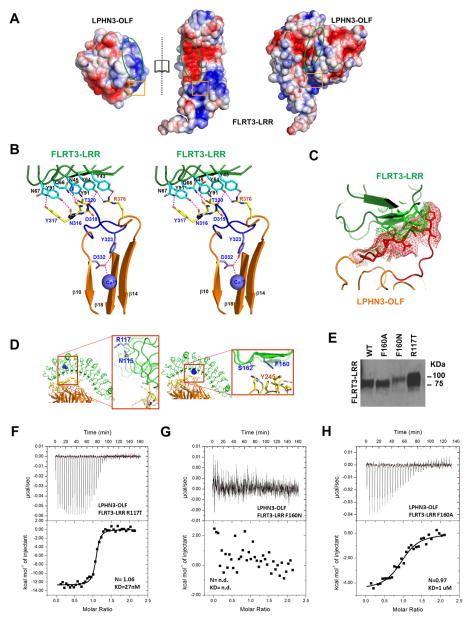
SUMMARY
Latrophilins (LPHNs) are adhesion-like G protein coupled receptors implicated in attention-deficit/hyperactivity disorder (ADHD). Recently, LPHN3 was found to regulate excitatory synapse number through trans interactions with fibronectin leucine-rich repeat transmembrane 3 (FLRT3). By isothermal titration calorimetry, we determined that only the olfactomedin (OLF) domain of LPHN3 is necessary for FLRT3 association. By multi-crystal native-SAD phasing, we determined the crystal structure of the OLF domain. This structure is a five-bladed β-propeller with a Ca2+ ion bound in the central pore, which is capped by a mobile loop that allows the ion to exchange with the solvent. The crystal structure of the OLF/FLRT3 complex shows that LPHN3-OLF in the closed state binds with high affinity to the concave face of FLRT3-LRR with a combination of hydrophobic and charged residues. Our study provides structural and functional insights into the molecular mechanism underlying the LPHN3/FLRT3 contribution to the development of glutamatergic synapses.
Graphical Abstract
INTRODUCTION
Latrophilins (LPHNs) are adhesion G protein-coupled receptors (adhesion GPCRs), the second largest class of GPCRs (Sudhof, 2001). LPHNs consists of three isoforms, LPHN1-3, expressed largely in the CNS (Ichtchenko et al., 1999). Common variants of the LPHN3 gene confer susceptibility to attention-deficit hyperactivity disorder (ADHD) (Arcos-Burgos et al., 2010), predict effectiveness of stimulant medication (Labbe et al., 2012) and impact behavioral and neurophysiological measures of cognitive response control (Fallgatter et al., 2013). A single point mutation in human LPHN3 was found in an ADHD patient (Domene et al., 2011). The mutation is in the olfactomedin domain (Ala247Ser, corresponding to mouse Ala313Ser) but the biological effects of this mutation are currently unknown.
Structurally, LPHNs are characterized by a seven-transmembrane region and a large N-terminal extracellular sequence containing the following domains: a rhamnose binding lectin (RBL) domain; an olfactomedin-like (OLF) domain followed by a Serine/Threonine rich domain that is O-linked glycosylated (O’Sullivan et al., 2014); a hormone binding (HR) domain; a GPCR Autoproteolysis INducing (GAIN) domain (Figure 1A). We recently reported that LPHN3 binds in vivo and in vitro to fibronectin leucine-rich repeat transmembrane 3 (FLRT3) to constitute a synaptic ligand-receptor pair that can modulate excitatory synapse number (O’Sullivan et al., 2012). There are three FLRT genes, FLRT1-3, encoding single-pass transmembrane proteins with one extracellular leucine-rich repeat (LRR) domain and a juxtamembrane fibronectin type 3 (FNIII) module (Lacy et al., 1999). Synaptic puncta density and cell based binding experiments using deletion of the OLF or the OLF-RBL domains prevented rescue of the synaptic phenotype, and suggested that whereas the OLF domain is required for interaction with FLRTs, both the RBL and OLF domains are involved in the interaction with teneurin 1 (TEN1), a cell surface protein involved in brain development. These data indicate that the extracellular domain of LPHN3 contains a distal ligand binding module situated on an elongated and potentially flexible stalk long enough to span the synaptic cleft, with differentiable FLRT3 and TEN1 binding sites (O’Sullivan et al., 2014).
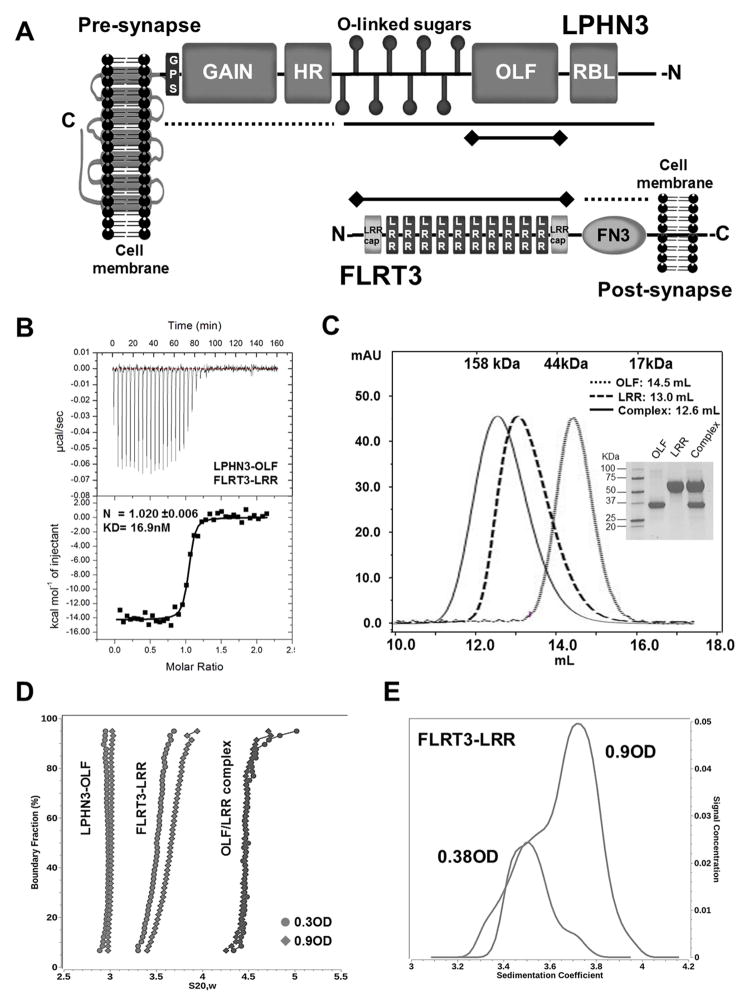
Figure 1. Characterization of LPHN3-OLF and FLRT3-LRR in solution.
(A), Schematic representation of LPHN3 and FLRT3 domain architecture. N- and C- designate N- and C-termini of the proteins. Dotted lines indicate constructs that did not bind, solid lines indicate constructs that bound and solid lines with terminal diamonds indicate constructs used in B. (B), ITC of LPHN3-OLF injected at 100μM into the cell containing the FLRT3-LRR domain at 10μM. (C), SEC of the purified LPHN3-OLF, FLRT3-LRR and their complex using a Superdex 200-10/300GL Column. Top margin shows the column calibration: 1, γ-globulin, 158KDa; 2, Ovalbumin, 44KDa; 3, myoglobin, 17KDa. OLF eluted with an apparent MW of ~30KDa; LRR at ~ 71KDa; the complex at ~90KDa as expected for a 1:1 stoichiometry. Inset, Coomassie blue staining SDS-PAGE of the purified proteins. (D), Sedimentation velocity of LPHN3-OLF, FLRT3-LRR and their complex, at two concentrations. Overlay of the diffusion corrected integral sedimentation coefficient distributions from the van Holde - Weischet analysis shows a single species with homogeneous distribution for LPHN3-OLF and the complex but significant self-association for FLRT-LRR. The frictional ratio of the monomer suggested a slightly non-globular shape and was determined to be 1.53 (1.48 and 1.58 with 95% confidence intervals). The dimer frictional ratio was found to be more elongated at 1.76 (1.74 and 1.78 with 95% confidence intervals). (E), The van Holde - Weischet analysis of the FLRT3-LRR concentrations reveal two distinct but overlapping species indicating a clear monomer-dimer equilibrium.
Olfactomedins (OLF) were first discovered in the frog olfactory neuroepithelium (Snyder et al., 1991). The OLF domain has a length of about 250 amino acids and it is present in at least 13 proteins in mammals (Tomarev and Nakaya, 2009). A growing body of evidence indicates that these proteins play important roles in normal development and disease in mammals, and mutations in some of these genes lead to profound pathologies in humans (Anholt, 2014). The most extensively studied OLF-containing protein is probably myocilin in which mutations are found in more than 10% of juvenile open-angle glaucoma cases and in 3–4% of patients with adult onset primary open-angle glaucoma (POAG) (Fingert et al., 1999; Stone et al., 1997). The available crystal structures of gliomedin and myocilin, two members of the OLF family reveal a 5-blade β-propeller fold and the presence of Ca2+ and/or Na+ in the central pore (Donegan et al., 2014; Han and Kursula, 2015). Recent findings on the extracellular domain of FLRT3 reveal that this protein has the tendency to homo-dimerize but a precise characterization of dimerization potential is currently unknown. Moreover, the study of FLRT2-LRR, alone and in complex with the cell surface receptor uncoordinated-5 D (UNC5D) revealed that FLRT2-UNC5D interaction controls neuronal radial migration whereas FLRT-FLRT interaction modulates spatial organization in the tangential axis (Seiradake et al., 2014). Subsequently, the same group solved the crystal structure of mouse LPHN3 lectin and olfactomedin-like domains, confirming that the OLF domain is a 5-bladed β-propeller containing a Ca2+ and a Na+ binding sites (Jackson et al., 2015).
Here we present the solution characterization of mouse LPHN3-OLF, FLRT3-LRR, the crystal structure of LPHN3-OLF alone in two different conformations, its complex with the LRR domain of FLRT3 and a characterization of the association including the effect of the Ser247Ala human mutation. Our structural analysis suggests that the high affinity interaction between LPHN3-OLF and FLRT3 is necessary for some aspect of the development of glutamatergic synapses.
RESULTS
Isothermal titration calorimetry (ITC) characterization of LPHN3 and FLRT3 Interactions
To determine which domain of FLRT3 and LPHN3 is directly involved in the association, we engineered different constructs for each protein (Figure 1A). The extracellular domain of LPHN3 was divided into three portions: the N-terminal half including the RBL, OLF, and the O-linked carbohydrate domains, the OLF domain alone, and the C-terminal half containing the HR-GAIN-GPS domains. The extracellular domain of FLRT3 was also split into two constructs, one containing the FN3 domain alone (FLRT3-FN3) and the other containing the ten LRR repeats between amino acids 29–386 (FLRT3-LRR). ITC experiments were performed to assess which construct retained the affinity of the full extracellular domain. Our data show that binding occurred between FLRT3-LRR and LPHN3-OLF (LPHN3 amino acids 199–495) (Figure 1B and Table 1) with an affinity similar to the one previously reported for the full length extracellular domains (O’Sullivan et al., 2012). Because no association was recorded using either the LPHN3-HR-GAINGPS construct or FLRT3-FN3 domains, we conclude that LPHN3-OLF and FLRT3-LRR represent the complete binding cassette of these proteins. ITC experiments in the presence of 1 mM EDTA or 2 mM Ca2+ showed that the binding affinity remains unchanged (data not shown), indicating that the interaction doesn’t require Ca2+ at the interface, as is the case of neuroligin/neurexin interaction (Fabrichny et al., 2007). The interaction data were interpreted by using the single site model fitting indicating that, in our conditions, the tendency of FLRT3-LRR to dimerize (see below) was abolished by the LPHN3-OLF/FLRT3-LRR association.
Table 1.
ITC binding experiments using various LPHN3 and FLRT3 constructs. For each pair, at least three experiments were recorded.
| Deletion constructs of LPHN3 and FLRT3 | ||||
|---|---|---|---|---|
| KD (nM) | LPHN3 | GAIN-HR | OLF-RBL | OLF |
| FLRT3 | 31±7.9 | No binding | 43.3±13.8 | n.d. |
| FN3 | No binding | n.d. | n.d. | n.d. |
| LRR | n.d. | n.d. | 34.2±7.5 | 16.9±15.9 |
| Amino-acid substitutions in LPHN3-OLF and FLRT3-LRR | ||||
|---|---|---|---|---|
| KD (nM) | LPHN3-OLF | LPHN3-OLF D332A | LPHN3-OLF Y323A | LPHN3-OLF A313S |
| FLRT3-LRR | 17 | 170 | No binding | 19 |
| FLRT3-LRR R117T | 27 | n.d. | n.d. | n.d. |
| FLRT3-LRR F160N | No binding | n.d. | n.d. | n.d. |
| FLRT3-LRR F160A | 1000 | n.d. | n.d. | n.d. |
Solution Characterization of the Protein Samples
Size exclusion chromatography (SEC)
The purified LPHN3-OLF domain, FLRT3-LRR and their complex were analyzed by SEC to ensure sample monodispersity for subsequent biophysical experiments and to help interpretation of the crystallography packing data. LPHN3-OLF and the OLF-LRR complex eluted as single peaks, indicating ideal monodispersity and apparent MWs consistent with their monomeric forms (Figure 1C). However, the elution profile of LRR alone appears larger than expected consistent with the fact that FLRT3-LRR undergoes a concentration-dependent oligomerization due to a low affinity homophilic interaction of the LRR domain of FLRT3 (Seiradake et al., 2014).
Analytical ultracentrifugation (AUC)
To obtain more quantitative data in solution on the oligomerization state of our samples we performed sedimentation velocity experiments (SV) at two loading concentrations (~0.3 and ~0.9 OD280). Diffusion corrected van Holde–Weischet sedimentation coefficient distributions of LPHN3-OLF and the OLF-LRR complex indicated single homogeneous species that did not change sedimentation distribution as a function of loading concentration (4.1 μM and 13.5 μM for LPHN3-OLF, and 2.4 μM and 7.5 μM for the OLF-LRR complex) (Figure 1D). These data indicate that LPHN3-OLF is a monomer with an estimated molecular weight of 37,195Da and 37,340Da for the low and high concentrations respectively, in line with the 36.6 kDa value calculated from the sequence of our expressed protein. The OLF-LRR complex behaves a 1:1 assembly with an estimated molecular weight of ~95KDa, a value compatible to the mass of the glycosylated complex (~81.9Da). To characterize the apparent dimerization of FRLT3-LRR, samples were measured in the AUC at 2 loading concentrations 0.38 and 0.92 OD 280, corresponding to 7.6 and 18.3 μM, respectively. The van Holde – Weischet analysis shows a strong mass action effect with a marked shift to a higher sedimentation coefficient distribution for the higher concentration (Figure 1E). Subsequently, to calculate dissociation constant for the monomer-dimer equilibrium, both concentrations were fitted with finite element solutions of the Lamm equation that models the reversible interaction for a monomer-dimer system (Cao and Demeler, 2008; Demeler et al., 2010). The fit resulted in virtually identical KD values for both fits, averaging 43.3 μM (39.7 – 46.9 μM with 95% confidence intervals). Taken together, these data indicate that when LPHN3-OLF associates with FLRT3-LRR, its weak dimerization is converted in the formation of a 1:1 complex.
The LPHN3-OLF crystal structure
In our experimental conditions, LPHN3-OLF crystallized in a hexagonal (P65) or an orthorhombic (C2221) lattice. The structure was solved by native-SAD phasing using the recently described multi-crystal phasing procedure (Liu et al., 2012; Liu et al., 2014) with the P65 crystal form. The resulting initial model was then used to position the LPHN3-OLF in the C2221 lattice by molecular replacement and both structures were completed and refined using 1.6 Å high resolution datasets (see Methods, Table 2).
Table 2. Crystallographic data statistics.
Values in parentheses are for highest-resolution shell.
| LPHN3-OLF | Complex OLF/LRR | |||
|---|---|---|---|---|
|
| ||||
| Native-SAD phasing | P65-form | C2221-form | ||
|
| ||||
| Data collection | ||||
| Beamline | NSLS X4A | NSLS X4C | CHESS F1 | CHESS F1 |
| Energy (keV)/Wavelength (Å) | 6.0/2.07 | 12.7/0.97 | 13.8/0.91 | 13.8/0.91 |
| Space group | P65 | P65 | C2221 | P3121 |
| Unit cell parameters | ||||
| a, b, c (Å) | 84.1, 84.1, 66.1 | 85.1, 85.1, 66.1 | 78.8, 96.8, 79.2 | 121.9, 121.9, 84.0 |
| α,β,γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 | 90.0, 90.0,120.0 |
| # of crystals | 2 | 1 | 1 | 1 |
| Bragg spacings (Å) | 40–2.60 (2.67–2.60) | 40–1.61 (1.65–1.61) | 48.4–1.6 (1.63–1.60) | 49.3–3.2 (3.41–3.19) |
| Rp.i.m.§ | 0.009 (0.257) | 0.018 (0.290) | 0.027 (0.379) | 0.055 (0.474) |
| <I/σ(I)> | 77.2 (2.2) | 30.5 (3.1) | 18.4 (1.9) | 9.4 (1.5) |
| Completeness (%) | 95.4 (60.4) | 99.1 (96.6) | 98.8 (86.9) | 99.7 (99.0) |
| CC1/2 | - | 1.00 (0.76) | 1.00 (0.66) | 0.96 (0.91) |
| Multiplicity | 92.9 (10.0) | 22.6 (19.7) | 6.6 (3.5) | 1.9 (1.9) |
| Phasing | ||||
| Number of sites | 5 | - | - | - |
| FOM (before/after density modification) | 0.293/0.649 | - | - | - |
| Refinement | ||||
| Resolution (Å) | 2.6 | 1.6 | 1.6 | 3.2 |
| R/Rfree# (%) | 14.5/21.5 | 16.5/19.0 | 17.7/19.6 | 25.7/33.7 |
| B factors (Å2) | ||||
| Protein | 31.5 | 21.0 | 21.6 | 153.9 |
| Heteroatoms | 30.6 | 25.8 | 26.1 | 226.9 |
| r.m.s.d. | ||||
| Bond length (Å) | 0.009 | 0.007 | 0.007 | 0.009 |
| Bond angles (°) | 1.14 | 1.13 | 1.07 | 1.1 |
| Ramachandran plot | ||||
| Favored regions (%) | 94.1 | 94.8 | 98.9 | 84.5 |
| Outliers (%) | 0 | 0 | 0 | 1.2 |
, where Ihkl,j is the intensity of the jth observation for unique hkl and 〈Ihkl〉 is the mean intensity for unique hkl.
, where Fobs and Fcalc are the observed and calculated structure factors, respectively. Rfree is calculated using a subset (10%) of the data excluded from the refinement.
In both crystal forms, the crystallized LPHN3-OLF construct polypeptidic chain was built in their respective electron density maps except from its N-terminal (linker, FLAG tag and Lys199) and C-terminal (463–495 and the 3C Protease residual recognition motif) fragments, as well as the loop 392–405 which could not be entirely modeled due to the lack of reliable definition of the position by the electron density maps for several residues (i.e. fragments 395–403 and 397–403 absent in the C2221 and the P65 form, respectively) (Table S1, related to Figure 2). Consistently, those three fragments are shown to be among the highest solvent-exposed portions of the protein by DXMS experiment (see below). The peptide 395-YEDDDNEAT-403 is most likely highly solvated on the five negatively charged Asp and Glu residues clustered in this small nine-residue fragment on the surface of the protein. As for the N- and C-terminal fragments, they likely have undergone cleavage during the crystallization process, as we found that the form of the protein that provided suitable crystals was shorter than the one present in freshly purified preparations used for AUC and DXMS (see Methods). A disulfide bond formed by Cys203 and Cys385 was found to be partially disrupted, most likely due to radiation damage from the X-ray beam.
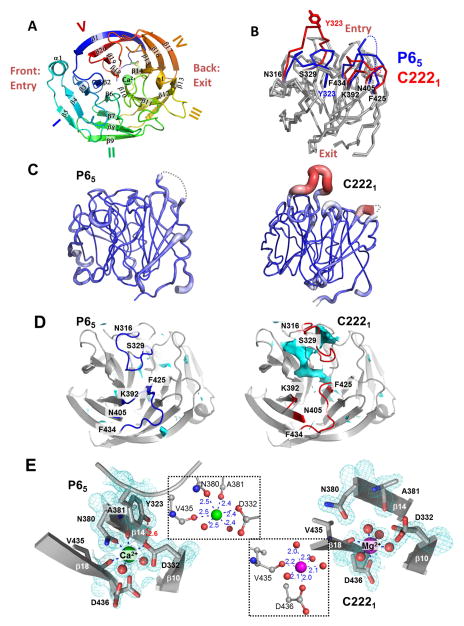
Figure 2. Crystal structure of LPHN3-OLF in P65 and C2221 crystal forms.
(A), Cartoon representation of the overall folding LPHN3-OLF, emphasizing its 5-bladed β-propeller folding. View from the “entry face” colored in rainbow mode from blue (N-terminus) to red (C-terminus). The Ca2+ ion, found in the central channel of the β-propeller, is represented in sphere mode. (B), Cα traces of the two superimposed LPHN3-OLF structures in P65 and C2221 crystal forms. The main structural differences are in the loops 316–329, 392–405, and 425–434 (red for the C2221 and blue for the P65 crystal form). The side chain of Tyr323 is represented to highlight a core stabilization of the loop 316–329 in the P65 form (“closed” conformation), versus the C2221 form (“open” conformation). (C), Comparison between the atomic thermal motions of the structure in both crystal forms along the backbone: from blue and thin for the lowest B-factor values to thick and red for the highest ones. The coloring/thickness of these cartoon representations is scaled in the range of 8.49 and 77.18 Å2, the minimal and maximal values of B-factors from both models. (D), Detailed views of solvent accessible surface of cavities and pockets (Cyan) in the P65 and C2221 crystal forms, highlighting an accessibility of the central pore in the C2221 form. (E), Metal ion binding in the central pore of the P65 and C2221 crystal forms and contoured with the 2mFo-DFc electron density map. Both panels summarize the protein residues and water molecules (red spheres) involved in the octahedric coordination of the observed Ca2+ (P65) or Mg2+ (C2221). The relevant inter-atomic distances (Å) are reported (insets). See also Figure S1).
Our LPHN3-OLF structure is very similar to several other OLF domain structures recently determined from LPHN3 (the mouse LPHN3 lectin and olfactomedin-like domains (LPHN3-RBL/OLF)) (Jackson et al., 2015), gliomedin (gliomedin-OLF) (Han and Kursula, 2015), myocilin (myocilin-OLF) (Donegan et al., 2014) and noelin (noelin-OLF) (Pronker et al., 2015) (Table S2, related to Figure 2). Consistently, our LPHN3-OLF structure displays a five-bladed β-propeller folding, i.e. twenty β-strands arranged in five four-stranded, highly twisted, anti-parallel β-sheets (blades) (Figure 2A). The blades are organized around a central conical pore ranging from approximately 6 Å at the “entry” face side to approximately 10 Å diameters at the “exit” face side. One specific feature of our LPHN3-OLF is the disordered 395–403 fragment, which was also disordered in the LPHN3-RBL/OLF structure (Jackson et al., 2015). A noticeable difference between LPHN3-OLF and gliomedin-OLF consists in the presence in the central pore of a Na+ atom instead of Ca2+ in the same position, showing that different metal ions can be found at this site in OLF domains (see below).
Comparison between the two crystal forms
LPHN3-OLF crystallized in two lattices – P65 and C2221. The major difference in the crystallization conditions was in the range of concentration of MgCl2 used (either 50 for the P65 form or 300 mM for the C2221 form, see Methods). When superimposed, the structures refined from the two lattices were very similar (r.m.s.d = 0.9 Å, 243 aligned Cα out of 254), with the exception of the loop 316–329 which is stabilized by the protein core in the P65 lattice (“closed” conformation) and projecting towards a symmetric neighbor in the C2221 lattice (“open” conformation) (Figure 2B). In the open state, this loop is highly flexible as illustrated by a dramatic increase of its thermal motion (B-factors), as compared to the rest of the protein and the closed conformation (Figure 2C). Indeed, the position of the loop in the open conformation was weakly defined by the electron density maps. Accompanying the movement of loop 316–329, two other neighboring loops (392–405 and 425–434) undergo a noticeable conformational change (Figure 2B). Interestingly, loop 392–405 includes the unmodeled fragment 397–403.
Consequently to the opening of the 316–329 loop (C2221), the β-propeller central cleft becomes solvent-accessible from the “entry” face (Figure 2D), with a total volume of accessible area of 1550 Å3, as opposed to the value of the closed state (P65) of only 220 Å3 of a buried cavity, disconnected from the surface of the protein. By examining the geometric characteristics of the coordination sphere of a heteroatom found in the central cleft, and exploiting the anomalous signal from the dataset collected at low energy through an f″ refinement (Liu et al., 2013), we identified a Ca2+in the closed state (Figure 2E). The shorter cation-oxygen distances within the octahedric coordination geometry of the open state (Figure 2E) clearly designate a Mg2+ ion (Harding, 2006) also validated by CheckMyMetal (Zheng et al., 2014). Hence, with the presence of 300 mM of Mg2+ used in crystallization, Ca2+ was replaced by Mg2+. In the closed state, the Ca2+ bridges three central cleft β-strands through the involvement of residue atoms in its coordination: Asp332 Oδ2 (β10), Asn380 Oδ1 (β14) and Val435 O (β18). Additionally, the loop 316–329 is involved in the binding of the calcium ion via a hydrogen bond between Tyr323 and Asp332. By contrast, the Mg2+ ion found in open state is coordinated only by residues from β18, Asp436 Oδ1 and Val435 O. Therefore, the opening of loop 316–329 and the concomitant disruption of the hydrogen bond between Tyr323 and Asp332 likely allows for a solvent accessibility of the central pore, which resulted in a displacement of the bound Ca2+ by Mg2+ at high concentration in the crystallization liquor.
The model refinement of the P65 crystal form indicated that the cation Ca2+ binding site was not fully occupied by the Ca2+, which was refined with 70% occupancy. Although partial exchange of Ca2+ by Mg2+ could have occurred in the P65 form, our crystallographic data are not sufficient to definitively conclude what proportion of Ca2+/Mg2+ composes the mixture. However, as the physiological concentration of Mg2+ in the extracellular fluids is 0.7–1mM (Bringhurst et al., 2012), the presence of Mg2+ in the C2221 crystal structure is most likely a crystallization artifact. Recently determined structures of myocilin-OLF (Donegan et al., 2014) and LPHN3-RBL/OLF (Jackson et al., 2015), contain the same Ca2+ binding site and a Na+ in the central pore that we could not identify from the electron density maps probably due to a partial occupancy (like Ca2+) as indicated by the alternative conformation of Asp436 (P65) (Figure 2E, Figure S1B, related to figure 2). Hence, a joint analysis of the two crystal forms of LPHN3-OLF and the LPHN3-RBL/OLF structure reveals that Ca2+ and Na+ both contribute to the conformational stability of the central pore residues, specifically Asp332 (Ca2+) and Asp436 (Na+). These ions also contribute to a core-stabilization of the loop 316–329 through the Ca2+- Asp332-Tyr323 interaction network. However, the partial occupancy of the ions in the P65 form and the availability of the C2221 open state where the Mg2+ occupancy reached 100 %, show that they are not absolutely required for the maintenance of the overall folding of LPHN3-OLF.
Hydrogen-deuterium exchange coupled with mass spectrometry (DXMS)
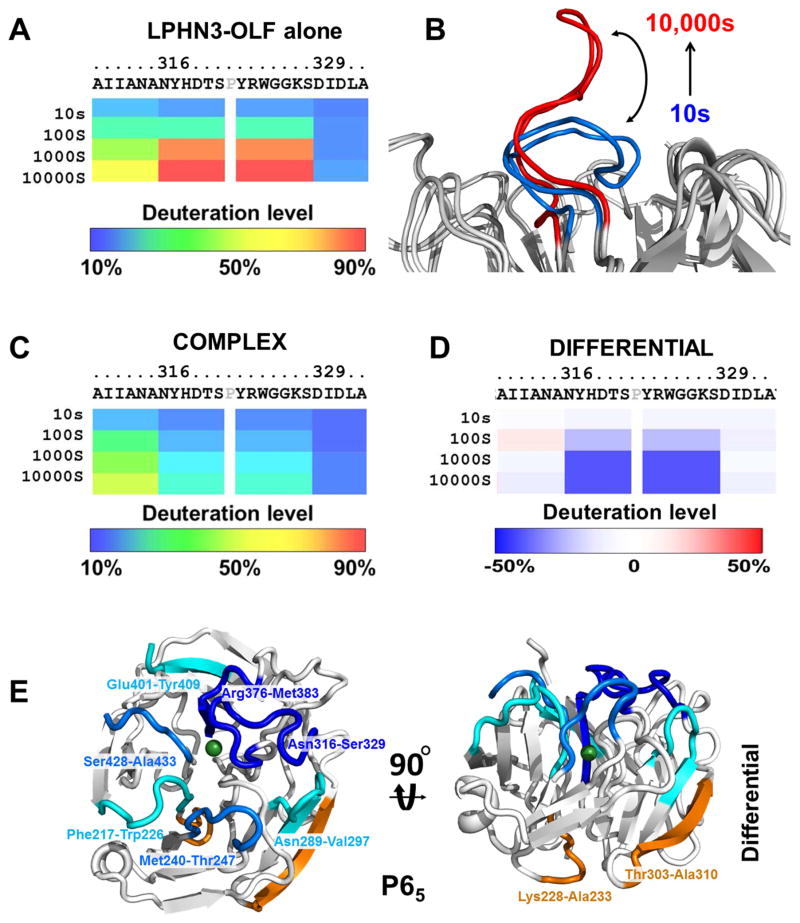
To obtain dynamic information on the individual structures and their complex, we used DXMS. After optimization of the mass spectrometry (MS) coverage of the individual proteins (data not shown), high resolution MS data we collected on LPHN3-OLF alone and bound to FLRT3-LRR. Deuteration levels of LPHN3-OLF alone show that the first ~20 amino acids (APSTDHLDYKDDDDKAAAKV) and the last ~40 amino acids (LDSRSGPVHHGQVSYISPPIHLDSELERPPVRGILEVLFQ) of the protein exchanged >90% deuteron in the first 10 seconds, indicating that these regions are fully solvent exposed and disordered. These 60 residues are clipped during crystallization trials and could not be built in our structure. Interestingly, among the LPHN3-OLF fragments that acquire deuteration over time (see supplemental information, Figure S2 and S3, related to figure 3), loop 316–329 is gradually deuterated and acquires >50% deuteration in 100s, reaching ~ 90 % deuteration after 10,000s (Figure 3A). This increase in deuteration level indicates a progressive solvent exposure of the amide hydrogens. In light of the crystallographic models, this strongly suggests that loop 316–329 is dynamic, as a result of a conformational motion between the closed and open state (Figure 3B). Therefore, our experiments show a propensity of the OLF central pore to be filled by solvent molecules, driven by a movement of loop 316–329.
Figure 3. DXMS analysis profiles of LPHN3-OLF, alone or bound to FLRT3-LRR domain.
(A), deuteration levels of the fragment 316–329 of LPHN3-OLF domain alone. (B), Schematics of the deuteration levels of loop 316–329 to indicate the deuteration over time. (C), Deuteration level of loop 316–329 of LPHN3-OLF in complex with FLRT3-LRR. (D), Difference in deuteration level of loop 316–329 between the free and bound LPHN3-OLF domain. The percentages of deuteration levels of each peptide fragment at various time points are shown as a heat map color-coded from blue (10%) to red (90%), as indicated at the bottom of each map. Each block under the protein sequence represents a peptide segment analyzed at each of the four time points (from top to bottom: 10, 100, 1,000, and 10,000s). Differential deuteration is shown in a color-coded map ranging from blue (−50%) to red (+50%), as indicated at the bottom of the panel. Proline residues and regions with no amide hydrogen exchange data available are colored in gray. (E), Structure of the entire P65 LPHN3-OLF color coded according to the differential deuteration map. Cyan to dark blue colors indicate increasing protection of LPHN3-OLF in complex with FLRT-LRR, orange indicate increased solvent accessibility. See also Figure S2, Figure S3.
Deuteration levels of LPHN3-OLF in complex with FLRT3-LRR should reveal whether some of the loops belonging to the binding interface become more solvent protected. In fact, LPHN3-OLF loop 316–329 in complex with FLRT3-LRR is significantly less deuterated than in the free protein (Figure 3C,D). These data suggest that loop 316–329 is stabilized in a solvent-inaccessible area of the complex (see below). To a lesser extent, fragment 396–400 also shows a high level of deuteration indicating a large degree of flexibility (>50% deuteration is 100s) consistent with the absence of this loop in the crystallographic models (Figure S2A,B, related to figure 3; Table S1, related to Figure 2). Other neighboring and superficial fragments show various degree of solvent protection (Phe217-Trp226; Met240-Thr247; Asn289-Val297; Arg376-Met383; Tyr429-Ala433; Glu401-Tyr409) whereas loops that are distant from the putative binding surface show higher deuteration exposure (Lys228-Ala233; Thr303-Ala310) (Figure 3E), in agreement with the interfacing area revealed by crystallography.
The LPHN3-OLF/FLRT3-LRR complex crystal structure
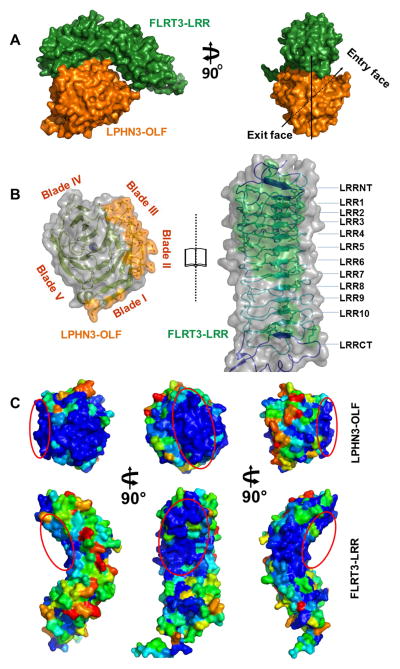
We have determined the structure of the 1:1 LPHN3-OLF/FLRT3-LRR complex at 3.2 Å resolution. Overall, the complex is assembled so that the concave face of the horseshoe-shaped solenoid FLRT3-LRR interacts with LPHN3-OLF, and the central pore axis of LPHN3-OLF is oriented ~45° relatively to the FLRT3-LRR long axis (Figure 4A). The LPHN3-OLF structure in complex with FLRT3-LRR enabled us to model four more residues in a disordered loop (400-NEAT-403) and five at the C-terminal end (463-DSRGP-467) not modeled in the free protein. The FLRT3-LRR interfacing residues are mainly provided by its N-terminal moiety and are part of the capping LRRNT and the first eight LRR motifs (Figure 4B; Table S3, related to Figure 4). All three predicted N-linked glycosylation sites of FLRT3-LRR are situated far from the interface and one N-Acetyl-glucosamine moiety could be modeled on Asn226. The LPHN3-OLF residues involved in the association belong to β-strands connecting loops projected by blades I to III towards the same region of the tertiary structure next to the “entry” face (4B; Table S3, related to Figure 4). The FLRT3-LRR remaining repeats and the capping LRRCT show only few residues (Arg275, Gln299, Lys326, Asn328 and Arg330) partly buried by the LPHN3-OLF fragment 261–267 (Table S3, related to Figure 4). The calculated total solvent-accessible area buried in the interface is 867 Å2 and this interface was computationally assigned as biologically relevant by the EPPIC server (data not shown).
Figure 4. Complex between LPHN3-OLF and FLRT3-LRR.
(A), Left panel, side view of the surface rendering of the LPHN3-OLF/FLRT3-LRR complex. Green, FLRT3-LRR, Orange, LPHN3-OLF. The view highlights the associating surfaces of the structure that is the N-terminal part of the concave surface of FLRT3-LRR and the lack of contact on the C-terminal part of it. Right panel, front view of the surface rendering of the complex to highlight the ~45°clockwise inclination of OLF through its central pore. (B), “Open book view” of the semi-transparent surface highlighting both ribbon diagram and side chains of the residues involved in the association and the main secondary elements. (C), Surface view of LPHN3-OLF (top) and FLRT3-LRR (bottom) highlighting the relative sequence conservation of the binding surface. Blue indicated high conservation and red indicates lowest conservation scores. Red ovals indicate the same binding surface in the different orientations. LPHN3-OLF was aligned with 20 other OLF domains of latrophilins and FLRT3-LRR was aligned with 73 unique sequences from CSI-BLAST.
To get better insights into the specificity of interaction, we mapped the surface of the two proteins according to their conservation score from the ConSurf server (Ashkenazy et al., 2010) (Figure 4C) using 73 LRRs and 20 LPHN-OLFs unique sequences. The analysis indicates that interfacing areas of both proteins are mainly composed of residues with high conservation scores. Interestingly, another highly conserved area proximal to the LPHN3-OLF/FLRT3-LRR interface was observed in both proteins (Figure 4C). In FLRT3-LRR this area matches the Unc5D/FLRT2 complex interface (Seiradake et al., 2014).
Mapping the electrostatic surface of the two proteins provides evidence on the observed binding mode: a cluster of negatively charged residues formed by Asp113, Asp136, Asp137 and Asp183 in FLRT3-LRR complements the positively charged Arg273, Arg292, Arg294 and in LPHN3-OLF (Figure 5A). Moreover, several hydrogen bonds and salt bridges are observed in the main interface (Table S3, related to Figure 4) added by hydrophobic and stacking contacts involving aromatic side chains, such as Phe160 from FLRT3-LRR observed in a π-stacking with Tyr245 from LPHN3-OLF (see below). Conversely, the C-terminal region of the concave face in FLRT3-LRR and the facing area in LPHN3-OLF do not seem to be in contact, probably because they are both positively charged and likely to repel each other (Figure 5A).
Figure 5. Analyses of the LPHN3-OLF/FLRT3-LRR association.
(A), Electrostatic surface of LPHN3-OLF and FLRT3-LRR in “open book view” to emphasize a charge complementarity within the main interfacing area of both partners (green oval) and a repulsive facing area (orange square). (B), Stereoview of the stabilization of the LPHN3-OLF loop 316–329 in the complex. Interfacing LPHN3-OLF and FLRT3-LRR residues are represented in yellow and cyan respectively, and the potential hydrogen bonds are shown as dashed magenta lines. Loop 316–329 (closed conformation) with the Tyr323, Asp332 residues and Ca2+ are in shown in purple. The central pore β-strands β10, β14 and β18 involved in the Ca2+ binding and the β13-β14 connecting loop that lies on top of the 316–329 loop are also shown in orange. (C), Loop 316–329 in the open state (C2221) of LPHN3-OLF from a superimposition with the complex structure. Red and green mashes represent the molecular surfaces of the residues that would display a steric clash. (D–H), Analyses of the binding between LPHN3-OLF/FLRT3-LRR by site-directed mutagenesis on FLRT3-LRR (D), Models showing the overall position of the mutations. (E), Western blot showing the FLRT3-LRR mutants used in this figure and their different migration pattern due to the additional glycosylation. (F), ITC experimental trace of the LPHN3-OLF injected at 100μM into the cell containing the FLRT3-LRR R117T control mutant at 10μM. (G), ITC experimental trace of the LPHN3-OLF injected at 100μM into the cell containing the FLRT3-LRR F160N mutant at 10μM. This mutant has an additional N-linked glycosylation site at the OLF binding surface that prevents the association. (H), ITC experimental trace of the LPHN3-OLF injected at 100uM into the cell containing the FLRT3-LRR F160A mutant at 10μM.
Within the complex, the LPHN3-OLF flexible 316–329 loop is stabilized in the closed state, via direct interactions with FLRT3-LRR established by Asn316, Tyr317, Asp319 and Thr320. The mobility of the entire loop is further restrained by the interfacing interactions established by Arg376 with Tyr43 and Tyr64 in FLRT3-LRR, which maintains the β13-β14 connecting loop on top of the 316–329 loop and contributes to lock its conformation (Figure 5B,C).
We next determined whether the association requires a significant conformational change of either of the two proteins, or it is of a rigid-body type. Superimposition of the LPHN3-OLF subunit with the P65 Ca2+-bound structure reveals an r.m.s.d. of 0.56 Å over 256 aligned Cα. The same procedure made with the FLRT3-LRR subunit with the recently determined structure (PDB ID 4V2E) (Seiradake et al., 2014) gives an overall r.m.s.d. of 1.20 Å for 321 aligned Cα, a value that falls to 0.96 Å when aligning the first 219 Cα (residues 30 to 248) which contain the main interfacing residues described earlier. This comparison indicates that the structures of LPHN3-OLF and FLRT3-LRR are rigid and undergo a rigid-body type interaction without any significant rearrangements, providing that the LPHN3-OLF is in the closed state.
Mutational analysis of the association
To test how the stabilization of the 316–329 loop affects the LPHN3-OLF/FLRT3-LRR association, we engineered an Asp332Ala and a Tyr323Ala that should disrupt the Ca2+-driven stabilization of this loop, and measured their interaction with FLRT3-LRR. All mutant proteins were first analyzed by SEC to reveal potential misfolding or oligomerization state abnormalities. As expected, the expression level of all mutants and their elution profile were identical to the ones measured for the WT proteins (data not shown). The mutant Asp332Ala showed a decreased affinity (KD ~170 nM) (Table 1), indicating that Asp332 is important by strengthening the closed state through the coordination of Ca2+ and the Asp332-Tyr323 hydrogen bond. Furthermore, the mutant Tyr323Ala showed no detectable binding in conditions similar to the wild type measurements, which strongly suggests that Tyr323 is key to maintain the “closed” state. Moreover, to confirm the binding mode between LPHN3-OLF and FLRT3-LRR revealed by the crystallographic study an N-linked glycosylation site was introduced in the FLRT3-LRR interfacing surface and compared to a glycosylated mutant outside of the binding interface. At the interfacing position 160 we substituted Phe160 with an Asn, to complete the canonical Nx(S/T) sequon. As a control mutant, the surface residue Asn115 was glycosylated by substituting Arg117 to a Thr (Table 1). The additional N-linked glycosylation was verified by SDS-PAGE showing slower migrating bands for the Phe160Asn and Arg117Thr mutants, compared to the Phe160Ala or the wild-type construct (Figure 5D,E and Table 1). The glycosylated Arg117Thr mutant had a measured dissociation constant KD of 27nM, typical of wild type values (Figure 5F and Table 1). In contrast, Phe160Asn mutant showed no detectable binding in similar conditions (Figure 5G and Table 1), indicating that introducing bulky sugar moieties in the interface observed in the crystal structure, severely impacted the LPHN3-OLF/FLRT3-LRR association. As the Phe160 side chain was found to establish a direct interaction with the LPHN3-OLF Tyr245 in the complex structure, an Ala mutant was also designed to get insights into the importance of this specific position for the affinity between the two partners. For the Phe160Ala mutant, a KD of ~1 μM was determined, suggesting that Phe160 is a key residue in the association and that the contact this residue establishes with LPHN3-OLF Tyr 245 is likely to be of major importance (Figure 5H and Table 1).
Finally, we analyzed the LPHN3-OLF mutation Ala313Ser reported in an ADHD patient (Domene et al., 2011). The mutant protein is expressed at levels similar to the WT protein and both hydrodynamic parameters and association with FLRT3-LRR were similar to the WT protein (Table 1). These data indicate that, Ala313, which is situated at a surface region distinct from the complex interface (~12 Å from the closest FLRT3-LRR residue), does not play an important role in the association with FRLT3-LRR.
DISCUSSION
We previously reported that LPHN3 binds in vivo and in vitro to fibronectin leucine-rich repeat transmembrane 3 (FLRT3) to constitute a synaptic ligand-receptor pair (O’Sullivan et al., 2012). Single particle electron microscopy of LPHN3 showed that its entire extracellular domain is organized into two globular regions separated by a semi-rigid domain that is O-linked glycosylated (O’Sullivan et al., 2014). However, these studies did not allow a detailed structural and functional characterization of the minimal domain necessary for the interaction with FLRT3. Here we show that only the OLF domain of LPHN3 is required for FLRT3-LRR binding and we determined precise dissociation constants and stoichiometry of the complex. As expected, the KD measured for this interaction (Table 1) are similar to the one determined for their entire extracellular domains (O’Sullivan et al., 2012), indicating that LPHN3-OLF alone is sufficient to bind FLRT3-LRR.
Characterization of the olfactomedin domain structure
Initial diffraction data were not sufficient to obtain a structure solution by molecular replacement approaches, most likely because at the time of our data analysis the closest five-bladed β-propeller homolog available had less than 13 % amino acid identity to our protein. Therefore, we successfully resorted to multi-crystal native SAD phasing (Liu et al., 2012), illustrating the robustness of using anomalous signal from light atoms such as sulfur for structure determination. Furthermore, we show how such signals can be critical to ascertain the nature of anomalous scatterers through the characteristic imaginary terms of their scattering factor (f″); the bound Ca2+ (P65) or Mg2+ (C2221) in our case. The crystal structure of the LPHN3-OLF shows that this domain has a five-blade β-propeller fold that is monomeric in solution. Despite the broad structural diversity of the β-propeller architecture, two features are often conserved amongst members of this fold: The four-stranded antiparallel β-sheets and the central pore where metal ions might also bind further stabilizing the structure (Chen et al., 2011).
Loop 316–329 mobility
In LPHN3-OLF, our crystallographic study designates loop 316–329 as one of the structural elements stabilized in presence of Ca2+ (the closed state), indirectly via a hydrogen bond between Asp332 and Tyr323. Moreover, this loop is involved in the binding interface with FLRT3-LRR as also shown by DXMS and in fact the open state of this loop is incompatible with the binding to FLRT3- LRR. However, loop 316–329 was found to be dynamic both in cristallo and in solution as shown by DXMS data of LPHN3-OLF alone (Figure 3A), which allows solvent exchange in the β-propeller central pore, evidenced by the C2221 crystal form. The high concentration of Mg2+ used for the crystallization of the C2221 crystal form led to a complete Ca2+/Mg2+ exchange in the central pore and to a stable enough conformation to be crystallized. Due to the well-established specific (de-)hydration properties of the Mg2+ ion as compared to Ca2+ (Maguire and Cowan, 2002), the ion coordination with Asp332 probably could not be established, maintaining loop 316–319 in an open state. Although loop 316–329 is dynamic under physiological condition, a Ca2+/Mg2+ exchange may rarely occurs in vivo due to relatively low concentration and the balance of these two ions in the extracellular fluids. These observations confer to the presence of Ca2+ in the central pore an important, albeit indirect, role in the complex formation. Some conformational motion of the myocilin-OLF equivalent loop B-10/C-11 has been reported when comparing the polyethylene glycol-containing wild type structure to the glycerol-containing form or to the SeMet derivative form of the E396D mutant (Donegan et al., 2014). Gliomedin-OLF appears to be different in this respect, since the LPHN3-OLF Tyr323-Asp332-Ca2+ trio structurally superimposes with Phe415-Asn423-Na+ (Figure S1D, related to figure 2), however, the gliomedin-OLF 407–420 loop (equivalent to LPHN3-OLF 316–329) is poorly conserved (Han and Kursula, 2015). The fact that Phe415 and the proximal Leu414 are involved in a core-buried hydrophobic cluster suggests that the entire loop is less dynamic. In general, solvent accessibility of the OLF β-propeller central cavity appears to be a shared feature as exhibited by the presence of a high density of bound water molecules in the available structures of OLF domains (Donegan et al., 2014; Han and Kursula, 2015; Jackson et al., 2015, Pronker et al., 2015, and this study). Furthermore, most of the Ca2+-binding residues in LPHN3-OLF are conserved – or conservatively substituted – in the OLF family, in particular those having their side chain involved in the ion coordination. Although our open-state structure was solved with higher-than-physiological amount of Mg2+, gliomedin-OLF (Han and Kursula, 2015) was crystallized with sub-physiological amount of Na+, indicating that Na+ likely exchanges in vivo.
The LPHN3-OLF/FLRT3-LRR complex
There is ample experimental evidence that LRR proteins tend to form quaternary structures with dimerization or multimerization surfaces (Scott et al., 2004; Seiradake et al., 2014). Although it is unclear whether these quaternary complexes are able to dissociate upon ligand binding (McEwan et al., 2006) our FLRT3-LRR/LPHN3-OLF complex is monomeric in solution, indicating that the tendency of FLRT3 to self-associate under concentrated condition (Figure 1D,E, (Seiradake et al., 2014)) is reversed upon binding with LPHN3-OLF. The curved structure of the LRR domain and the exposed β-sheet on the concave side form a large binding surface, which makes the LRR domain a very effective protein-binding motif (Kobe and Kajava, 2001). Although ligand-binding through LRR domains concave surfaces is common, it is not exclusive: for example, UNC5D binds to one side of FLRT2 (Seiradake et al., 2014). It is worth noting that the LRR-OLF interface area appears relatively small with respect to the high affinity of the interaction (Chen et al., 2013). Therefore, beyond the solvent-inaccessibility of interface residues, some key structural features, such as the strong electrostatic surface complementary between the concave face of FLRT3-LRR and globular LPHN3-OLF, are at play in the stability of the OLF-LRR association. Our mutational analyses point to the FLRT3-LRR Phe160. Indeed, its central position within the interface, its high level of buried area (Table S3, related to Figure 4) and the observed affinity decrease in Phe160Ala make this position as a “hot spot” for the interaction (Bogan and Thorn, 1998). Regarding the ADHD-linked mutation, this residue is not conserved in the LPHN family and the Ala313 Ser mutation does not affect FLRT3 binding, suggesting that it may simply represent a benign SNP.
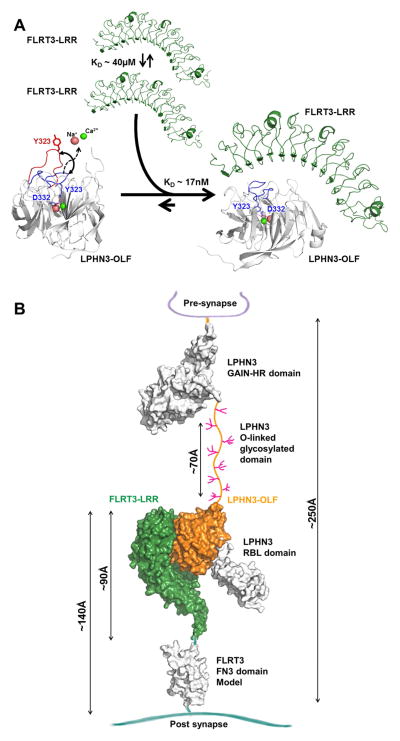
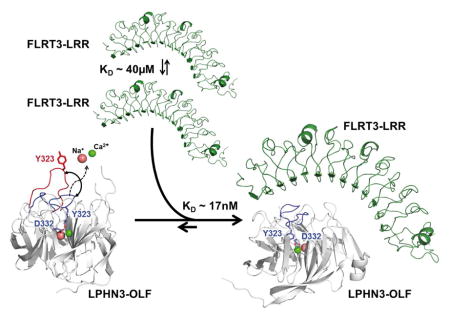
Taken together, an overall association model emerges from our studies: In the unbound state, the LRR motif of FLRT3 can dimerize with low affinity (~40μM) and LPHN3-OLF loop 316–329 is dynamic in the seconds to minutes time scale. Once these domains associate, there would be strong stabilization of the LPHN3-OLF domain in the close conformation and the disruption of the FLRT3 dimerization toward a high affinity LPHN3-OLF/FLRT3-LRR complex (Figure 6A). LPHN3 is a presynaptic protein containing an OLF domain that through the central O-linked glycosylation domain the OLF domain can reach the LRR of postsynaptic FLRT3 across the synaptic cleft, where they can interact (Figure 6B) (O’Sullivan et al., 2014).
Figure 6. Overall model of the association in the context of the synapse.
(A), FLRT-LRR alone tends to weakly dimerize and LPHN3-OLF alone has loop 316-329 that is mobile possibly allowing and Ca2+ and Na+ exchange. The high affinity of the association maintains LPHN3-OLF loop 316–329 closed and dissociates the low-affinity homo-dimerization of FLRT3-LRR. (B), LPHN3 is presynaptic and it binds through the OLF domain to the LRR domain of FLRT3, a post-synaptic protein. With the exception of the central O-linked glycosylated stalk domain drawn between the GAIN domain and the OLF domain, the rest of extracellular domain of LPHN3 has been determined using crystallography. FLRT3 FN3 domain is represented as homology model using other FN3 domains as template. Approximate dimensions are shown in Angstrom (Å).
It was recently reported that LPHN3-OLF also binds FLRT2 with low nM affinity and that neurons were repelled by the presence of LPHN3-OLF in the growth matrix of a stripe assay (Jackson et al., 2015). Moreover, overlay of the coordinates of the published mouse FLRT2-LRR in complex with the first Ig domain of UNC5D (PDB ID: 4V2C) (Seiradake et al., 2014) with our complex suggest that a trimeric complex composed by FLRT2 or FLRT3-LRR/LPHN 3-OLF/UNC5D-Ig1 is possible. Owing to this intricate recognition code between FLRTs and LPHN3, which may generate adhesion or repulsion, the structural characterization of the presented complex will be important to inform testable mechanistic-based hypotheses in the biology of synapse formation and in vivo axonal attraction and repulsion.
EXPERIMENTAL PROCEDURES
Cloning and expression of FLRT3 and LPHN3
The extracellular domains of mouse LPHN3 and mouse FLRT3 and their shorter constructs were cloned as C-terminal Fc-fusion proteins into a modified pCMV6-XL4 plasmid and were expressed as soluble entities in the cell culture medium of stably transfected HEK293 GnTI- cells and maintained in 10%FBS in Dulbecco’s Modified Eagle Medium (DMEM).
Protein purification
Proteins were affinity purified using Protein-A resin using in 50mM TrisHCl pH 7.4, 150mM NaCl, 1 mM EDTA (TNE), and subsequently cleaved with 3C protease to remove the Fc fragment. Proteins were further purified by size exclusion chromatography in 50mM Tris-HCl pH 7.4 and 150mM NaCl (TN) and concentrated depending of the downstream experiment.
ITC
Experiments were performed with a MicroCaliTC200 system, purified proteins were buffer-exchanged in 50 mM Na-HEPES pH 7.0, 150 mM NaCl, 1 mM EDTA (or 2mM CaCl2). The protein solution in the syringe was added to the cell in a series of multiple injections at 25°C. The raw ITC data was processed and fitted using a single site model using the ORIGIN software provided by GE MicroCal.
AUC experiments - were performed on a Beckman Optima XL-I analytical ultracentrifuge. All samples were measured at 35 krpm and scanned at 281 nm and data were acquired in intensity mode. All samples were measured in a buffer containing 50 mM TRIS-HCl, pH 7.4; 150 mM NaCl, and 1 mM EDTA. Data were analyzed with UltraScan-III ver. 3.1, release 1964.
Protein crystallography
Crystals of LPHN3-OLF were made by under-oil microbatch method by mixing 1 μL of protein with an equal volume of crystallization solution containing [15–25]% polyethylene glycol 8000, [50–300] mM MgCl2 and 0.1 M TAPS pH 9.0. C2221 crystal forms were harvested from 50 mM MgCl2 containing conditions whereas P65 crystal forms grew in 300 mM MgCl2 containing conditions. Crystals of the complex were prepared by vapor diffusion in hanging drops by mixing 1 μL of a 1:1.2 molar ratio mixture of LPHN3-OLF and FLRT3-LRR with an equal volume of crystallization solution containing 6 % polyethylene glycol 3350 and 0.2 M NaNO3. Complete data sets were collected from individual crystals under a cryogenic stream at 100K. All diffraction data sets were processed by XDS and SCALA. For the LPHN3-OLF structure Native-SAD phasing was performed first by substructure solution with SHELXD and subsequent SAD phasing with PHENIX module PHASER, using four diffraction datasets collected with a 6 keV X-Ray beam. The subsequent 2.6 Å resolution initial model was further refined using Phenix.refine against separated collected P65 and C2221 data at 1.6 Å resolution, after a molecular replacement with the PHENIX module PHASER for the latter. The complex structure was solved by molecular replacement using the PHENIX module PHASER using the P65 structure of LPHN3-OLF and the recently determined structure of FLRT3-LRR (PDB ID 4V2E) as starting models. Subsequent refinement of the structure was performed using BUSTER.
DXMS
Comparative deuterium exchange mass spectrometry studies were carried out with LPHN3- OLF alone and with LPHN3-OLF/FLRT3 complex by mixing these proteins at 1:1 molar ratio. The exchange reactions were quenched at various time points by addition of 18μl of ice-cold 0.8% formic acid, 0.8M GuHCl, 16.6% glycerol for a final pH of 2.5. Peptide identification was done by the aid of Proteome Discoverer software (ThermoFisher).
More details can be found in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
The olfactomedin domain of LPHN3 alone is sufficient for FLRT3 binding
The crystal structures of LPHN3-OLF alone and in complex with FLRT3-LRR are presented.
LPHN3-OLF shows an open and a close conformation and Ca2+ bound in the central pore
The high affinity binding is based on electrostatic and hydrophobic interactions
Acknowledgments
We thank Randy Abramowitz at National Synchrotron Light Source (NSLS) beamlines X4A and X4C and the MacCHESS staff for their assistance in data collection, and the superb support provided by Virgil Schirf (CAUMA) and others of the staff at the Texas Advanced Computing Center at the University of Texas at Austin. This work was supported by National Institutes of Health grants MH092906 and grant # 67038 from the Robert Wood Johnson Foundation to the Child Health Institute of New Jersey to D.C., GM107462 to W.A.H., NIH R01AI081982, R01GM020501, R01AI101436 to S.L.. The development of the UltraScan software is supported by National Science Foundation grant NSF-ACI-1339649 to B.D.. Supercomputer time allocations were provided through National Science Foundation grant TG-MCB070039N to B.D. X4 beamlines are supported by the New York Structural Biology Center at the NSLS of Brookhaven National Laboratory, a DOE facility. CHESS is supported by the NSF & NIH/NIGMS via NSF awards DMR-0936384 and DMR-1332208, and the MacCHESS resource is supported by NIGMS award GM-103485. We thank other members of the Comoletti Lab and especially Ian Hagee for his excellent technical support during protein expression and purification.
Footnotes
PROTEIN DATA BANK ACCESSION CODES
The atomic coordinates and associated structure factors were deposited in the Protein Data Bank under the accession codes 4RMK (P65) and 4RML (C2221) for the LPHN3-OLF structures and 4YEB for the LPHN3-OLF/FLRT3-LRR complex structure.
AUTHOR CONTRIBUTION
F.R., Q.L., F.M., F.B., S.v.D., S.L., D.L., performed all experiments, F.R., B.D., W.A.H. and D.C. wrote the manuscript.
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anholt RR. Olfactomedin proteins: central players in development and disease. Frontiers in cell and developmental biology. 2014;2:6. doi: 10.3389/fcell.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcos-Burgos M, Jain M, Acosta MT, Shively S, Stanescu H, Wallis D, Domene S, Velez JI, Karkera JD, Balog J, et al. A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Molecular psychiatry. 2010;15:1053–1066. doi: 10.1038/mp.2010.6. [DOI] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic acids research. 2010;38:W529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. Journal of molecular biology. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- Bringhurst FR, DM, Krane SM, Kronenberg HM. Bone and Mineral Metabolism in Health and Disease. In: Longo DL, FA, Kasper DL, Hauser SL, Jameson JL, Loscalzo J, editors. Harrison’s Principles of Internal Medicine. 18. New York: McGraw-Hill; 2012. [Google Scholar]
- Cao W, Demeler B. Modeling analytical ultracentrifugation experiments with an adaptive space-time finite element solution for multicomponent reacting systems. Biophysical journal. 2008;95:54–65. doi: 10.1529/biophysj.107.123950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Chan NL, Wang AH. The many blades of the beta-propeller proteins: conserved but versatile. Trends in biochemical sciences. 2011;36:553–561. doi: 10.1016/j.tibs.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Chen J, Sawyer N, Regan L. Protein-protein interactions: general trends in the relationship between binding affinity and interfacial buried surface area. Protein science : a publication of the Protein Society. 2013;22:510–515. doi: 10.1002/pro.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeler B, Brookes E, Wang R, Schirf V, Kim CA. Characterization of reversible associations by sedimentation velocity with UltraScan. Macromolecular bioscience. 2010;10:775–782. doi: 10.1002/mabi.200900481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domene S, Stanescu H, Wallis D, Tinloy B, Pineda DE, Kleta R, Arcos-Burgos M, Roessler E, Muenke M. Screening of human LPHN3 for variants with a potential impact on ADHD susceptibility. American journal of medical genetics Part B, Neuropsychiatric genetics. 2011;156B:11–18. doi: 10.1002/ajmg.b.31141. [DOI] [PubMed] [Google Scholar]
- Donegan RK, Hill SE, Freeman DM, Nguyen E, Orwig SD, Turnage KC, Lieberman RL. Structural basis for misfolding in myocilin-associated glaucoma. Human molecular genetics. 2014 doi: 10.1093/hmg/ddu730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrichny IP, Leone P, Sulzenbacher G, Comoletti D, Miller MT, Taylor P, Bourne Y, Marchot P. Structural analysis of the synaptic protein neuroligin and its beta-neurexin complex: determinants for folding and cell adhesion. Neuron. 2007;56:979–991. doi: 10.1016/j.neuron.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallgatter AJ, Ehlis AC, Dresler T, Reif A, Jacob CP, Arcos-Burgos M, Muenke M, Lesch KP. Influence of a latrophilin 3 (LPHN3) risk haplotype on event-related potential measures of cognitive response control in attention-deficit hyperactivity disorder (ADHD) European neuropsychopharmacology. 2013;23:458–468. doi: 10.1016/j.euroneuro.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingert JH, Heon E, Liebmann JM, Yamamoto T, Craig JE, Rait J, Kawase K, Hoh ST, Buys YM, Dickinson J, et al. Analysis of myocilin mutations in 1703 glaucoma patients from five different populations. Human molecular genetics. 1999;8:899–905. doi: 10.1093/hmg/8.5.899. [DOI] [PubMed] [Google Scholar]
- Han H, Kursula P. The Olfactomedin Domain from Gliomedin Is a beta-Propeller with Unique Structural Properties. The Journal of biological chemistry. 2015;290:3612–3621. doi: 10.1074/jbc.M114.627547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding MM. Small revisions to predicted distances around metal sites in proteins. Acta crystallographica Section D, Biological crystallography. 2006;62:678–682. doi: 10.1107/S0907444906014594. [DOI] [PubMed] [Google Scholar]
- Ichtchenko K, Bittner MA, Krasnoperov V, Little AR, Chepurny O, Holz RW, Petrenko AG. A novel ubiquitously expressed alpha-latrotoxin receptor is a member of the CIRL family of G-protein-coupled receptors. The Journal of biological chemistry. 1999;274:5491–5498. doi: 10.1074/jbc.274.9.5491. [DOI] [PubMed] [Google Scholar]
- Jackson VA, del Toro D, Carrasquero M, Roversi P, Harlos K, Klein R, Seiradake E. Structural basis of latrophilin-FLRT interaction. Structure. 2015;23:774–781. doi: 10.1016/j.str.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Current opinion in structural biology. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- Labbe A, Liu A, Atherton J, Gizenko N, Fortier ME, Sengupta SM, Ridha J. Refining psychiatric phenotypes for response to treatment: contribution of LPHN3 in ADHD. American journal of medical genetics Part B, Neuropsychiatric genetics. 2012;159B:776–785. doi: 10.1002/ajmg.b.32083. [DOI] [PubMed] [Google Scholar]
- Lacy SE, Bonnemann CG, Buzney EA, Kunkel LM. Identification of FLRT1, FLRT2, and FLRT3: a novel family of transmembrane leucine-rich repeat proteins. Genomics. 1999;62:417–426. doi: 10.1006/geno.1999.6033. [DOI] [PubMed] [Google Scholar]
- Liu Q, Dahmane T, Zhang Z, Assur Z, Brasch J, Shapiro L, Mancia F, Hendrickson WA. Structures from anomalous diffraction of native biological macromolecules. Science. 2012;336:1033–1037. doi: 10.1126/science.1218753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Guo Y, Chang Y, Cai Z, Assur Z, Mancia F, Greene MI, Hendrickson WA. Multi-crystal native SAD analysis at 6 keV. Acta crystallographica Section D, Biological crystallography. 2014;70:2544–2557. doi: 10.1107/S1399004714013376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Liu Q, Hendrickson WA. Robust structural analysis of native biological macromolecules from multi-crystal anomalous diffraction data. Acta crystallographica Section D, Biological crystallography. 2013;69:1314–1332. doi: 10.1107/S0907444913001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire ME, Cowan JA. Magnesium chemistry and biochemistry. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine. 2002;15:203–210. doi: 10.1023/a:1016058229972. [DOI] [PubMed] [Google Scholar]
- McEwan PA, Scott PG, Bishop PN, Bella J. Structural correlations in the family of small leucine-rich repeat proteins and proteoglycans. Journal of structural biology. 2006;155:294–305. doi: 10.1016/j.jsb.2006.01.016. [DOI] [PubMed] [Google Scholar]
- O’Sullivan ML, de Wit J, Savas JN, Comoletti D, Otto-Hitt S, Yates JR, 3rd, Ghosh A. FLRT proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron. 2012;73:903–910. doi: 10.1016/j.neuron.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan ML, Martini F, von Daake S, Comoletti D, Ghosh A. LPHN3, a presynaptic adhesion-GPCR implicated in ADHD, regulates the strength of neocortical layer 2/3 synaptic input to layer 5. Neural development. 2014;9:7. doi: 10.1186/1749-8104-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronker MF, Bos TG, Sharp TH, Thies-Weesie DM, Janssen BJ. Olfactomedin-1 has a V-shaped disulfide-linked tetrameric structure. JBC. 2015 doi: 10.1074/jbc.M115.653485. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott PG, McEwan PA, Dodd CM, Bergmann EM, Bishop PN, Bella J. Crystal structure of the dimeric protein core of decorin, the archetypal small leucine-rich repeat proteoglycan. PNAS. 2004;101:15633–15638. doi: 10.1073/pnas.0402976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiradake E, del Toro D, Nagel D, Cop F, Hartl R, Ruff T, Seyit-Bremer G, Harlos K, Border EC, Acker-Palmer A, et al. FLRT structure: balancing repulsion and cell adhesion in cortical and vascular development. Neuron. 2014;84:370–385. doi: 10.1016/j.neuron.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder DA, Rivers AM, Yokoe H, Menco BP, Anholt RR. Olfactomedin: purification, characterization, and localization of a novel olfactory glycoprotein. Biochemistry. 1991;30:9143–9153. doi: 10.1021/bi00102a004. [DOI] [PubMed] [Google Scholar]
- Stone EM, Fingert JH, Alward WL, Nguyen TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A, Nichols BE, et al. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–670. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. alpha-Latrotoxin and its receptors: neurexins and CIRL/latrophilins. Annual review of neuroscience. 2001;24:933–962. doi: 10.1146/annurev.neuro.24.1.933. [DOI] [PubMed] [Google Scholar]
- Tomarev SI, Nakaya N. Olfactomedin domain-containing proteins: possible mechanisms of action and functions in normal development and pathology. Molecular neurobiology. 2009;40:122–138. doi: 10.1007/s12035-009-8076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Chordia MD, Cooper DR, Chruszcz M, Muller P, Sheldrick GM, Minor W. Validation of metal-binding sites in macromolecular structures with the CheckMyMetal web server. Nature protocols. 2014;9:156–170. doi: 10.1038/nprot.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.