Abstract
A genome-wide differential expression of long noncoding RNAs (lncRNAs) was identified in blood specimens of autism spectrum disorder (ASD). A total of 3929 lncRNAs were found to be differentially expressed in ASD peripheral leukocytes, including 2407 that were upregulated and 1522 that were downregulated. Simultaneously, 2591 messenger RNAs (mRNAs), including 1789 upregulated and 821 downregulated, were also identified in ASD leukocytes. Functional pathway analysis of these lncRNAs revealed neurological pathways of the synaptic vesicle cycling, long-term depression and long-term potentiation to be primarily involved. Thirteen synaptic lncRNAs, including nine upregulated and four downregulated, and 19 synaptic mRNAs, including 12 upregulated and seven downregulated, were identified as being differentially expressed in ASD. Our identification of differential expression of synaptic lncRNAs and mRNAs suggested that synaptic vesicle transportation and cycling are important for the delivery of synaptosomal protein(s) between presynaptic and postsynaptic membranes in ASD. Finding of 19 lncRNAs, which are the antisense, bi-directional and intergenic, of HOX genes may lead us to investigate the role of HOX genes involved in the development of ASD. Discovery of the lncRNAs of SHANK2-AS and BDNF-AS, the natural antisense of genes SHANK2 and BDNF, respectively, indicates that in addition to gene mutations, deregulation of lncRNAs on ASD-causing gene loci presents a new approach for exploring possible epigenetic mechanisms underlying ASD. Our study also opened a new avenue for exploring the use of lncRNA(s) as biomarker(s) for the early detection of ASD.
Introduction
Autism spectrum disorder (ASD) has a reported prevalence of 1 in 68 children in the United States.1 ASD is a grouping of lifelong neurodevelopmental disorders, characterized by impairments in reciprocal social interaction and communication, and the presence of stereotypical behaviors, interests or activities. The etiology of ASD is not yet well understood. Although mutations of many genes, including NLGN3, NLGN4, NRXN1, SHANK2, SHANK3 and PTCHD1, have been associated with ASD,2, 3 metabolic, infectious, inflammatory and other environmental factors have also been implicated in the pathogenesis of ASD.4, 5, 6, 7, 8, 9 We previously determined that hypermethylation of the ENO2 gene is present in 15% of children with ASD,10 indicating that epigenetic factor(s) may contribute to the etiology of ASD.3, 11 In addition, as transcriptional and posttranscriptional regulators, both microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) have been reported to be involved in ASD as well as in many other neurological disorders.12, 13, 14, 15, 16, 17, 18, 19, 20, 21 Overexpression and knockdown studies have shown that lncRNAs have important roles in regulating a variety of processes, including splicing,22 transcription,23 localization24 and the organization of subcellular compartments.25, 26, 27, 28, 29, 30, 31 Underscoring the importance of lncRNAs' regulatory roles is their emergence as essential components in the etiology of many disorders, and of complex diseases in particular, for which genetic and environmental interactions have key roles.31, 32, 33, 34
LncRNAs are a subset of RNA molecules greater than 200 nt in length that are transcribed but not translated. They may be positioned in genomic sequences as antisense, intronic and large intergenic noncoding RNAs (ncRNAs), as well as at promoter-associated and untranslated regions, which function as cis or trans regulators. LncRNAs can function as translational and posttranslational regulators of brain development and differentiation, and are associated with various human brain disorders.16, 17, 18, 19, 20, 21, 35, 36, 37, 38, 39, 40 LncRNAs have been reported to be involved in many complex diseases, including neurodegenerative and psychiatric diseases, cardiovascular disease, immune dysfunction and auto-immunity, carcinogenesis and reproductive diseases.41, 42, 43, 44, 45, 46, 47, 48, 49, 50 Deregulation of lncRNAs is becoming recognized as a major feature of many types of diseases. Importantly, cancer-associated lncRNAs may serve as diagnostic or predictive biomarkers and provide targets for new therapeutic strategies for selective silencing.51 Among 168 human diseases that have been found to be associated with lncRNAs, and that are recorded in the lncrnadisease (http://cmbi.bjmu.edu.cn/lncrnadisease) database, neurological diseases, cardiovascular diseases and cancers account for 8.3%, 10.7% and 40.5%, respectively.52
Altered lncRNA levels have been identified in ASD brains.21, 53, 54, 55 In one study of ~33 000 annotated lncRNAs and 30 000 messenger RNA (mRNA) transcripts from the postmortem prefrontal cortex and cerebellar tissues of two ASD and two control subjects, over 200 differentially expressed lncRNAs were detected. These differentially expressed lncRNAs in the ASD subjects were enriched for genomic regions containing genes related to neurodevelopment and neuropsychiatric diseases. Comparison of differences in the expression of mRNAs between the prefrontal cortex and the cerebellum within individual ASD brains showed more transcriptional homogeneity than within control brains. This finding was also true of the lncRNA transcriptome.55 Abnormalities in mRNA expression in ASD have also been observed in peripheral blood mononuclear cells, which are safely and easily assayed in infants and offer the potential of a peripheral blood-based, early biomarker panel to detect risk for ASD in infants and toddlers.56 We undertook this study to determine whether lncRNAs are differentially expressed in the blood of individuals with ASD, rather than in ASD brains. Our positive findings may open a new approach to investigate potential epigenetic mechanisms underlying ASD and to explore biomarker identifications for possible clinical screening and diagnosis of ASD.
Materials and methods
Ethics statement
The Hospital Ethics Committee reviewed and approved the research project. Informed consent was obtained from the parents of the participating children. All the material and data were previously de-identified and coded, and were anonymous to the investigators.
Subjects
Twenty-five pairs of gender- and age-matched Chinese ASD and control children were recruited for this discovery study at their first-time clinical visit before any clinical laboratory studies, intervention or medication. The children with ASD were clinically diagnosed by means of DSM-IV criteria and did not have epilepsy, any physical disabilities or family history of ASD. The controls were phenotypically and developmentally normal children who were undergoing an annual health checkup. There were 17 pairs of boys and eight pairs of girls, 3–5 years of age, in both the ASD and control groups. Lymphocytes were isolated from 3 to 5 ml of peripheral blood specimens of the Caucasian participants and stored at −70 °C until total RNA was extracted with a Qiagen Mini kit (Qiagen, Valencia, CA, USA). In addition, total RNAs, isolated from 10 lymphoblast cell lines derived from Caucasian children (seven boys and three girls, aged 3 to 8 years) with ASD, were subjected to the validation study. There were no Caucasian control samples used for the validation.
Microarray hybridization
The Arraystar Human LncRNA Array v2.0 (www.arraystar.com), which detects genome-wide lncRNAs and mRNAs simultaneously, was used for this study. This array covers 33 045 lncRNAs and 30 218 mRNAs that were identified from authoritative data sources, including RefSeq, UCSC Knowngenes and Ensembl. RNA labeling and array hybridizations were performed according to the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technologies, Santa Clara, CA, USA) with minor modifications. Briefly, mRNA was purified from total RNA after the removal of ribosomal RNA with the mRNA-ONLY Eukaryotic mRNA Isolation Kit (Epicentre, Omaha, NE, USA). Each sample was amplified and transcribed into fluorescent complementary RNA (cRNA) along the entire length of the transcripts without a 3′ bias, utilizing the random priming method. The labeled cRNAs were purified using an RNeasy Mini Kit (Qiagen). The concentration and specific activity of the labeled cRNAs (pmol Cy3 per μg cRNA) were measured with NanoDrop ND-1000. One microgram of each labeled cRNA was fragmented by adding 5 μl of 10 × blocking agent and 1 μl of 25 × fragmentation buffer, and then heating the mixture to 60 °C for 30 min. Finally, 25 μl 2 × GE of hybridization buffer was added to dilute the labeled cRNA. Fifty microliters of hybridization solution was dispensed onto the gasket slide and assembled with the lncRNA expression microarray slide. The slides were incubated for 17 h at 65 °C in an Agilent hybridization oven. The hybridized arrays were washed, fixed and scanned by using the Agilent DNA Microarray Scanner (Agilent Technologies). Agilent Feature Extraction software (version 11.0.1.1) was used to analyze the acquired array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX v12.1 software package (Agilent Technologies). After normalization of the raw data, lncRNAs and mRNAs that had flags (‘All Targets Value') were chosen for further data analysis. Differentially expressed lncRNAs and mRNAs between the two groups with statistical significance were identified through volcano plot filtering. Hierarchical clustering was performed using the Agilent GeneSpring GX software (Version 12.1). Both ‘GO analysis' and ‘Pathway analysis' were performed with the DAVID program (http://david.abcc.ncifcrf.gov), in which analysis of gene ontology (GO) and KEGG PATHWAY was conducted. The results were also analyzed using the genetic and molecular interaction software GeneMANIA,57, 58 an algorithm to determine the relationship between these mRNAs. The bio-functions and canonical pathways associated with our data were generated by using the core-analysis option in Ingenuity Pathway Analysis (Ingenuity Systems; http://www.ingenuity.com).
Quantitative real-time PCR analysis
The total RNA extracted from leukocytes or from lymphoblasts was used to synthesize cDNA. The expression levels of lncRNAs and of lncRNA-targeted mRNAs were determined by quantitative real-time PCR. Quantitative PCR reactions (the primer sequences used in quantitative PCR are listed in Supplementary Table S1) were performed by the ABI7900 system (Life Technologies, Grand Island, NY, USA) and SYBR green dye SuperArray PCR master mix (SABiosciences, Frederick, MD, USA). mRNA of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control for quantitative analysis of lncRNA or mRNA. The lncRNA or mRNA values were normalized to GAPDH levels. For each lncRNA or mRNA, triple reactions were analyzed simultaneously, and the result was reported as the relative expression calculated relative to this control. All the data were given in terms of relative expression of the mean±S.E. (n=10). The data were subjected to one-way analysis of variance followed by an unpaired, two-tailed t-test. Differences were considered significant at P<0.05.
Results
Differential expression profiles of lncRNA and mRNAs
A total of 3929 lncRNAs were identified as differentially expressed in Chinese ASD peripheral blood cells, including 2407 that were upregulated and 1522 that were downregulated. Among these, intergenic lncRNAs were the most common (accounting for 43%), followed by natural antisense (19%), intronic antisense (12%), exon sense-overlapping (9%), bi-directional (5%) and intron sense-overlapping (4%). Five percent of identified lncRNAs belong to uncharacterized groups (Supplementary Table S2). Simultaneously, 2610 mRNAs, including 1789 upregulated and 821 downregulated, that were differentially expressed in ASD blood cells genome-wide were also identified. The entire data set has been deposited in a public domain (DataDryad.org, DOI: doi:10.5061/dryad.d8f84).
Functional pathways derived from lncRNAs–mRNAs
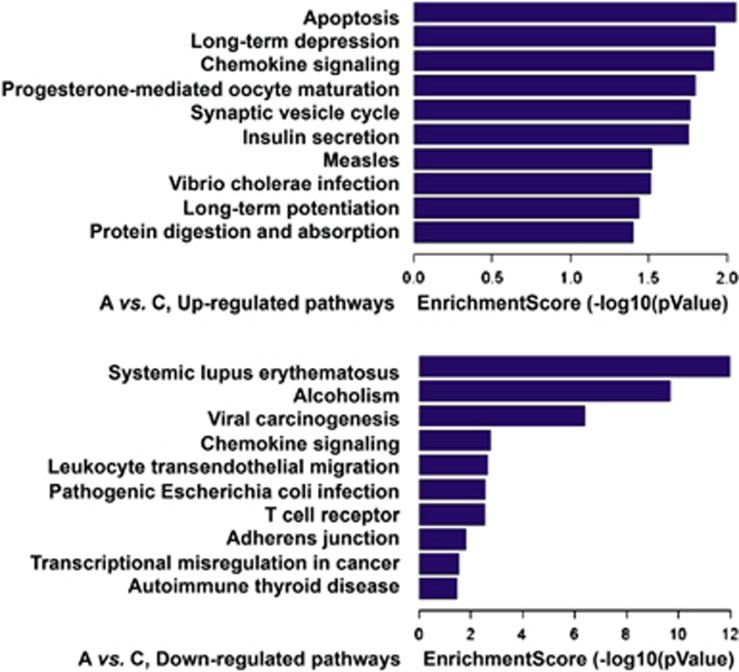
The gene loci where the lncRNAs are localized were subjected to pathway and gene ontology analysis. A total of 13 pathways derived from upregulated lncRNAs and 14 from downregulated lncRNAs were identified as being significant in ASD group (P<0.05). The 10 pathways with the highest enrichment score (Figure 1) showed that downregulated lncRNA loci were mainly involved in infection and inflammatory pathways. However, three pathways that are related to neurological regulation—the long-term depression, the synaptic vesicle cycling and the long-term potentiation pathways—were characterized from the upregulated lncRNA loci. The plot of enrichment score shows the relevant level of probability of the involvement of differentially expressed lncRNAs in the pathway. The range-axis in the upregulated pathways is lower than in the downregulated, suggesting that the pathogenic impact of lncRNAs in downregulated pathways is heavier than in upregulated pathways and the downregulated pathways are more likely involved in ASD than in upregulated pathways.
Figure 1.
Metabolic pathways characterized from the lncRNAs differentially expressed in ASD: The top-10 score of up- and downregulated pathways were characterized with KEGG functional analysis. Three P-values, the EASE-score, Fisher P-value and Hypergeometric P-value were integrated for the analysis. The bar plot shows the top Enrichment Score [−log10(P-value)] value of the significant enrichment pathway. The higher Enrichment Score indicates that more lncRNA molecules are involved in this pathway. ASD, autism spectrum disorder; lncRNA, long noncoding RNA.
Differential expression of synaptic lncRNAs and mRNAs
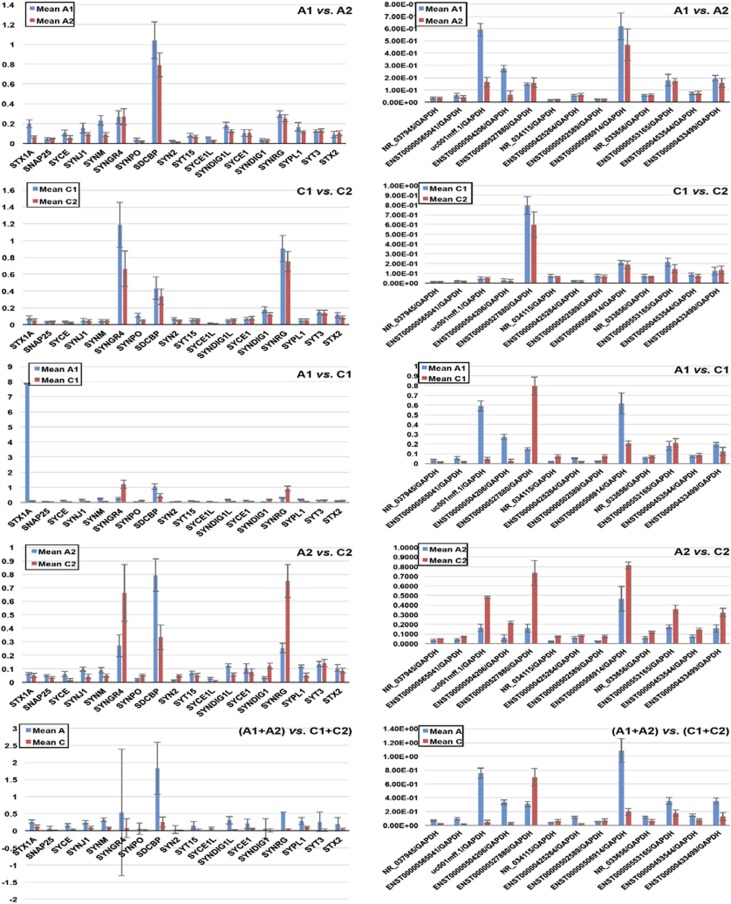
Thirteen synaptic lncRNAs (Table 1A), including nine upregulated and four downregulated, and 19 synaptic mRNAs (Table 1B), including 12 upregulated and seven downregulated, were identified as being differentially expressed in children with ASD. Among the upregulated lncRNAs, six were intronic antisense, two exon sense-overlapping and one natural antisense. Among the downregulated lncRNAs, three were natural antisense and one intronic antisense. To validate the differential expression of the synaptic lncRNAs and mRNAs identified from the microchip-based discovery study, the lncRNAs and mRNAs were subjected to quantitative analysis by quantitative PCR. Before the differential expression between the ASD and the control groups was analyzed, inter-group comparisons were made between the lymphocytes in a Chinese population and the lymphoblasts in a Caucasian population (A1 vs A2, C1 vs C2). As shown in Table 2A, the expression of lncRNAs NR_037945 (STX16), ENST00000565041 (SYNGR3), ENST00000425264 (SLC18A2), ENST00000502589 (SV2C) and ENST00000453544 (SNAP25) showed no significant difference between the Chinese and the Caucasian populations in both the ASD and the control groups; expression of ENST00000527880 (SYP), NR_034115 (STXBP5), NR_033656 (STX8) and ENST00000553165 (SYT1) showed no significant difference between the Chinese ASD groups; and expression of uc001mff.1 (SYT9), ENST00000504206 (SYT9), ENST00000506914 (SYT15) and ENST00000433499 (STXBP5) showed no significant difference between the two control groups. The differential expression of all lncRNAs, except ENST00000453544 (locus SNAP25) in the Caucasian ASD subjects and ENST00000553165 (SYT1) in both the Chinese and Caucasian ASD subjects, was statistically significant between the ASD and control groups. However, there was no statistical significance (P>0.05) observed between the males and females, nor between different age groups, in ASD or controls. No significant difference in the expression of three mRNAs—NGR4, SYNDIG1 and STX2—was evident between the Chinese and Caucasian ASD subjects. Expression of the mRNAs SYNJ1, SDCBP, SYPL1, SYNM and SYNDIG1L was not significantly different between the Chinese and Caucasian control groups. Expressions of all mRNAs, except SYCE1 in the Caucasian population and STX2 and SYT3 in both populations, were determined to be significantly different between the ASD and control groups in both the populations (Table 2B). To integrate the genome-wide-expressed lncRNAs with the synaptic mRNAs, we were able to draw the networks between the lncRNAs and mRNAs (Figure 2). This helps associate differential gene expression with gene ontology, biological pathway and the regulatory functions of the lncRNAs.
Table 1A. Synaptic lncRNAs differentially expressed in ASD (discovery).
| LncRNA name | P-value | Fold change | Regulation | LncRNA symbol | Chromosome | LncRNA strand | Relationship | Associated gene name | Associated gene strand | Associated protein |
|---|---|---|---|---|---|---|---|---|---|---|
| ENST00000506914 | 6.22808E−10 | 2.986859 | Up | RP11-38L15.3 | Chr10 | + | Intronic antisense | SYT15 | − | Synaptotagmin-15 isoform a |
| NR_037945 | 4.65294E−09 | 2.772749 | Up | STX16-NPEPL1 | Chr20 | + | Exon sense-overlapping | STX16 | + | Syntaxin-16 isoform |
| ENST00000565041 | 3.29675E−09 | 2.453277 | Up | AC005606.14 | Chr16 | − | Intronic antisense | SYNGR3 | + | Synaptogyrin-3 |
| NR_033656 | 3.16257E−08 | 2.318933 | Up | STX8 | Chr17 | − | Exon sense-overlapping | STX8 | − | Syntaxin-8 |
| ENST00000553165 | 1.09728E−09 | 2.007777 | Up | RP1-78O14.1 | Chr12 | − | Intronic antisense | SYT1 | + | Synaptotagmin-1 |
| ENST00000453544 | 2.61069E−09 | 2.291905 | Up | RP5-839B4.7 | Chr20 | − | Intronic antisense | SNAP25 | + | Synaptosomal-associated protein 25 isoform |
| ENST00000425264 | 2.99156E−08 | 2.407836 | Up | RP11-501J20.5 | Chr10 | − | Natural antisense | SLC18A2 | + | Synaptic vesicular amine transporter |
| uc001mff.1 | 1.14979E−11 | 3.098403 | Up | AK128569 | Chr11 | − | Intronic antisense | SYT9 | + | Synaptotagmin-9 |
| ENST00000504206 | 9.3421E−13 | 2.766950 | Up | CTD-2516F10.2 | Chr11 | − | Intronic antisense | SYT9 | + | Synaptotagmin-9 |
| ENST00000502589 | 8.16875E−10 | 2.804515 | Down | RP11-466P24.2 | Chr5 | − | Intronic antisense | SV2C | + | Synaptic vesicle glycoprotein 2C |
| ENST00000527880 | 5.19056E−22 | 3.546924 | Down | SYP-AS1 | ChrX | + | Natural antisense | SYP | + | Synaptic vesicle glycoprotein 2C |
| ENST00000433499 | 1.12117E−10 | 2.567277 | Down | STXBP5-AS1 | Chr6 | − | Natural antisense | STXBP5 | − | Synaptophysin |
| NR_034115 | 3.93428E−12 | 3.163574 | Down | STXBP5-AS1 | Chr6 | − | Natural antisense | STXBP5 | + | Syntaxin-binding protein 5 isoform |
Abbreviations: ASD, autism spectrum disorder; lncRNA, long noncoding RNA.
Table 1B. Synaptic mRNAs differentially expressed in ASD (discovery).
| Gene symbol | P-value | Fold change | Regulation | LncRNA name | Chromosome | Gene strand | Protein |
|---|---|---|---|---|---|---|---|
| SYNDIG1L | 2.07E−10 | 2.4465995 | Up | ENST00000331628 | chr14 | − | Synapse differentiation-inducing 1-like |
| SYNJ1 | 1.05E−09 | 2.0476525 | Up | NM_203446 | chr21 | − | Synaptojanin-1 isoform b |
| SYCE1 | 2.58E−09 | 2.0756986 | Up | ENST00000303903 | chr10 | − | Synaptonemal complex central element protein 1 |
| SYCE1L | 2.63E−07 | 2.1141472 | Up | NM_001129979 | chr16 | + | Synaptonemal complex central element protein 1-like |
| SYCE2 | 1.76E−10 | 2.7053528 | Up | NM_001105578 | chr19 | − | Synaptonemal complex central element protein 2 |
| SYPL1 | 9.68E−09 | 2.6788418 | Up | NM_006754 | chr7 | − | Synaptophysin-like protein 1 isoform a |
| SNAP25 | 7.44E−11 | 2.2339847 | Up | NM_130811 | chr20 | + | Synaptosomal-associated protein 25 isoform |
| SYT15 | 8.02E−07 | 2.0689712 | Up | ENST00000374325 | chr10 | − | Synaptotagmin XV |
| SYT3 | 7.75E−09 | 2.019983 | Up | NM_001160329 | chr19 | − | Synaptotagmin-3 |
| SDCBP | 4.00E−09 | 3.5665565 | Up | ENST00000260130 | chr8 | + | Syndecan binding protein (syntenin) |
| SYNM | 7.08E−11 | 2.8931897 | Up | NM_015286 | chr15 | + | Synemin isoform B |
| STX1A | 6.43E−10 | 2.8111503 | Up | NM_004603 | chr7 | − | Syntaxin-1A isoform 1 |
| SYNDIG1 | 1.17E−11 | 2.1523478 | Down | NM_024893 | chr20 | + | Synapse differentiation-inducing gene protein 1 |
| SYN2 | 8.58E−09 | 2.0109727 | Down | NM_133625 | chr3 | + | Synapsin-2 isoform IIa |
| SYNGR4 | 4.79E−06 | 2.0704043 | Down | NM_012451 | chr19 | + | Synaptogyrin-4 |
| SYNJ1 | 6.97E−12 | 2.1072302 | Down | NM_003895 | chr21 | − | Synaptojanin-1 isoform a |
| SYNPO | 1.59E−13 | 2.8214178 | Down | NM_001109974 | chr5 | + | Synaptopodin isoform B |
| SYNRG | 3.89E−17 | 2.5995717 | Down | NM_198882 | chr17 | − | Synergin gamma isoform 3 |
| STX2 | 6.70E−10 | 2.1167502 | Down | NM_194356 | chr12 | − | Syntaxin-2 isoform 2 |
Abbreviations: ASD, autism spectrum disorder; lncRNA, long noncoding RNA; mRNA, messenger RNA.
Table 2A. Synaptic lncRNAs differentially expressed in ASD (validation).
| NR_037945 | ENST00000565041 | ENST00000425264 | ENST00000502589 | ENST00000453544 | ENST00000527880 | NR_034115 | NR_033656 | ENST00000553165 | uc001mff.1 | ENST00000504206 | ENST00000506914 | ENST00000433499 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 vs A2 | P | 0.4559 | 0.0688 | 0.1267 | 0.6145 | 0.6630 | 0.3911 | 0.3517 | 0.3925 | 0.7064 | 0.0000 | 0.0000 | 0.0110 | 0.0128 |
| t | 0.762 | 1.935 | 1.601 | 0.513 | 0.443 | 0.896 | 0.966 | 0.876 | 0.385 | 20.982 | 16.173 | 2.829 | 2.764 | |
| C1 vs C2 | P | 0.2169 | 0.0862 | 0.5017 | 0.2529 | 0.272 | 0.0009 | 0.013 | 0.0369 | 0.0021 | 0.9356 | 0.3177 | 0.1648 | 0.6184 |
| t | 1.28 | 1.815 | 0.686 | 1.181 | 2.405 | 3.949 | 2.756 | 2.265 | 3.595 | 0.082 | 1.028 | 1.448 | 0.507 | |
| A1 vs C1 | P | 0.0001 | 0.0002 | 0.0000 | 0.0000 | 0.0193 | 0.0000 | 0.0000 | 0.0056 | 0.1089 | 0.0000 | 0.0000 | 0.0000 | 0.0001 |
| t | 5.792 | 5.646 | 14.119 | 11.704 | 2.569 | 22.538 | 12.138 | 3.197 | 1.687 | 31.772 | 26.686 | 11.679 | 4.858 | |
| A2 vs C2 | P | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.1370 | 0.0000 | 0.0000 | 0.0297 | 0.7743 | 0.0000 | 0.0000 | 0.0000 | 0.0003 |
| t | 9.768 | 9.6730 | 16.1620 | 13.3310 | 1.5660 | 25.8450 | 17.5950 | 2.3720 | 0.2910 | 26.2790 | 22.3500 | 12.1380 | 4.4780 |
Abbreviations: ASD, autism spectrum disorder; A1, Chinese ASD; A2, Caucasian ASD; C1, Chinese control; C2, Caucasian control; lncRNA, long noncoding RNA.
Table 2B. Synaptic mRNAs differentially expressed in ASD (validation).
| SYCE1 | SYNGR4 | SYNDIG1 | SNAP25 | STX2 | SYT3 | SYT15 | SYNRG | SYNJ1 | SDCBP | SYPL1 | STX1A | SYCE2 | SYNM | SYNPO | SYN2 | SYCE1L | SYNDIG1L | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 vs A2 | P | 0.9354 | 0.8693 | 0.4318 | 0.3603 | 0.2745 | 0.2609 | 0.0707 | 0.0118 | 0.0041 | 0.0023 | 0.0019 | 0.0000 | 0.0004 | 0.0000 | 0.0009 | 0.0000 | 0.0000 | 0.0000 |
| t | 0.0820 | 0.1670 | 0.8040 | 0.9510 | 1.1270 | 1.1610 | 1.9210 | 2.8000 | 3.6080 | 3.5410 | 3.6370 | 10.0110 | 4.3530 | 8.7390 | 3.9480 | 5.6170 | 7.9180 | 5.9400 | |
| C1 vs C2 | P | 0.4557 | 0.0001 | 0.0008 | 0.0813 | 0.0320 | 0.7826 | 0.4860 | 0.0239 | 0.3098 | 0.0748 | 0.9628 | 0.0035 | 0.0007 | 0.5587 | 0.0000 | 0.0006 | 0.0057 | 0.5068 |
| t | 0.7620 | 4.8350 | 4.0040 | 1.8470 | 2.3250 | 0.2800 | 0.7110 | 2.4660 | 1.0450 | 1.8910 | 0.0470 | 3.3600 | 4.0750 | 0.5510 | 7.9550 | 4.1700 | 3.1400 | 0.6770 | |
| A1 vs C1 | P | 0.0055 | 0.0000 | 0.0000 | 0.0146 | 0.1238 | 0.0511 | 0.0027 | 0.0000 | 0.0001 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| t | 3.1560 | 10.4850 | 11.4310 | 2.7870 | 1.6140 | 2.0900 | 3.4830 | 11.9790 | 6.1330 | 8.4260 | 8.0720 | 7.7860 | 7.2810 | 11.8300 | 8.6720 | 9.6360 | 12.7730 | 13.4130 | |
| A2 vs C2 | P | 0.4620 | 0.0001 | 0.0000 | 0.0001 | 0.0816 | 0.5086 | 0.0076 | 0.000 | 0.0000 | 0.0000 | 0.0000 | 0.0155 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| t | 2.1410 | 5.4670 | 13.0950 | 4.9060 | 1.8440 | 0.6740 | 3.0030 | 12.478 | 6.9610 | 9.5700 | 11.8830 | 2.6730 | 6.4040 | 5.5550 | 10.0800 | 10.4070 | 6.8990 | 11.9880 | |
| A vs C | P | 0.0090 | 0.0000 | 0.0000 | 0.0000 | 0.8327 | 0.0910 | 0.0015 | 0.000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| t | 3.1670 | 14.3460 | 15.3330 | 6.6640 | 0.2140 | 1.8380 | 3.7420 | 17.277 | 7.6950 | 11.4130 | 12.1930 | 7.1870 | 8.8100 | 13.0440 | 11.0910 | 15.1610 | 19.9780 | 19.7310 |
Abbreviations: ASD, autism spectrum disorder; A1, Chinese ASD; A2, Caucasian ASD; C1, Chinese control; C2, Caucasian control; mRNA, messenger RNA.
Figure 2.
Validation of lncRNAs: qRT-PCR was applied to validate differentially expressed lncRNAs (right panels) and mRNAs (left panels) between the ASD and the control groups. Other than the gene symbol, each lncRNA was labeled with its name that can be matched to the symbol in Table 3. The height of each bar, measured by mean±s.d., represents the relative expression level. ASD, autism spectrum disorder; lncRNA, long noncoding RNA; qRT-PCR, quantitative real-time PCR.
Association of lncRNAs with autistic genes
Genetic and genomic studies have revealed that a substantial proportion of ASD risk resides in high-impact rare variation, ranging from chromosome abnormalities, single-nucleotide variation, copy-number variation to gene mutations.59 To match our lncRNA results to these gene loci, 19 lncRNAs were found to be associated with ASD genes (Table 3A). Among these lncRNAs, seven were natural antisense (AS); six, intronic antisense; three, bi-directional; two, intron sense-overlapping; and one, intergenic. Twelve of the 19 lncRNAs associated with ASD genes were homeobox or homeobox-related genes, including nine that were upregulated and three that were downregulated, followed by four brain-derived neurotrophic factor (BDNF) isoforms. Interestingly, differential expression of mRNAs for HOXA and HOXB was also identified in our microarray-based discovery study of the ASD patients (Table 3B). However, no mRNAs, which are the targets of BDNF-AS and of the intronic AS of SHANK2—a membrane of the SHANK gene family and its gene mutations found in ASD patients (http://autism.mindspec.org/GeneDetail/SHANK2)—were identified.
Table 3A. LncRNAs associated with autistic genes.
| LncRNA name | P-value | Fold change | LncRNA symbol | LncRNA strand | Relationship | Associated gene accession | Associated gene name | Associated gene strand | Associated protein |
|---|---|---|---|---|---|---|---|---|---|
| Upregulated | |||||||||
| ENST00000454594 | 9.88E−08 | 2.25 | HOTTIP | + | Intronic antisense | NM_000522 | HOXA13 | − | Homeobox protein Hox-A13 |
| ENST00000476204 | 2.06E−11 | 3.41 | HOXB-AS3 | + | Intronic antisense | NM_002147 | HOXB5 | − | Homeobox protein Hox-B5 |
| ENST00000477144 | 6.89E−11 | 2.09 | HOXB-AS3 | + | Natural antisense | NM_018952 | HOXB6 | − | Homeobox protein Hox-B6 |
| ENST00000476204 | 2.06E−11 | 3.41 | HOXB-AS3 | + | Intronic antisense | NM_018952 | HOXB6 | − | Homeobox protein Hox-B6 |
| ENST00000462131 | 4.89E−10 | 2.45 | HOXB-AS4 | + | Intergenic | ||||
| ENST00000452365 | 2.85E−10 | 2.79 | HOXD-AS1 | − | Bidirectional | NM_024501 | HOXD1 | + | Homeobox protein Hox-D1 |
| ENST00000430404 | 4.212E−04 | 2.97 | DLX6-AS1 | − | Intronic antisense | NM_005222 | DLX6 | + | Homeobox protein DLX6 |
| ENST00000558292 | 1.73E−13 | 3.71 | AC108449.2 | + | Intron sense-overlapping | NM_001135726 | HMBOX1 | + | Homeobox-containing protein 1 |
| ENST00000558292 | 1.73E−13 | 3.71 | AC108449.2 | + | Intron sense-overlapping | NM_024567 | HMBOX1 | + | Homeobox-containing protein 1 |
| ENST00000454594 | 8.43E−09 | 2.27 | RP11-231K24.2 | + | Bidirectional | NM_021570 | BARX1 | − | Homeobox protein BarH-like 1 |
| NR_033314 | 9.454E−08 | 3.30 | BDNF-AS | + | Natural antisense | NM_170735 | BDNF | − | Brain-derived neurotrophic factor isoform a preproprotein |
| uc009yim.3 | 7.610E−09 | 2.71 | BDNF-AS1 | + | Natural antisense | NM_001143805 | BDNF | − | Brain-derived neurotrophic factor isoform a preproprotein |
| ENST00000307548 | 6.98E−09 | 2.62 | SHANK2-AS3 | + | Intronic antisense | NM_012309 | SHANK2 | − | SH3 and multiple ankyrin repeat domains protein 2 isoform 1 |
| Downregulated | |||||||||
| ENST00000521231 | 6.06E−10 | 3.73 | HOXA-AS3 | + | Natural antisense | NM_024014 | HOXA6 | − | Homeobox protein Hox-A6 |
| ENST00000526796 | 3.83E−08 | 2.09 | RP11-687M24.8 | − | Intronic antisense | NM_022062 | PKNOX2 | + | Homeobox protein PKNOX2 |
| ENST00000526796 | 1.33E−06 | 2.53 | RP11-687M24.5 | − | Bidirectional | NM_022062 | PKNOX2 | + | Homeobox protein PKNOX2 |
| NR_033315 | 1.57E−12 | 2.11 | BDNF-AS | + | Natural antisense | NM_001143810 | BDNF | − | Brain-derived neurotrophic factor isoform e |
| uc009yix.3 | 1.77E−17 | 2.60 | BDNF-AS1 | + | Natural antisense | NM_001143805 | BDNF | − | Brain-derived neurotrophic factor isoform a preproprotein |
| ENST00000481143 | 2.82E−07 | 2.15 | DMD-AS1 | + | Natural antisense | NM_004014 | DMD | − | Dystrophin Dp116 isoform |
Abbreviations: ASD, autism spectrum disorder; BDNF, brain-derived neurotrophic factor; lncRNA, long noncoding RNA.
Table 3B. Differential expression of mRNAs encoded by autistic genes.
| Gene symbol | P-value | Fold change | Regulation | Gene accession | ccdsID | Chromosome | Gene strand | Protein |
|---|---|---|---|---|---|---|---|---|
| HOXA1 | 6.8332E−14 | 2.2217333 | Down | NM_153620 | CCDS5402.2 | Chr7 | − | Homeobox protein Hox-A1 isoform b |
| HOXA4 | 8.80085E−16 | 2.0322213 | Down | NM_002141 | CCDS5405.1 | Chr7 | − | Homeobox protein Hox-A4 |
| HOXB2 | 1.45676E−15 | 2.055423 | Down | NM_002145 | CCDS11527.1 | Chr17 | − | Homeobox protein Hox-B2 |
| HOXB4 | 2.47971E−07 | 2.034943 | Down | NM_024015 | CCDS11529.1 | Chr17 | − | Homeobox protein Hox-B4 |
| HOXB8 | 1.37338E−09 | 2.101062 | Down | ENST00000239144 | CCDS11533.1 | Chr17 | − | Homeobox B8 |
Abbreviation: mRNA, messenger RNA.
Discussion
Differentially expressed lncRNAs represent a new potential biomarker category for the early detection of ASD
Previous studies have reported that lncRNAs were aberrantly expressed in brain tissues and associated with ASD.56 However, brain tissue cannot be used as clinical material for early screening or for diagnostic purposes. In addition, there is no specific biomarker at present that can be applied in clinical practice, owing to the genetic heterogeneity of ASD,10 although efforts have been undertaken to characterize blood mRNA profiles.60, 61, 62, 63 Use of a panel of lncRNAs that are specifically associated with ASD phenotypes and are differentially expressed in ASD peripheral blood would be valuable and practical. In this study, we presented metabolic pathways (Figure 1) that define peripheral blood lncRNAs that are differentially expressed in ASD. Among these, synaptic vesicle cycling, long-term depression and long-term potentiation are neurologically related pathways. LncRNAs that are differentially expressed in ASD and have been identified in these pathways include IGF1, mGluR1, CRFR1, IGF1R, NMDAR and VDCC, which are localized to cell membranes; and Ras, G protein, PLC, IP3R, PKG, ERK1, ERK2 and PP2A, which are in the cytoplasm and involved in signal transduction for long-term depression and long-term potentiation. The differentially expressed lncRNAs involved in the synaptic vesicle cycling pathway are Rab3A, Munc13, Syntaxin, SNAP25, Clathrin, V-ATPase and trans-SNARE complex. Expression of all these genes is found with various technology platforms (www.genecards.org). It is not clear whether the lncRNAs and mRNAs identified in the peripheral lymphocytes are identical to those expressed in neuronal cells or whether they reflect ASD brain functions.
Although lncRNAs have been determined to associate with transcriptional regulation in neuronal development and diseases,17 applying gene differential expression profile analysis of peripheral bloods for brain disorders presents a challenge to convince that the differential expression profile in the peripheral bloods may be relevant to that in brain tissues. In fact, there is no way to obtain human brain tissue for routine screening or diagnostic analysis in clinical practice. The eQTL gene transcripts identified from brains was demonstrated as being stably expressed in peripheral bloods.64, 65 Our earlier study also determined that the brain gene ENO2 showed differentially expressed methylation in peripheral bloods.10 Therefore, to analyze differential expression profile in blood may open a new approach to explore applying differential expression profile in blood as a biomarker for brain diseases.
The differential expression we found demonstrates the potential for lncRNAs to be applied as clinical biomarkers. Replication of our study with larger samples and various ethnic backgrounds will be needed. Indeed, we noted differences in the lncRNAs when comparing the Chinese and Caucasian populations (Table 2A). These differences suggest that there exist, at certain gene loci, inter-population and inter-condition differences in gene expression. A similar finding of the influence of different ethnic backgrounds was also observed in the mRNAs (Table 2B).
Synaptic lncRNAs may regulate synaptic vesicle transportation
Among the 13 synaptic lncRNAs, three were lncRNAs, which resided at the genes for synaptic vesicle proteins (Table 1A). The gene SLC18A2, a member of the vesicular monoamine transporter family, encodes a vesicular monoamine transporter of cytosolic monoamines into synaptic vesicles, using the proton gradient maintained across the synaptic vesicular membrane. Its proper function is essential for the proper activity of monoaminergic systems, which have been implicated in several human neuropsychiatric disorders, including brain dopamine–serotonin vesicular transport disease and cocaine dependence.66 The gene SV2C encodes synaptic vesicle glycoprotein 2C, which has a role in the control of regulated secretion in neural and endocrine cells, selectively enhances low-frequency neurotransmission, and positively regulates vesicle fusion by maintaining the readily releasable pool of secretory vesicles.67 SYP is a gene that encodes the synaptic protein synaptophysin, an integral membrane protein of small synaptic vesicles in the brain and endocrine cells, which is a transporter and a calcium ion–binding protein. This protein may also bind cholesterol and is thought to direct targeting of vesicle-associated membrane protein 2 (synaptobrevin) to intracellular compartments.68 Mutations in this gene are associated with X-linked mental retardation (www.researchgate.net/publication/12901506_XLMR_database).69 In addition to these three synaptic vesicle proteins, the genes STX8 and STX16 are also involved in synaptic vesicle metabolism. The STX8 gene is involved in protein trafficking from early to late endosomes via vesicle fusion and exocytosis. It encodes a vesicle trafficking protein that functions in the early secretory pathway, possibly by mediating retrograde transport from cis-Golgi membranes to the endosome reticulum.70, 71 The STX16 gene is a member of the t-SNARE (target-SNAP receptor) family. Proteins in this family are found on cell membranes and serve as the targets for V-SNARES (vesicle-SNAP receptors), permitting specific synaptic vesicle docking and fusion. A disease associated with STX8 includes visual epilepsy, and diseases associated with STX16 are pseudohypoparathyroidism type 1b and pseudohypoparathyroidism.72, 73
HOX genes are likely to be deregulated in ASD
Several studies have demonstrated that lncRNAs can function in the regulation of in vivo transcription. An lcnRNA dubbed linc-HOXA1 RNA has been found to repress Hoxa1 expression. Knockdown of linc-HOXA1 increases transcription of the Hoxa1 gene that is located some 50 kb adjacent to the linc-HOXA1.74 HOXA cluster antisense RNA 2 (HOXA-AS2) is an lncRNA located between the HOXA3 and HOXA4 genes in the HOXA cluster. HOXA-AS2 is an apoptosis repressor in all trans retinoic acid-treated NB4 promyelocytic leukemia cells.75 Its transcript is expressed in NB4 promyelocytic leukemia cells and human peripheral blood neutrophils, and expression is increased in NB4 cells treated with all trans retinoic acid. The all trans retinoic acid induction of HOXA-AS2 suppresses all trans retinoic acid-induced apoptosis.75 The HOTAIR (Hox transcript antisense RNA) gene contains 6232 bp and encodes a 2.2- kb lncRNA. Its source DNA is located within a HOXC gene cluster. Recently, differential expression of HOTAIR has been determined to be associated with cancer metastasis and possibly to represent an independent prognostic factor.76 The 5′ end of HOTAIR interacts with the polycomb-group protein Polycomb repressive complex 2 and as a result regulates chromatin state. It is required for gene-silencing of the HOXD locus by Polycomb repressive complex 2 and the 3′ end of HOTAIR interacts with the histone demethylase LSD1.77 In our study, we identified several lncRNAs of the HOX genes (HOXA13, HOXB5, HOXB6 and HOXD1) and HOX-related genes (DLX6, HMBOX1 and BARX1) from leukocytes derived from ASD patients (Table 3A). These findings led us to hypothesize that the differentially expressed lncRNAs of HOX genes and HOX-related genes, referred to as lncHOXs, could represent a new set of biomarkers for ASD.
The variety of identified lncRNAs suggests that lncRNAs' regulatory functions involved in ASD may have various epigenetic mechanisms.
LncRNAs have been recognized as transcriptional and posttranscriptional regulators.28, 29, 30, 31, 32, 33 They may function to activate gene transcription by binding a transcriptional factor to the promoter region to signal or guide transcription. They may prevent miRNA from binding to the target gene, may suppress gene transcription by decoying the transcription factor away from the promoter, or may be a chromatin modifier by bringing a chromatin enzyme onto chromatin to form a complex and thereby modify histones.28, 29, 30, 31, 32, 33 Usually, if the lncRNA is the antisense of a gene, the lncRNA likely functions as the cis-suppressor to inhibit gene transcription. This could be the case for both the natural and intronic antisense lncRNAs that we identified (Tables 1A and 3A). To further understand their molecular mechanisms, transgenic models created by introducing extragenic lncRNA to generate a knockout, knockin or knockdown model at the cellular and/or animal level could be investigated. Such a transgenic model could provide phenotype(s) to mimic ASD. So far, there is little evidence that details the molecular and pathogenic mechanisms of the bi-directional and intergenic lncRNAs involved in ASD and other neurological diseases. In the present study, we identified three bi-directional and one intergenic lncRNA, and observed that all are located within HOX loci (Table 3A). A better understanding of the molecular mechanisms will clarify how these lncRNAs are involved in regulating gene expression in ASD and other neurological conditions.
In conclusion, we have profiled here the differential expression of lncRNAs and mRNAs in ASD peripheral leukocytes and have identified important clusters that may be associated with this disorder. Our findings suggest the importance of synaptic lncRNAs, which are likely involved in synaptic vesicle transportation and cycling and thus would be important for the delivery of synaptosomal protein(s) between presynaptic and postsynaptic membranes. LncRNAs that are the antisense of the HOX genes may be related to ASD. This finding may open a new approach to investigate the pathogenic mechanisms of the HOX genes in the development of ASD. Identification of the lncRNAs of SHANK2-AS and BDNF-AS indicate that in addition to gene mutation, de-regulation of lncRNAs on ASD-causing gene loci may represent a new category of, and allow exploration of, the epigenetic mechanisms involved in ASD. Further investigation with larger sample sizes may validate the use of lncRNAs as biomarkers for early detection of ASD.
Acknowledgments
We thank the autism and control subjects and their parents for making this study feasible. This study was supported in part by the National ‘973' program (2012CB517905) granted from the Chinese Ministry of Science and Technology, and funding from the Shanghai Municipal Department of Science and Technology (2009JC1412600), Peking University Center of Medical Genetics and the New York State Office for People With Developmental Disabilities (OPWDD).
Author contributions
YW and WJ were responsible for the procurement of specimens. YW, WJ and JZ participated in the sample preparations and quantitative studies. XZ and MF carried out the statistical analysis. XT and PW performed bioinformatics analysis. WTB and NZ drafted the manuscript. NZ conceived the study and was responsible for the research design, data analysis and coordination. All the authors read and approved the final manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Centers for Disease Control and Prevention. MMWR Surveill Summ 2014; 63: 1–21.
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet 2008; 9: 341–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhong N. Clinical and genetic heterogeneity of autismCooper DN, Chen JM. Mutations in Human Genetic Disease. Intech: Croatia, Anchor, 2012, pp 217–232. [Google Scholar]
- Akshoomoff N, Pierce K, Courchesne E. The neurobiological basis of autism from a developmental perspective. Dev Psychopath 2002; 14: 613–634. [DOI] [PubMed] [Google Scholar]
- Institute of Medicine of the National Academies. Workshop Proceedings. The National Academies Press: Washington, DC, USA, 2007. [Google Scholar]
- Pardo CA, Vargas DL, Zimmerman AW. Immunity, neuroglia and neuroinflammation in autism. Int Rev Psychiatry 2005; 17: 485–495. [DOI] [PubMed] [Google Scholar]
- Bailey A, Phillips W, Rutter M. Autism: towards an integration of clinical, genetic, neuropsychological, and neurobiological perspectives. J Child Psychol Psychiatry 1996; 37: 89–126. [DOI] [PubMed] [Google Scholar]
- Yan GD, Lian YY, Wang Y, Huang W, Zou XB, Zhong N. Copy number variation analysis of a Chinese Han family with autism spectrum disorder. J Chinese Med Genet 2010; 27: 654–658. [DOI] [PubMed] [Google Scholar]
- Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res 2011; 1380: 42–77. [DOI] [PubMed] [Google Scholar]
- Wang Y, Fang Y, Zhang F, Xu M, Zhang J, Yan J et al. Hypermethylation of the enolase gene (ENO2) in autism. Eur J Pediatr 2014; 173: 1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschou M, Paraskevopoulou MD, Vlachos IS, Koukouraki P, Hatzigeorgiou AG, Doxakis E. miRNA regulons associated with synaptic function. PLoS One 2012; 7: e46189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanna P, Vernes S. A direct molecular link between the autism candidate gene RORa and the schizophrenia candidate MIR137. Sci Rep 2014; 4: 3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlakha YK, Saini N. Brain microRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Mol Cancer 2014; 13: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Yu D, Gu QH, Yang Y, Tu K, Zhu J et al. miR-191 and miR-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nat Commun 2014; 5: 3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barca-Mayo O, Pietri De, Tonelli D. Convergent microRNA actions coordinate neocortical development. Cell Mol Life Sci 2014; 71: 2975–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Devondervoort II, Gordebeke PM, Khoshab N, Tiesinga PH, Buitelaar JK, Kozicz T et al. Long non-coding RNAs in neurodevelopmental disorders. Front Mol Neurosci 2013; 6: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BS, Blackshaw S. Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. Front Genet 2014; 5: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori C, Wahlestedt C. Involvement of long noncoding RNAs in diseases affecting the central nervous system. RNA Biol 2012; 9: 860–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petazzi P, Sandoval J, Szczesna K, Jorge OC, Roa L, Sayols S et al. Dysregulation of the long non-coding RNA transcriptome in a Rett syndrome mouse model. RNA Biol 2013; 10: 1197–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori C, Peschansky VJ, Barbouth D, Mehta A, Silva JP, Wahlestedt C. Comprehensive analysis of the transcriptional landscape of the human FMR1 gene reveals two new long noncoding RNAs differentially expressed in Fragile X syndrome and Fragile X-associated tremor/ataxia syndrome. Hum Genet 2014; 133: 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson B, Campbell DB. Contribution of long noncoding RNAs to autism spectrum disorder risk. Int Rev Neurobiol 2013; 113: 3559. [DOI] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010; 39: 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev 2006; 20: 1470–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham A, Orth A, Batalov S, Peters EC, Wen BG, Aza-Blanc P et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 2005; 309: 1570–1573. [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009; 33: 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauss JL, Sun T. Regulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and function. Neuroscience 2013; 235: 200–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009; 136: 629–641. [DOI] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011; 43: 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science 2012; 338: 1435–1439. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012; 81: 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013; 152: 1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenoglio C, Ridolfi E, Galimberti D, Scarpini E. An emerging role for long non-coding RNA dysregulation in neurological disorders. Int J Mol Sci 2013; 14: 20427–20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung JTY, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics 2013; 193: 651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res 2012; 40: 6391–6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D et al. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS One 2011; 6: e23356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soreq L, Guffanti A, Salomonis N, Simchovitz A, Israel Z, Bergman H et al. Long non-coding RNA and alternative splicing modulations in Parkinson's leukocytes identified by RNA sequencing. PLoS Comput Biol 2014; 10: e1003517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G. Integrating the roles of long and small non-coding RNA in brain function and disease. Mol Psychiatry 2014; 19: 410–416. [DOI] [PubMed] [Google Scholar]
- Wu P, Zuo X, Deng H, Liu X, Liu L, Ji A. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res Bull 2013; 97: 6980. [DOI] [PubMed] [Google Scholar]
- Ng SY, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet 2013; 29: 461–468. [DOI] [PubMed] [Google Scholar]
- Tan L, Yu JT, Hu N, Tan L. Non-coding RNAs in Alzheimer's disease. Mol Neurobiol 2013; 47: 382–393. [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 2009; 10: 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annilo T, Kepp K, Laan M. Natural antisense transcript of natriuretic peptide precursor A (NPPA): structural organization and modulation of NPPA expression. BMC Mol Biol 2009; 10: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci 2010; 3: ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Onc 2011; 18: 1243–1250. [DOI] [PubMed] [Google Scholar]
- Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011; 71: 6320–6326. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Horlings HM, Wong DJ, Tsai MC et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, He Y, Xi B, Zheng J, Zeng X, Cai Q et al. LncRNAs expression in preeclampsia placenta reveals the potential role of LncRNAs contributing to preeclampsia pathogenesis. PLoS One 2013; 8: e81437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo XC, Shi QX, Ya Gu, Pan J, Wang LL, Hua MF et al. LncRNA pathway involved in premature preterm rupture of membrane (PPROM): an epigenomic approach to study the pathogenesis of reproductive disorders. PLoS One 2013; 8: e79897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cao Q, Ge J, Liu C, Ma Y, Meng Y et al. LncRNA-regulated infection and inflammation pathways associated with pregnancy loss: genome wide differential expression of lncRNAs in early spontaneous abortion. Am J Reprod Immunol 2014; 72: 359–375. [DOI] [PubMed] [Google Scholar]
- Shen C, Zhong N. Long non-coding RNAs: the epigenetic regulators involved in the pathogenesis of reproductive disorder. Am J Reprod Immunol 2014; 73: 95–108. [DOI] [PubMed] [Google Scholar]
- Shi X, Sun M, Liu H, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett 2013; 339: 159–166. [DOI] [PubMed] [Google Scholar]
- Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X et al. LncRNA disease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res 2013; 41: D983–D986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerin T, Ramanathan A, Rivas K, Grepo N, Coetzee GA, Campbell DB. A noncoding RNA antisense to moesin at 5p14.1 in autism. Sci Transl Med 2012; 4: 128ra40. [DOI] [PubMed] [Google Scholar]
- Velmeshev D, Magistri M, Faghihi MA. Expression of non-protein-coding antisense RNAs in genomic regions related to autism spectrum disorders. Mol Autism 2013; 4: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziats MN, Rennert OM. Aberrant expression of long noncoding RNAs in autistic brain. J Mol Neurosci 2013; 49: 589–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt SJ, Tsuang MT, Winn M, Chandler SD, Collins M, Lopez L et al. Blood-based gene expression signatures of infants and toddlers with autism. J Am Acad Child Adolesc Psychiatry 2012; 51: 934–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010; 38: W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberi K, Franz M, Rodriguez H, Montojo J, Lopes CT, Bader GD et al. GeneMANIA prediction server 2013 update. Nucleic Acids Res 2013; 41: W115–W122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet 2014; 94: 677–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enstrom AM, Lit L, Onore CE, Gregg JP, Hansen RL, Pessah IN et al. Altered gene expression and function of peripheral blood natural killer cells in children with autism. Brain Behav Immun 2009; 23: 124–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuszek G, Talebizadeh Z. Autism Genetic Database (AGD): a comprehensive database including autism susceptibility gene-CNVs integrated with known noncoding RNAs and fragile sites. BMC Med Genet 2009; 10: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong SW, Collins CD, Shimizu-Motohashi Y, Holm IA, Campbell MG, Lee IH et al. Characteristics and predictive value of blood transcriptome signature in males with autism spectrum disorders. PLoS One 2012; 7: e49475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong SW, Shimizu-Motohashi Y, Campbell MG, Lee IH, Collins CD, Brewster SJ et al. Peripheral blood gene expression signature differentiates children with autism from unaffected siblings. Neurogenetics 2013; 14: 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemandez DG, Nalls MA, Moore M, Chong S, Dillman A, Trabzuni D et al. Integration of GWAS SNPs and tissue specific expression profiling reveal discrete eQTLs for human traits in blood and brain. Neurobiol Dis 2012; 47: 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinska AJ, Service S, Choi QW, DeYoung J, Grujic O, Kong SY et al. Identification of brain transcriptional variation reproduced in peripheral blood: an approach for mapping brain expression traits. Hum Mol Genet 2009; 18: 4415–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Zhao Y, Chung CY, Zhou Y, Xiong N, Glatt CE et al. High regulatability favors genetic selection in SLC18A2, a vesicular monoamine transporter essential for life. FASEB J 2010; 24: 2191–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit RM, Frey D, Hilbert M, Kevenaar JT, Wieser MM, Stirnimann CU et al. Structural basis for recognition of synaptic vesicle protein 2C by botulinum neurotoxin A. Nature 2014; 505: 108–111. [DOI] [PubMed] [Google Scholar]
- Ozçelik T, Lafreniere RG, Archer BT 3rd, Johnston PA, Willard HF, Francke U et al. Synaptophysin: structure of the human gene and assignment to the X chromosome in man and mouse. Am J Hum Genet 1990; 47: 551–561. [PMC free article] [PubMed] [Google Scholar]
- Chiurazzi P, Hamel BCJ, Neri G. XLMR genes: update 2000. Eur J Hum Genet 2001; 9: 71–81. [DOI] [PubMed] [Google Scholar]
- Thoreau V, Bergès T, Callebaut I, Guillier-Gencik Z, Gressin L, Bernheim A et al. Molecular cloning, expression analysis, and chromosomal localization of human syntaxin 8 (STX8). Biochem Biophys Res Commun 1999; 257: 577–583. [DOI] [PubMed] [Google Scholar]
- He Y, Linder ME. Differential palmitoylation of the endosomal SNAREs syntaxin 7 and syntaxin 8. J Lipid Res 2009; 50: 398–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Bremnes B, Rønning E, Aasland R, Stenmark H. Syntaxin-16, a putative Golgi t-SNARE. Eur J Cell Biol 1998; 75: 223–231. [DOI] [PubMed] [Google Scholar]
- Amessou M, Fradagrada A, Falguières T, Lord JM, Smith DC, Roberts LM et al. Syntaxin 16 and syntaxin 5 are required for efficient retrograde transport of several exogenous and endogenous cargo proteins. J Cell Sci 2007; 120: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Cabili MN, Rinn J, Raj A. linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev 2013; 27: 1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhang X, Frazão JB, Condino-Neto A, Newburger PE. HOX antisense lincRNA HOXA-AS2 is an apoptosis repressor in all trans retinoic acid treated NB4 promyelocytic leukemia cells. J Cell Biochem 2013; 114: 2375–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chen S, Yang G, Gu F, Li M, Zhong B et al. Long noncoding RNA HOTAIR as an independent prognostic marker in cancer: a meta-analysis. PLoS One 2014; 9: e105538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329: 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.