanov S, Chudakov DM. Chromophore-assisted light inactivation (CALI) using the phototoxic fluorescent protein KillerRed. Nat Protoc 2006, 1: 947–953. [DOI] [PubMed] [Google Scholar]
- 40.Shay JW, Roninson IB. Hallmarks of senescence in carcinogenesis and cancer therapy. Oncogene 2004, 23: 2919–2933. [DOI] [PubMed] [Google Scholar]
- 41.d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003, 426: 194–198. [DOI] [PubMed] [Google Scholar]
- 42.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol Cell 2004, 14: 501–513. [DOI] [PubMed] [Google Scholar]
- 43.Makarov VL, Lejnine S, Bedoyan J, Langmore JP. Nucleosomal organization of telomere-specific chromatin in rat. Cell 1993, 73: 775–787. [DOI] [PubMed] [Google Scholar]
- 44.Tommerup H, Dousmanis A, de Lange T. Unusual chromatin in human telomeres. Mol Cell Biol 1994, 14: 5777–5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benetti R, Garcia-Cao M, Blasco MA. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat Genet 2007, 39: 243–250. [DOI] [PubMed] [Google Scholar]
- 46.Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, Eguia R, et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol 2005, 7: 420–428. [DOI] [PubMed] [Google Scholar]
- 47.Garcia-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet 2004, 36: 94–99. [DOI] [PubMed] [Google Scholar]
- 48.Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol 2006, 8: 416–424. [DOI] [PubMed] [Google Scholar]
- 49.Galati A, Magdinier F, Colasanti V, Bauwens S, Pinte S, Ricordy R, Giraud-Panis MJ, et al. TRF2 controls telomeric nucleosome organization in a cell cycle phase-dependent manner. PLoS One 2012, 7: e34386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pisano S, Leoni D, Galati A, Rhodes D, Savino M, Cacchione S. The human telomeric protein hTRF1 induces telomere-specific nucleosome mobility. Nucleic Acids Res 2010, 38: 2247–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, Jenuwein T, Blasco MA. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J Cell Biol 2007, 178: 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 2008, 452: 492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palacios JA, Herranz D, De Bonis ML, Velasco S, Serrano M, Blasco MA. SIRT1 contributes to telomere maintenance and augments global homologous recombination. J Cell Biol 2010, 191: 1299–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Canudas S, Houghtaling BR, Bhanot M, Sasa G, Savage SA, Bertuch AA, Smith S. A role for heterochromatin protein 1gamma at human telomeres. Genes Dev 2011, 25: 1807–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol 2010, 17: 1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bartocci C, Diedrich JK, Ouzounov I, Li J, Piunti A, Pasini D, Yates JR III, et al. Isolation of chromatin from dysfunctional telomeres reveals an important role for Ring1b in NHEJ-mediated chromosome fusions. Cell Rep 2014, 7: 1320–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peuscher MH, Jacobs JJ. DNA-damage response and repair activities at uncapped telomeres depend on RNF8. Nat Cell Biol 2011, 13: 1139–1145 [DOI] [PubMed] [Google Scholar]
- 58.Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet 2007, 8: 299–309. [DOI] [PubMed] [Google Scholar]
- 59.Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 2000, 406: 641–645. [DOI] [PubMed] [Google Scholar]
- 60.Armanios M. Syndromes of telomere shortening. Annu Rev Genom Hum Genet 2009, 10: 45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Armanios M, Alder JK, Parry EM, Karim B, Strong MA, Greider CW. Short telomeres are sufficient to cause the degenerative defects associated with aging. Am J Hum Genet 2009, 85: 823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willeit P, Willeit J, Mayr A, Weger S, Oberhollenzer F, Brandstatter A, Kronenberg F, et al. Telomere length and risk of incident cancer and cancer mortality. JAMA 2010, 304: 69–75. [DOI] [PubMed] [Google Scholar]