Abstract
Resistance to RAF- and MEK-targeted therapy is a major clinical challenge1–4. RAF and MEK inhibitors are initially but only transiently effective in some but not all patients with BRAF gene mutation and are largely ineffective in those with RAS gene mutation because of resistance5–14. Through a genetic screen in BRAF-mutant tumor cells, we show that the Hippo pathway effector YAP (encoded by YAP1) acts as a parallel survival input to promote resistance to RAF and MEK inhibitor therapy. Combined YAP and RAF or MEK inhibition was synthetically lethal not only in several BRAF-mutant tumor types but also in RAS-mutant tumors. Increased YAP in tumors harboring BRAF V600E was a biomarker of worse initial response to RAF and MEK inhibition in patients, establishing the clinical relevance of our findings. Our data identify YAP as a new mechanism of resistance to RAF- and MEK-targeted therapy. The findings unveil the synthetic lethality of combined suppression of YAP and RAF or MEK as a promising strategy to enhance treatment response and patient survival.
Oncogenic activation of RAF-MEK-ERK (mitogen-activated protein kinase (MAPK) pathway) signaling frequently occurs in human cancers, often through somatic activating mutations in BRAF or RAS genes. MAPK pathway–targeted therapies (RAF and MEK inhibitors) have been deployed in patients with BRAF- and RAS-mutant tumors and have been demonstrated to have clinical efficacy in melanoma and non–small cell lung cancer (NSCLC) harboring BRAF V600E1–4, but responses are variable, incomplete and transient because of resistance1–4. Furthermore, some patients with BRAF V600E–mutant melanoma or NSCLC and almost all patients with BRAF V600E–mutant colorectal or thyroid cancer do not initially respond to BRAF inhibitor therapy1–4,8–15. Similarly, MAPK pathway inhibition with MEK inhibitor therapy is largely ineffective in individuals with mutant RAS because of primary resistance5–7,16,17. Thus, there is an urgent need to uncover the molecular targets that limit the response to RAF- and MEK-targeted therapy in both BRAF- and RAS-mutant tumors to develop new therapeutic strategies to enhance treatment response and patient survival.
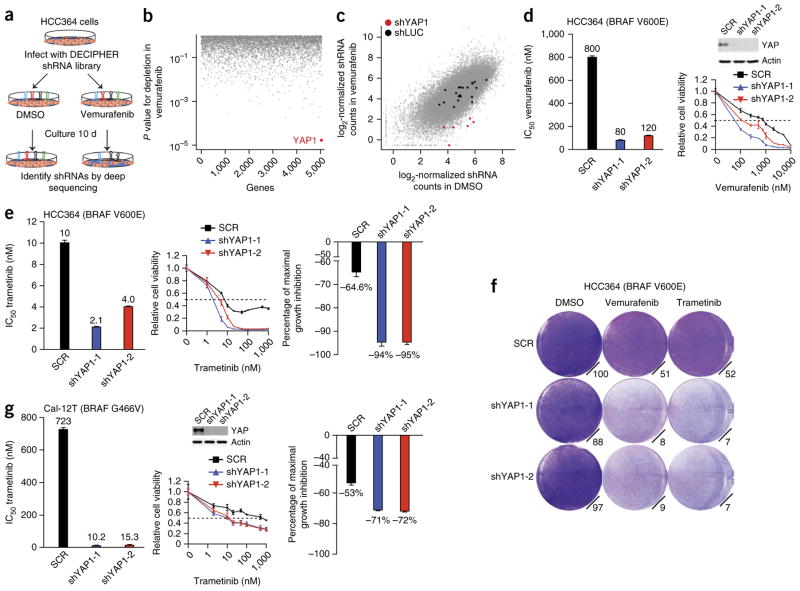
To uncover new genetic modifiers of the response to RAF- targeted therapy in human cancer, we conducted a pooled short hairpin RNA (shRNA) screen in human NSCLC cells harboring BRAF V600E (HCC364 cells) that are dependent on oncogenic BRAF for growth11. Our goal was to identify genes that, when silenced, enhanced the response to RAF inhibitor. We screened 27,500 shRNAs targeting 5,046 signaling components (Supplementary Table 1). After infecting HCC364 cells with lentiviruses expressing the shRNA library and subjecting them to selection, we treated the cells with the selective BRAF inhibitor vemurafenib or with vehicle control (Fig. 1a). We quantified the abundance of each barcoded hairpin to identify shRNAs that were selectively depleted during treatment with vemurafenib but not vehicle (Fig. 1a), as described previously12,18. The Hippo signaling pathway component YAP1 was the best-scoring hit in the screen, as all six YAP1-targeted shRNAs present in the screening library were depleted during treatment with vemurafenib but not vehicle (Fig. 1b,c, Supplementary Fig. 1 and Supplementary Table 2). We therefore hypothesized that the encoded YAP protein is a new determinant of the response to RAF inhibitor and that YAP inhibition might enhance the efficacy of RAF-targeted therapy.
Figure 1.
A pooled shRNA screen in BRAF-mutant human lung cancer cells identifies new modifiers of the RAF inhibitor response including YAP. (a) Summary of the pooled shRNA screening strategy in BRAF-mutant human lung cancer cells. (b,c) Primary screen data showing gene targets (b) and shRNAs depleted specifically upon vemurafenib treatment (c), highlighting YAP1 in red. shYAP1, shRNA to YAP1; shLUC, shRNA to the luciferase gene. (d) Validation of the effects of YAP1 knockdown on sensitivity to vemurafenib in HCC364 BRAF-mutant lung cancer cells (both IC50 and cell viability results are shown). The inset shows the effects of each YAP1 shRNA by immunoblot for YAP protein expression. SCR, scrambled control shRNA. Data are shown as means ± s.e.m. (n = 3 biological replicates). (e) Validation of the effects of YAP1 knockdown on sensitivity to trametinib in HCC364 BRAF-mutant lung cancer cells (IC50, cell viability and maximal growth inhibition results are shown). Data are shown as means ± s.e.m. (n = 3 biological replicates). (f) Effects of YAP1 knockdown on sensitivity to vemurafenib and trametinib in HCC364 BRAF-mutant lung cancer cells (cell growth by crystal violet staining assays is shown, with quantification for each condition relative to cells expressing the scrambled control shRNA treated with DMSO control). (g) Effects of YAP1 knockdown on sensitivity to trametinib in Cal-12T BRAF-mutant (non-V600E) lung cancer cells (IC50, cell viability and maximal growth inhibition results are shown). Data are shown as means ± s.e.m. (n = 3 biological replicates).
We used independent shRNAs to knock down YAP1 in HCC364 cells. YAP1 silencing enhanced sensitivity to vemurafenib with little effect in vehicle-treated cells, confirming the initial screening results (Fig. 1d,f, Supplementary Fig. 1 and Supplementary Table 3). As BRAF activates MEK and MEK inhibitor monotherapy has incomplete efficacy in patients with BRAF V600E–mutant tumors1,3, we tested whether YAP1 silencing enhanced the response to MEK inhibitor in HCC364 cells. YAP1 knockdown enhanced sensitivity to the MEK inhibitor trametinib in this system (Fig. 1e,f and Supplementary Table 3). YAP1 suppression enhanced not only sensitivity to trametinib (IC50, half-maximal inhibition concentration) but also the degree to which maximal growth inhibition was achieved by MEK inhibition (Fig. 1e and Supplementary Table 4). These effects of YAP1 silencing were specific to targeted inhibition of RAF-MEK signaling, as YAP1 knockdown had no effect on sensitivity to cytotoxic chemotherapy (Supplementary Fig. 2). We found that the transcriptional output of YAP is likely critical for regulation of the response to RAF- and MEK-targeted therapy, as silencing either of the Hippo-YAP pathway transcription factor effectors TEAD2 and TEAD4 (encoding TEA domain (TEAD) family members 2 and 4)19,20 phenocopied the effects of YAP1 suppression on sensitivity to RAF and MEK inhibitors in HCC364 cells (Supplementary Fig. 2). Moreover, we observed nuclear YAP expression in these BRAF-mutant cells in cellular fractionation studies (Supplementary Fig. 3). We further found that stable overexpression of either YAP1 or its paralog TAZ19 substantially decreased sensitivity to vemurafenib and trametinib in HCC364 cells (Supplementary Fig. 4).
As MEK inhibitor therapy is more effective than RAF inhibitor therapy in some tumor cells with non-V600E forms of mutant BRAF11, we tested whether YAP1 silencing enhanced sensitivity to trametinib in Cal-12T human NSCLC cells that exhibit MEK-ERK activation but harbor a BRAF mutation encoding a G466V substitution. YAP1 depletion enhanced the efficacy of the MEK inhibitor in Cal-12T cells, indicating that the effects of YAP1 suppression in response to MEK inhibitor are not restricted to V600E forms of mutant BRAF (Fig. 1g and Supplementary Tables 3 and 4). Collectively, these data demonstrate that YAP modulates the response to targeted inhibition of RAF signaling in human NSCLC models.
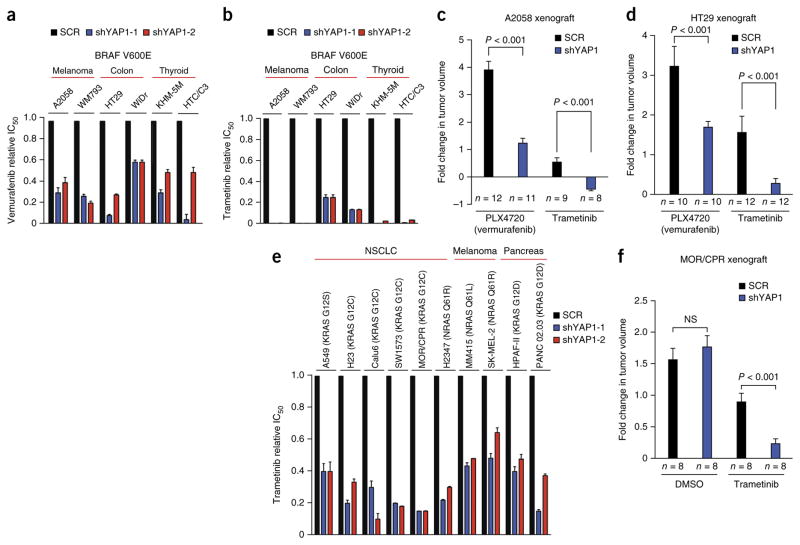
We next investigated whether YAP regulates the response to targeted inhibition of BRAF signaling in other BRAF-mutant tumor histologies, using human melanoma, colon and thyroid cancer cell lines with endogenous BRAF mutation encoding the V600E substitution. YAP1 suppression enhanced the efficacy of both vemurafenib and trametinib in the A2058 and WM793 melanoma cell lines, the HT29 and WiDr colon cancer cell lines, and the KHM-5M and HTC/C3 thyroid cancer cell lines, all harboring BRAF V600E, without significantly affecting vehicle-treated cells (Fig. 2a,b, Supplementary Figs. 3, 5 and 6, and Supplementary Tables 3 and 4). We again observed nuclear YAP expression in these other BRAF-mutant cell lines (Supplementary Fig. 3). Extending our in vitro findings, we found that silencing YAP1 enhanced the response to not only vemurafenib but also trametinib in vivo in A2058 melanoma xenografts, without significantly affecting tumor growth in vehicle-treated tumors (Fig. 2c and Supplementary Fig. 7). YAP1 suppression led to tumor regression upon trametinib treatment in this in vivo system (Fig. 2c and Supplementary Fig. 7). Additionally, we found that YAP1 silencing enhanced the response to RAF and MEK inhibitors in vivo in HT29 colon cancer xenografts harboring BRAF V600E, without significantly affecting tumor growth in vehicle-treated tumors (Fig. 2d and Supplementary Fig. 7). These data show that YAP1 suppression enhances RAF and MEK inhibitor efficacy in many BRAF-mutant tumor types, not only in cells with intrinsic sensitivity but also in those with intrinsic resistance to monotherapy with RAF or MEK inhibitor (Fig. 2, Supplementary Figs. 5, 6 and 7, and Supplementary Tables 3 and 4). YAP1 silencing even overcame intrinsic resistance to RAF and MEK inhibitors in A2058 and KHM-5M cells in which no effect from inhibition of epidermal growth factor receptor (EGFR), which can promote resistance to RAF inhibitor in some tumors with BRAF V600E8,12, was observed (Supplementary Fig. 8). These data indicate a distinct and broad role for YAP in promoting resistance to RAF and MEK inhibition across a wide spectrum of BRAF-mutant tumors.
Figure 2.
YAP regulates the response to RAF and MEK inhibitors in multiple BRAF-mutant tumor types. (a,b) Effects of YAP1 knockdown on sensitivity to vemurafenib (a) and trametinib (b) in the indicated BRAF-mutant cell lines, shown as relative IC50 (data are shown as means ± s.e.m. for all cell viability data; n = 3 biological replicates). (c) Effects of YAP1 knockdown (shYAP1-1) on the efficacy of vemurafenib (PLX4720) and trametinib in A2058 melanoma xenografts encoding BRAF V600E (data are shown as means ± s.e.m.; n = 8–12 tumors/group). (d) Effects of YAP1 knockdown on the efficacy of vemurafenib (PLX4720) and trametinib in HT29 colon cancer xenografts encoding BRAF V600E (data are shown as means ± s.e.m.; n = 8–12 tumors/group). (e) Effects of YAP1 knockdown on sensitivity to trametinib in the indicated RAS-mutant tumor cell lines, shown as relative IC50 (data are shown as means ± s.e.m. for all cell viability data; n = 3 biological replicates). (f) Effects of YAP1 knockdown on the efficacy of trametinib in MOR/CPR RAS-mutant NSCLC xenografts (data are shown as means ± s.e.m.; n = 8–12 tumors/group). NS, not significant; P < 0.05 for IC50 differences between each YAP1 shRNA and scrambled shRNA control in a, b and e, Student t test.
We next explored whether YAP regulates the response to MAPK pathway inhibition in tumor cells with oncogenic RAS, which drives tumor growth, in part, through MEK-ERK signaling21. No effective targeted therapies exist for patients with tumors having mutant RAS, with MEK inhibitor therapy exhibiting limited efficacy5–7,16,17. We investigated whether YAP1 suppression could enhance the response to MEK inhibitor in RAS-mutant tumors. Silencing YAP1 enhanced the efficacy of trametinib in multiple KRAS- and NRAS-mutant human NSCLC, melanoma and pancreatic adenocarcinoma models and across several distinct mutant alleles of RAS genes (Fig. 2e, Supplementary Figs. 5, 9 and 10, and Supplementary Tables 3 and 4). YAP1 suppression enhanced the response to trametinib, as measured by IC50 across these models, but had only modest effects on maximal growth inhibition upon MEK inhibitor treatment in RAS-mutant melanoma and pancreatic adenocarcinoma cells (Fig. 2e and Supplementary Fig. 5). We again observed nuclear YAP expression in these RAS-mutant cell lines (Supplementary Fig. 3). We confirmed these findings in vivo by demonstrating that YAP1 silencing enhanced the response to trametinib in MOR/CPR xenograft tumors encoding KRAS G12C (Fig. 2f and Supplementary Fig. 7). These data identify combined inhibition of YAP and MEK as a promising strategy to enhance treatment response in patients with mutant RAS. Our findings extend recent studies indicating that YAP regulates KRAS oncogene dependence in some tumor types22–24 by establishing that YAP1 suppression enhances the response to MEK inhibitor in multiple tumor histologies and functions across several different forms of oncogenic MAPK signaling.
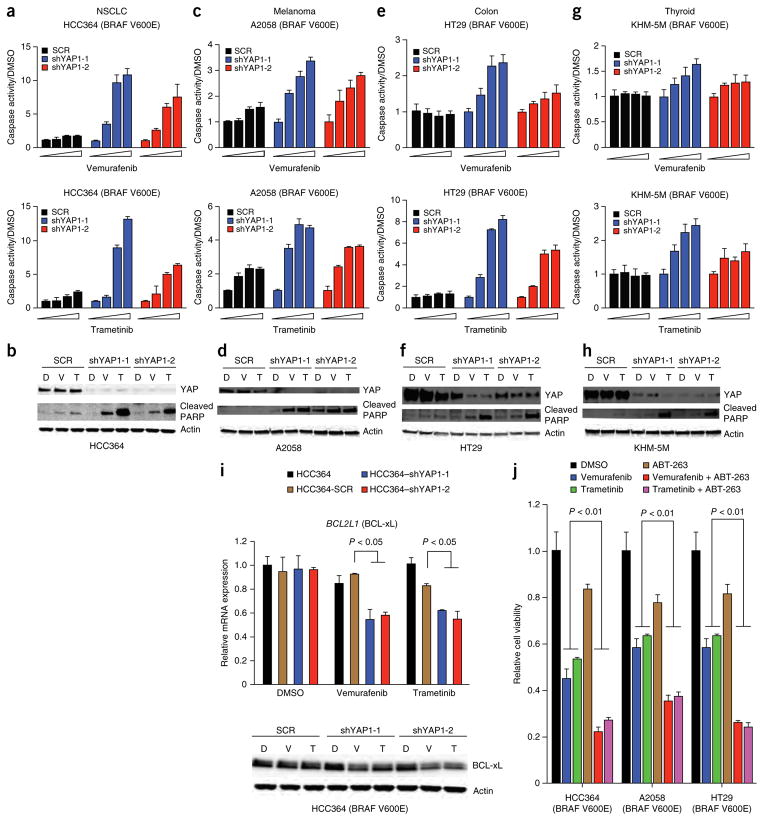
We next investigated the mechanism through which YAP regulates the response to RAF- and MEK-targeted therapy. As YAP1 silencing profoundly impaired cell viability, specifically upon treatment with RAF or MEK inhibitor, and previous work indicates that YAP can regulate apoptosis22,24–26, we reasoned that suppression of YAP together with RAF-MEK signaling might be synthetically lethal. Indeed, we found that YAP1 knockdown promoted apoptosis, as measured by both the induction of caspase-3 and caspase-7 activity and PARP cleavage, upon treatment with either RAF or MEK inhibitor that alone was insufficient to induce apoptosis in NSCLC (Fig. 3a,b), melanoma (Fig. 3c,d), colon cancer (Fig. 3e,f and Supplementary Fig. 11) and thyroid cancer (Fig. 3g,h) models harboring BRAF V600E. YAP1 silencing also enhanced apoptosis upon treatment with MEK inhibitor, which by itself did not induce cell death in RAS-mutant NSCLC models, albeit more modestly than in many BRAF-mutant models (Supplementary Fig. 11). These findings show the synthetic lethality of combined suppression of YAP and RAF-MEK signaling.
Figure 3.
Synthetic lethality and synergistic induction of apoptosis with concurrent inhibition of YAP and oncogenic MAPK signaling. (a–h) Effects of YAP1 knockdown on apoptosis induced by treatment with vemurafenib (top) or trametinib (bottom) in HCC364 BRAF-mutant lung cancer cells as measured by caspase-3 and caspase-7 activation (a) and the levels of cleaved PARP by immunoblot analysis (b); in A2058 BRAF-mutant melanoma cells as measured by caspase-3 and caspase-7 activation (c) and the levels of cleaved PARP by immunoblot analysis (d); in HT29 BRAF-mutant colon cancer cells as measured by caspase-3 and caspase-7 activation (e) and the levels of cleaved PARP by immunoblot analysis (f); and in KHM-5M BRAF-mutant thyroid cancer cells as measured by caspase-3 and caspase-7 activation (g) and the levels of cleaved PARP by immunoblot analysis (h). Data are shown as means ± s.e.m.; n = 3 biological replicates. Caspase activation was normalized to levels in cells treated with DMSO vehicle. D, DMOS; V, vemurafenib; T, trametinib. (i) Effects of YAP1 suppression and treatment with vemurafenib or trametinib on BCL-xL levels in HCC364 cells. Levels of BCL-xL were measured by quantitative RT-PCR (top) and immunoblot analysis (bottom). Data are shown as means ± s.e.m.; n = 3 biological replicates. Comparisons were to levels in cells expressing scrambled shRNA and treated with DMSO. (j) Effects of pharmacological inhibition of BCL-xL using ABT-263 on sensitivity to vemurafenib or trametinib in HCC364 BRAF-mutant lung cancer cells, A2058 BRAF-mutant melanoma cells and HT29 BRAF-mutant colon cancer cells. Data are shown as means ± s.e.m.; n = 3 biological replicates. Comparisons were to levels in corresponding cells treated with DMSO. P values are indicated for statistical analysis.
We reasoned that YAP might enhance the expression of an antiapoptotic factor to promote survival and resistance to RAF and MEK inhibitors. YAP can transcriptionally upregulate the expression of specific antiapoptotic components, including the BCL2 family member protein BCL-xL (encoded by BCL2L1) in some cell types27. Therefore, we hypothesized that YAP might control the threshold for apoptosis induction during RAF- and MEK-targeted therapy by promoting BCL-xL expression as a parallel survival input in tumor cells with oncogenic BRAF. Indeed, YAP1 suppression resulted in decreased expression of BCL-xL, specifically in the context of treatment with either vemurafenib or trametinib in HCC364 NSCLC, A2058 melanoma, HT29 colon cancer and KHM-5M thyroid cancer cells (Fig. 3i and Supplementary Fig. 12). These data show that YAP and RAF-MEK signaling function in parallel to regulate BCL-xL levels, which might ensure that the threshold for the induction of apoptosis is achieved only with inhibition of both YAP and RAF-MEK signaling.
We confirmed the relevance of BCL-xL function downstream of YAP, establishing that BCL-xL overexpression rescued the effect of YAP1 silencing on the response to RAF and MEK inhibitors in HCC364 cells (Supplementary Fig. 13). These data indicate that BCL-xL is a critical effector by which YAP promotes resistance to RAF or MEK inhibition. Consistent with these observations and those of others28, pharmacological BCL-xL inhibition using ABT-263 (navitoclax) enhanced the efficacy of treatment with either RAF or MEK inhibitor in several tumor cell lines harboring BRAF V600E, including NSCLC (HCC364), melanoma (A2058) and colon cancer (HT29) models (Fig. 3j). Treatment with BCL-xL inhibitor also enhanced sensitivity to trametinib in RAS-mutant NSCLC cells (Supplementary Fig. 11). These effects of ABT-263 treatment were phenocopied by treatment with another BCL-xL inhibitor, TW37 (Supplementary Fig. 11). These data indicate that YAP acts as a parallel survival input via BCL-xL to promote resistance to RAF and MEK inhibitors, extending recent findings linking BCL-xL with the response to MEK inhibitor in some KRAS-mutant tumors29,30.
To further explore the synthetic lethal relationship between YAP and RAF or MEK inhibition, we conducted unbiased transcriptional profiling in HCC364 cells harboring BRAF V600E in which YAP and MEK were suppressed individually or concurrently. These profiling data showed few significant changes in gene expression with silencing of YAP1 alone (adjusted P value < 0.05 and at least 40% decreased or at least 66% increased expression, compared to the control) (Supplementary Table 5), consistent with our functional studies indicating that YAP1 silencing has a weaker impact on cell viability (Fig. 1 and Supplementary Fig. 1). Treatment with trametinib led to differential expression of more genes, consistent with the important role of BRAF-MEK signaling in these cells (Supplementary Table 5). Silencing YAP1 together with trametinib treatment similarly led to differential expression of a substantial number of genes, with evidence of coregulation of some genes, including BCL2L1 (Supplementary Table 5). Pathway analysis of the genes that were significantly altered specifically by combined inhibition of YAP and MEK (such as BCL2L1) indicated an enrichment of genes involved in apoptosis (Supplementary Fig. 14). These findings offer additional insight into the role of BCL-xL and the synthetic lethality of simultaneous suppression of YAP and RAF-MEK signaling.
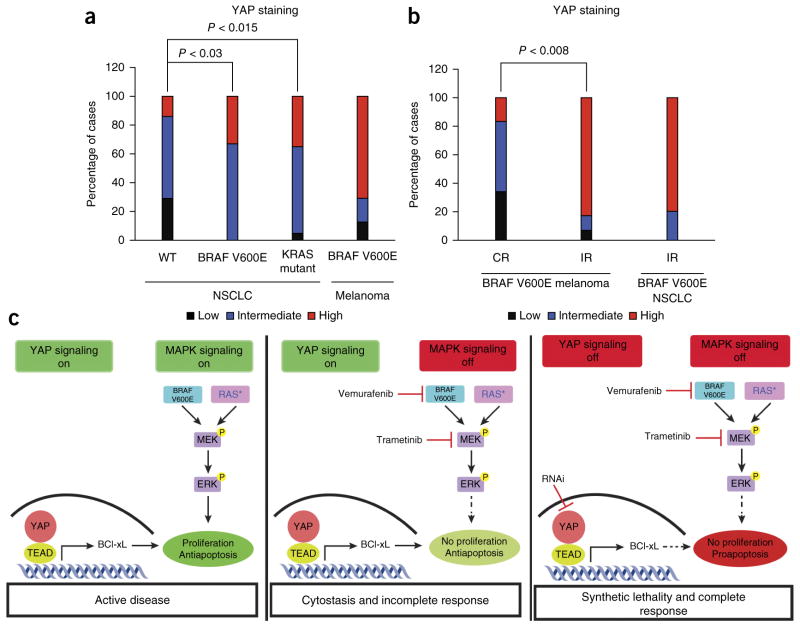
We next sought to determine the clinical relevance of our findings. We hypothesized that YAP might be upregulated in some BRAF- or RAS-mutant human tumors and that this upregulation might promote resistance to RAF and MEK inhibitors in patients. We therefore measured YAP levels in primary human tumor specimens obtained from patients with NSCLC (n = 13) or melanoma (n = 35) encoding BRAF V600E, from patients with KRAS-mutant NSCLC (n = 23) or from patients with NSCLC with wild-type BRAF and KRAS (n = 14). We observed high levels of YAP in the majority of the tumors encoding BRAF V600E (NSCLC and melanoma) and the KRAS-mutant NSCLC tumors and lower levels of YAP in tumors with wild-type BRAF and KRAS (NSCLC), as measured by immunohistochemistry using an antibody to YAP (Fig. 4a and Supplementary Fig. 15). These data show that YAP is upregulated in some BRAF- and RAS-mutant human tumors. As MEK inhibitor monotherapy is largely ineffective in patients with KRAS-mutant tumors17, these findings suggest that increased YAP levels in these tumors might contribute to primary resistance to MEK inhibition.
Figure 4.
Increased YAP levels in human tumor specimens encoding BRAF V600E is a biomarker of worse response to RAF inhibitor in patients. (a) Quantification of the levels of YAP in human NSCLC (n = 13) and melanoma (n = 29) specimens encoding BRAF V600E, KRAS-mutant NSCLC specimens (n = 23) and human tumor specimens with wild-type BRAF and KRAS (WT; NSCLC; n = 14), as measured by immunohistochemistry with a validated antibody to YAP and scoring of the staining as low, intermediate or high. (b) Quantification of YAP levels in melanoma or NSCLC specimens encoding BRAF V600E from patients with either a confirmed complete response (melanoma CR, n = 6) or an incomplete response (melanoma IR, n = 29; NSCLC IR, n = 5; incomplete response includes either a partial response or stable disease by RECIST criteria) to initial treatment with a BRAF inhibitor (vemurafenib, dabrafenib or LGX818; n = 16) or to combined RAF and MEK inhibitor treatment (dabrafenib and trametinib or LGX818 and MEK162; n = 19) (P = 0.008 distinguishing the proportion of complete response from incomplete response among melanomas with high YAP levels). YAP immunohistochemistry analysis was conducted as in a. (c) YAP functions via BCL-xL as a parallel input to suppress apoptosis and promote survival, protecting BRAF- and RAS-mutant tumor cells from death (left). RAF- and MEK-targeted therapy is therefore ineffective or cytostatic, resulting in an incomplete tumor response (middle). YAP suppression lowers the threshold for the induction of apoptosis upon RAF or MEK inhibition, promoting a complete response (right). P, phosphorylation; RAS*, mutant RAS.
We next assessed whether YAP expression was inversely correlated with the initial response to RAF and MEK inhibition in patients with BRAF V600E (n = 35). We measured YAP levels by immunohistochemistry in melanoma specimens harboring BRAF V600E from patients (n = 35) with either a confirmed complete response or an incomplete response (including either a partial response or stable disease by RECIST (Response Evaluation Criteria in Solid Tumors) criteria) to monotherapy with a BRAF inhibitor (vemurafenib, dabrafenib or LGX818; n = 16) or to combined RAF and MEK inhibitor treatment (dabrafenib and trametinib or LGX818 and MEK162; n = 19). Patients with melanoma harboring BRAF V600E who had a complete response to therapy exhibited lower YAP expression in the pretreatment tumor samples (Fig. 4b and Supplementary Table 6). Conversely, patients who had an incomplete response to therapy had higher YAP expression in the baseline tumor samples (Fig. 4b and Supplementary Table 6). We also analyzed YAP expression in NSCLC tumors with BRAF V600E obtained from patients (n = 5) before investigational treatment with dabrafenib, which resulted in an incomplete response in each patient. All but one of these pretreatment NSCLC specimens exhibited high YAP expression (Fig. 4b and Supplementary Table 6). Taken together, these data suggest that increased YAP levels are a biomarker of decreased response to RAF or MEK inhibitor in patients with BRAF-mutant tumors.
We also assessed YAP levels in melanomas obtained from patients at the time of progression on RAF or MEK inhibitor therapy after an initial response to test the hypothesis that YAP might contribute to acquired resistance. We examined YAP expression by immunohistochemistry in an additional 32 paired melanoma specimens with BRAF V600E obtained from 16 patients both before RAF or MEK inhibitor treatment and upon the development of acquired resistance. As the majority of the pretreatment melanoma specimens encoding BRAF V600E from patients harbored high baseline YAP levels (Supplementary Table 7), we examined whether YAP levels were higher at the time of progression in the subset of patients whose pretreatment tumors had low or intermediate YAP levels (25%, 4/16 cases; Supplementary Fig. 15 and Supplementary Table 7). We found increased YAP levels in each paired tumor obtained at acquired resistance in comparison to the matched pretreatment specimen (Supplementary Fig. 15 and Supplementary Table 7). These findings further suggest that increased YAP levels might limit the clinical efficacy of RAF and MEK inhibitors.
All together, our findings unveil the synthetic lethality of concurrent inhibition of YAP and RAF-MEK signaling (Fig. 4c) and augment findings indicating an emerging role for YAP in tumorigenesis across several different tumor types22–24,31,32. The data show unanticipated functional cross-talk between YAP and RAF-MEK signaling. Our findings uncover a new, promising polytherapeutic strategy to enhance the efficacy of RAF and MEK inhibitors and survival in patients with a broad range of BRAF- and RAS-mutant tumors.
ONLINE METHODS
Cell lines and culture reagents
The HCC364 cell line was kindly provided by D. Solit (Memorial Sloan Kettering Cancer Center). MM415 and SKMEL-2 cells were kindly provided by B. Bastian (University of California, San Francisco). WM793, HPAF-II and PANC 02.03 cells were kindly provided by E. Collisson (University of California, San Francisco). CAL-12T, A2058, HT29, WiDr, A549, Calu-1, H23, SW1573 and H2347 cells were purchased from the American Type Culture Collection (ATCC). HTC-C3 cells were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DMSZ), Germany. KHM-5M cells were purchased from the Japanese Collection of Research Bioresources (JCRB). MOR/CPR cells were purchased from Sigma. Cells were maintained at 37 °C in a humidified atmosphere at 5% CO2, grown in RPMI 1640 supplemented with 10% FBS, 100 IU/ml penicillin and 100 μg/ml streptomycin. All cell lines used tested negative for mycoplasma.
Compounds
Vemurafenib, trametinib, TW37 and ABT-263 were purchased from Sellekchem. PLX4720 was kindly provided by Plexxikon.
shRNA screen with the DECIPHER shRNA library
Lentiviral plasmids encoding shRNAs including the Cellecta DECIPHER shRNA library human module 1 are described online at the company’s website. shRNA lentiviruses were generated from HEK293FT cells according to the manufacturer’s protocol (Cellecta). HCC364 cells were infected with lentiviral supernatant containing shRNAs with a multiplicity of infection (MOI) of 0.3. After 48 h, the infection rate was measured by flow cytometry, and cells were replated with medium containing 1 μg/ml puromycin. Seventy-two hours after the addition of puromycin, 27 million cells were frozen for further analysis, and 27 million cells were replated in the presence and absence of 1 μM or 3 μM vemurafenib; the medium was refreshed every 3 d for 10 d. Genomic DNA was isolated, and shRNA inserts were retrieved according to the manufacturer’s protocol (Cellecta). Indexes and adaptors for deep sequencing were incorporated into PCR primers. Sample quantification was performed on a Bioanalyzer (Agilent Technologies) to ensure that samples were pooled at the same quantity. Deep sequencing was performed using the Illumina HiSeq 2500 platform at the Center for Advanced Technology of the University of California, San Francisco. shRNA barcodes were segregated and deconvoluted from each sequencing read. P values for each gene were calculated as follows. For each gene G with k barcodes, each with shRNA count c in the control condition and d in the experimental condition, a test statistic r(G) was computed as the second lowest ranked value of
Subsequently, a P value was computed on the basis of r(G) as
where
and s(k) is the second lowest ranked value of k randomly chosen values t from all barcodes in all genes and N is the number of permutation trials performed. For this sample, we set N at 10,000. Individual shRNAs used for the validation experiments were purchased from Sigma.
Cell viability assays
Cells (3,000–5,000) were plated per well in 96-well plates 24 h before drug treatment. The number of viable cells was determined 72 h after the initiation of drug treatment using CellTiter-Glo luminescent cell viability reagent according to the manufacturer’s protocol (Promega). Each assay consisted of four replicate wells and was repeated at least three times. Data are expressed as percentage of the cell viability of control cells. Data were graphically displayed using GraphPad Prism version 5.0 for Mac (GraphPad Software). IC50 values were calculated as 50% of growth inhibition as measured by the cell viability assays. Maximal growth inhibition was calculated as the maximum percentage of growth inhibition achieved upon trametinib treatment from 0.1 nM to 1,000 nM, as measured by the cell viability assays.
Apoptosis assays
Cells (7,500–10,000) were plated per well in 96-well white plates 24 h before drug treatment. The proportion of apoptotic cells was determined 24 h after the initiation of drug treatment using Caspase3/7-Glo luminescent assay reagent according to the manufacturer’s protocols (Promega). Each assay consisted of four replicate wells and was repeated at least three times. Data are expressed as percentage of the cell viability of control cells. Data were graphically displayed using GraphPad Prism version 5.0 for Mac (GraphPad Software).
Quantitative PCR
Total RNA was collected from cultured cells using the RNeasy kit (Qiagen). cDNA was synthesized with SuperScript III reverse transcriptase using random hexamer primers (Invitrogen), and RT-PCR was performed on a QuantStudio instrument with TaqMan probes (Life Technologies), using the following program: holding at 50 °C for 2 min and polymerase activation at 95 °C for 20 s, followed by 40 cycles of amplification (95 °C for 1 s, 60 °C for 20 s). TBP expression was used as an internal reference to normalize input cDNA. Ratios of the expression level of each gene to that of the reference gene were then calculated.
Protein blot analysis
Cells (200,000) were seeded per well in 6-well plates 24 h before drug treatment, and whole-cell lysates were prepared using RIPA buffer (10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.1% sodium deoxycholate, 0.1% SDS, 140 mM NaCl) supplemented with protease inhibitor and phosphatase inhibitor (Roche) and clarified by sonication and centrifugation. Nuclear and cytosol fractionation was performed using NE-PER nuclear and cytoplasmic extraction reagents (Thermo). Equal amounts of protein were separated by 4–15% SDS-PAGE and were transferred onto nitrocellulose membranes (Bio-Rad) for protein blot analysis. Membranes were incubated with primary antibody overnight, washed and incubated with secondary antibody. Membranes were exposed using either a fluorescence system (Li-Cor) or a chemiluminescent reagent; images were captured, and bands were quantified using an ImageQuant LAS 4000 instrument (GE Healthcare).
Antibodies
Antibodies to YAP/TAZ (8418), Parp (9542) and Bcl-xL (2764) were purchased from Cell Signaling Technology. Antibodies to YAP (sc-101199) and GAPDH (sc-59540) for protein blotting were purchased from Santa Cruz Biotechnology. The antibody to YAP (sc-15407) for immunohistochemistry was purchased from Santa Cruz Biotechnology. The antibody to β-actin (A2228) was purchased from Sigma-Aldrich.
Tumor xenograft study
Each indicated cell line (A2058, HT29 and MOR/CPR) was infected with lentiviruses encoding either scrambled control shRNA or YAP1 shRNA was injected subcutaneously into the left and right posterior flanks of 7-week-old immunodeficient NOD-SCID female mice (Charles River). Tumor formation was measured twice a week, and tumor volumes were calculated by length × width × height. When the tumor reached approximately 200–300 mm3, the mice were randomized for drug treatments (PLX4720, 50 mg/kg; trametinib, 1 mg/kg). The fold change in tumor volume was normalized to the tumor volume when treatments were initiated for each tumor. The duration of treatment was 14 d for A2058 and MOR/CPR and 21 d for HT29 tumor xenografts. All mouse experiments were performed in accordance with protocols approved by the University of California at San Francisco Institutional Animal Care and Use Committee. Sample size was not predetermined.
Immunohistochemical analyses of human tumor specimens
All specimens were acquired from individuals with NSCLC and melanoma under the auspices of institutional review board (IRB)-approved clinical protocols at each hospital in which informed consent was obtained. BRAF and KRAS mutation status was assessed by established clinical DNA sequencing assays. Immunohistochemistry for YAP (H-125, Santa Cruz Biotechnology) was conducted on formalin-fixed, paraffin-embedded tumor sections as previously described33,34. Statistical significance was assessed and is reported as the P value from the χ2 test, with P < 0.05 considered significant.
RNA deep sequencing and analysis
RNA from each of the indicated cell lines was extracted by RNeasy kit (Qiagen). In total, 2 μg of total RNA was used for deep sequencing library preparation using Illumina TruSeq sample prep kits according to the manufacturer’s protocols. Sequencing libraries with different indices were pooled and sequenced in paired-end format to a length of 100 bp using the HiSeq 2500 platform at the Center for Advanced Technology at the University of California, San Francisco. Reads were aligned against NCBI Build 37 (hg19) of the human genome using NCBI Ensembl transcript annotation (version 75) with RSEM35, which also yielded gene-level quantification of expression.
Differential expression and pathway analysis
Differential gene expression analysis was performed with DESeq36 among three sets of conditions: (i) YAP1 knockdown using two independent validated shRNAs, (ii) shSCR (scrambled shRNA control) and trametinib (100 nM), and (iii) YAP1 knockdown using two independent shRNAs and trametinib (100 nM), each compared to the control condition (shSCR and DMSO). Pathway analysis (gene set enrichment) was performed on genes significantly differentially expressed (q < 0.05) that were exclusive to the YAP1 knockdown and trametinib treatment condition, using MSigDB37.
Supplementary Material
Acknowledgments
We thank G. Bollag and P. Lin (Plexxikon) for providing PLX4720. We thank J. Weissman and members of the Bivona laboratory for critical review of the manuscript. The authors acknowledge funding support (to T.G.B.) from the following sources: US National Institutes of Health (NIH) Director’s New Innovator Award (DP2CA174497), the Howard Hughes Medical Institute (Collaborative Innovation Award), the Doris Duke Charitable Foundation, the American Lung Association, the National Lung Cancer Partnership, the Addario and Van Auken Private Foundations, the Sidney Kimmel Foundation for Cancer Research and the Searle Scholars Program. The authors also acknowledge funding from La Caixa Foundation (to R.R.) and from US NIH grant R01CA131261 (to M.M.).
Footnotes
Accession codes. Transcriptome analysis (RNA sequencing) of HCC364 cells in the context of YAP and MEK suppression, individually or concurrently, GSE64550.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
L.L., E.C., A.J.S., V.O., D.N., J.J.Y., E.P., X.L., M.M.W., L.P. and E.A.C. conducted all experiments and/or analyzed data. L.C. and S.A. conducted the analysis of the shRNA screening data. M.M. provided cell lines and analyzed data. N.K., M.G.C., J.L.M., J.L.R., J.M.S.T., F.B., C.M.R., A.A., E.R., I.O., E.C., E.M.-C., J.C., A.M., R.R., D.T.F., Z.A.C., K.T.F. and J.A.W. provided clinical samples and data. All authors contributed to the design of experiments and to data analysis and interpretation. T.G.B., L.L. and A.J.S. wrote the manuscript with input from all coauthors.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Flaherty KT, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flaherty KT, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367:107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 4.Sosman JA, et al. Survival in BRAF V600–mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adjei AA, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol. 2008;26:2139–2146. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jänne PA, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 2013;14:38–47. doi: 10.1016/S1470-2045(12)70489-8. [DOI] [PubMed] [Google Scholar]
- 7.Migliardi G, et al. Inhibition of MEK and PI3K/mTOR suppresses tumor growth but does not cause tumor regression in patient-derived xenografts of RAS-mutant colorectal carcinomas. Clin Cancer Res. 2012;18:2515–2525. doi: 10.1158/1078-0432.CCR-11-2683. [DOI] [PubMed] [Google Scholar]
- 8.Fordyce SL, et al. Genetic diversity among pandemic 2009 influenza viruses isolated from a transmission chain. Virol J. 2013;10:116. doi: 10.1186/1743-422X-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roth AD, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 10.Planchard D, et al. Interim results of phase II study BRF113928 of dabrafenib in BRAF V600E mutation–positive non–small cell lung cancer (NSCLC) patients. J Clin Oncol. 2013;31(suppl) abstract 8009. [Google Scholar]
- 11.Lin L, et al. Mapping the molecular determinants of BRAF oncogene dependence in human lung cancer. Proc Natl Acad Sci USA. 2014;111:E748–E757. doi: 10.1073/pnas.1320956111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prahallad A, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 13.Corcoran RB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montero-Conde C, et al. Relief of feedback inhibition of HER3 transcription by RAF and MEK inhibitors attenuates their antitumor effects in BRAF-mutant thyroid carcinomas. Cancer Discov. 2013;3:520–533. doi: 10.1158/2159-8290.CD-12-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudin CM, Hong K, Streit M. Molecular characterization of acquired resistance to the BRAF inhibitor dabrafenib in a patient with BRAF-mutant non-small-cell lung cancer. J Thorac Oncol. 2013;8:e41–e42. doi: 10.1097/JTO.0b013e31828bb1b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 17.Young A, et al. Ras signaling and therapies. Adv Cancer Res. 2009;102:1–17. doi: 10.1016/S0065-230X(09)02001-6. [DOI] [PubMed] [Google Scholar]
- 18.Bivona TG, et al. FAS and NF-κB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471:523–526. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong W, Guan KL. The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev Biol. 2012;23:785–793. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapoor A, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158:185–197. doi: 10.1016/j.cell.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, et al. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci Signal. 2014;7:ra42. doi: 10.1126/scisignal.2005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shao DD, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Vigneron AM, Ludwig RL, Vousden KH. Cytoplasmic ASPP1 inhibits apoptosis through the control of YAP. Genes Dev. 2010;24:2430–2439. doi: 10.1101/gad.1954310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenbluh J, et al. β-catenin–driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell. 2012;151:1457–1473. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frederick DT, et al. Clinical profiling of BCL-2 family members in the setting of BRAF inhibition offers a rationale for targeting de novo resistance using BH3 mimetics. PLoS ONE. 2014;9:e101286. doi: 10.1371/journal.pone.0101286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corcoran RB, et al. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell. 2013;23:121–128. doi: 10.1016/j.ccr.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan N, et al. Bcl-2/Bcl-xL inhibition increases the efficacy of MEK inhibition alone and in combination with PI3 kinase inhibition in lung and pancreatic tumor models. Mol Cancer Ther. 2013;12:853–864. doi: 10.1158/1535-7163.MCT-12-0949. [DOI] [PubMed] [Google Scholar]
- 31.Feng X, et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated Rho GTPase signaling circuitry. Cancer Cell. 2014;25:831–845. doi: 10.1016/j.ccr.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu FX, et al. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell. 2014;25:822–830. doi: 10.1016/j.ccr.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall CA, et al. Hippo pathway effector Yap is an ovarian cancer oncogene. Cancer Res. 2010;70:8517–8525. doi: 10.1158/0008-5472.CAN-10-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.