Abstract
Auditory information is initially processed in the cochlear nuclei before being relayed to the brain. The cochlear nuclei are subdivided into dorsal, anterior ventral, and posterior ventral domains, each containing several subtypes of neurons that are thought to play discrete roles in the processing of sound. However, the ontogeny of these neurons is poorly understood, and this gap in knowledge hampers efforts to understand the basic neural circuitry of this nucleus. Here, we reveal that Bhlhb5 is expressed in both excitatory (unipolar brush cells) and inhibitory neurons (cartwheel cells) of the DCN during development. To gain genetic access to Bhlhb5-expressing neurons in the DCN, we generated a Bhlhb5::flpo knockin allele. Using an intersectional genetic strategy, we labeled cartwheel cells, thereby providing proof of concept that subpopulations of Bhlhb5-expressing neurons can be genetically targeted. Moreover, fate-mapping experiments using this allele revealed that Bhlhb5 is required for the proper development of the DCN, since mice lacking Bhlhb5 showed a dramatically diminished number of neurons, including unipolar brush and cartwheel cells. Intriguingly, the Bhlhb5::flpo allele also genetically labels numerous other regions of the nervous system that process sensory input, including the dorsal horn, the retina, and the nucleus of the lateral olfactory tract, hinting at a more general role for Bhlhb5 in the development of neurons that mediate sensory integration.
Keywords: Dorsal cochlear nucleus, Bhlhb5, Bhlhe22, Cre recombinase, Flp recombinase
1. Introduction
The dorsal cochlear nucleus (DCN) is a brainstem auditory nucleus where acoustic information is first processed and integrated with somatosensory and cochlear input. One of the functions of this nucleus is to help localize sound in the vertical plane so that an organism can determine whether the acoustic source is from above or below, thereby helping an organism evade predators or locate prey. In addition, the DCN may also contribute to our experience of painfully loud sound, an aversive sensation that evolved to protect hearing from acoustic assault. When acoustic damage occurs repeatedly, this trauma can cause ringing in the ear due to the false perception of sound, a pathological condition known as tinnitus (Axelsson and Ringdahl, 1989; Shargorodsky et al., 2010; Maes et al., 2013). Importantly, recent studies suggest that tinnitus is caused, at least in part, by hyper-activity within the dorsal cochlear nucleus due to decreased inhibition (Wang et al., 2009, 2011; Middleton et al., 2011). These physiological and pathophysiological roles underscore the importance of understanding the neural circuitry of the DCN, which remains poorly defined. In this regard, the development of tools that allow cell type-specific genetic control would be extremely useful.
Elegant fate-mapping studies have revealed that the DCN arises largely from progenitors within rhombomere 5 of the auditory lip (Farago et al., 2006; Nichols and Bruce, 2006). These neurons migrate to form a laminar structure comprised of three layers: the molecular layer, the fusiform cell layer, and the deep layer (Osen, 1969; Ryugo and Willard, 1985; Hackney et al., 1990). Within these layers, there are eight major morphological classes of neurons that have been identified. Previous work has revealed that the excitatory subtypes (granule cell, unipolar-brush cells, giant cells, and fusiform cells) arise from Atoh1-expressing progenitors, whereas the inhibitory subtypes (golgi cells, superficial-stellate cells, cartwheel cells and tuberculo-ventral cells) arise from Ptf1a-expressing progenitors (Fujiyama et al., 2009). However, the transcriptional programs within the DCN that mediate terminal differentiation and neuronal connectivity are poorly understood.
To determine what role these different cell types play in the function of the DCN, we need tools that allow cell-type specific genetic manipulation. We find that a key factor in the development of the DCN is Bhlhb5 (also known as Bhlhe22), a basic helix-loop-helix transcription factor that is related to Atonal (Ross et al., 2003). In many regions of the nervous system, Bhlhb5 is selectively expressed in early post-mitotic neurons as they undergo terminal differentiation and establish synaptic connections (Ross et al., 2010). Using genome-wide CHIP-seq analysis, we previously showed that Bhlhb5 binds to a specific DNA consensus motif (CATATGNTNT), which contains a canonical E-box (underlined). In addition, we provided evidence that Bhlhb5 recruits a specific PR/SET domain containing co-repressor, Prdm8, to mediate the repression of its target genes (Ross et al., 2012). Thus, Bhlhb5 is a neural-specific transcriptional repressor that is predominantly expressed during development.
At the cellular level, Bhlhb5 has been shown to be important for the proper development and/or survival of a number of neural cell types. For instance, Bhlhb5 is required for the survival of a population of inhibitory neurons in the dorsal horn of the spinal cord that function to inhibit itch (Ross et al., 2010; Kardon et al., 2014). Bhlhb5 is also expressed during the development of excitatory neurons in the dorsal telencephalon, where it shows an area-specific pattern of expression (Joshi et al., 2008; Ross et al., 2012). Loss of Bhlhb5 results in an almost complete absence of major axon tracts in the telencephalon including the corpus collosum, hippocampal commissure, anterior commissure, and the corticospinal tract (Ross et al., 2012). Finally, Bhlhb5 has been shown to play an important role in the development of the retina, where it is expressed in both type II cone bipolar cells and a specific subset of amacrine cells (Feng et al., 2006). These examples highlight the idea that Bhlhb5 plays a key role in the development of neurons in which it is expressed. However, the expression of Bhlhb5 in many regions of the nervous system is poorly characterized, and its possible function in these regions is unknown.
Here, we show that Bhlhb5 is expressed in a subset of DCN neurons during its development, including unipolar brush cells and cartwheel cells. To gain genetic access to these cells, we generated a Bhlhb5::flpo allele, and find that this allele causes recombination in subsets of Bhlhb5-expressing neurons. Using this allele, we compare the efficiencies of Cre and Flpo in a mouse. In addition, we show that the Bhlhb5::flpo allele can be coupled with other Cre lines to mark neurons at the genetic intersection, as illustrated by the targeting of cartwheel cells. Finally, our fate mapping studies reveal that Bhlhb5 is a critical factor in the ontogeny and survival of DCN neurons, which show dramatic reduction in cell number in its absence. In addition to marking cells within the DCN, the Bhlhb5::flpo allele described here may be a useful tool for the study of sensory integration in other regions of the nervous system, such as the retina, the dorsal horn, and the nucleus of the lateral olfactory tract.
2. Methods
2.1. Animal husbandry
Bhlhb5::cre mice (also called Bhlhb5-cre) and Bhlhb5 null mice were generated as previously described (Ross et al., 2010) and were maintained on a mixed C57bl/6.129J background. Bhlhb5::flpo mice were generated as described below and genotyped using the following primers: F: GGTGAATCCAAGCAAGATAAACGG; R: CAGCACAGGTAGGTCAGCTC.
RCE::dual mice were obtained from Gordon Fishell (Sousa et al., 2009). RCE::FRT and RCE::LoxP mice were obtained by mating RCE::dual mice with either EIIa-cre mice (Jackson Labs) or ROSA26Flpo mice (Jackson Labs), respectively. Mice were given free access to food and water and housed under standard laboratory conditions. The use of animals was approved by the Institutional Animal Care and Use Committee of the University of Pittsburgh.
2.2. Generation of Bhlhb5::flpo knockin mouse
Targeting vectors were constructed from 129s6/SvEvTAC mouse genomic fragments, which were amplified by PCR and sequenced. For the Bhlhb5::flpo knockin construct, the coding region of the Bhlhb5 gene was replaced with the coding region for Flpo re-combinase by fusion PCR, which was confirmed by restriction digest and sequencing. The fusion product was then introduced into a targeting vector that contained 5′ and 3′ homologous arms. In addition, the construct featured a LoxP-flanked PGK-neomycin positive selection cassette (Neo) downstream to the 3′UTR, as well as a diptheria toxin negative selection cassette (DTA) 3′ to the genomic fragment. The targeting construct was directly sequenced in its entirety prior to its use in gene targeting. Linearized targeting vectors were then electroporated into sv129 mouse ES cells, and G418-resistant clones were screened for homologous recombination at the Bhlhb5 locus by PCR. Primers were used to amplify the 5′ arm (F: TAAAAGACCCGTCCCTTGTG and R: GTTCACGATGTCGAAGCTCA) and the 3′ arm (F: GGGGAACTTCCT-GACTAGGG and R: CTTGTGAAGCCTGCAAAACA). Two positive clones were transfected with a Cre recombinase-expressing plas-mid, and proper removal of the LoxP-flanked neomycin cassette was confirmed by PCR. Confirmed ES cells were injected into C57BL/6 blastocysts and implanted into pseudopregnant females to generate chimeric offspring. More details are available upon request.
2.3. Immunohistochemistry
For immunocytochemistry, post-natal mice were fixed with 4% paraformaldehyde in PBS by intracardial perfusion and brains and/or spinal cords were dissected; embryonic mice were drop fixed. Tissues were post-fixed for 1 h to overnight at 4 °C, washed extensively with PBS, cryopreserved in 30% sucrose in PBS overnight, embedded in OTC, and frozen. Sections were cut at 20 µm on a cryostat and placed on slides. For immunostaining, sections were blocked in 10% goat serum and 0.25% triton-X in PBS for 1 h at RT. Sections were incubated with primary antibodies in block overnight at 4 °C. Slides were washed 4 × 5 min in PBS containing 0.1% triton-X. Detection was carried out using Alexa-fluor secondary antibodies diluted 1:500 in blocking solution. Sections were counterstained with the nuclear dye Hoechst 33342 (1:10,000) for one minute at room temperature. Sections were washed, as above, and coverslipped. The primary antibodies used were: chicken anti-GFP (1:2000, GFP-1020, Aves, Tigard OR), rabbit anti-GFP (1:2000, A11122, Life technologies, Grand Island, NY), rat anti-Bhlhb5 (1:2000, Ross et al., 2010), mouse anti-NeuN (1:1000, MAB 377, Millipore, Billerica, MA), mouse anti-Pax6 (1:200, DSHB, Iowa City, IA), rabbit anti-Pax2 (1:1000, 71–6000, Life technologies, Grand Island, NY), mouse anti-CamKIIα (1:500; 05–532 EMD Millipore, Billerica, MA), rabbit anti-Tbr2 (1:2000; generous gift from Robert Hevner (Englund et al., 2005)) and mouse anti-parvalbumin (1:1000, MAB1572, Chemicon).
2.4. Imaging
All sections reacted with fluorescent antibodies were examined with either a Nikon A1R confocal microscope (Nikon; Tokyo, Japan) or an Olympus BX53 florescence microscope (Olympus; Tokyo, Japan). Images from confocal microscope were generated by single focal plane observation through a 10×, 20×, or 60× oil-immersion lens.
2.5. Quantification of cells in DCN
To generate control Bhlhb5flpo/+ and Bhlhb5flpo/− mice for quantification of neurons in the DCN, heterozygous Bhlhb5flpo/+; RCEFRT/FRT were mated with heterozygous Bhlhb5 null mice and littermate offspring were used for analysis. Pairs of littermate mice were perfused and the brains were sectioned in the coronal plane. Twenty-micron sections were collected on slides in a series of 10 (such that sections on a given slide are ~200 µm apart). Sections were co-stained with antibodies to GFP and NeuN and counter-stained with Hoechst. Sections from each pair were matched based on landmarks. The sections containing the largest segment of the DCN were imaged on a confocal microscope using a 60× objective, and images (at least 4 × 4) were stitched together to give a image containing the entire DCN. The DCN was outlined using Nikon software and the number of cells in each channel was quantified by a person who was blind to genotype.
3. Results
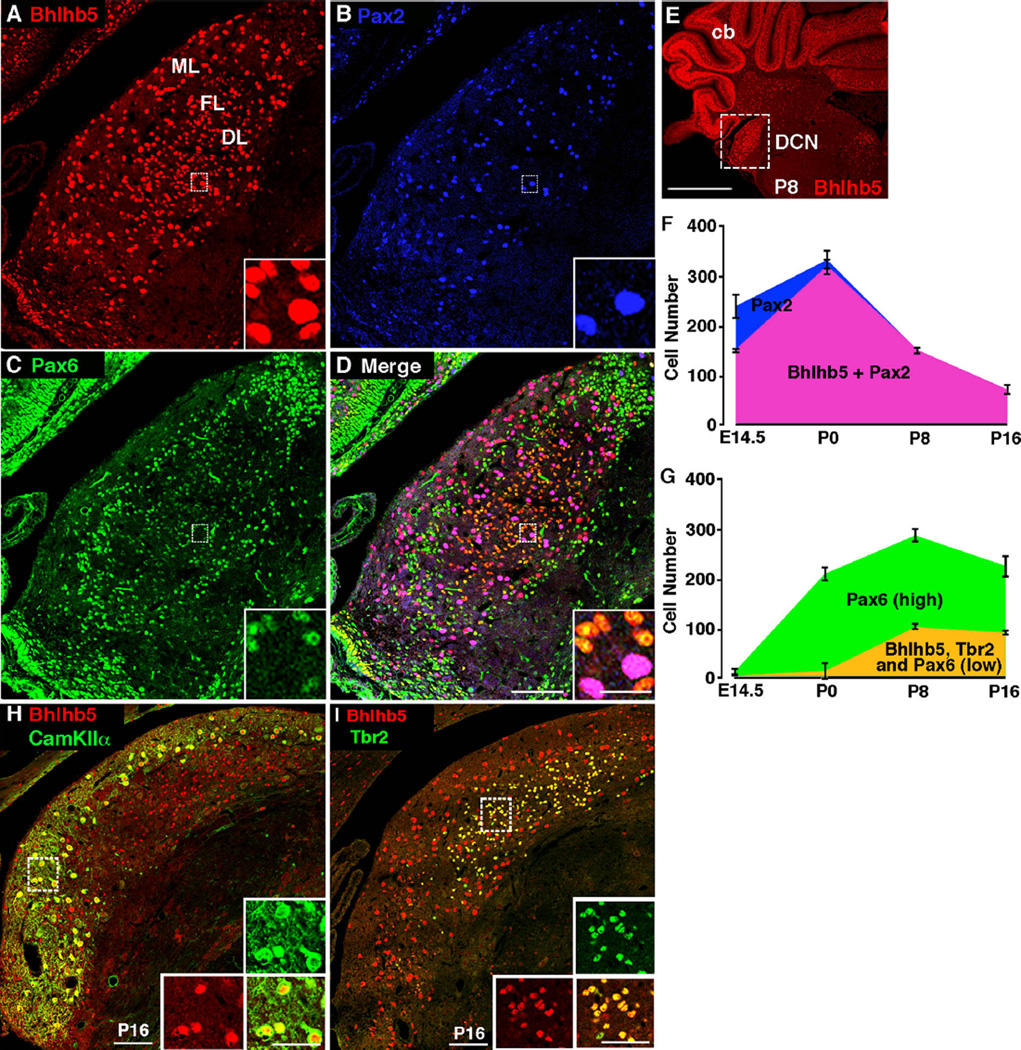
Bhlhb5 has been found to play a key role in the development of every neuronal subtype that has been examined in detail thus far (Feng et al., 2006; Joshi et al., 2008; Ross et al., 2010; Ross et al., 2012). We therefore set out to perform a more thorough characterization of its expression pattern using immunohistochemical analysis throughout embryonic and early post-natal development. For these studies, we used Bhlhb5-directed antibodies that we had previously generated and validated in Bhlhb5−/− mice (Ross et al., 2010). While numerous regions of the nervous system showed transient expression of Bhlhb5 during embryonic development, the DCN stood out due to prolonged expression of Bhlhb5 in the post-natal mouse and a high prevalence of Bhlhb5-expressing neurons within this nucleus. Bhlhb5 is first detectable in the developing dorsal cochlear nucleus at E12.5 where it is expressed at low levels in newly post-mitotic neurons (data not shown). Co-staining experiments reveal that Bhlhb5 is co-expressed with the neuronal marker, MAP2, but not astrocyte marker, ALDH1L1, or the oligodendrocyte marker, Olig2 (Fig. S1). We found that all Bhlhb5-positive cells co-stained with either Pax2 or Pax6 during early postnatal development (e.g., P8), and these two populations were found to be completely non-overlapping (Fig. 1A–D). We therefore used these markers to do a time-course analysis of Bhlhb5 expression. These efforts revealed that there is an early phase of Bhlhb5 expression, peaking at P0, in which Bhlhb5 is predominantly co-expressed with Pax2 (Fig. 1F). In contrast, from P8–P16 the majority of Bhlhb5-positive cells co-express Pax6 (Fig. 1G). These findings suggest that there are two overlapping waves of Bhlhb5 expression in at least two distinct cell types.
Fig. 1.
Bhlhb5 is expressed in the dorsal cochlear nucleus during development. (A–D) Coronal section from mouse at P8 immunostained with antibodies directed against Bhlhb5 (red), Pax2 (blue) and Pax6 (green), as indicated. All Bhlhb5-expressing cells either express either Pax2 or Pax6. Scale bar = 100 µm; inset scale bar = 20 µm; ML, molecular layer; FL, fusiform layer; DL deep cell layer. (E) Coronal brain hemi-section stained with Bhlhb5; boxed inset of DCN is shown in A–D. Scale bar = 500 µm; cb, cerebellum. (F) Quantification of the number of Pax2-expressing cells that do (pink) or do not (blue) co-express Bhlhb5 from E14.5 – P16 (data are average +/− SEM, n = 3). (G) Quantification of the number of Pax6-expressing cells in the DCN that do (orange) or do not (green) co-express Bhlhb5 from E14.5–P16. Bhlhb5-expressing cells showed a relatively low level of Pax6 expression and also co-expressed Tbr2 (data are average +/− SEM, n = 3). (H–I) Coronal section from mouse at P16 immunostained with antibodies directed against Bhlhb5 (red), CamKIIα (green) and Tbr2 (green) as indicated. Boxed area is shown in insets (H: molecular layer; I: fusiform-deep cell layers). Scale bar = 100 µm; inset scale bar = 50 µm. All images are single confocal optical sections.
Next, we investigated the identity of the Pax2- and Pax6-expressing subsets of Bhlhb5 neurons in the DCN. Many of the Pax2-positive subset co-expressed CamKIIα, a marker of cartwheel cells (Ochiishi et al., 1998; Takaoka et al., 2005). In particular, at P16 almost all Bhlhb5-expressing cells in the molecular cell layer co-label with CamKIIα (Fig. 1H). In contrast, the majority of Bhlhb5-expressing cells in the fusiform layer co-stained with Pax6 (Fig. 1D). Although, Pax6 is frequently used as a marker of granule cells, we had reason to suspect that the Pax6-expressing subset of Bhlhb5 neurons were not granule cells. Specifically, these Bhlhb5-positive cells were found exclusively in the fusiform layer and deep layers and expressed lower levels of Pax6 relative to the presumptive granule cells in the molecular layer (Fig. 1C and D). Given their location, we hypothesized that these small cells within the fusiform cell and deep layers might be unipolar brush cells. Consistent with this idea, we found that all cells that co-express Bhlhb5 and Pax6 also express Tbr2 (Fig. 1I), a marker of unipolar brush cells (Dino and Mugnaini, 2008). Taken together, these data indicate that Bhlhb5 is expressed in the DCN during embryonic and post-natal development, where it is expressed by Tbr2-positive unipolar brush cells as well as in Pax2-positive neurons that include cartwheel cells.
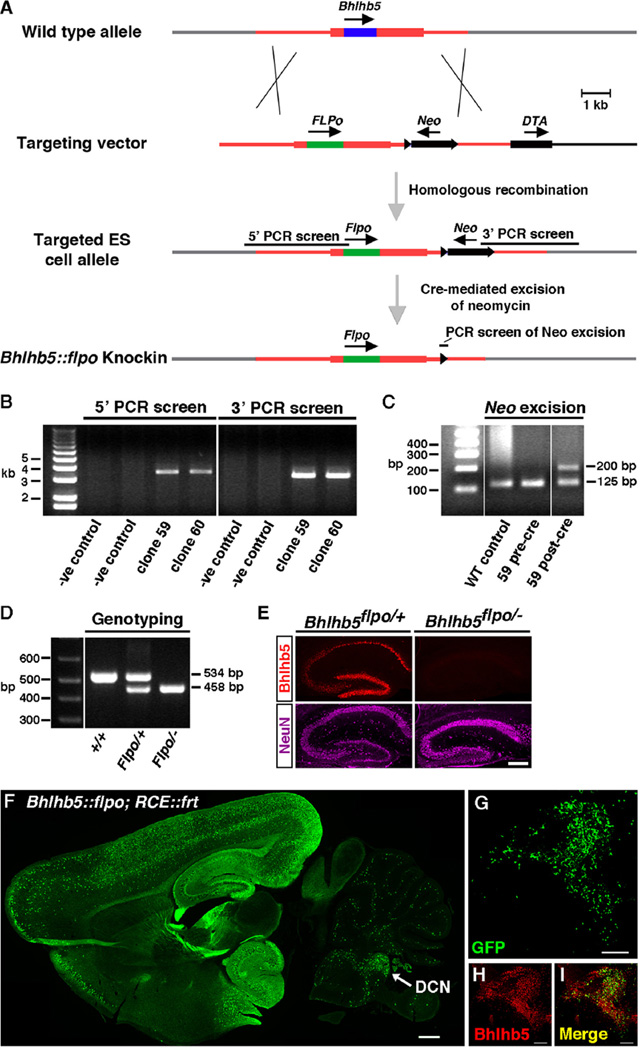
Since Bhlhb5 is widely and persistently expressed in the DCN, we hypothesized it is important in the development and function of this nucleus. To visualize and manipulate Bhlhb5-expressing cells in the DCN and elsewhere in the nervous system, we generated a Bhlhb5::flpo knockin allele in which the recombinase Flpo was targeted to the endogenous Bhlhb5 locus. Specifically, we precisely replaced the coding region of the Bhlhb5 gene with that of Flpo recombinase (Fig. 2A). (Flpo is a codon-optimized and thermostable derivative of Flp that has been optimized for high activity in mammalian cells (Dymecki et al., 2010)). To screen for ES cells in which homologous recombination had occurred, we used a PCR-based approach to amplify the 5′ and 3′ arms (Fig. 2B). Upon identification of correct clones, we removed the neomycin selection cassette via Cre-mediated excision (Fig. 2C). Routine genotyping by PCR gave rise to products of the anticipated size in wild-type, heterozygous, and Bhlhb5::flpo mutant mice (Fig. 2D). To further validate that Flpo was targeted correctly to the endogenous locus, we performed immunohistochemical analysis of hippocampal sections from mice that were either heterozygous for Bhlhb5::flpo (Bhlhb5flpo/+) or Bhlhb5::flpo mutant mice (Bhlhb5ftpo/−). As expected, whereas robust expression of Bhlhb5 was observed in the hippocampus of heterozygous mice, Bhlhb5ftpo/− mutant mice showed no Bhlhb5 expression (Fig. 2E). Together, these findings indicate that the Bhlhb5::flpo line is a true knockin into the endogenous Bhlhb5 locus that abrogates Bhlhb5 expression.
Fig. 2.
Generation of a Bhlhb5::flpo knockin mouse. (A) Targeting strategy to replace the Bhlhb5 open reading frame with that of flpo recombinase. Open reading frames are indicated with arrows and dark shaded regions within rectangles for Bhlhb5 (blue) and Flpo (green). LoxP sites are indicated with black triangles. PCR products are marked with black bars. Neo, neomycin positive selection cassette; DTA, diphtheria toxin negative selection cassette. (B) PCR-based screening for homologous recombination at the 5′ and 3′ arms. PCR products of appropriate size are observed upon screening clones 59 and 60. (C) PCR analysis of DNA from WT control, clone 59 before cre-mediated removal of neomycin (pre-cre) and after (post-cre). A larger PCR product is observed upon removal of neomycin due to additional bases associated with the remaining LoxP site in the targeted allele. Genotyping of genomic DNA isolated from tails of mice that are wild type (+/+), heterozygous for the Bhlhb5::flpo allele (Flpo/+), or mutants harboring one copy of the Bhlhb5::flpo allele and one copy of the Bhlhb5 null allele (Flpo/−) shows PCR products of the expected size. This PCR amplifies the junction between the 3′ UTR and the coding region of either the Bhlhb5 gene of the flpo recombinase gene. (E) Immunohistochemistry for Bhlhb5 (red) or NeuN (magenta) in coronal sections of the hippocampus from heterozygous mice with one copy of the Bhlhb5::flpo allele (Bhlhb5flpo/+) or mutant mice with one copy of the Bhlhb5::flpo allele and one copy of the null allele (Bhlhb5flpo/−) shows the loss of Bhlhb5 protein in the hippocampus of Bhlhb5fIp0/− mice, indicating correct targeting of the Bhlhb5::flpo allele into the endogenous Bhlhb5 locus. Scale bar = 50 µm (F) Immunohistochemical analysis of sagittal sections from a Bhlhb5::flpo; RCE::FRT mice at P8 using antibodies against GFP to visualize cells in which recombination has occurred. Arrow indicates the DCN as well as the granular cell domain. Single optical section; Scale bar = 200 µm. (G–I) Magnified view of DCN/granule cell domain showing numerous cells expressing GFP (green) and Bhlhb5 (red) as indicated. Scale bar = 100 µm.
To investigate whether the Bhlhb5::flpo causes recombination in Bhlhb5-expressing cells, we crossed mice with the Bhlhb5::flpo allele with mice harboring the RCE::FRT reporter. This particular reporter is one in which the Gt(ROSA)26Sor locus is targeted with a CAG promoter, driving the expression of eGFP following a STOP sequence that is flanked by FRT sites (Sousa et al., 2009). Immunohistochemical analysis of sections from double transgenic Bhlhb5::flpo; RCE::FRT mice at P8 revealed widespread expression of eGFP in the hippocampus and the cortex—a pattern that faithfully recapitulates the expression pattern of Bhlhb5 in postnatal mice (Fig. 2F). Importantly, we also saw extensive Flpo-mediated recombination within cells of the DCN (Fig. 2G). This recombination was largely selective to this nucleus, since there was very little recombination in either the anteroventral or posteroventral regions of the cochlear nucleus (Fig. S2). Co-staining of these sections with Bhlhb5-directed antibodies revealed that many Bhlhb5-expressing cells show eGFP expression and, conversely, that the vast majority of eGFP-positive cells co-express Bhlhb5.
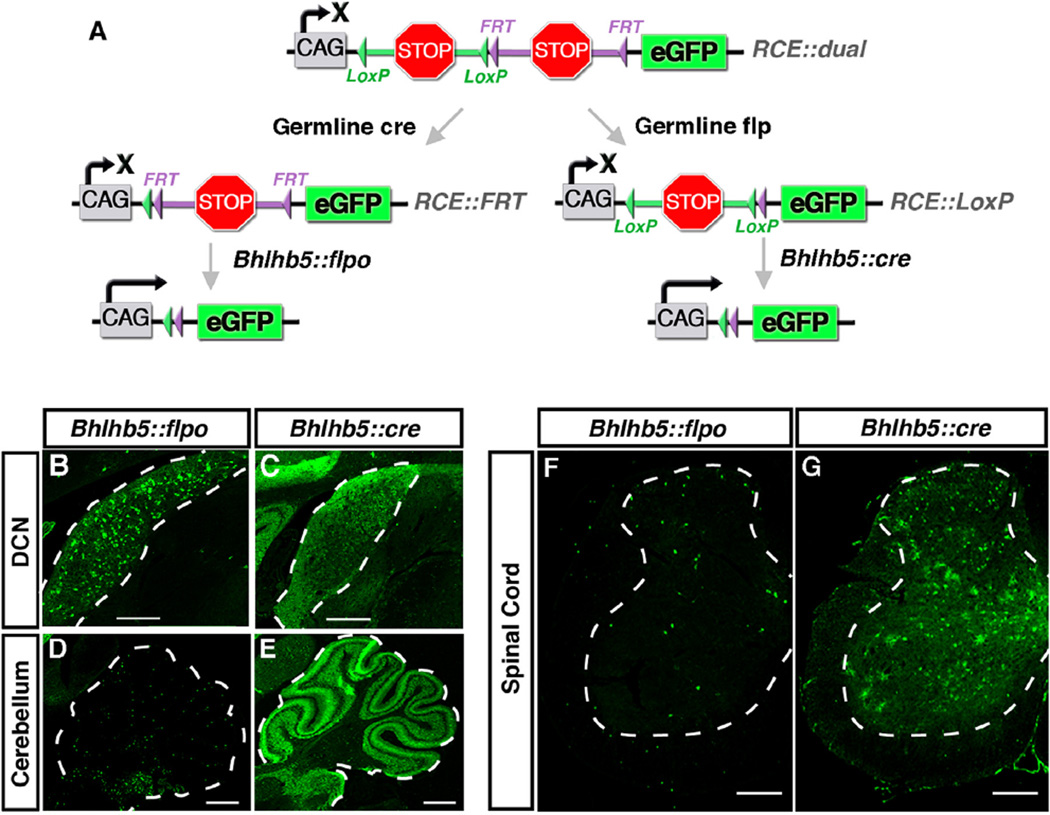
Because the Bhlhb5::cre and Bhlhb5::flpo alleles are identical in design except that they express different recombinases, these alleles gave us the opportunity to directly compare Cre and Flpo activity in mice. Flpo is known to be somewhat less efficient than Cre, and so it seemed likely that these two alleles would cause somewhat different recombination patterns (Dymecki et al., 2010). For a fair comparison, it was important to analyze recombination activity—at least to the extent possible—using the same reporter. Thus, we utilized two derivative versions of the RCE allele (Sousa et al., 2009). Specifically, we began with a dual responsive reporter (RCE::dual), and then converted it to make it either responsive to Flp alone (RCE::FRT) or responsive to Cre alone (RCE::LoxP) via germline removal of one of the two stop cassettes (Fig. 3A). Thereafter, the activity of the Bhlhb5::flpo allele was analyzed using the RCE::FRT reporter, whereas the activity of the Bhlhb5::cre allele was analyzed using the RCE::LoxP reporter. These experiments revealed that the Bhlhb5::flpo allele consistently shows more restricted expression than the Bhlhb5::cre allele (Fig. 3B–G). For instance, the Bhlhb5::flpo allele causes recombination in a subset of neurons in the DCN and cerebellum, whereas the Bhlhb5::cre allele causes recombination in virtually all cells of these regions (Fig. 3B–E). Similarly, in the spinal cord, more selective GFP expression is observed following Flpo-mediated recombination relative to that observed with Cre (Fig. 3F and G). Thus, the pattern of recombination that we observed using these two alleles suggested that Cre appears to cause recombination in all of the neurons that have ever expressed Bhlhb5. In contrast, the Flpo allele appears to cause recombination in neurons that show persistent expression of Bhlhb5. These data clearly illustrate that Flpo is significantly less active than Cre in mammalian cells. As a result, this allele generally marks cells in which the expression of the Bhlhb5 gene is prolonged.
Fig. 3.
Comparison of Bhlhb5::flpo and Bhlhb5::cre alleles. (A) Strategy to compare the recombination from Bhlhb5::flpo and Bhlhb5::cre alleles using the RCE family of reporters. The RCE::dual allele contains both FRT-flanked and LoxP-flanked stop sequences. Upon cre-mediated removal of a stop sequence, a Flp-dependent reporter is generated (RCE::FRT) that can be used to visualize recombination from the Bhlhb5::flpo allele. Alternatively, upon Flp-mediated removal of a stop cassette in the RCE::dual allele, a cre-dependent reporter is generated (RCE::LoxP) that can be used to visualize recombination from the Bhlhb5::cre allele. (B) Coronal sections of Bhlhb5::flpo; RCE::FRT mice at P12 immunostained for GFP reveals restricted recombination a subset of cells in the DCN. Scale bar = 100 µm (C) Coronal sections of adult Bhlhb5::cre; RCE::LoxP mice at P12 immunostained for GFP reveals widespread recombination throughout the DCN. Scale bar = 100 µm (D–E) Sagittal sections from mice at P12 showing GFP expression in DCN and cerebellum from Bhlhb5::flpo; RCE::FRT (E) and Bhlhb5::cre; RCE::LoxP (F) mice. Scale bar = 200 µm. (F–G) Transverse sections showing GFP expression in the lumbar spinal cord from adult Bhlhb5::flpo; RCE::FRT (F) and Bhlhb5::cre; RCE::LoxP (G) mice. Scale bar = 100 µm.
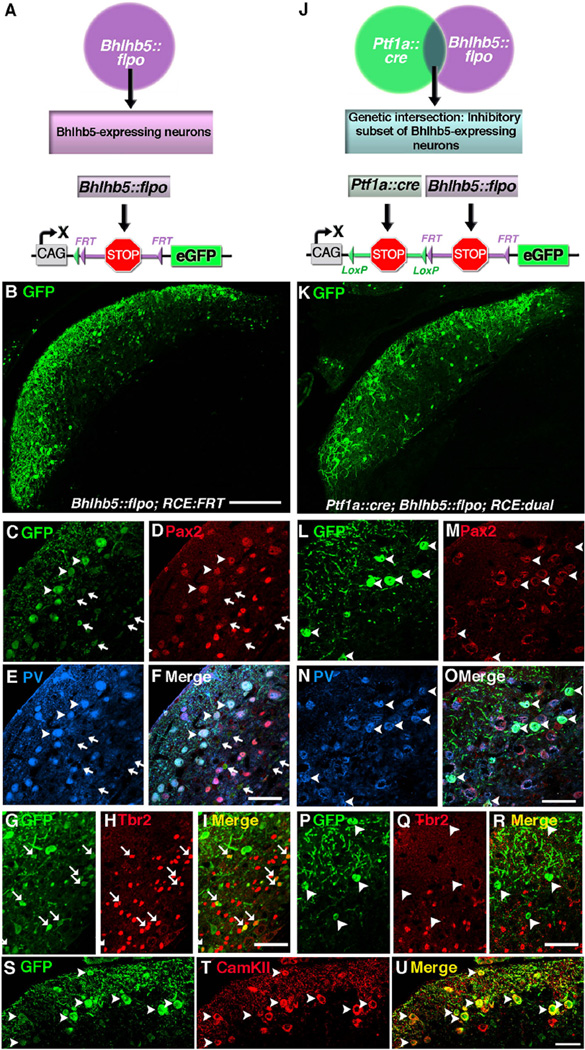
Intersectional genetic approaches give us a way to label subsets of neurons that express two different genes of interest. With the Bhlhb5::flpo tool in hand, we were now positioned to test the feasibility of using this strategy to label subsets of Bhlhb5-expressing neurons. Here, we coupled the Ptf1a::cre allele, which is selective to inhibitory neurons (Fujiyama et al., 2009), to the Bhlhb5::flpo allele in order to selectively label the inhibitory subset of Bhlhb5-expressing cells in the DCN. This allowed us to compare the recombination pattern using RCE reporters that were either singly responsive (i.e., to Bhlhb5::flpo; Fig. 4A) or dually responsive (i.e., to Bhlhb5::flpo and Ptf1a::cre; Fig. 4J). Using this strategy, we found that the Bhlhb5::flpo allele caused eGFP expression in at least two cell types. The first are medium-sized, Pax2-positive neurons whose cell bodies are found predominantly between the fusiform cell and the molecular layers and that co-express parvalbumin (Fig. 4B–F); arrowheads). In addition, we found a population of small neurons that are found predominantly in the fusiform and deep layers that lack both Pax2 and parvalbumin expression (Fig. 4B–F); arrows). However, these small neurons co-express Tbr2 (Fig. 4G–I; arrows), a marker of unipolar brush cells (Dino and Mugnaini, 2008).
Fig. 4.
Dual intersectional strategy distinguishes subpopulations of Bhlhb5-expressing cells: cartwheel and unipolar brush neurons. (A) Schematic illustrating the use of the Bhlhb5::flpo with RCE::FRT to label Bhlhb5-expressing neurons. (B–I) Coronal sections of the DCN from an adult Bhlhb5::flpo; RCE::FRT mouse immunostained with antibodies against GFP, Pax2, parvalbumin (PV), or Tbr2, as indicated. (J) Schematic illustrating the use of Bhlhb5::flpo, Ptf1a::cre and RCE::dual to label a population of neurons at the genetic intersection, namely the inhibitory subset of Bhlhb5-expressing neurons. (L–R) Coronal sections of the DCN from an adult Ptf1a::cre; Bhlhb5::flpo; RCE::dual mouse immunostained with antibodies against GFP, Pax2, parvalbumin (PV), or Tbr2, as indicated. (S–U) Coronal sections of the DCN from an adult Bhlhb5::flpo; RCE::FRT mouse immunostained with antibodies against GFP, CamKIIα, as indicated. Images are single confocal optical plane of coronal sections from six- to eight-week old mice. Arrowheads and arrows indicate examples of double-labeled inhibitory and excitatory neurons, respectively. Scale bars: B, K = 200 µm; all others = 50 µm.
Consistent with this interpretation, when we used the inter-sectional genetic strategy to label the inhibitory subset of Bhlhb5-expressing neurons selectively (Fig. 4J and K), we found that all GFP positive neurons expressed Pax2 and parvalbumin (Fig. 4L–O); arrowheads) and the recombined cells no longer included Tbr2-expressing neurons (Fig. 4P–R); arrows). Finally, to confirm that the inhibitory subset of Bhlhb5-expressing neurons consists predominantly of cartwheel cells, we co-stained sections from mice with the Bhlhb5::flpo allele with CamKIIα;, a cartwheel cell marker (Ochiishi et al., 1998; Takaoka et al., 2005) and found that the majority of cartwheel cells were recombined by the Bhlhb5::flpo allele (Fig. 4S–U). Together, these findings indicate that the Bhlhb5::flpo allele labels inhibitory cartwheel cells as well as excitatory unipolar brush cells. In addition, our studies illustrate that the Bhlhb5::flpo allele together with genetic intersectional strategies can be used to target a specific subsets of Bhlhb5-expressing cells in the DCN.
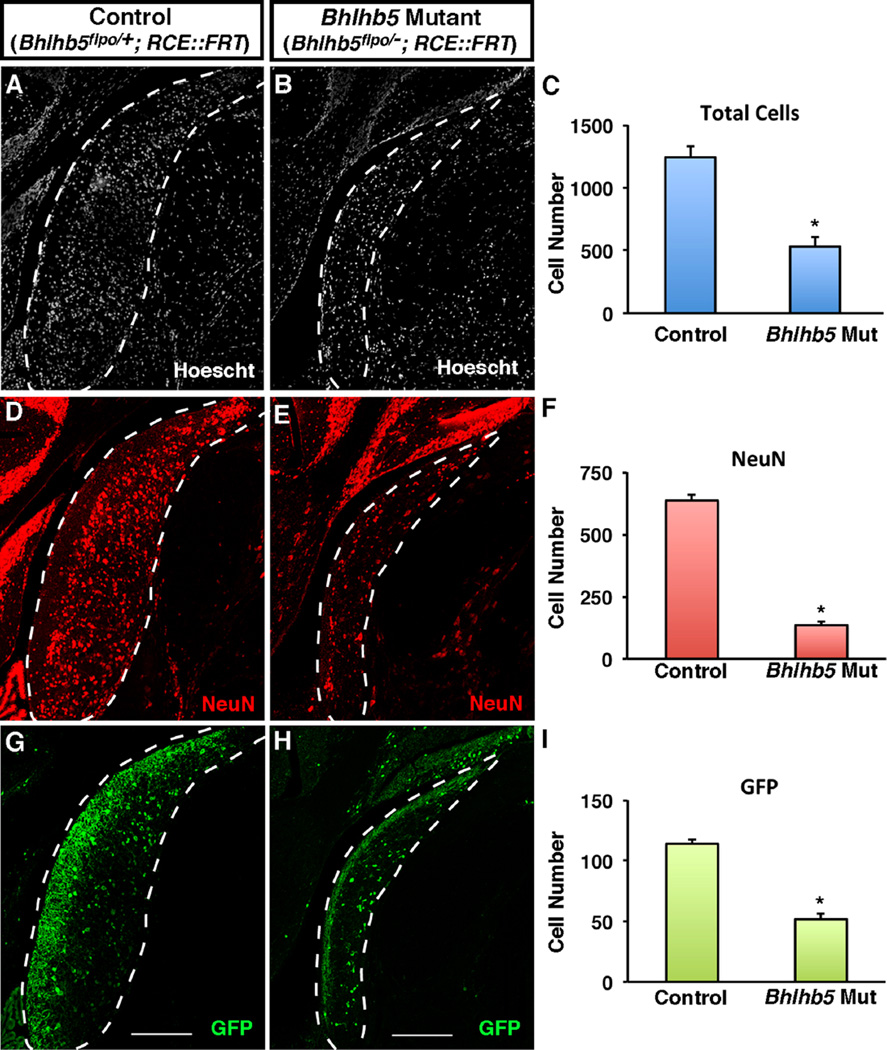
Given that Bhlhb5 shows prolonged expression in the DCN, we hypothesized that this transcription factor may play a key role in the development of this nucleus. To investigate this idea, we used immunohistochemical and fate-mapping approaches to compare the DCN in control and Bhlhb5 mutant mice. Upon loss of Bhlhb5, we observed a dramatic reduction of the DCN, which showed a ~50% reduction in total cell number (Fig. 5A–C). This cell loss could be almost completely accounted for by the finding that Bhlhb5 mutant mice showed an 80% decrease in the number of NeuN positive neurons (Fig. 5D–F). Approximately 20% of neurons in the DCN are genetically labeled using the Bhlhb5::flpo allele, and these neurons also showed a significant decrease in total number in the DCN of Bhlhb5 mutant mice (Figs. 5G–I and S3). In contrast, there was no obvious defect in the AVCN or the PVCN, which appeared grossly normal in size (Fig. S3). Thus, immunohistochemical and fate-mapping studies reveal an essential role for Bhlhb5 for the proper development of the DCN.
Fig. 5.
Mice lacking Bhlhb5 show a reduced cell number in the DCN. (A and B) Coronal sections from adult mice showing the DCN in control (Bhlhb5flpo/+; RCE::FRT) or Bhlhb5 mutant (Bhlhb5flpo/−; RCE::FRT) mice stained with Hoechst to mark nuclei. (C) Quantification of (A–B) showing a significant reduction in the total cell number in the dorsal cochlear nucleus in Bhlhb5 mutant mice relative to controls. (D–F) Immunostaining (D,E) and quantification (F) of NeuN-positive neurons in the DCN showing a significant loss of neurons in Bhlhb5 mutant mice relative to controls. (G–I) Immunostaining (G–H) and quantification (I) of GFP-expressing neurons, again showing a significant decrease in the absence of Bhlhb5. Representative confocal optical sections are shown. Scale bar = 200 µm. For quantification, n = 4 pairs of control/mutant littermates. Data are presented as mean + SEM and * indicates a significant difference relative to controls (P < 0.05, t-test). All quantification was conducted blind to genotype.
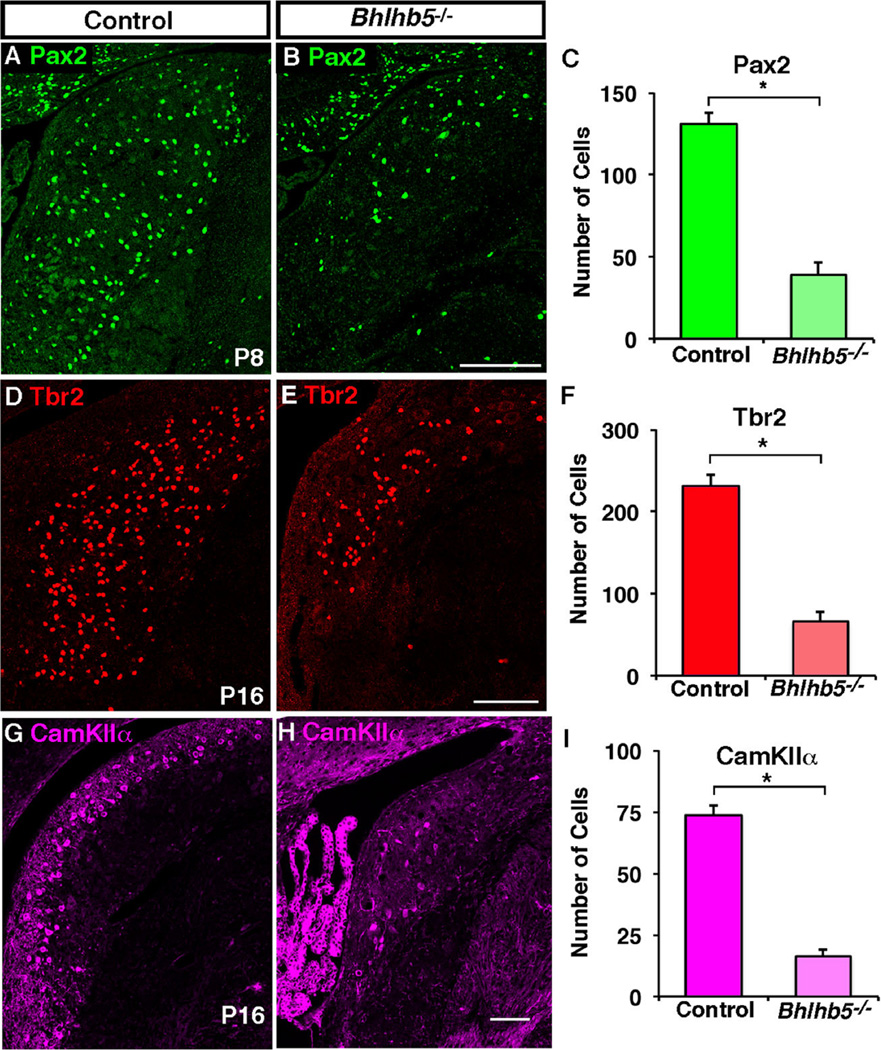
To gain further insight into the timing of cell loss in the DCN of mice that lack Bhlhb5, we compared the expression of genetically marked cells in control and Bhlhb5 mutant littermates at a variety of developmental time points (E14.5, P0, P8 and P16). A clear loss of genetically labeled cells in the DCN of Bhlhb5 mutant mice was not observed during embryonic development or at birth. In contrast, by P8 the cell loss in the Bhlhb5 mutant mice was evident. To address which specific cell types are lost in the absence of Bhlhb5, we performed immunostaining with cell-type specific markers at P8 and P16. These experiments revealed a significant decrease in Pax2-, Tbr2-, and CamKIIα-expressing neurons in the DCN of Bhlhb5 mutant mice relative to control littermates (Figs. 6 and S4). Together these findings suggest that a large fraction of Bhlhb5-expressing cells die in the absence of this transcription factor, including the majority of unipolar brush cells and cartwheel cells.
Fig. 6.
Pax2-, Tbr2- and CamKIIα-expressing neurons are reduced in the DCN of Bhlhb5 mutant mice. (A–B) Coronal sections from mice at P8 showing the DCN in control (Bhlhb5flpo/+; RCE::FRT) or Bhlhb5 mutant (Bhlhb5flpo/−; RCE::FRT) mice stained with antibodies to Pax2. (C) Quantification of (A–B) showing a significant reduction in the number of Pax2-expressing cells in the dorsal cochlear nucleus in Bhlhb5 mutant mice relative to controls. (D–F) Immunostaining (D, E) and quantification (F) of Tbr2-positive neurons in the DCN of P16 mice showing a significant loss of neurons in Bhlhb5 mutant mice relative to controls. (G–I) Immunostaining (G–H) and quantification (I) of CamKIIα-expressing neurons, again showing a significant decrease in the absence of Bhlhb5. Representative confocal optical sections are shown. Scale bar = 200 µm. For quantification, n = 3–4 DCN from control/mutant littermate pairs. Data are presented as mean + SEM and * indicates a significant difference relative to controls (P < 0.05, t-test). All counts were conducted blind to genotype.
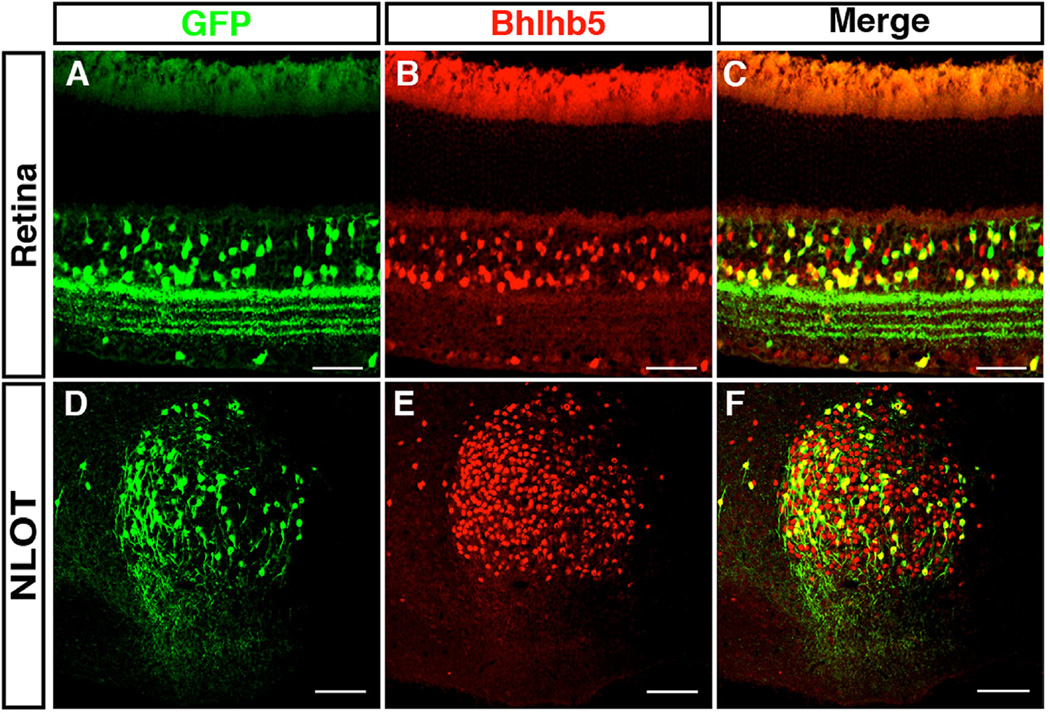
The DCN is where auditory information is first processed and integrated (Oertel and Young, 2004). In this regard, it is noteworthy that Bhlhb5 also shows prolonged expression in many other regions of the nervous system that have an analogous role in information integration. For instance, Bhlhb5 shows prolonged expression in spinal cord interneurons that are involved in integrating somatosensory input (Ross et al., 2010; Kardon et al., 2014). We also find that Bhlhb5 shows persistent expression in particular subtypes of bipolar and amacrine cells in the retina (Fig. 7A), which are involved in processing visual input, as well as in neurons of the nucleus of the lateral olfactory tract (Fig. 7B), which are involved in integrating olfactory input. Importantly, the Bhlhb5::flpo allele labels the Bhlhb5-expressing neurons in these structures (Figs. 7B and 6F), and may therefore be a useful tool in the study of a variety of types of sensory integration.
Fig. 7.
Bhlhb5 and Bhlhb5::flpo mark subsets of neurons in the retina and the nucleus of the lateral olfactory tract. (A–C) Transverse sections from retina of Bhlhb5::flpo; RCE::FRT that are co-stained with antibodies to Bhlhb5 (red) and GFP (green). Scale = 50 µm (D–F) Sagittal sections of the brain of Bhlhb5::flpo; RCE::FRT mouse at P8 showing the nucleus of the lateral olfactory tract (NLOT) stained with antibodies to Bhlhb5 (red) and GFP (green). Scale bar = 50 µm.
4. Discussion
Here we uncover an essential requirement for Bhlhb5 in the development of the DCN, and show that there is dramatic cell loss in mice that lack this transcription factor. In addition, we characterize the Bhlhb5::flpo knock-in allele, which genetically labels a subset of Bhlhb5-expressing cells, likely those that show persistent expression of Bhlhb5. Through a direct comparison of the recombination efficiencies, we provide clear evidence that Flpo is less active than Cre in mammalian cells. In addition, we provide proof of concept that the Bhlhb5::flpo allele can be coupled with Cre alleles to label subsets of Bhlhb5-expressing neurons. This tool may be useful not only in the DCN but also in the retina and nucleus of the lateral olfactory tract, three regions of the nervous system that are involved in sensory integration.
One of the interesting aspects of our study is that we are able to compare Cre and Flpo activities. To our knowledge, this is the first direct comparison between Cre and Flpo alleles that are expressed from the same genetic locus. We find that the Bhlhb5::cre allele and the Bhlhb5::flpo allele show distinct patterns of recombination, likely due to the fact that Cre is a very active enzyme in mammalian cells, whereas Flpo is significantly less active (Dymecki et al., 2010). As a result, the Bhlhb5::cre allele appears to label all cells that have ever expressed Bhlhb5, even if the expression is low and/or transient, while the Bhlhb5::flpo allele mainly appears to label cells that show high expression of Bhlhb5 over an extended period. Consistent with this idea, in regions of the nervous system such as the retina, which continue to express Bhlhb5 throughout the life of the animal, we eventually get almost complete recombination within the Bhlhb5-expressing population. In contrast, we see only moderate recombination in the DCN, where Bhlhb5 is expressed for approximately three weeks. In particular, just ~20% of neurons become recombined with the Bhlhb5::flpo allele in the DCN despite the fact that approximately 40% of neurons transiently express this transcription factor at E14. Intriguingly, the majority of recombined cells appear to be one of two cell types: cartwheel and unipolar brush cells, hinting at a particularly important role for Bhlhb5 in these cells.
Our study reveals that Bhlhb5 has a widespread role for the survival of cells in the DCN, since a large proportion of cells (~50% of total) are missing in the Bhlhb5 mutant mouse. At this stage, it is not possible to determine whether this effect is entirely due to a cell autonomous requirement for Bhlhb5, or whether cells that do not express this transcription factor are also affected by its absence. In any event, we speculate that auditory information is aberrantly integrated in Bhlhb5 mutant mice, given the degree of cell loss that is observed. In this regard, it is noteworthy to consider that Bhlhb5 mutant mice develop abnormally elevated itch due to loss of inhibitory neurons in the dorsal spinal cord (Ross et al., 2010; Kardon et al., 2014). Whether these mice also develop tinnitus due to loss of inhibitory neurons in the DCN is an interesting question to be addressed in future studies.
Although our study is focused on the DCN, we find that Bhlhb5 shows persistent expression in several areas of the nervous system that are necessary for sensory processing and integration. For instance, Bhlhb5 is required for the survival of B5-I neurons in the spinal cord that function to inhibit itch (Ross et al., 2010; Kardon et al., 2014). This transcription factor is also required for the survival of type II cone bipolar cells and a subset of amacrine cells in the retina (Feng et al., 2006). Here, we show that Bhlhb5 is required for the survival of neurons in the DCN, and we reveal that this transcription factor also expressed in the nucleus of the olfactory tract (NLOT), a nucleus that integrates olfactory input. These observations raise the possibility that Bhlhb5 plays a conserved function in differentiation of neurons that are involved in sensory integration.
Supplementary Material
Acknowledgments
This research was supported by a grant from the American Hearing Research Foundation to S. E. R. Additional support came from NIH grants R01 AR063772 and R21 AR064445 to S. E. R, and NIDCD 013048 to M. E. R. Part of this work was supported by a grant from the Rita Allen Foundation to S. E. R who is a Rita Allen Foundation Pain Scholar, as well as the Richard King Mellon Institute for Pediatric Research at the University of Pittsburgh to S. M. M. The Bhlhb5::flpo line was generated with help from the Mouse Gene Manipulation Facility of the Boston Children's Hospital Intellectual and Developmental Disabilities Research Center (IDDRC), which is supported by NIHP30-HD 18655. We thank Stephanie Buerk for help with figures. We also thank Robert Hevner for the Tbr2 antibody (Englund et al., 2005).
Footnotes
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2016.04.028.
References
- Axelsson A, Ringdahl A. Tinnitus—a study of its prevalence and characteristics. Br. J. Audiol. 1989;23:53–62. doi: 10.3109/03005368909077819. [DOI] [PubMed] [Google Scholar]
- Dino MR, Mugnaini E. Distribution and phenotypes of unipolar brush cells in relation to the granule cell system of the rat cochlear nucleus. Neuroscience. 2008;154:29–50. doi: 10.1016/j.neuroscience.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymecki SM, Ray RS, Kim JC. Mapping cell fate and function using recombinase-based intersectional strategies. Methods Enzym. 2010;477:183–213. doi: 10.1016/S0076-6879(10)77011-7. [DOI] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci.: Off. J. Soc. Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farago AF, Awatramani RB, Dymecki SM. Assembly of the brainstem cochlear nuclear complex is revealed by intersectional and subtractive genetic fate maps. Neuron. 2006;50:205–218. doi: 10.1016/j.neuron.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Feng L, Xie X, Joshi PS, Yang Z, Shibasaki K, Chow RL, Gan L. Requirement for Bhlhb5 in the specification of amacrine and cone bipolar subtypes in mouse retina. Development. 2006;133:4815–4825. doi: 10.1242/dev.02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama T, Yamada M, Terao M, Terashima T, Hioki H, Inoue YU, Inoue T, Masuyama N, Obata K, Yanagawa Y, Kawaguchi Y, Nabeshima Y, Hoshino M. Inhibitory and excitatory subtypes of cochlear nucleus neurons are defined by distinct bHLH transcription factors, Ptf1a and Atoh1. Development. 2009;136:2049–2058. doi: 10.1242/dev.033480. [DOI] [PubMed] [Google Scholar]
- Hackney CM, Osen KK, Kolston J. Anatomy of the cochlear nuclear complex of guinea pig. Anat. Embryol. 1990;182:123–149. doi: 10.1007/BF00174013. [DOI] [PubMed] [Google Scholar]
- Joshi PS, Molyneaux BJ, Feng L, Xie X, Macklis JD, Gan L. Bhlhb5 regulates the postmitotic acquisition of area identities in layers II–V of the developing neocortex. Neuron. 2008;60:258–272. doi: 10.1016/j.neuron.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon AP, Polgar E, Hachisuka J, Snyder LM, Cameron D, Savage S, Cai X, Karnup S, Fan CR, Hemenway GM, Bernard CS, Schwartz ES, Nagase H, Schwarzer C, Watanabe M, Furuta T, Kaneko T, Koerber HR, Todd AJ, Ross SE. Dynorphin acts as a neuromodulator to inhibit itch in the dorsal horn of the spinal cord. Neuron. 2014;82:573–586. doi: 10.1016/j.neuron.2014.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes IH, Cima RF, Vlaeyen JW, Anteunis LJ, Joore MA. Tinnitus: a cost study. Ear Hear. 2013;34:508–514. doi: 10.1097/AUD.0b013e31827d113a. [DOI] [PubMed] [Google Scholar]
- Nichols DH, Bruce LL. Migratory routes and fates of cells transcribing the Wnt-1 gene in the murine hindbrain. Dev. Dyn.: Publ. Am. Assoc. Anat. 2006;235:285–300. doi: 10.1002/dvdy.20611. [DOI] [PubMed] [Google Scholar]
- Ochiishi T, Yamauchi T, Terashima T. Regional differences between the immunohistochemical distribution ofCa2+/calmodulin-dependent protein kinase II alpha and beta isoforms in the brainstem of the rat. Brain Res. 1998;790:129–140. doi: 10.1016/s0006-8993(98)00058-4. [DOI] [PubMed] [Google Scholar]
- Oertel D, Young ED. What’s a cerebellar circuit doing in the auditory system? Trends Neurosci. 2004;27:104–110. doi: 10.1016/j.tins.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Osen KK. Cytoarchitecture of the cochlear nuclei in the cat. J. Comp. Neurol. 1969;136:453–484. doi: 10.1002/cne.901360407. [DOI] [PubMed] [Google Scholar]
- Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- Ross SE, Mardinly AR, McCord AE, Zurawski J, Cohen S, Jung C, Hu L, Mok SI, Shah A, Savner EM, Tolias C, Corfas R, Chen S, Inquimbert P, Xu Y, McInnes RR, Rice FL, Corfas G, Ma Q, Woolf CJ, Greenberg ME. Loss of inhibitory interneurons in the dorsal spinal cord and elevated itch in Bhlhb5 mutant mice. Neuron. 2010;65:886–898. doi: 10.1016/j.neuron.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SE, McCord AE, Jung C, Atan D, Mok SI, Hemberg M, Kim TK, Salogiannis DM, Hu L, Cohen S, Lin Y, Harrar D, McInnes RR, Greenberg ME. Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron. 2012;73:292–303. doi: 10.1016/j.neuron.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryugo DK, Willard FH. The dorsal cochlear nucleus of the mouse: a light microscopic analysis of neurons that project to the inferior colliculus. J. Comp. Neurol. 1985;242:381–396. doi: 10.1002/cne.902420307. [DOI] [PubMed] [Google Scholar]
- Shargorodsky J, Curhan GC, Farwell WR. Prevalence and characteristics of tinnitus among US adults. Am. J. Med. 2010;123:711–718. doi: 10.1016/j.amjmed.2010.02.015. [DOI] [PubMed] [Google Scholar]
- Sousa VH, Miyoshi G, Hjerling-Leffler J, Karayannis T, Fishell G. Characterization of Nkx6–2-derived neocortical interneuron lineages. Cereb. Cortex. 2009;19(Suppl 1):i1–i10. doi: 10.1093/cercor/bhp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka Y, Setsu T, Misaki K, Yamauchi T, Terashima T. Expression of reelin in the dorsal cochlear nucleus of the mouse. Brain Res. Dev. Brain Res. 2005;159:127–134. doi: 10.1016/j.devbrainres.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Wang H, Brozoski TJ, Turner JG, Ling L, Parrish JL, Hughes LF, Caspary DM. Plasticity at glycinergic synapses in dorsal cochlear nucleus of rats with behavioral evidence of tinnitus. Neuroscience. 2009;164:747–759. doi: 10.1016/j.neuroscience.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Brozoski TJ, Caspary DM. Inhibitory neurotransmission in animal models of tinnitus: maladaptive plasticity. Hear. Res. 2011;279:111–117. doi: 10.1016/j.heares.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.