Abstract
Dissimilatory sulfate reduction in peatlands is sustained by a cryptic sulfur cycle and effectively competes with methanogenic degradation pathways. In a series of peat soil microcosms incubated over 50 days, we identified bacterial consortia that responded to small, periodic additions of individual fermentation products (formate, acetate, propionate, lactate or butyrate) in the presence or absence of sulfate. Under sulfate supplementation, net sulfate turnover (ST) steadily increased to 16–174 nmol cm–3 per day and almost completely blocked methanogenesis. 16S rRNA gene and cDNA amplicon sequencing identified microorganisms whose increases in ribosome numbers strongly correlated to ST. Natively abundant (⩾0.1% estimated genome abundance) species-level operational taxonomic units (OTUs) showed no significant response to sulfate. In contrast, low-abundance OTUs responded significantly to sulfate in incubations with propionate, lactate and butyrate. These OTUs included members of recognized sulfate-reducing taxa (Desulfosporosinus, Desulfopila, Desulfomonile, Desulfovibrio) and also members of taxa that are either yet unknown sulfate reducers or metabolic interaction partners thereof. Most responsive OTUs markedly increased their ribosome content but only weakly increased in abundance. Responsive Desulfosporosinus OTUs even maintained a constantly low population size throughout 50 days, which suggests a novel strategy of rare biosphere members to display activity. Interestingly, two OTUs of the non-sulfate-reducing genus Telmatospirillum (Alphaproteobacteria) showed strongly contrasting preferences towards sulfate in butyrate-amended microcosms, corroborating that closely related microorganisms are not necessarily ecologically coherent. We show that diverse consortia of low-abundance microorganisms can perform peat soil sulfate reduction, a process that exerts control on methane production in these climate-relevant ecosystems.
Introduction
Peatlands currently store about one-third of all terrestrial carbon (Limpens et al., 2008), which is predicted to be partially released as the greenhouse gases CO2 and CH4 because of climate change (Hodgkins et al., 2014; Schuur et al., 2015). In these water-saturated soils, steep gradients in redox conditions sustain a complex network of biogeochemical processes (Limpens et al., 2008), with organic matter degradation being carried out by different functional guilds of microorganisms. This diversity of simultaneously occurring organic carbon degradation pathways is favored by frequently fluctuating concentrations of electron acceptors (Schmalenberger et al., 2007; Küsel et al., 2008; Knorr and Blodau, 2009) and donors (Schmalenberger et al., 2007; Küsel et al., 2008; Wüst et al., 2009). A changing water table that steadily shifts the oxic–anoxic interface (Knorr et al., 2009; Reiche et al., 2009) and complex flow paths of infiltrating and exfiltrating water create distinct spatial and temporal patterns (hot spots and hot moments) of various biogeochemical processes, making peatlands very dynamic ecosystems (Jacks and Norrström, 2004; Knorr et al., 2009; Knorr and Blodau, 2009; Frei et al., 2012).
Below the water table, where anoxic conditions persist, the processes of fermentation and subsequent methanogenesis are competing with energetically more favorable degradation pathways such as anoxic respiration coupled to nitrate, Fe(III), humic matter or sulfate reduction (Loy et al., 2004; Beer et al., 2008; Hamberger et al., 2008; Küsel et al., 2008; Drake et al., 2009; Keller et al., 2009; Knorr and Blodau, 2009; Knorr et al., 2009; Reiche et al., 2009; Wüst et al., 2009; Hunger et al., 2011; Pester et al., 2012b). The importance of sulfate reduction for peatland biogeochemistry is often neglected because of the low prevailing sulfate concentrations (μm range). This is contrasted by a highly active but cryptic sulfur cycle that rapidly reoxidizes reduced sulfur species and thus sustains sulfate reduction rates that are as high as in sulfate-rich marine surface sediments (Pester et al., 2012b). Here, (bio)geochemical sulfur reoxidation can be either coupled to the reduction of O2 at the oxic–anoxic interphase or to the reduction of Fe(III) (Hansel et al., 2015) and/or humic matter (Heitmann and Blodau, 2006; Yu et al., 2015) under completely anoxic conditions. As sulfate-reducing microorganisms (SRM) generally outcompete methanogens and syntrophically associated fermenters (Muyzer and Stams, 2008), they exert an important intrinsic control function on CH4 production in peatlands (Gauci et al., 2004, 2005; Gauci and Chapman, 2006).
Microbial communities in soils are typically composed of a large number of species (e.g., Roesch et al., 2007; Serkebaeva et al., 2013). Few of these taxa are very or moderately abundant (arbitrarily classified at ⩾1% and ⩾0.1% relative abundance, respectively). The remaining majority of species have an individual relative abundance of <0.1% and are summarized as the rare biosphere (Pedrós-Alió, 2012; Lynch and Neufeld, 2015). Because of its species richness and genetic diversity, the rare biosphere functions as a microbial seed bank for the recruitment of dormant cells to become active and numerically dominant under favorable environmental conditions. For example, singular disturbance events such as oil spills resulted either in the recruitment of specific taxa (Teira et al., 2007) or even widespread changes of the total microbial community (Newton et al., 2013). Furthermore, periodic environmental filtering through seasonal changes led to reoccurring temporal shifts between rare and abundant population sizes in marine bacterioplankton species (Campbell et al., 2011; Vergin et al., 2013; Alonso-Sáez et al., 2015). Dormant microorganisms may also be introduced to environments by transport mechanisms such as ocean currents (Müller et al., 2014) or wind (Hervàs and Casamayor, 2009; Hervàs et al., 2009), where they stay inactive and eventually undergo taphonomy (Lynch and Neufeld, 2015). However, there is increasing evidence that the rare biosphere is not just a seed bank but also harbors active populations. Seasonal patterns in marine bacterial and archaeal plankton revealed that many taxa that displayed reoccurring annual abundance changes were actually rare biosphere members and even during their bloom periods never reached abundant numbers (Campbell et al., 2011; Hugoni et al., 2013; Alonso-Sáez et al., 2015). In addition, microorganisms with a specialized metabolism often fulfill gatekeeper functions in an ecosystem while sustaining low-abundance populations (Lynch and Neufeld, 2015), for example, N2-fixing microorganisms in the ocean (Großkopf et al., 2012) or nitrifiers in wastewater treatment plants (Wang et al., 2014). The reasons for their constantly low abundance are not yet resolved but most likely vary between different taxa and may include a decreased efficiency in the competition for resources, predation and viral attack, or an energy metabolism sustaining little growth (Lynch and Neufeld, 2015).
Recognized SRM are typically members of the rare biosphere in peatlands (Loy et al., 2004; Costello and Schmidt, 2006; Dedysh et al., 2006; Kraigher et al., 2006; Pester et al., 2010; Steger et al., 2011; Tveit et al., 2013). Previous research showed that rare peatland Desulfosporosinus spp. have the potential for high cell-specific sulfate reduction rates (Pester et al., 2010) that are at the upper limit of cell-specific rates observed for pure cultures (Detmers et al., 2001). This provided indirect evidence that rare biosphere members could contribute substantially to biogeochemical cycling in peatlands. In the same peatland, a high diversity of deep-branching lineages of the dsrAB genes (subunit A and B of the dissimilatory sulfite reductase) was identified previously (Loy et al., 2004; Schmalenberger et al., 2007; Pester et al., 2010; Steger et al., 2011). Although dsrAB generally serve as functional marker genes for SRM (Müller et al., 2015), the taxonomic identity of these novel lineages and their function with regard to sulfur and carbon metabolism remain unclear (Pester et al., 2012b; Müller et al., 2015). Here, we set up controlled incubation experiments supplemented with single substrates and sulfate to simulate sulfate-reducing hot spots and gain insights into the discrepancy between the low abundance of SRM and the high sulfate reduction rate regularly observed in peatlands.
Materials and methods
Peat microcosms
The acidic peatland Schlöppnerbrunnen II is located in Germany (50°07′54.8″N, 11°52′51.8″E). Main site characteristics and soil core sampling are described in the Supplementary Methods. Peat soil microcosms were set up by mixing 30 g of fresh soil (10–20 cm depth) with 60 ml of filter-sterilized (0.2 μm) and anoxic peat water in 250 ml sterile glass bottles. Microcosms were sealed under 100% N2 with butyl rubber septa and incubated for 53 days in the dark at 14 °C, which is within the observed air temperature range of this peatland (Schmidt et al., 2015). Microcosms were periodically amended from the first day of incubation with formate, acetate, propionate, lactate or butyrate, or incubated without any external substrate (Supplementary Figure S1). Added substrates generally did not exceed a final concentration of 100–200 μm (Supplementary Figure S2a). A side aspect of the formate additions was its potential to target H2-using microorganisms as well. Interconversion of formate to H2+CO2 is at thermodynamic equilibrium (−4 kJ mol–1) and has been documented in peat soil incubations of the analyzed peatland (Hunger et al., 2011). Half of the substrate-amended and -unamended microcosms additionally received periodic amendments of sulfate (Supplementary Figure S1). These microcosms were initially spiked with sulfate to a final concentration of 190–387 μm, and, thereafter, were periodically amended with small amounts of sulfate equivalent to 79–161 μm (Supplementary Figure S2b). Triplicate microcosms were set up per incubation condition (36 microcosms in total) and periodically sampled for substrate/sulfate concentrations directly after amendment (Supplementary Figure S1). Identical microcosms were set up to measure CO2 and CH4 production (Supplementary Methods).
Quantification of sulfate and substrate turnover
Sulfate and substrate concentrations at days 0, 4, 7, 11, 14, 18, 21, 25, 28, 35, 42 and 49 were determined by capillary electrophoresis (P/ACE MDQ; Beckman Coulter, Brea, CA, USA) directly after amendment using the CEofix Anions 5 Kit (Analis, Suarlée, Belgium). To obtain substrate-specific sulfate turnover (ST) rates, we determined how much sulfate was used in the substrate- and sulfate-amended microcosms in comparison with the sulfate-amended controls without external substrate. This served to discriminate against sulfate dissimilation caused by SRMs using endogenous substrates and sulfate assimilation in general. The calculation was performed by subtracting average sulfate concentrations of the sulfate-amended no-substrate controls from the individual substrate- and sulfate-amended microcosms at each corresponding time point. A nonparametric regression was fitted to the obtained values in R (R Core Team, 2015), and the slope of the regression curve was used to calculate net ST rates. Sulfate concentrations were previously converted from μm to nmol cm–3 of fresh peat soil using the measured soil water content (78% Supplementary Methods) and the bulk density of peat soil (0.29 g cm–3; Goldberg et al., 2008) to allow comparison with results obtained in other studies (e.g., Knorr et al., 2009; Knorr and Blodau, 2009).
Amplicon sequencing and qPCR
After 5, 26 and 50 days of incubation, total nucleic acids were extracted from soil samples, purified, separated into RNA and DNA fractions and quantified (Supplementary Methods). Complete removal of DNA in RNA samples was verified with a quantitative PCR (qPCR) assay targeting the 16S rRNA genes of most Bacteria and Archaea (Supplementary Methods). The V4 region of the 16S rRNA gene and its cDNA was amplified as described previously (Caporaso et al., 2011) and sequenced in three Illumina MiSeq runs at the Joint Genome Institute (genome portal projects 1 016 201, 1 016 203 and 1 031 338) and deposited at NCBI (SRA277144, SRA277986 and SRA277988). All reads were subjected to quality control (iTagger; https://bitbucket.org/berkeleylab/jgi_itagger/), UPARSE clustering (including de novo chimera filtering) (Edgar, 2013), singleton filtering (iTagger) and UCHIME chimera filtering (Edgar et al., 2011) (Supplementary Methods). This resulted in 10.0 and 11.3 million high-quality reads from DNA and RNA samples, respectively, which formed a total of 7435 species-level operational taxonomic units (OTUs) (97% sequence identity). The library size varied between 17 795–323 368 and 46 764–341 948 reads per DNA and RNA sample, respectively, with a median of 84 629 and 93 245 reads (Supplementary Figure S3a). Taxonomic identity was assigned with the Ribosomal Database Project (RDP) classifier 2.9 and the RDP 16S rRNA training set 10 using a confidence threshold of 0.5 (Wang et al., 2007). Relative abundances obtained from 16S rRNA gene samples were corrected for copy number bias using rrnDB 4.3.3 (Stoddard et al., 2015) in R (R Core Team, 2015), resulting in estimated relative genome abundance values for each OTU (Supplementary Methods). This was previously shown to improve diversity and abundance estimates (Kembel et al., 2012). For selected OTUs, the taxonomic assignment was verified by phylogenetic tree reconstruction as outlined in the Supplementary Methods. qPCR assays targeting 16S rRNA genes or cDNA of the genus Desulfosporosinus, most Bacteria and Archaea, and selected dsrA variants were performed as described in the Supplementary Methods. Reliability of relative abundance shifts in our amplicon sequencing approach was verified by a mock community analysis as internal control (Supplementary Figure S4; Herbold et al., 2015).
Statistical analysis
Differential abundance of OTUs between individual treatments and time points was tested in R (R Core Team, 2015) using the package edgeR (Robinson et al., 2010) based on the recommendations of McMurdie and Holmes (2014). Pairwise comparisons were performed by testing independently for the effect of sulfate, substrate and incubation time (Supplementary Methods). Each test included three replicates per treatment, with the exception of the propionate- and sulfate-amended microcosms, where one replicate was excluded from all analyses because of its inconsistent ST (Figure 1). OTUs entering statistical analysis required >10 reads in at least two out of six samples per pairwise comparison. Significantly responding OTUs were assigned to sulfate-stimulated or -deterred conditions with respect to a given substrate and response time. For association network analysis, regularized log transformation was applied to OTU abundance data to stabilize variance heterogeneity, as implemented in the DESeq2 package (Love et al., 2014). Resulting variance homogeneity was verified with the Brown–Forsythe test (P-value>0.1) in R (R Core Team, 2015). Transformed OTU abundances of responsive OTUs were correlated pairwise to each other and to ST using Pearson's correlation. Significant (false discovery rate-corrected P-value<0.05) OTU–ST and OTU–OTU pairwise correlations were used for network construction.
Figure 1.
Substrate-specific net ST rates in sulfate-stimulated microcosm. Individual microcosms are depicted by different symbols (and are consistent in all figures that show triplicate data). Solid symbols indicate time points where sulfate concentrations were measured, and open symbols indicate time points selected for 16S rRNA (gene) amplicon sequencing. Minor negative values are because of variations caused by manual sulfate addition to individual microcosms in comparison with the corresponding no-substrate controls and as such can be neglected. Sulfate pools were never depleted in any sulfate-amended microcosm.
Physiological characterization of Telmatospirillum siberiense
Telmatospirillum siberiense 26-4b1T (DSM-18240; Sizova et al., 2007) was incubated anoxically with DSMZ medium 1126 (DSMZ, Braunschweig, Germany). Sulfate was added to the medium to a final concentration of 1 mm. Anaerobic growth at 28 °C was inferred from measuring optical density (600 nm) in parallel incubations in medium supplemented with different carbon sources, that is, citrate (positive control for growth of T. siberiense), formate, propionate, lactate or butyrate (5 mm each). DNA was extracted from biomass grown on the citrate-amended medium with the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) and tested for the presence of the dsrAB genes with the primer pairs DSR190F/DSR916R (dsrA), DSR1762F/DSR2107R (dsrB) and DSR190F/DSR2107R (both) (Pelikan et al., 2015).
Results
Fermentation products differ in their impact on ST
Anoxic peat microcosms were set up to reflect naturally occurring concentrations of substrates and generally did not exceed 100–200 μm (Supplementary Figure S2a). For this purpose, microcosms were periodically amended with small amounts of formate, acetate, propionate, lactate or butyrate—substrates that represent typical organic carbon degradation intermediates in peatlands (Schmalenberger et al., 2007; Küsel et al., 2008; Limpens et al., 2008). Microcosms without external substrate served as controls for the impact of endogenous substrates. To mimic a sulfate reduction hot spot in the peat (Jacks and Norrström, 2004; Knorr and Blodau, 2009; Knorr et al., 2009; Frei et al., 2012), half of the microcosms were periodically amended with sulfate in the lower μm range (accumulating over time to a maximum of ~0.5 to 2 mm depending on the substrate supplemented in parallel; Supplementary Figure S2b). In sulfate-stimulated incubations, supplemented substrates were completely turned over after each addition. The same was true for incubations without external sulfate with the exception of butyrate-amended microcosms, where butyrate started to accumulate slightly after 30 days (Supplementary Figure S2a).
Net ST increased over time but differed substantially between different substrates. Highest ST was observed for butyrate at the end of the incubations (averaging 174 nmol cm–3 per day), followed by propionate, lactate, acetate and formate (averaging 16 nmol cm–3 per day) in decreasing order (Figure 1). In formate- and propionate-amended microcosms, one replicate each showed a clear deviation in ST from the other two, being either substantially higher or lower, respectively. As the peat soil had not been homogenized before separation into individual microcosms, this likely reflects the spatially heterogeneous distribution of the microbiota in the peat soil matrix. ST increased overincubation time in each of the individual substrate-amended microcosms. Little ST at early time points is possibly due to a presence of alternative, energetically more favorable electron acceptors such as nitrate, Fe(III) or humic matter (Thauer et al., 1977) as observed previously in this peatland (Küsel et al., 2008; Knorr and Blodau, 2009). The presence of such alternative electron acceptors was corroborated by a parallel microcosm setup, where over the total incubation time of 27 days only a minor carbon flow towards methanogenesis (0.1–0.2%) was observed in incubations not amended with sulfate (Supplementary Table S1). In sulfate-stimulated microcosms, CH4 production was reduced by 83–100% as compared with controls without additional sulfate (based on the time frame between 18 and 27 days; Supplementary Figure S2c). In the same parallel microcosm setup, sulfate amendment did not alter total CO2 production (Supplementary Figure S2c), but shifted 43–100% of organic carbon mineralization towards sulfate reduction at the very end of the incubation time (assuming that equivalents of acetate are completely oxidized to two CO2 with the eight released reducing equivalents being used to reduce sulfate to sulfide, detailed in Supplementary Methods and Results).
Only low-abundance OTUs responded to sulfate stimulation
We used changes in relative genome abundance (i.e., relative 16S rRNA gene abundance corrected for rrn operon copy numbers) per OTU to estimate microbial growth. In addition, the protein synthesis potential of an OTU population was determined by calculating its relative ribosome abundance (i.e., relative 16S rRNA cDNA abundance). The abundant community (OTUs with ⩾0.1% relative genome abundance) of the native soil was dominated by members of the Acidobacteria, Alphaproteobacteria, Actinobacteria and Planctomycetes (Supplementary Figure S3b). None of these abundant OTUs was stimulated by sulfate in any of the treatments. In contrast, 18 low-abundance OTUs each responded positively (P-value<0.05) to either sulfate-stimulated or -unstimulated conditions under different substrate scenarios (Figure 2a). Most of these OTUs stayed below or reached levels slightly above 0.1% relative genome abundance throughout the incubation period. Only Telmatospirillum OTU0029 exceeded 1% relative genome abundance towards the end of the incubation period in butyrate incubations without sulfate amendments (Figure 2b).
Figure 2.
OTUs responding to individual carbon degradation intermediates. (a) OTUs responding positively to sulfate stimulation (+Sulfate) and/or unamended conditions (−Sulfate) at the 16S rRNA cDNA (i.e., ribosome) or gene (i.e., genome) level. OTUs 0062+0273, 0346 or 0029+0062 exhibited both +Sulfate and −Sulfate responses in propionate-, lactate- or butyrate-amended microcosms, respectively (stronger response indicated). Taxonomic classification indicates phyla (classes for Proteobacteria) and the highest taxonomic level resolved by the RDP classifier. OTUs without RDP classification at class level (marked with asterisks) were assigned tentative classifications by the SINA online classifier (Pruesse et al., 2012) using a minimum identity threshold of 0.75. (b) Relationship of relative genome and ribosome abundance per responsive OTU at time points with significant abundance shifts. Lines connect replicate microcosms for each OTU (abundances of zero not shown). White circles represent 10 000 randomly selected data points (>0.0005% relative abundance).
In accordance to the high ST observed under butyrate, propionate and lactate, 5–11 OTUs responded positively to sulfate stimulation under each of these substrates, whereas none responded significantly to the addition of formate or acetate. Responsive OTUs were affiliated with taxa containing known SRM (Desulfosporosinus, Desulfomonile, Desulfopila and Desulfovibrio) and also to taxa not known to harbor SRM. The latter were affiliated to Alphaproteobacteria, Acidobacteria and Gammaproteobacteria, and tentatively also to candidate phylum TM6, Fibrobacteres, ‘Parcubacteria' (candidate phylum OD1; Rinke et al., 2013) and Verrucomicrobia (Figure 2a). Our stringent statistical approach did not reveal any OTUs responding positively to sulfate stimulation with formate. However, although ST only increased minimally in two of the formate-amended microcosms, it clearly increased in the third microcosm (Figure 1). Interestingly, these differences in ST were mirrored in concurrent response patterns at the ribosome level for several OTUs that responded also positively to the sulfate addition under other substrates (Desulfosporosinus OTU0051, Telmatospirillum OTU0062, Desulfomonile OTU0144, Desulfopila OTU0256, Magnetospirillum OTU0339 and unclassified Rhodospirillaceae OTU0577; Figure 3 and Supplementary Figures S5a–e).
Figure 3.
Temporal changes in relative ribosome and genome abundance of the most abundant Desulfosporosinus OTU (0051) in incubations with different substrates. Solid lines and symbols depict sulfate-stimulated microcosms, whereas dashed lines and open symbols depict controls without additional sulfate. Diagonal crosses depict the abundance in the native soil, which was sampled from parallel peat soil subsamples and plotted at day 0 in all panels. Halos around symbols indicate significantly higher abundance in the sulfate-stimulated microcosms as compared with their respective controls and day 5.
Low-abundance bacteria differ in their response strategy towards favorable conditions
The majority of OTUs with significantly increased ribosome abundance showed only a weak and some even no significant increase in genome abundance over a period of 50 days (Figure 2a). These relative abundance shifts were not obscured by changes in absolute microbial abundance as the number of bacterial and archaeal 16S rRNA genes cm–3 soil remained constant throughout all incubations as measured by qPCR (Supplementary Figure S6a). Members of the genus Desulfosporosinus (Supplementary Figure S7a) were major responders to the addition of sulfate and substrate at the ribosome level but apparently did not grow. Two Desulfosporosinus OTUs were detected with a relative genome abundance of 0.003% (OTU0051) and 0.001% (OTU0228) in the native soil. Their relative genome abundance was slightly higher at the first measured time point in the microcosms (day 5, on average 0.022% and 0.009%, respectively), which could simply be due to the different soil subsamples used to characterize the native peat soil community and for setting up the microcosms. Alternatively, this might be explained by germination of a Desulfosporosinus sub-population (Stackebrandt, 2014), as the nucleic acids extraction from spores is less efficient than from active cells. Thereafter, the relative genome abundance of both OTUs did not change significantly over 50 days of incubation, as confirmed with a qPCR assay targeting the genus Desulfosporosinus (Supplementary Figure S6b). Also here, no growth was observed at a median abundance of 1.2 × 105 genomes cm–3 soil with one small outlier under lactate at day 50 that reached an abundance of 6.9 × 105 genomes cm–3 soil. In contrast, the relative ribosome abundance of both OTUs increased significantly (P-value<0.001) from 0.30% to 3.69% (OTU0051) and 0.03% to 0.48% (OTU0228) between days 5 and 50 in microcosms with butyrate and sulfate amendment (Figure 3 and Supplementary Figure S5f). The more abundant Desulfosporosinus OTU0051 also responded in the same manner to additions of lactate and propionate. These findings were corroborated by qPCR analysis, which showed that the ribosome per genome ratio of the genus Desulfosporosinus increased from 2600–7300 to 57 000–84 000 throughout the incubation in sulfate-stimulated microcosms amended with propionate, lactate or butyrate (Supplementary Figure S6b). Such high cellular ribosome contents have been reported for exponentially growing pure cultures (Bremer and Dennis, 1996) but so far have not been observed in low-abundance microorganisms keeping a steady population size. Among recognized SRM OTUs, Desulfosporosinus OTU0051 correlated best with substrate-specific ST (Supplementary Figure S8) and dominated the average relative ribosome abundance of this metabolic group (Figure 2b).
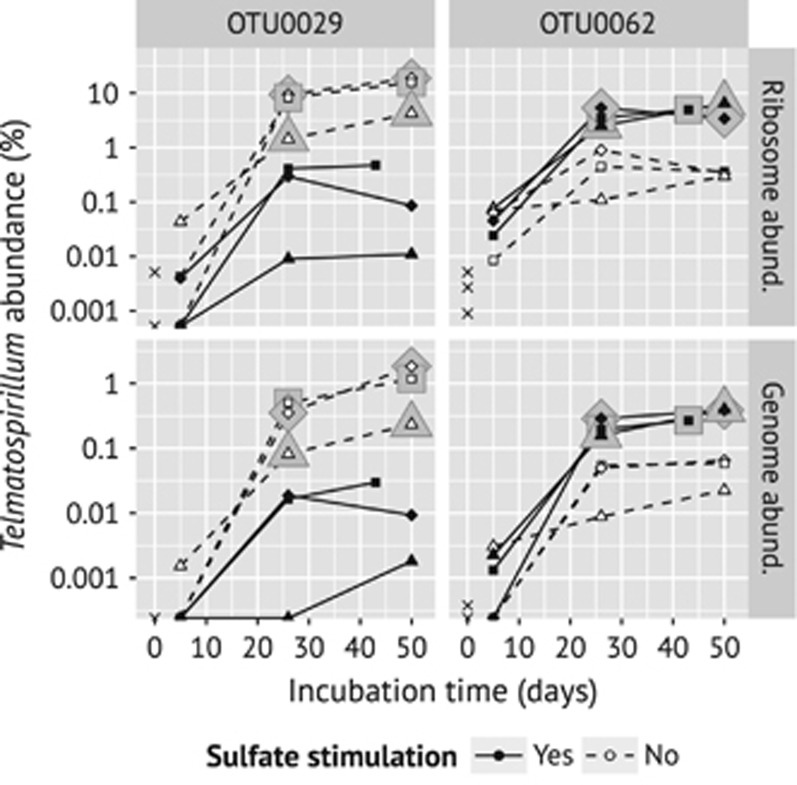
In contrast to Desulfosporosinus and other slow- or non-growing taxa, OTUs of the genus Telmatospirillum (Supplementary Figure S7b) responded with both increased relative ribosome and genome abundance (Supplementary Figure S5a). The two most abundant Telmatospirillum OTUs were standing out among the responsive OTUs that are not affiliated with recognized SRM because of the magnitude of their response as well as their ecologically incoherent behavior, that is, they showed an opposite response to sulfate under butyrate amendment. Although OTU0062 responded stronger in sulfate-stimulated as compared with -unstimulated microcosms, the opposite response was observed for OTU0029 (Figure 4). OTU0029 dominated the response under butyrate without sulfate amendment, reaching at the end of the incubation an average relative genome abundance of 1.1% and an average relative ribosome abundance of 12.9%. In comparison, in butyrate incubations with sulfate amendment OTU0062 reached an average relative genome abundance of 0.4% and an average relative ribosome abundance of 5.0%.
Figure 4.
Temporal changes in relative ribosome and genome abundance of the two most abundant Telmatospirillum OTUs (0029 and 0062) in butyrate-amended incubations. Solid lines and symbols depict sulfate-stimulated microcosms, whereas dashed lines and open symbols depict controls without additional sulfate (note the logarithmic scale on the y axis). Diagonal crosses depict the abundance in the native soil. Halos around symbols indicate significantly higher abundance in the unstimulated or sulfate-stimulated microcosms, respectively, as compared with their respective controls and day 5. Both Telmatospirillum OTUs showed a significant increase in relative ribosome abundance over time, irrespective of sulfate stimulation. Data points drawn directly on the x axis represent relative abundance values of zero.
To assess the possibility that Telmatospirillum OTU0062 could represent a so far unrecognized SRM, we performed growth experiments with T. siberiense, which is the only cultured species of this genus (97.2% sequence similarity). T. siberiense was previously reported to ferment various dicarboxylic acids, pyruvate and glucose under anoxic conditions. It was further tested negative for sulfate reduction, but only with acetate as electron donor (Sizova et al., 2007). Here, we show that T. siberiense is also unable to grow anaerobically with either formate, propionate, lactate or butyrate in the presence of sulfate as electron acceptor (Supplementary Figure S9). In addition, all attempts to amplify the SRM marker genes dsrAB from DNA extracts of T. siberiense using specific PCR assays failed.
Discussion
Low-abundance microorganisms drive a biogeochemically relevant process
In this study, we performed a series of controlled microcosm experiments that constituted a window into naturally fluctuating biogeochemical conditions in peat soils favorable for sulfate reduction. We show that SRM were clearly involved in the degradation of the intermediate fermentation products propionate, lactate and butyrate. It is possible that at least some of these substrates were not directly used by SRM but initially degraded by secondary fermenters to acetate, formate or H2/CO2 with SRM thriving on the latter. However, lower ST under formate and acetate as compared with propionate, lactate and butyrate rather indicated a direct utilization of these C3 and C4 compounds (Figure 1).
Responsive OTUs affiliated with ST were all composed of low-abundance OTUs in the native soil, most of which stayed below or around 0.1% relative genome abundance throughout the incubation period and were permanent members of the rare biosphere in our experiments. The only major exceptions were Desulfomonile OTU0144 and Telmatospirillum OTUs 0029 and 0062, which increased in sulfate-amended incubations to maximum relative genome abundances of 0.26–0.46% (Supplementary Figures S5a and b). As all three OTUs were of low abundance in the native soil, they classify as conditionally rare biosphere members (Lynch and Neufeld, 2015). Some responsive OTUs were affiliated to the sulfate-reducing genera Desulfovibrio, Desulfomonile and Desulfopila (Deltaproteobacteria), and Desulfosporosinus (Firmicutes), and showed a strong correlation in their response to ST (r=0.71–0.94; Supplementary Figure S8). In addition, several responsive OTUs not related to recognized SRM were also strongly associated with ST and/or SRM (Supplementary Figure S8). Here, two OTUs were standing out, with Telmatospirillum OTU0062 correlating strongly under butyrate to ST (r=0.87) as well as to all recognized SRM (r=0.90–0.96) and Rhodospirillaceae OTU0577 correlating to ST under propionate, lactate and butyrate (r=0.83–0.89). In the light of the large diversity of novel dsrAB variants, which have been repeatedly detected in the analyzed peatland (Loy et al., 2004; Pester et al., 2010; Steger et al., 2011), we speculated whether these OTUs may represent so far unrecognized SRM (detailed in Supplementary Results). Because Telmatospirillum OTU0062 had the highest 16S rRNA identity to a cultivated species, we used the type species T. siberiense as a case example to evaluate this hypothesis. However, T. siberiense tested negative for growth under sulfate-reducing conditions with all five substrates investigated in this study (Sizova et al., 2007; and this study, Supplementary Figure S9) and for the presence of dsrAB. Hence, it seems more likely that Telmatospirillum OTU0062, and possibly also Rhodospirillaceae OTU0577, fermented the provided substrates, whereas SRM made syntrophic use of the formed formate and/or H2. However, this would not explain why both OTUs also responded to formate in the one microcosm that showed pronounced ST. As such, it may also be possible that these OTUs represent anaerobic sulfur oxidizers. As sulfide oxidation in wetlands can be coupled to the reduction of Fe(III) (Hansel et al., 2015) or potentially also humic matter (Pester et al., 2012b), Telmatospirillum OTU0062 and Rhodospirillaceae OTU0577 may represent microorganisms that are responsible for this so far missing link in the cryptic sulfur cycle, either by directly being involved in catalyzing these reactions or using elemental sulfur resulting from chemical oxidation of sulfide with Fe(III) compounds. Recently, correlation of sulfate-reducing activity and anaerobic CH4 oxidation was reported for three North American wetlands (Segarra et al., 2015). We did not observe any evidence for the presence and activity of typical microorganisms mediating sulfate-dependent anaerobic CH4 oxidation in our incubations, despite the good coverage of the used primers for ANME archaea (ANME-1: 77% ANME-2a/2b: 89% ANME-2c: 76% and ANME-3: 82% Quast et al., 2013) and the detection of related archaea within the orders Methanosarcinales (five OTUs) and Methanocellales (five OTUs).
A novel mechanism to display metabolic activity
The relationship between non-growth activity and cellular rRNA content is not understood but important for the interpretation of environmental rRNA data to characterize microbial communities (Blazewicz et al., 2013). Our results provide strong evidence that a rare Desulfosporosinus species can follow an ecological strategy to increase its cellular rRNA content while maintaining its population size over a period of 50 days. As Desulfosporosinus OTU0051 always correlated best in its ribosome response to ST among all responsive SRM (r=0.89–0.94) and contributed the highest relative ribosome abundance of this metabolic group, our results strongly suggest that this increase in protein synthesis potential was also translated into metabolic activity. All microorganisms have to divide at some time in their lives to avoid extinction. Population growth of a Desulfosporosinus species was observed in a DNA-stable isotope probing study of the same peatland after a longer period of incubation (73 days) (Pester et al., 2010). Direct comparisons between the two studies are difficult because of the different incubation conditions; that is, peat soil microcosm incubations with a substrate mixture of formate, acetate, lactate and propionate, and two starvation phases in the previous study (Pester et al., 2010). Whether the addition of a substrate mixture, especially in the presence of acetate that is a commonly supplemented building block for biomass production in anoxic cultivation media, was responsible for the observed growth remains uncertain, but may explain the different growth response as compared with this study. The previous study also showed that Desulfosporosinus stayed rare in snapshot samples of the native soil spanning over a time period of 3 years. The combined results of this and the previous study thus indicate that rare peatland Desulfosporosinus species (i) maintain a stable, low-abundance population in the studied peatland and (ii) respond to favorable environmental conditions first by increasing their ribosome content and metabolic activity and only after prolonged time by growth. This is supported by the fact that Desulfosporosinus species contain an exceptionally high number of ribosomal (rrn) operons in their genomes (8–10 copies; Pester et al., 2012a), which would enable them to react very quickly to favorable conditions by an increase in ribosomes. This strategy would make sense in the highly fluctuating conditions of peatlands in space and time (Jacks and Norrström, 2004; Knorr et al., 2009; Reiche et al., 2009; Frei et al., 2012) and would be beneficial in escaping grazing pressure or viral attack (Lynch and Neufeld, 2015). In addition, such a strategy may be widespread in different habitats as suggested by the high ribosome to genome ratios of certain rare biosphere members in a 3-year survey of marine bacterioplankton (Campbell et al., 2011) and in a snapshot analysis of a hypersaline lake sediment (Weigold et al., 2015).
Rare biosphere members are either considered as conditionally rare taxa that eventually grow to large population sizes upon favorable conditions or as permanently rare taxa that express rapid, periodic small abundance changes upon activity (Lynch and Neufeld, 2015). Here, we provide conclusive evidence that the Desulfosporosinus species in the studied peatland soil is prototypical of a new strategy of rare biosphere members, namely to postpone growth and remain at a constant low population size while maintaining increased metabolic activity over prolonged time spans. As Desulfosporosinus species are members of the rare biosphere in permafrost soil, natural wetlands and rice paddies worldwide (Supplementary Figure S10), their sulfate-reducing activity and ecological strategy may have a widespread effect on ecosystem functioning in these environments. In conclusion, our results underscore the importance of the rare microbial biosphere not only as a seed bank of dormant microorganisms but also as an active mediator of biogeochemical processes that buffer against climate change.
Acknowledgments
This research was financially supported by the Austrian Science Fund (FWF, P23117-B17 to MP and AL), the US Department of Energy (CSP605 to MP and AL), the German Research Foundation (DFG, PE 2147/1-1 to MP) and the European Union (FP7-People-2013-CIG, Grant No. PCIG14-GA-2013-630188 to MP). The work conducted by the Joint Genome Institute was supported by the Office of Science of the US Department of Energy under Contract No. DE-AC02-05CH11231. We are grateful to Martin Huemer and Norbert Bittner for technical support during qPCR analysis and field sampling, respectively. We additionally thank Ilias Lagkouvardos for performing analyses with the integrated microbial NGS platform, Craig Herbold for help with the taxonomic classification and the staff of the Joint Genome Institute for amplicon library preparation, sequencing and standard bioinformatics support.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alonso-Sáez L, Díaz-Pérez L, Morán XAG. (2015). The hidden seasonality of the rare biosphere in coastal marine bacterioplankton. Environ Microbiol 17: 3766–3780. [DOI] [PubMed] [Google Scholar]
- Beer J, Lee K, Whiticar M, Blodau C. (2008). Geochemical controls on anaerobic organic matter decomposition in a northern peatland. Limnol Oceanogr 53: 1393–1407. [Google Scholar]
- Blazewicz SJ, Barnard RL, Daly RA, Firestone MK. (2013). Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J 7: 2061–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer H, Dennis PP. (1996) Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B et al (eds), Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press: Washington, DC, USA, pp 1553–1569. [Google Scholar]
- Campbell BJ, Yu L, Heidelberg JF, Kirchman DL. (2011). Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci USA 108: 12776–12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108: 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Schmidt SK. (2006). Microbial diversity in alpine tundra wet meadow soil: novel Chloroflexi from a cold, water-saturated environment. Environ Microbiol 8: 1471–1486. [DOI] [PubMed] [Google Scholar]
- Dedysh SN, Pankratov TA, Belova SE, Kulichevskaya IS, Liesack W. (2006). Phylogenetic analysis and in situ identification of Bacteria community composition in an acidic Sphagnum peat bog. Appl Environ Microbiol 72: 2110–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmers J, Brüchert V, Habicht KS, Kuever J. (2001). Diversity of sulfur isotope fractionations by sulfate-reducing prokaryotes. Appl Environ Microbiol 67: 888–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake HL, Horn MA, Wüst PK. (2009). Intermediary ecosystem metabolism as a main driver of methanogenesis in acidic wetland soil. Environ Microbiol Rep 1: 307–318. [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996–998. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei S, Knorr K-H, Peiffer S, Fleckenstein JH. (2012). Surface micro-topography causes hot spots of biogeochemical activity in wetland systems: a virtual modeling experiment. J Geophys Res Biogeosci 117: G00N12. [Google Scholar]
- Gauci V, Chapman SJ. (2006). Simultaneous inhibition of CH4 efflux and stimulation of sulphate reduction in peat subject to simulated acid rain. Soil Biol Biochem 38: 3506–3510. [Google Scholar]
- Gauci V, Dise N, Blake S. (2005). Long-term suppression of wetland methane flux following a pulse of simulated acid rain. Geophys Res Lett 32: L12804. [Google Scholar]
- Gauci V, Matthews E, Dise N, Walter B, Koch D, Granberg G et al. (2004). Sulfur pollution suppression of the wetland methane source in the 20th and 21st centuries. Proc Natl Acad Sci USA 101: 12583–12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg SD, Knorr K-H, Gebauer G. (2008). N2O concentration and isotope signature along profiles provide deeper insight into the fate of N2O in soils. Isotopes Environ Health Stud 44: 377–391. [DOI] [PubMed] [Google Scholar]
- Großkopf T, Mohr W, Baustian T, Schunck H, Gill D, Kuypers MMM et al. (2012). Doubling of marine dinitrogen-fixation rates based on direct measurements. Nature 488: 361–364. [DOI] [PubMed] [Google Scholar]
- Hamberger A, Horn MA, Dumont MG, Murrell JC, Drake HL. (2008). Anaerobic consumers of monosaccharides in a moderately acidic fen. Appl Environ Microbiol 74: 3112–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel CM, Lentini CJ, Tang Y, Johnston DT, Wankel SD, Jardine PM. (2015). Dominance of sulfur-fueled iron oxide reduction in low-sulfate freshwater sediments. ISME J 9: 2400–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitmann T, Blodau C. (2006). Oxidation and incorporation of hydrogen sulfide by dissolved organic matter. Chem Geol 235: 12–20. [Google Scholar]
- Herbold CW, Pelikan C, Kuzyk O, Hausmann B, Angel R, Berry D et al. (2015). A flexible and economical barcoding approach for highly multiplexed amplicon sequencing of diverse target genes. Front Microbiol 6: 731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervàs A, Camarero L, Reche I, Casamayor EO. (2009). Viability and potential for immigration of airborne bacteria from Africa that reach high mountain lakes in Europe. Environ Microbiol 11: 1612–1623. [DOI] [PubMed] [Google Scholar]
- Hervàs A, Casamayor EO. (2009). High similarity between bacterioneuston and airborne bacterial community compositions in a high mountain lake area. FEMS Microbiol Ecol 67: 219–228. [DOI] [PubMed] [Google Scholar]
- Hodgkins SB, Tfaily MM, McCalley CK, Logan TA, Crill PM, Saleska SR et al. (2014). Changes in peat chemistry associated with permafrost thaw increase greenhouse gas production. Proc Natl Acad Sci USA 111: 5819–5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugoni M, Taib N, Debroas D, Domaizon I, Jouan Dufournel I, Bronner G et al. (2013). Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc Natl Acad Sci USA 110: 6004–6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger S, Schmidt O, Hilgarth M, Horn MA, Kolb S, Conrad R et al. (2011). Competing formate- and carbon dioxide-utilizing prokaryotes in an anoxic methane-emitting fen soil. Appl Environ Microbiol 77: 3773–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks G, Norrström A-C. (2004). Hydrochemistry and hydrology of forest riparian wetlands. For Ecol Manage 196: 187–197. [Google Scholar]
- Keller JK, Weisenhorn PB, Megonigal JP. (2009). Humic acids as electron acceptors in wetland decomposition. Soil Biol Biochem 41: 1518–1522. [Google Scholar]
- Kembel SW, Wu M, Eisen JA, Green JL. (2012). Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLoS Comput Biol 8: e1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorr K-H, Blodau C. (2009). Impact of experimental drought and rewetting on redox transformations and methanogenesis in mesocosms of a northern fen soil. Soil Biol Biochem 41: 1187–1198. [Google Scholar]
- Knorr K-H, Lischeid G, Blodau C. (2009). Dynamics of redox processes in a minerotrophic fen exposed to a water table manipulation. Geoderma 153: 379–392. [Google Scholar]
- Kraigher B, Stres B, Hacin J, Ausec L, Mahne I, Van Elsas JD et al. (2006). Microbial activity and community structure in two drained fen soils in the Ljubljana Marsh. Soil Biol Biochem 38: 2762–2771. [Google Scholar]
- Küsel K, Blöthe M, Schulz D, Reiche M, Drake HL. (2008). Microbial reduction of iron and porewater biogeochemistry in acidic peatlands. Biogeosciences 5: 1537–1549. [Google Scholar]
- Limpens J, Berendse F, Blodau C, Canadell JG, Freeman C, Holden J et al. (2008). Peatlands and the carbon cycle: from local processes to global implications—a synthesis. Biogeosciences 5: 1475–1491. [Google Scholar]
- Love MI, Huber W, Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy A, Küsel K, Lehner A, Drake HL, Wagner M. (2004). Microarray and functional gene analyses of sulfate-reducing prokaryotes in low-sulfate, acidic fens reveal cooccurrence of recognized genera and novel lineages. Appl Environ Microbiol 70: 6998–7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MDJ, Neufeld JD. (2015). Ecology and exploration of the rare biosphere. Nat Rev Microbiol 13: 217–229. [DOI] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. (2014). Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10: e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, Stams AJM. (2008). The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol 6: 441–454. [DOI] [PubMed] [Google Scholar]
- Müller AL, de Rezende JR, Hubert CRJ, Kjeldsen KU, Lagkouvardos I, Berry D et al. (2014). Endospores of thermophilic bacteria as tracers of microbial dispersal by ocean currents. ISME J 8: 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller AL, Kjeldsen KU, Rattei T, Pester M, Loy A. (2015). Phylogenetic and environmental diversity of DsrAB-type dissimilatory (bi)sulfite reductases. ISME J 9: 1152–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton RJ, Huse SM, Morrison HG, Peake CS, Sogin ML, McLellan SL. (2013). Shifts in the microbial community composition of gulf coast beaches following beach oiling. PLoS One 8: e74265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrós-Alió C. (2012). The rare bacterial biosphere. Ann Rev Mar Sci 4: 449–466. [DOI] [PubMed] [Google Scholar]
- Pelikan C, Herbold CW, Hausmann B, Müller AL, Pester M, Loy A. (2015). Diversity analysis of sulfite- and sulfate-reducing microorganisms by multiplex dsrA and dsrB amplicon sequencing using new primers and mock community-optimized bioinformatics. Environ Microbiol e-pub ahead of print 2 December 2015; doi:10.1111/1462-2920.13139. [DOI] [PubMed]
- Pester M, Bittner N, Deevong P, Wagner M, Loy A. (2010). A ‘rare biosphere' microorganism contributes to sulfate reduction in a peatland. ISME J 4: 1591–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pester M, Brambilla E, Alazard D, Rattei T, Weinmaier T, Han J et al. (2012. a). Complete genome sequences of Desulfosporosinus orientis DSM765TDesulfosporosinus youngiae DSM17734TDesulfosporosinus meridiei DSM13257T, and Desulfosporosinus acidiphilus DSM22704T. J Bacteriol 194: 6300–6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pester M, Knorr K-H, Friedrich MW, Wagner M, Loy A. (2012. b). Sulfate-reducing microorganisms in wetlands—fameless actors in carbon cycling and climate change. Front Microbiol 3: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2015) R: A Language and Environment For Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. Available at: http://www.r-project.org/ accessed 15 February 2016.
- Reiche M, Hädrich A, Lischeid G, Küsel K. (2009). Impact of manipulated drought and heavy rainfall events on peat mineralization processes and source-sink functions of an acidic fen. J Geophys Res Biogeosci 114: G02021. [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD et al. (2007). Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1: 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalenberger A, Drake HL, Küsel K. (2007). High unique diversity of sulfate-reducing prokaryotes characterized in a depth gradient in an acidic fen. Environ Microbiol 9: 1317–1328. [DOI] [PubMed] [Google Scholar]
- Schmidt O, Horn MA, Kolb S, Drake HL. (2015). Temperature impacts differentially on the methanogenic food web of cellulose-supplemented peatland soil. Environ Microbiol 17: 720–734. [DOI] [PubMed] [Google Scholar]
- Schuur EAG, McGuire AD, Schädel C, Grosse G, Harden JW, Hayes DJ et al. (2015). Climate change and the permafrost carbon feedback. Nature 520: 171–179. [DOI] [PubMed] [Google Scholar]
- Segarra KEA, Schubotz F, Samarkin V, Yoshinaga MY, Hinrichs K-U, Joye SB. (2015). High rates of anaerobic methane oxidation in freshwater wetlands reduce potential atmospheric methane emissions. Nat Commun 6: 7477. [DOI] [PubMed] [Google Scholar]
- Serkebaeva YM, Kim Y, Liesack W, Dedysh SN. (2013). Pyrosequencing-based assessment of the Bacteria diversity in surface and subsurface peat layers of a northern wetland, with focus on poorly studied phyla and candidate divisions. PLoS One 8: e63994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova MV, Panikov NS, Spiridonova EM, Slobodova NV, Tourova TP. (2007). Novel facultative anaerobic acidotolerant Telmatospirillum siberiense gen. nov. sp. nov. isolated from mesotrophic fen. Syst Appl Microbiol 30: 213–220. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E. (2014) The emended family Peptococcaceae and description of the families Desulfitobacteriaceae, Desulfotomaculaceae, and Thermincolaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds), The Prokaryotes—Firmicutes and Tenericutes. Springer: Berlin Heidelberg, Germany, pp 285–290. [Google Scholar]
- Steger D, Wentrup C, Braunegger C, Deevong P, Hofer M, Richter A et al. (2011). Microorganisms with novel dissimilatory (bi)sulfite reductase genes are widespread and part of the core microbiota in low-sulfate peatlands. Appl Environ Microbiol 77: 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard SF, Smith BJ, Hein R, Roller BRK, Schmidt TM. (2015). rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res 43: D593–D598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teira E, Lekunberri I, Gasol JM, Nieto-Cid M, Álvarez-Salgado XA, Figueiras FG. (2007). Dynamics of the hydrocarbon-degrading Cycloclasticus bacteria during mesocosm-simulated oil spills. Environ Microbiol 9: 2551–2562. [DOI] [PubMed] [Google Scholar]
- Thauer RK, Jungermann K, Decker K. (1977). Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev 41: 100–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tveit A, Schwacke R, Svenning MM, Urich T. (2013). Organic carbon transformations in high-Arctic peat soils: key functions and microorganisms. ISME J 7: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergin KL, Done B, Carlson Ca, Giovannoni SJ. (2013). Spatiotemporal distributions of rare bacterioplankton populations indicate adaptive strategies in the oligotrophic ocean. Aquat Microb Ecol 71: 1–13. [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhang X-X, Lu X, Liu B, Li Y, Long C et al. (2014). Abundance and diversity of bacterial nitrifiers and denitrifiers and their functional genes in tannery wastewater treatment plants revealed by high-throughput sequencing. PLoS One 9: e113603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigold P, Ruecker A, Loesekann-Behrens T, Kappler A, Behrens S. (2015). Ribosomal tag pyrosequencing of DNA and RNA reveals ‘rare' taxa with high protein synthesis potential in the sediment of a hypersaline lake in Western Australia. Geomicrobiol J e-pub ahead of print 1 July 2015; doi:10.1080/01490451.2015.1049304.
- Wüst PK, Horn MA, Drake HL. (2009). Trophic links between fermenters and methanogens in a moderately acidic fen soil. Environ Microbiol 11: 1395–1409. [DOI] [PubMed] [Google Scholar]
- Yu Z-G, Peiffer S, Göttlicher J, Knorr K-H. (2015). Electron transfer budgets and kinetics of abiotic oxidation and incorporation of aqueous sulfide by dissolved organic matter. Environ Sci Technol 49: 5441–5449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.